Abstract
The wealth of genomic technologies has enabled biologists to rapidly ascribe phenotypic characters to biological substrates. Central to effective biological investigation is the operational definition of the process under investigation. We propose an elucidation of categories of biological characters, including disease relevant traits, based on natural endogenous processes and experimentally observed biological networks, pathways and systems rather than on externally manifested constructs and current semantics such as disease names and processes. The Ontological Discovery Environment (ODE) is an Internet accessible resource for the storage, sharing, retrieval and analysis of phenotype-centered genomic data sets across species and experimental model systems. Any type of data set representing gene-phenotype relationships, such quantitative trait loci (QTL) positional candidates, literature reviews, microarray experiments, ontological or even meta-data, may serve as inputs. To demonstrate a use case leveraging the homology capabilities of ODE and its ability to synthesize diverse data sets, we conducted an analysis of genomic studies related to alcoholism. The core of ODE’s gene-set similarity, distance and hierarchical analysis is the creation of a bipartite network of gene-phenotype relations, a unique discrete graph approach to analysis that enables set-set matching of non-referential data. Gene sets are annotated with several levels of metadata, including community ontologies, while gene set translations compare models across species. Computationally derived gene sets are integrated into hierarchical trees based on gene-derived phenotype interdependencies. Automated set identifications are augmented by statistical tools which enable users to interpret the confidence of modeled results. This approach allows data integration and hypothesis discovery across multiple experimental contexts, regardless of the face similarity and semantic annotation of the experimental systems or species domain.
Keywords: homology, combinatorial algorithms, microarray, ontology
Introduction
High-thoughput molecular biology provides a means to rapidly associate underlying molecular pathways and other substrates to biological structures and functions. These associations are used to characterize phenotypes and in a limited way, to define the relations among them. There are numerous methodologies for empirical creation and analysis of gene-sets from this type of data. In contrast, defining biologically meaningful categories of phenotypes, particularly those which share a common mechanism is problematic due to the often subjective and phenomenological description of such categories.
Working from the top down, ontology development efforts develop and impose a knowledge structure on biology. Phenotype ontologies such as the Mammalian Phenome Ontology (MPO) [1] and the Phenotype And Trait Ontology (PATO) [2] are projects designed to organize higher order phenotypes based on construct knowledge. Both make use of formalized processes for describing relations pioneered by the Gene Ontology Consortium [3]. These and other existing ontology development strategies often do not allow for the description of explicit structure and relationship among defined phenotypes. In the case of behavior, for example, there is limited shorthand to describe the essential categories of complex characteristics mediated by shared biological pathways. This is in contrast to biochemical pathways which are often more-well worked, though even the humble biochemical pathway becomes exquisitely complex as pathway members expand beyond reaction enzymes to the tremendous array of associated gene products involved in transport, anchoring, aggregation, synthesis and other processing of enzymes and substrates. Furthermore, it is challenging to compactly define and unify sets of processes that have different external manifestations of common internal processes. It then becomes vital to implement an approach that discovers the natural organizations of related behavioral processes as a reflection of underlying empirically-derived gene sets using dynamic points of intersection. Lastly, existing paradigms rely on prior knowledge or relevant gene groupings to describe new relationships successfully. For many new or largely uncharacterized genomic features, this is a significant problem. By constructing hierarchical ontologies from known gene-phenotype relationships, ODE breaks from existing constructs by separating the naturally occurring gene-network from the a priori concept structure of the ontology.
The automated and semi-automated creation and analysis of gene sets is a well-developed area enabling rapid development and interpretation of empirical data. This data is often synthesized and grouped through category matching approaches, wherein new empirical data is intersected with known, curated functional annotations for groups of genes. The most widely supported effort of this sort is the Gene Ontology [3] annotation effort which uses carefully curated experimental data from functional studies of each gene-phenotype association. Other pathway databases such as the Kyoto Encyclopedia of Genes and Genomes (KEGG) [4], GenMAPP [5], and the Biocarta collection contain gene set annotations largely based on known systems and pathways. Highly curated data banks and tools for pathway reconstruction, such as Ingenuity’s Pathway Analysis package (Ingenuity Systems, Mountain View, CA), can be used to construct and annotate gene networks. Indeed, numerous tools have been described for the analysis of various category representations [6–9]. While these tools are often an invaluable aid for distilling and interpreting gene lists and pathways resulting from differential expression analysis, they suffer from a few limitations. Most notably, these include the need for cross-species data integration, and the need to understand, identify and analyze a highly granular and uncharacterized set of related biological processes underlying the broad disease constructs that are assessed through various experimental methods. Analysis of cross-species convergence of gene-phenotype associations, termed ‘convergent functional genomics,’ has been profitably employed in an analysis of bipolar disorder across species in several experimental contexts [10].
From a genome perspective, there have been many attempts to produce convergent analysis of phenome expression on genome scales, covering a variety of species including mouse, rat, human, and yeast [11–16]. Although each such example provides forward thinking approaches to cross-experimental data integration, the methodology of these existing efforts focuses on the creation of comprehensive ontologies of narrow domains, or on the mapping of high-throughput data to existing ontologies. These approaches often preclude the set-set comparison on non-referential data across diverse experimental domains or between species. Current mapping efforts to facilitate large scale phenotype interoperability are encouraging [17–19], but suffer from the challenges inherent to the lofty goals of structuring and describing compactly knowledge of all of biological function.
