Abstract
Although cancers have altered glucose metabolism, termed the Warburg effect that describes the increased uptake and conversion of glucose to lactate by cancer cells under adequate oxygen tension, changes in the metabolism of glutamine and fatty acid have also been documented. The MYC oncogene, which contributes to the genesis of many human cancers, encodes a transcription factor, c-Myc, that links altered cellular metabolism to tumorigenesis. c-Myc regulates genes involved in the biogenesis of ribosomes and mitochondria, and regulation of glucose and glutamine metabolism. With E2F1, c-Myc induces genes involved in nucleotide metabolism and DNA replication, and microRNAs that homeostatically attenuate E2F1 expression. With the hypoxia inducible transcription factor HIF-1, ectopic c-Myc cooperatively induces a transcriptional program for hypoxic adaptation. Myc regulates gene expression either directly, such as glycolytic genes including lactate dehydrogenase A (LDHA), or indirectly, such as repression of microRNAs miR-23a/b to increase glutaminase (GLS) protein expression and glutamine metabolism. Ectopic MYC expression in cancers, therefore, could concurrently drive aerobic glycolysis and/or oxidative phosphorylation to provide sufficient energy and anabolic substrates for cell growth and proliferation in the context of the tumor microenvironment. Collectively, these studies indicate that Myc-mediated altered cancer cell energy metabolism could be translated for the development of new anti-cancer therapies.
Background
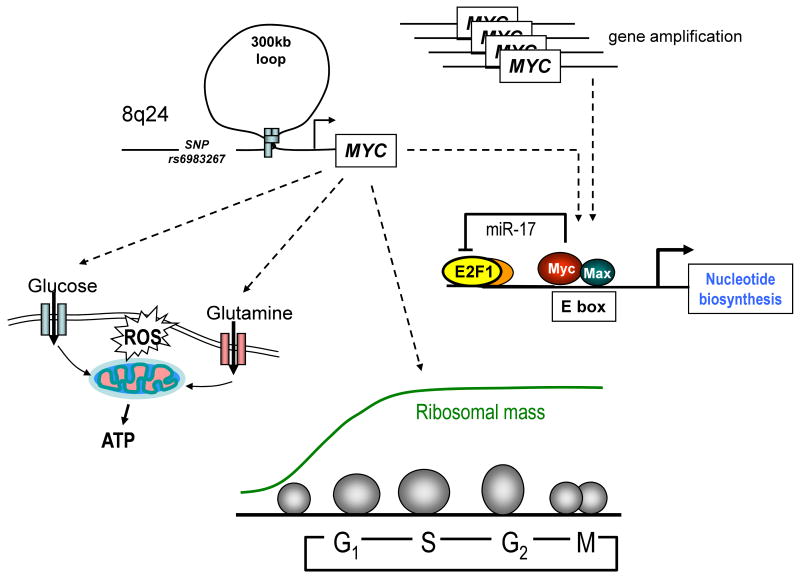
The MYC gene has long been known to be altered by chromosomal translocations and gene amplification in human cancers. In addition, common single nucleotide polymorphisms (SNPs) on human chromosome 8q24, which predispose to colon, breast, prostate and bladder cancers, have been implicated in deregulated MYC expression (1). The region containing 8q24 SNP rs6983267 confers increased cancer risk (odds ratio ∼1.5) and is located >300 kb away from MYC (2). This SNP contributes to a consensus TCF4 binding site and has features of a long-distance regulatory sequence or enhancer that is tethered to the MYC promoter via DNA looping. Therefore, this SNP could increase MYC expression through TCF4 generated by activation of the WNT signaling pathway. In this regard, subtle changes in deregulated MYC expression can have a profound effect on tumorigenesis in animal models(3). MYC is, hence, central to the genesis of most commonly occurring human cancers (Figure 1).
Figure 1. Altered MYC expression and Myc function.
Cancer predisposing SNP r6983267 located >300 kb away is shown tethered to the MYC promoter, altering its expression. MYC gene amplification, which is found in some human cancers, is also illustrated. Myc in turn regulates energy metabolism and ribosomal biogenesis, which provides the bulk of cellular mass through the cell cycle. As cells enter S phase, Myc together with E2F1 regulates nucleotide metabolism with a negative regulatory loop involving miR-17 that attenuates E2F1 protein levels for safe passage through DNA replication.
Given the central role of MYC in many human cancers, it is critical that the c-Myc (herein termed Myc) transcription factor be well-understood, particularly regarding its role in stem cell maintenance and tumorigenesis (4-6). Myc is a helix-loop-helix leucine zipper transcription factor that dimerizes with its partner protein Max to bind specific DNA sequences and transactivates genes. The Myc-Max heterodimer can also repress gene expression through complex formation with the transcription factor Miz1 (5). In addition to its role in cancer, Myc is one of four transcription factors that collectively can re-program differentiated adult cells back to a pluripotent stem cell state (7).
In addition to its role in cancer, Myc also plays an important role in normal cell physiology. The key distinction between physiological and oncogenic Myc function is whether MYC expression is regulated by normal circuitries, such as growth factor signaling that occurs when cells enter into the cell cycle and proliferate for tissue repair or whether, as in cancers, MYC activation can be short-circuited by genetic alterations, permitting deregulated Myc expression to alter transcription that no longer responds to external cues, particularly negative regulatory ones (8). Ectopic Myc expression is normally kept in check by its activation of p53 or Arf, which triggers apoptosis, senescence or cell cycle arrest. In this regard, Myc mediated lymphomagenesis requires p53 or Arf loss of function (9). It has been of major interest to understand the transcriptional program, particularly, downstream of deregulated Myc expression. To this end, many laboratories have recently used high throughput methods to map the Myc target gene network (4, 5).
Intriguingly, multiple studies have demonstrated that Myc can directly bind the promoters of thousands of genes – as high as 30% of all known genes, but only a fraction of the bound genes actually respond (up-regulated or down-regulated) to Myc (10-13). Myc-mediated transcriptional regulation requires other cooperating transcription factors to regulate target genes (10, 11, 14). For example, Myc cooperates with other stem cell transcription factors to regulate genes found in embryonic stem cells (15). Myc cooperates with E2F1 to regulate genes involved in nucleotide metabolism and with the hypoxia inducible factor 1 (HIF-1) to regulate genes involved in glucose metabolism (11, 16, 17).
The key lesson learned from lower organisms and mammals is that while Myc regulates a myriad of genes, a conserved core set of Myc target genes appears to be involved in ribosomal and mitochondrial biogenesis, energy metabolism, and regulation of cell cycle (Figure 1). Direct mapping of dMyc target genes in Drosophila identified CDK4 (18), which was also independently confirmed in mammalian cells as a direct Myc target, thereby linking Myc to cell cycle regulation across species (19). Mutant flies with diminished dMyc function have small cell and body size that pheno-copies mutants with loss of ribosomal protein function, linking Myc to ribosomal biogenesis (20). The finding that Myc could regulate transcription mediated by RNA polymerases I (for rRNA transcription) and III (for tRNA and small RNAs transcription), in addition to RNA Pol II, further demonstrates its role in biogenesis of ribosomes (4, 5, 21). Studies in multiple mammalian systems document a role for Myc in the regulation of genes involved in mitochondrial biogenesis and function, such that increased Myc function is associated with enhanced mitochondrial mass and function (22-25). These studies collectively indicate a key role for Myc in organellar biogenesis that is required for energy production, biosynthesis and cell growth.