We present The Ontological Discovery Environment (ODE) as a Web-based software environment that extracts existing phenomenologically-driven complex trait genomic analysis, and integrates it with a simultaneous analysis of instances (gene-trait associations) and ontologies (classes of genes and traits). In this way, ODE provides and analyzes articulations between gene space and phenome space [20]. ODE addresses the challenge of phenome mapping by accumulating gene-phenotype knowledge through data integration and hypothesis driven discovery across multiple labs and multiple experimental contexts. Emergent discovery in this software environment relies on user-submitted and publicly available gene sets associated with various species and phenotypes, and integrates them using categorical metadata, such as homology. In this way, ODE seeks to define the ontology of complex biological processes, such as behavior, based on intrinsic biological entities, rather than external phenotypic manifestations, which are often subject to historical and cultural biases. The collection of unique ODE tools builds a shared biological architecture of apparently distinct processes, enabling recognition of biological function in health and disease.
ODE’s novel approach to gene set analysis also incorporates computation-critical aspects of genome-scale discovery. This is a particularly pressing issue because classification and assessment of the phenome space is theoretically unbounded. Recent Bayesian network approaches have made significant contributions to our understanding of cross-domain synthesis but do not offer robust information about local relationships [19] needed for granular analysis. Since set relationships are discrete structures that can naturally be described as finite simple graphs, graph algorithms can be harnessed to interpret and analyze the enormous correlation matrices that arise in the study of transcriptomic and other sorts of -omic data. Bipartite graph representations of gene-phenotype associations are a discrete combinatorial approach that shows promise in preserving information while escaping constrained semantics as demonstrated by clustering of disease phenotype and genes in a fixed data set [21]. In particular, by representing each gene list as a phenotype vertex connected to vertices representing each gene on the list in a bi-partite graph, ODE provides data integration while maintaining substructure relationships of nested gene-set clusters. The creation of emergent phenome ontologies as presented here addresses these computational demands in large part by exploiting novel mathematical tools, such as fixed-parameter tractability [22], and by employing innovative implementations of combinatorial algorithms we have synthesized for supercomputers at our disposal [23]. Consequently, by leveraging high performance computing, ODE is uniquely positioned to provide phenome models in genome-scale space.
Gene sets, the primary input to the analyses, may be empirically defined or dynamically created within ODE’s repository of gene relationships. Multiple tools are available to perform integrative, gene-centered analysis, and provide confidence metrics for model structure and data aggregation. ODE’s tools include gene set clustering, pairwise Jaccard Similarity and Distance Analysis, Hypergeometric tests, and a highly efficient biclique method for constructing a map of the gene-centered, empirical phenome. Visualization of the resultant phenotypes can then be seen in real time and used for iterative testing and gene set creation. By integrating this approach into a web-based software system, we facilitate the analysis and interpretation of sets of genomic results, enabling comparison, intersection and integration of convergent data from several species and many experiment types, including mutant analyses, genome wide association studies, microarray experiments and virtually any other genomic data type.
Results and Discussion
The ODE environment uses bipartite graphs to dynamically create phenotype relationship diagrams to enable users to produce new knowledge about phenotype similarity and the underlying gene interconnectivity. Indeed, any type of data set representing gene-phenotype relationships, such quantitative trait loci (QTL), literature reviews, microarray experiments and ontological annotations, may be used as the foundation to create self-describing phenotype hierarchical graphs. To demonstrate a use case leveraging the homology underpinnings of ODE and its ability to synthesize information from various data sets, we conducted an analysis of alcoholism related behaviors in several model systems.
The initial data set includes genes from mouse strains selected for their functional abilities after acute ethanol exposure, called high and low acute functional tolerance or HATF2 and LAFT2, respectively [24]. A second set of genes that are differentially expressed in response to acute ethanol in two mouse strains, C57BL/6 and DBA/2 [25] is added. Cross-homology functionality is demonstrated by the inclusion of a differential gene expression analysis in rats after traumatic induced brain injury [26]. Finally, to bring in genes associated with differing states of complex behavior, a set of bipolar disorder candidate genes derived from a mouse differential expression study are included [10]. Each of these data sets are publicly available and pre-loaded into ODE as part of a large library of experimental data currently included as part of the environment, which currently includes data from Mus Musculus, Homo Sapiens, Drosophila Melanogaster, Rattus Norvegicus and Danio Rerio. This library also includes data from the Kyoto Encyclopedia of Gene and Genomes (KEGG), Gene Ontology (GO), and phenotypic alleles table of the Mouse Genome Informatics database, which consists of all of the Mammalian Phenotype Ontology terms and the mutant alleles to which these terms are associated, and results from many published genetic and genomic studies entered by users of our Web-based software system.