Clinical Translational Advances
The Role of Myc in Cancer Energy Metabolism
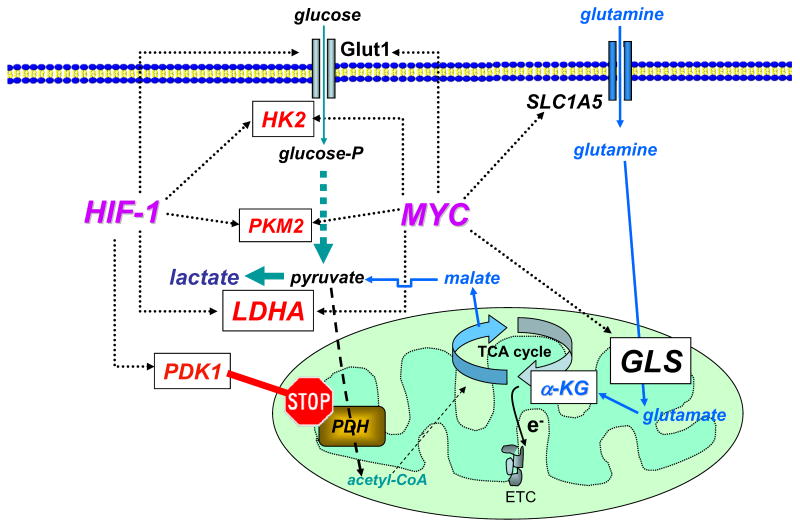
Deregulated MYC expression is found in many commonly occurring human cancers including colon, breast, prostate and bladder cancer. It is estimated that increased MYC expression contributes to the cause of at least 40% of all human cancers (www.myccancergene.org). Early studies established Myc as a transcription factor through the identification of the transactivation domain and specific DNA binding domain (26-28). Subsequent studies of Myc target genes focused on its role in the regulation of the cell cycle, since the genesis of cancers was thought to reside mainly in a deregulated cell cycle machinery. In fact, Myc appears to play a role in DNA replication distinct from its role as a transcription factor (29). The link between Myc and regulation of glucose metabolism was first established when an early unbiased screen for Myc target genes uncovered lactate dehydrogenase A (LDHA) among 20 other putative Myc target genes (30, 31). LDHA converts pyruvate, which is derived from glucose through glycolysis or other sources, to lactate (Figure 2).
Figure 2. Myc and HIF-1 regulate glucose metabolism and stimulate the Warburg effect.
Myc and HIF-1 are depicted to regulate (dotted lines) genes involved in glucose metabolism (glucose transporter Glut1, hexokinase 2 (HK2), pyruvate kinase M2 (PKM2), lactate dehydrogenase A (LDHA), and pyruvate dehydrogenase kinase 1 (PDK1)), favoring the conversion of glucose to lactate (glycolysis). Myc is also depicted to stimulate glutamine metabolism through the regulation of transporters (SLC1A5) and glutaminase (GLS). Glutamine is shown converted to α-ketoglutarate (α-KG) for catabolism through the tricarboxylic acid (TCA) cycle to malate, which is transported into the cytoplasm and converted to pyruvate and then to lactate (glutaminolysis). High energy electrons (e-) from the TCA cycle is shown transported with the electron transport chain (ETC). PDH = pyruvate dehydrogenase.
Many other glucose metabolism genes were subsequently documented to be directly regulated by Myc. Chief among these are the glucose transporter GLUT1, hexokinase 2 (HK2), phosphofructokinase (PFKM), and enolase 1 (ENO1) (32-34). Myc is hence able to stimulate genes that increase the transport of glucose, its catabolism to trioses and pyruvate and ultimately to lactate (Figure 2). Because glycolytic genes are also directly responsive to the hypoxia inducible factor, HIF-1, the interplay between Myc and HIF was documented through genes that could be regulated by both transcription factors (17). Collectively, these studies suggest that HIF-1 transactivates glucose transporter and glycolytic genes in common with Myc; HIF-1 transactivates these genes under hypoxic conditions (anaerobic glycolysis), while Myc regulates the same set of genes under non-hypoxic conditions. These observations imply that Myc could contribute to the Warburg effect (aerobic glycolysis) or the ability to convert glucose to pyruvate and in turn to lactate even under adequate oxygen tension.
The ability of Myc to transactivate genes involve in glycolysis under normal oxygen tension begged the question of whether pyruvate, which is converted to lactate by LDHA, could also be converted to acetyl-CoA and oxidized by increased Myc-mediated mitochondrial biogenesis. In this regard, Li et al. documented that genes involved in mitochondrial biogenesis and function are statistically over-represented amongst Myc target genes (23). They further document through gain-of-function and loss-of-function analysis of Myc, that mitochondrial mass and function correlate with Myc function, a link that was suggested earlier and has since been corroborated by additional studies (22, 24, 25).
Myc not only induces genes that convert glucose to lactate but also those conferring mitochondrial oxidation of substrates via the TCA cycle. What then would happen to glucose metabolism in hypoxic Myc-transformed cancer cells? In this regard, Kim et al. screened for genes that might be in common to Myc and HIF-1 under hypoxic conditions (32, 33, 35). They found that PDK1, encoding pyruvate dehydrogenase kinase 1, was robustly transactivated by HIF-1 and further increased by Myc (Figure 2). They further documented that PDK1, which blocks the conversion of pyruvate to acetyl-CoA by phosphorylation of pyruvate dehydrogenase, inhibits mitochondrial oxidation of pyruvate under hypoxic conditions (35). These observations suggest that Myc could stimulate glucose oxidation and lactate production in normoxia. Under hypoxia, Myc collaborates with HIF-1 to induce PDK1, thereby suppressing mitochondrial respiration and favoring the conversion of glucose to lactate. Decreased expression of PDK1 mediated by siRNA under hypoxic conditions was accompanied by cell death due to oxidative stress from continued mitochondrial respiration under low oxygen tension. Concurrent studies of Papandreou et al. (36) corroborated the significance of these observations, which are underscored by the fact that inhibition of PDK1 by dichloroacetate decrease tumor growth in a xenograft model of lung carcinoma (37).
Because Myc is able to induce mitochondrial biogenesis and oxygen consumption in human B cels, Gao et al. sought to determine the effects of Myc on their mitochondrial proteome (38). Through proteomic analysis of mitochondria from high Myc expressing human B lymphocytes as compared to control lymhocytes, mitochondrial glutaminase (GLS) was among 7 proteins identified with >10-fold induction by Myc. Further analyses revealed that unlike glutamine transporters (ASCT2 and SLC7A25), which are direct Myc target genes, GLS protein level was induced by Myc through direct suppression of microRNAs, miR-23a and miR-23b, which targets the GLS mRNA 3′-UTR. GLS is the first enzyme that converts glutamine to glutamate, which is in turn converted to α-ketoglutarate for further metabolism in the TCA cycle (Figure 2). It is notable that Myc-overexpressing human cell lines were found to be dependent on glutamine, such that its withdrawal triggered apoptosis (39). Furthermore, independent studies by Wise et al. also documented that Myc induces genes involved in glutamine metabolism to confer glutamine-addiction (40). With adequate oxygen tension, α-ketoglutarate derived from glutamine could then be oxidized through the TCA cycle, indicating that Myc can induce glutamine oxidation concurrently with aerobic glycolysis. Studies of a glioblastoma cell line indicate that glutamine can also be converted to lactate through glutaminolysis (41), which was recognized several decades ago as a major mode of glutamine metabolism in certain cell lines (42).
Glutaminolysis involves lactate production from the conversion of glutamine to α-ketoglutarate, which is in turn catabolized through part of the TCA cycle to malate and then transported out of the mitochondrion (Figure 2). Cytoplasmic malic enzyme converts malate to pyruvate with the concomitant production of NADPH from NADP+(43). Pyruvate is then converted to lactate by LDHA. While a portion of lactate is produced from glutamine in selected cancer cell lines in vitro (41, 44), the context as to when glutaminolysis plays a role in cancer metabolism in vivo remains to be established. Notwithstanding these caveats, Myc is able to induce the expression of genes involved in both glycolysis and glutaminolysis with LDHA being critical for both processes.
Cancer Cell Metabolism and Therapeutic Opportunities
In addition to the induction of glucose and glutamine metabolic enzyme genes, Myc also induces genes involved in nucleotide metabolism and polyamine synthesis. With E2F1, Myc regulates genes involved in nucleotide metabolism and DNA replication, as well as microRNAs that homeostatically attenuate E2F1 expression for safe passage of cells through S phase (11, 45-47). Interference with this microRNA (miR-17 cluster) circuitry results in DNA replication stress (48). Nucleotide metabolism has been a key target for cancer therapeutics over the last five decades, culminating in many agents, such as 5-fluorouracil and nucleosides that are part of the current therapeutic armamentarium. The new understanding that an important oncogene directly regulates multiple metabolic pathways suggests that new opportunities exist, particularly in altered cancer energy metabolism.