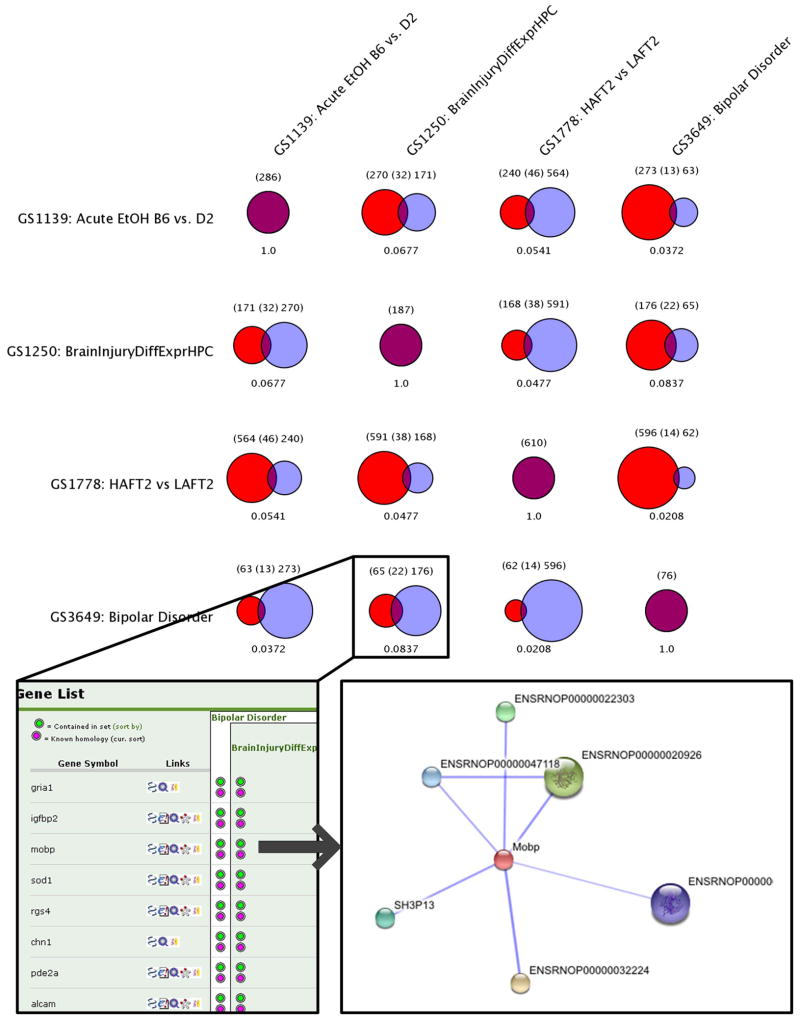
The ODE function, Jaccard Similarity (Figure 1), is one of several ODE tools for pairwise comparison of diverse gene sets. This analysis uses Jaccard’s positive match correlations to identify statistically similar gene sets. A complete pairwise Venn diagram display reveal 11 genes at the intersection of bipolar disorder [26] and traumatic brain injury [25], 13 genes at the intersection of bipolar disorder and acute ethanol response, and 14 genes at the intersection of bipolar disorder and acute functional tolerance to alcohol. All other pairwise intersections are populated.
Figure 1. Jaccard Similarity.
When combined with PhISH, this technique demonstrates the significant overlap of gene sets between comparison groups. This provides a rapid means to identify and organize sets of interest. The Jaccard results may be used to identify common genes of interest (inset) that, in turn, may be linked to external resources, such as STRING.
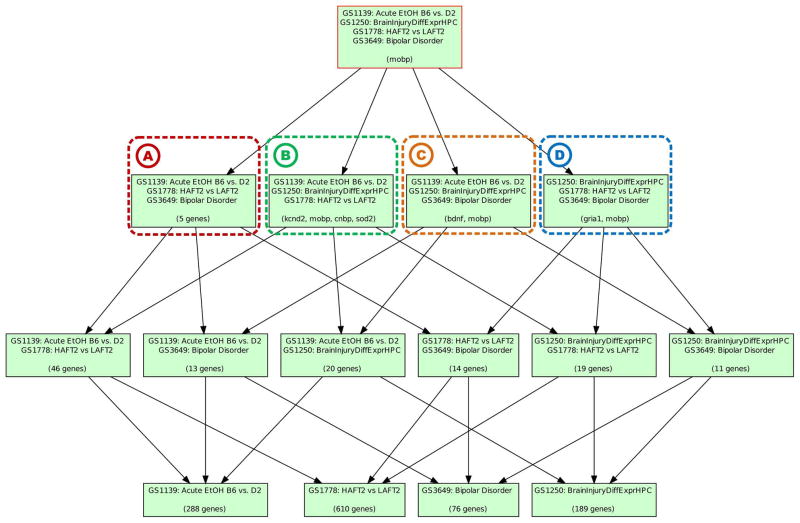
To integrate these data sets, an analysis of higher order intersections was performed using the PhISH tool, which enumerates and illustrates all intersections. Results of the PhISH analysis of these data sets (Figure 2) highlight gene-phenotype relationships based on empirically derived heterogeneous data sets. The hierarchical distribution of intersections demonstrate a separation of genes into distinct categories that reflect underlying phenotypic states; genes involved in neural function, oxidative stress, depression, or mania emerge as a part of the empirically created ontology. In the root node a genetic singularity converges on mobp, a gene with demonstrated increased levels in schizophrenia patients with a history of substance abuse [27].
Figure 2. PhISH representation of gene-phenotype interactions.
Genes sets representing a variety of phenotype states, including ethanol exposure in the mouse, GS1139 [25] and GS1778 [24], mouse bipolar candidate genes, GS1250 [10], and differential gene expression in the rat following traumatic brain injury, GS3649 [26], were analyzed using bipartite association matrices, and organized in a PhISH diagram. The resulting intersections demonstrate a separation of genes into distinct categories that reflect initial phenotype sets. Genes involved in (A) neural function, (B) oxidative stress, (C) depression-related, and (D) mania-related emerge as a part of the empirically created ontology. In the root node, genes converge on the gene mobp, a gene with demonstrated increased levels in schizophrenic patients with a history of substance abuse [27]. Significance of the network is ascertained through a permutation test of two parameters. The phenotype parsimony is normal and non-significant due to the presence of all combinations of phenotypes (p=1.0, n=50,000). The second measurement determines if there is more gene overlap in node intersections than expected by random chance. This is significant since, given a few thousand genes, there are more overlaps than predicted during permutation analysis (p=5.99988 · 10−5, n=50,000).
Significance of the tree is ascertained by examining phenotype parsimony and node overlap parameters. After permutation testing the parsimony value, which is reflected in the shape of the tree, is found to be normal and non-significant due to the presence of all combinations of phenotypes (p=1.0, n=50,000). The second measurement determines if there is more gene overlap in node intersections than expected by random chance. This is significant since, given multiple permutation tests, there are more observed overlaps than expected (p=5.99988 · 10−5, n=50,000).
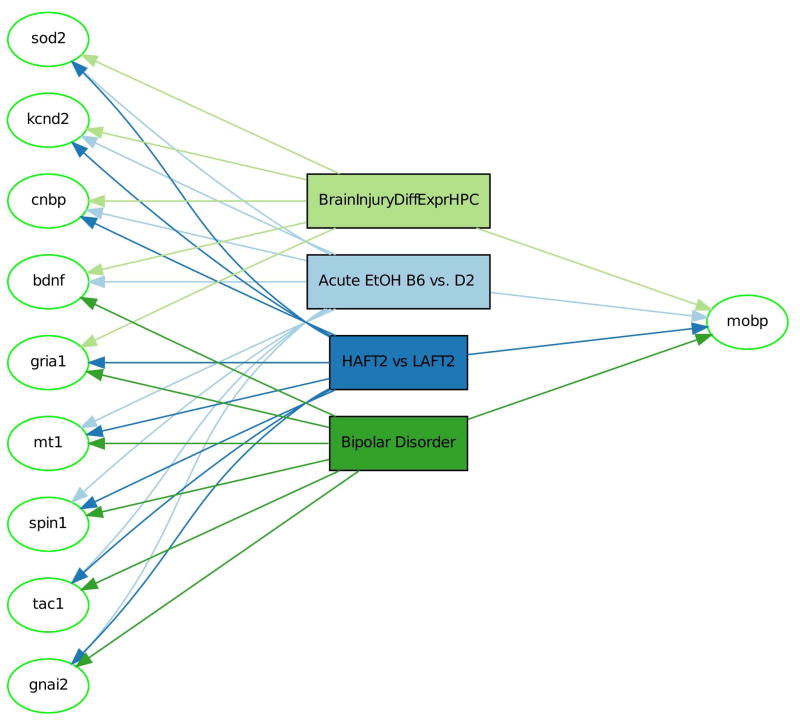
Interactive visualization of the gene-phenotype association bi-partite graph (Figure 3) reveals highly connected (high-degree) gene nodes, and the pattern of gene-phenotype aggregation. A degree threshold can be set to filter out low-degree nodes, i.e. those genes which are connected to only a small number of phenotypes. Selection of a gene node can be used to perform a search for additional connected phenotypes.
Figure 3. Overlap Visualization.
Genes are laid out from left to right in increasing overlap with data sets. Each gene is hyperlinked to search for related data sets.
ODE creates an environment in which data from existing, phenomenologically-driven genomic analysis can be integrated for a simultaneous and seamless analysis of instances (genes – traits associations) and ontologies (classes of genes and traits). Using ODE, a natural organization of complex traits such as basal and alcohol related behavioral processes may be elucidated, thereby reflecting common biological substrates for the relevant behaviors. By integrating genome-wide empirical associations, new information may be added to known pathways and novel relations may be revealed. The goal is not biochemical reaction or interaction analysis, but rather, to ask fundamental questions about the relations among behavioral processes such as stress response and alcohol consumption, or learning and addiction. Thus, the arbitrary and incomplete nature of experimental pathway data is not an impediment. By making use of a “gene and gene product parts list” that is empirically associated with a phenotype, common components can be identified and used to identify relations among any process. The relations of common components form a rational ontology, and can be identified through strictly empirical approaches. This enables well-studied biological and behavioral constructs to be mapped to actual biological processes, pathways and systems.
The ODE has numerous applications. The tool can be used for convergent validation of experimental results, validation of biological assays as metrics of related phenotypes, translational analysis for validation of animal models and treatments designed to mimic human disease and identification of candidate genes from among a list of positional candidates found in quantitative trait locus analysis and linkage analysis. Links to other resources from inside the tool facilitate annotation and aggregation of additional information around discovered networks. This interactive environment with features for storage and sharing of interim results can support integration of diverse data across interdisciplinary collaborative efforts. Indeed, ODE-associated tools may be extended to include alternative methods to test associations between disparate sources using a variety of statistical tests, such as edge permutation and node label permutation tests [28].
A property of phenome ontology we find exciting is its ability to create ontologies that can be mapped, linked and aligned. Previous attempts at ontological alignment have focused on semantic equalities [29]. These approaches, however, are subject to lexical and data prejudice. Using inter-species homology translations, along with a consequent mapping of a variety of annotations, will enable empirically based ontology alignments and, perhaps, a convergence of the vast numbers of community ontologies being created. Through the process of ontological discovery from empirical observation, we believe that a fundamental reclassification of disease based on biological substrate, rather than external manifestation will one day be possible. This will enable biologists and clinicians to define the effects of genetic diversity, environmental perturbation and points of therapeutic intervention in terms of the functional processes underlying diverse mechanisms of disease rather than in terms of the often convergent outputs of these diverse perturbations.
Methods and Materials
Data Structure and Interoperability
ODE’s organizing metaphor is the gene and the subsequent superset of gene-sets and sets of gene-sets. Consequently, ODE accepts gene sets generated through any methodology dedicated to gene-network creation. For example, gene sets may defined from public microarray data including the Genome Institute of Novartis tissue specific gene expression data [30], MGI tables of phenotypic alleles, Gene Network’s genetic correlation to gene expression [31], literature associations obtain via text mining using bibliographic similarity based approaches [32] or Latent-Semantic Indexing [33], and even hand curated NCBI’s Gene Reference Into Function. A higher order and somewhat less empirical class of gene lists comes from numerous literature reviews and hypothesis-based studies in which researchers have compiled gene lists involved in various behavioral constructs including pain [34], aggression [35], alcohol specific [25], and drug abuse [36], among others. In addition, GAGGLE integration through FireGoose [37] enables bi-directional ODE interface with MeV, R, Cytoscape or other sites such as DAVID [38], STRING [39], or KEGG [4]. Novel gene sets are also dynamically generated as a function of the analysis tools, iteratively optimized by users, and edited to create new sets of genes.
The software environment attempts to alleviate data incompatibility through the collection of metadata and pooling community gene annotation information. Metadata is collected during gene upload, using a web-based form designed to maximize free-form, ontological, and publication-centric information. For example, a PubMed ID (PMID) is sufficient to extract published information associated with the data set of interest and asynchronous tree menus allow users to assign multiple observations from community ontologies [3, 40] that may be used to describe their data. The use of existing OBOs means that metadata is extensible to any number of emerging ontologies and allows gene sets to be searched via a variety of biologically-relevant relationships. Plasticity in ontology metadata also allows the ontological alignment between different organisms, community ontology efforts, and experimental data sets.
Gene identifiers used in upload can come from a variety of databases, which are filtered based on the species and identifier type provided by the user during upload. The ODE upload process maps uploaded genes to the species’ reference database identifier (i.e. HGNC, MGI, RGD, etc.). If there is no reference identifier, the next most unique identifier is used (typically Entrez or Ensembl identifiers). This process ensures that ambiguous gene symbols from different species are kept distinct in the database. During analysis, gene name collisions across species are avoided by feeding unique ODE GENE IDs to the analysis tools. Homology relations are established using Homologene tables, though other mappings can be easily incorporated into the software. Once complete, the results are post-processed for on-screen display to add gene names.
To insulate against rapid changes in the underlying technology, the ODE web interface is built on standardized and open source middleware and server-side development tools. Database interaction and HTML production is handled by PHP 5 and CSS. Dynamic client-side objects are achieved through javascript and asynchronous client-server interactions (AJAX), where appropriate, and web security is offered through https protocols. Pages are served using Apache Server 2.0 [41], while dynamic database accessibility is provided by PostgreSQL v 8.0, an object-relational active database that provides lightweight, but robust, data consistency. Data interoperability with the ODE environment is further enhanced through the use of XML. The modular implementation of ODE’s web interface allows dynamic access to multiple tools in isolation of the set-enabled data structures paradigm. Documentation and tutorials are available on the site in the form of a quick-start guide, interactive help, and a narrated movie demonstration.
Analysis Tools
Phenome Interdependency and Similarity Hierarchy
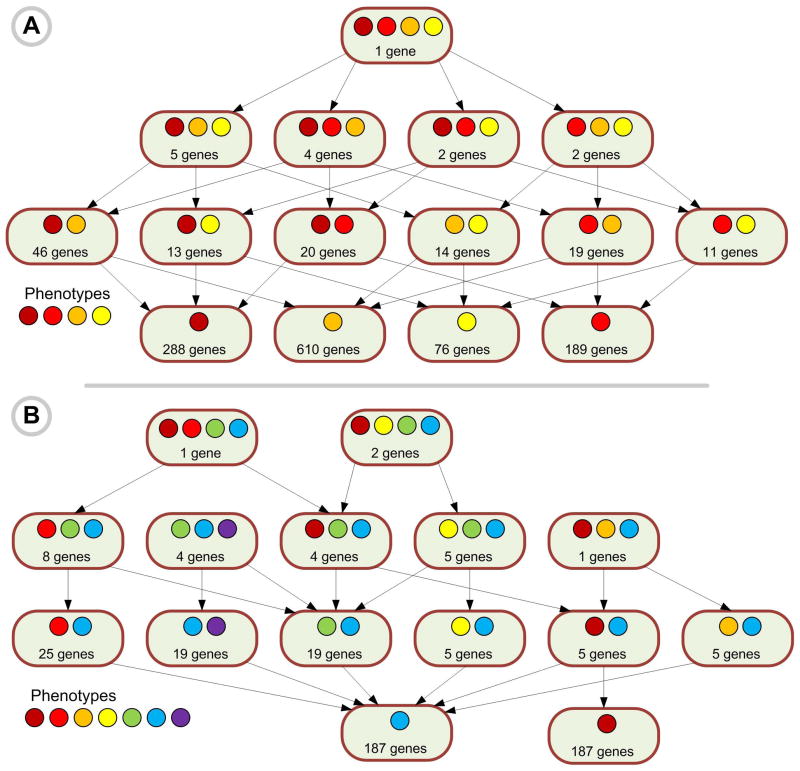
The ultimate goal of ODE is to construct empirically-derived phenome ontologies based on user-submitted and dynamically-generated sets of genes, displayed by the ODE as a Phenome Interdependency and Similarity Hierarchy (PhISH). Creating a PhISH graph is computationally challenging but solvable due to recent advances in algorithms for bipartite graph analysis [42]. Briefly, phenotype supersets are defined by common connections to a gene or genes (Figure 4). These sets reside in the root node of an is-a hierarchy for the classification of phenotypes. Subsets are defined by connections to additional genes. These child nodes are associated with the same biological networks as the parent node, but are also connected to additional genes. Node splitting rules based on similarity, and stopping rules based on node size, are applied to limit the growth and density of the tree. To enhance the multi-domain integration of divergent data types, this approach using bipartite graphs employs discrete associations, of which types and thresholds may be defined by the user.
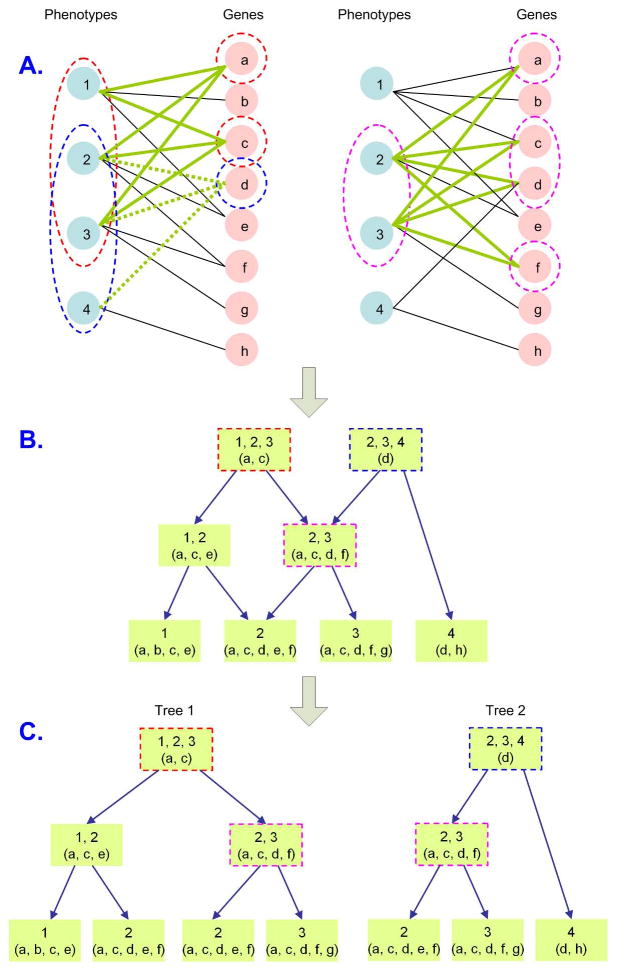
Figure 4. Creation of Phenome Interdependency and Similarity Hierarchy (PhISH).
Phenotype supersets are defined by common connections to a gene or genes. These sets reside in the root node of an is-a hierarchy for the classification of phenotypes. Subsets are defined by connections to additional genes. These child nodes are associated with the same biological networks as the parent node, but are also connected to additional genes. Node splitting rules based on similarity, and stopping rules based on node size, are applied to limit the growth and density of the tree. (A) Gene-phenotype bipartite graph and three maximal bicliques, (B) Representation of the Phenome Map as a DAG of all maximal bicliques, (C) Decomposition of the DAG into trees for each root (node of indegree 0).
Information Condensation
The automated and semi-automated creation of models requires algorithms that ensure users the ability to rapidly gauge the context and confidence of results. We recognize that the literature describing statistical significance of network relationships within fixed data sets remains unresolved, and attempt to provide qualifying, if not deterministic, measurements of dynamic result sets. This is achieved by measuring characteristics representative of information aggregation occurring at the level of genes and phenotypes and applying permutation tests or other metrics to determine the chance occurrence of similar results. For example, the goal of phenome information aggregation in a bipartite graph or biclique is to minimize the number of intersections present, meaning that a large number of phenotypes were reduced to a limited set of categories based on shared biological substrates. In practical terms this is viewed as the parsimony of the phenome map, represented by (Eq. 1.1) and (Eq. 1.2) where Phenotypes is number of genes in an input set.
| Eq. 1.1 |
| Eq. 1.2 |
Here, larger values reflect the greater aggregation or condensation of phenotypes. From this perspective, a single root containing all phenotypes is an optimal result with maximal aggregation. According to (Eq. 1.2) it is apparent that even the addition of a single disjoint phenotype substantially reduces parsimony. Figure 5 demonstrates how parsimony is a generalization of the PhISH diagram shape, where irregular graph distributions have lower phenotype aggregation values and may be assigned probability values based on permutation tests.
Figure 5. Phenotype Condensation.
A single root containing all phenotypes is an optimal result of PhISH analysis with maximal aggregation, but may produce a tree with a non-random distribution of nodes. This schematic depicts how phenotype aggregation is a generalization of the PhISH diagram shape. (A) When all phenotypes are represented, the tree is not-significantly more aggregated than chance trees when compared to a randomized background with the same edge density in a permutation test (p=0.99994, n=50,000). (B) Irregular graph distributions have lower phenotype aggregation values and may be more unusual after permutation testing (p = 0, n = 50,000)
PhISH Permutation Tests
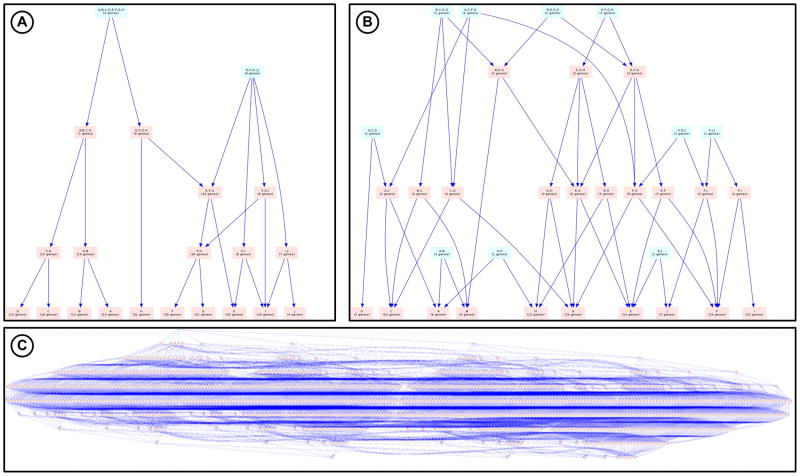
Permutation tests were performed to place gene aggregation and phenotype aggregation into statistical context and to determine how the topology of the PhISH diagram deviates from random [43]. Here, genes and phenotypes are shuffled within the information set, keeping the same overall density of gene-phenotype connections. Simulations against randomized data sets have a two-fold benefit. First, it enables assessment of the impact of false positive and false negative information on the resulting graph. The addition of false positive gene-phenotype associations adds links and nodes, connecting non-overlapping pairs of phenotypes, condensing two 2-phenotype nodes into a single 3-phenotype node, for example. In general this produces a taller tree that approaches the maximal phenotype aggregation value of a regular tree where all combinations of phenotypes are represented. Adding false negatives breaks links and removes nodes, deconstructing a tree into the minimal aggregation of all input phenotypes represented by a completely disjoint tree. These effects of permutation testing are described for a synthetic data set in Figure 6. Secondly, permuting a known data set n number of times produces a distribution of phenotype aggregation values allowing the probability measurement of the observed values.
Figure 6. Implications of false positive and false negative information.
These images show a synthetic dataset with high overlap between 10 sets taken from a possible 1000 genes. (A) shows the original PhISH diagram. After simulating 50% false negatives in a graph of the same size and density, the resulting PhISH diagram (B) has multiple root nodes (split from the original root) and overall smaller phenotype overlap due to the lack of gene aggregation. (C) shows the diagram after simulating 50% false positives in a graph of the same size and density, which results in a much wider diagram with many nodes due to the large number of combinations of gene aggregation possible from such a high overlap.
Another property of interest is overlap, or the density of gene-phenotype associations. This is calculated per node and aggregated across the entire tree. Based on the density of intersections of any sets of genes, we compute the exact probability of obtaining a result of higher or lower overlap. The scores of individual bicliques (Eq. 2.1) are combined across all sets in the entire tree (Eq. 2.1), where Geneschildren is the number of genes in the union of all children of a biclique node. Either result is desirable depending on the user’s goal of identifying common or unique substrates.
| Eq. 2.1 |
| Eq. 2.2 |
Set Analysis Tools
ODE’s analysis tools build on maturing approaches to set analysis, specifically, on a variant of the binomial or hypergeometric test to determine whether members of each category are over-represented among a list of genes. ODE adds the Jaccard positive-match coefficient as a metric of set similarity, because this measure is not upwardly biased by a high rate of true negative results found in comparison of sparse sets. GoTree Machine was among the first to use a reference set [9] to estimate whether category members were over-represented among a list of genes relative to possible representation from the set of genes considered. Newer tools, such as ErmineJ, take advantage of the entire vector of gene expression values rather than forcing the gene set to have a categorical representation [7]. Both standalone and web-based tools exist, but most of them simply allow an identification of relations to a single user entered gene set, or a limited group of gene sets, with a very limited set of functions facilitating union and intersection analysis. For example, existing tools allow one to ask questions such as, “Does this set of genes differentially expressed in response to stressors correspond to any known pathways or categories?” In contrast, ODE tool variants expand upon this approach to include matching sets of sets to other sets of sets, for example, by asking “Do stress related gene sets have any common relationships with alcohol consumption related gene sets?” Using hypergeometric, Jaccard similarity and distance, and fisher tests produces a high-level view of the landscape of gene relationships represented in the test set and, while not required to construct PhISH graphs, these gene set similarity matrices provide inputs to clustering methods and act as filters for empirical ontology classifications.
Acknowledgments
This work is a project of the Integrative Neuroscience Initiative on Alcoholism and is supported by NIH U01AA13499, U24AA13513.
Abbreviations
- ODE
Ontological Discovery Environment
- PhISH
Phenome Integration and Similarity Hierarchy
- MPO
Mammalian Phenome Ontology
- PATO
Phenotype and Trait Ontology
- QTL
Quantitative Trait Locus
- KEGG
Kyoto Encyclopedia of Gene and Genomes
Footnotes
Availability: http://ontologicaldiscovery.org
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith CL, Goldsmith CA, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005;6(1):R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gkoutos GV, Green EC, Mallon AM, Hancock JM, Davidson D. Building mouse phenotype ontologies. Pac Symp Biocomput. 2004:178–189. doi: 10.1142/9789812704856_0018. [DOI] [PubMed] [Google Scholar]
- 3.Ashburner M, Ball CA, Blake JABD, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28 (1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31(1):19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 6.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4(10):R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee HK, Braynen W, Keshav K, Pavlidis P. ErmineJ: Tool for functional analysis of gene expression data sets. BMC Bioinformatics. 2005;6(1):269. doi: 10.1186/1471-2105-6-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H, Hu ZZ, Wu CH. DynGO: a tool for visualizing and mining of Gene Ontology and its associations. BMC Bioinformatics. 2005;6:201. doi: 10.1186/1471-2105-6-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5(1):16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le-Niculescu H, McFarland MJ, Mamidipalli S, Ogden CA, Kuczenski R, Kurian SM, Salomon DR, Tsuang MT, Nurnberger JI, Jr, Niculescu AB. Convergent Functional Genomics of bipolar disorder: from animal model pharmacogenomics to human genetics and biomarkers. Neurosci Biobehav Rev. 2007;31(6):897–903. doi: 10.1016/j.neubiorev.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bogue MA, Grubb SC, Maddatu TP, Bult CJ. Mouse Phenome Database (MPD) Nucleic Acids Res. 2007;35(Database issue):D643–649. doi: 10.1093/nar/gkl1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-Ricaud L, Warringer J, Ericson E, Glaab K, Davidsson P, Nilsson F, Kemp GJ, Nerman O, Blomberg A. PROPHECY--a yeast phenome database, update 2006. Nucleic Acids Res. 2007;35 (Database issue):D463–467. doi: 10.1093/nar/gkl1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuya H, Yoshikawa S, Heida N, Toyoda T, Wakana S, Shiroishi T. Phenosite: a web database integrating the mouse phenotyping platform and the experimental procedures in mice. J Bioinform Comput Biol. 2007;5(6):1173–1191. doi: 10.1142/s0219720007003168. [DOI] [PubMed] [Google Scholar]
- 14.Muilu J, Peltonen L, Litton JE. The federated database--a basis for biobank-based post-genome studies, integrating phenome and genome data from 600,000 twin pairs in Europe. Eur J Hum Genet. 2007;15(7):718–723. doi: 10.1038/sj.ejhg.5201850. [DOI] [PubMed] [Google Scholar]
- 15.Toyoda T, Mochizuki Y, Player K, Heida N, Kobayashi N, Sakaki Y. OmicBrowse: a browser of multidimensional omics annotations. Bioinformatics. 2007;23(4):524–526. doi: 10.1093/bioinformatics/btl523. [DOI] [PubMed] [Google Scholar]
- 16.van Driel MA, Bruggeman J, Vriend G, Brunner HG, Leunissen JA. A text-mining analysis of the human phenome. Eur J Hum Genet. 2006;14(5):535–542. doi: 10.1038/sj.ejhg.5201585. [DOI] [PubMed] [Google Scholar]
- 17.Kim WK, Krumpelman C, Marcotte EM. Inferring mouse gene functions from genomic-scale data using a combined functional network/classification strategy. Genome Biol. 2008;9(Suppl 1):S5. doi: 10.1186/gb-2008-9-s1-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pena-Castillo L, Tasan M, Myers CL, Lee H, Joshi T, Zhang C, Guan Y, Leone M, Pagnani A, Kim WK, et al. A critical assessment of Mus musculus gene function prediction using integrated genomic evidence. Genome Biol. 2008;9 (Suppl 1):S2. doi: 10.1186/gb-2008-9-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Li X, Su H, Chen H, Galbraith DW. A framework of integrating gene relations from heterogeneous data sources: an experiment on Arabidopsis thaliana. Bioinformatics. 2006;22 (16):2037–2043. doi: 10.1093/bioinformatics/btl345. [DOI] [PubMed] [Google Scholar]
- 20.Oti M, Huynen MA, Brunner HG. Phenome connections. Trends Genet. 2008;24(3):103–106. doi: 10.1016/j.tig.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104(21):8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Downey RG, Fellows MR. Parameterized Complexity. Springer-Verlag; 1999. [Google Scholar]
- 23.Zhang Y, Abu-Khzam FN, Baldwin NE, Chesler EJ, Langston MA, Samatova NF. Genome-Scale Computational Approaches to Memory-Intensive Applications in Systems Biology. Super Computing. 2005 [Google Scholar]
- 24.Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, et al. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci U S A. 2006;103(16):6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RW, Miles MF. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25(9):2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matzilevich DA, Rall JM, Moore AN, Grill RJ, Dash PK. High-density microarray analysis of hippocampal gene expression following experimental brain injury. J Neurosci Res. 2002;67 (5):646–663. doi: 10.1002/jnr.10157. [DOI] [PubMed] [Google Scholar]
- 27.Mitkus SN, Hyde TM, Vakkalanka R, Kolachana B, Weinberger DR, Kleinman JE, Lipska BK. Expression of oligodendrocyte-associated genes in dorsolateral prefrontal cortex of patients with schizophrenia. Schizophr Res. 2008;98(1–3):129–138. doi: 10.1016/j.schres.2007.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balasubramanian R, LaFramboise T, Scholtens D, Gentleman R. A graph-theoretic approach to testing associations between disparate sources of functional genomics data. Bioinformatics. 2004;20(18):3353–3362. doi: 10.1093/bioinformatics/bth405. [DOI] [PubMed] [Google Scholar]
- 29.Marquet G, Mosser J, Burgun A. A method exploiting syntactic patterns and the UMLS semantics for aligning biomedical ontologies: The case of OBO disease ontologies. Int J Med Inform. 2007;76 (Suppl 3):S353–361. doi: 10.1016/j.ijmedinf.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 30.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99(7):4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chesler EJ, Wang J, Lu L, Qu Y, Manly KF, Williams RW. Genetic correlates of gene expression in recombinant inbred strains: a relational model system to explore neurobehavioral phenotypes. Neuroinformatics. 2003;1(4):343–357. doi: 10.1385/NI:1:4:343. [DOI] [PubMed] [Google Scholar]
- 32.Blaschke C, Valencia A. Automatic ontology construction from the literature. Genome Inform Ser Workshop Genome Inform. 2002;13:201–213. [PubMed] [Google Scholar]
- 33.Homayouni R, Heinrich K, Wei L, Berry MW. Gene clustering by latent semantic indexing of MEDLINE abstracts. Bioinformatics. 2005;21(1):104–115. doi: 10.1093/bioinformatics/bth464. [DOI] [PubMed] [Google Scholar]
- 34.Belfer I, Wu T, Kingman A, Krishnaraju RK, Goldman D, Max MB. Candidate gene studies of human pain mechanisms: methods for optimizing choice of polymorphisms and sample size. Anesthesiology. 2004;100(6):1562–1572. doi: 10.1097/00000542-200406000-00032. [DOI] [PubMed] [Google Scholar]
- 35.Maxson SC. Searching for candidate genes with effects on an agonistic behavior, offense, in mice. Behav Genet. 1996;26(5):471–476. doi: 10.1007/BF02359751. [DOI] [PubMed] [Google Scholar]
- 36.Kreek MJ, Nielsen DA, LaForge KS. Genes associated with addiction: alcoholism, opiate, and cocaine addiction. Neuromolecular Med. 2004;5(1):85–108. doi: 10.1385/NMM:5:1:085. [DOI] [PubMed] [Google Scholar]
- 37.Bare JC, Shannon PT, Schmid AK, Baliga NS. The Firegoose: two-way integration of diverse data from different bioinformatics web resources with desktop applications. BMC Bioinformatics. 2007;8:456. doi: 10.1186/1471-2105-8-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherman BT, Huang da W, Tan Q, Guo Y, Bour S, Liu D, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics. 2007;8:426. doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25(11):1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fielding RT, Kaiser G. The Apache HTTP Server Project. IEEE Internet Computing. 1997;1(4):88–90. [Google Scholar]
- 42.Zhang Y, Chesler E, Langston M. On finding bicliques in bipartite graphs: a novel algorithm with application to the integration of diverse biological data types. HICSS: 2008; Big Island, Hawaii. 2008 doi: 10.1186/1471-2105-15-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]