A number of small organic molecules have been reported to target glycolysis, but none to date has been shown to have specific molecular targets. For example, 3-bromopyruvate, a highly active akylating agent, is reported to target HK2, but to date little biochemical evidence is available to support this claim (49). In fact, recent studies have implicated GAPDH as a potential target of 3-bromopyruvate (50). Although 2-deoxyglucose can be phosphorylated by HK2 and in turn inhibit HK2, it has non-glycolytic effects (51). Other attractive targets for therapy include pyruvate kinase M2 (PKM2), which converts phosphoenolpyruvate to pyruvate (52-54) and LDHA, because three independent studies have demonstrated that loss of LDHA function results in dramatically diminished cellular transformation or xenograft tumor growth (31, 55, 56).
Upon its identification as a direct Myc target in 1997, anti-sense mediated suppression of LDHA expression in several human lymphoid tumor cell lines markedly decreased soft agar colony growth (31). This study also suggested that LDHA is necessary for adaptation to hypoxia resulting from growth of spheroid cellular masses in soft agar. Indeed, Fantin et al. later demonstrated that stable interference RNA (shRNA) mediated knock-down of LDHA expression in mouse mammary tumor cells resulted in prolonged survival of tumor inoculated animals as compared with those inoculated with control cells (55). They also documented that decreased LDHA expression was associated with increased mitochondrial respiration, but neither the effect on reactive oxygen species (ROS) production nor the mechanism for cellular toxicity due to lowered LDHA expression was reported. Similar to PDK1 inhibition, which enhances pyruvate oxidation in hypoxia and ROS, lowered LDHA expression could also result in oxidative stress and ensuing apoptosis. More recently, shRNA mediated knock-down of LDHA in a lung carcinoma xenograft model was also documented to inhibit tumor xenograft growth (56). In aggregate, these studies provide proof-of-concept that targeting LDHA could be a fruitful avenue, particularly since humans genetically lacking LDHA are viable and normal except for exercise-induced myoglobinuria. Our preliminary studies (Le and Dang, unpublished observations) suggest that small organic molecules capable of inhibiting human LDHA (57) could inhibit in vivo xenograft tumor growth of human B lymphoid tumor and pancreatic cancer cells, paving the way for further development of therapeutic LDHA inhibitors.
As discussed, glutamine metabolism is an important pathway regulated by Myc and glutaminase was documented to be required for the proliferation of human B lymphoid tumor cells and the prostate PC3 cancer cell lines, suggesting that it could be a key target for therapy (38). Furthermore, anti-sense reduction of glutaminase expression diminishes the in vivo tumorigenicity of Erhlich ascites tumor (58, 59). In this regard, it is notable that the anti-leukemic effect of L-asparaginase in childhood acute lymphocytic leukemia is due to the associated glutaminase activity, which diminishes circulating glutamine levels and deprives leukemic cells of a major energy and anabolic substrate (60). Unfortunately, 6-diazo-5-oxo-l-norleucine (DON) and acivicin, which are both glutamine analogs, have been met with significant central nervous system (CNS) side effects (61). Glutamine is taken up by neurons and converted by glutaminase to glutamate, a major neurotransmitter, which is released into the synaptic cleft and then rapidly cleared to prevent prolonged, toxic neuronal stimulation. In this regard, specific inhibitors of glutaminase or glutamate dehydrogenase may prove to be more useful for non-CNS cancers if the inhibitors could not cross the blood brain barrier (62).
Conclusions
The molecular biology revolution has taken modern cancer biology from the discovery of oncogenes and tumor suppressors back to the past with direct links of these genetic alterations to altered cancer cell metabolism, which was first described by Otto Warburg over 80 years ago. In this regard, Myc-mediated alterations in glucose and glutamine metabolism provide fertile ground for the development of a new class of anti-cancer drugs, which are likely to emerge in the clinics over the next 5 to 10 years.
Acknowledgments
We apologize for the omission of primary references due to space limitation. We thank Lawrence Gardner, Linda Lee and Peng Sun for comments. Our original work is supported by the Leukemia Lymphoma Society, the National Cancer Institute, and National Institutes of Health.
References
- 1.Wokolorczyk D, Gliniewicz B, Sikorski A, et al. A range of cancers is associated with the rs6983267 marker on chromosome 8. Cancer Res. 2008;68:9982–6. doi: 10.1158/0008-5472.CAN-08-1838. [DOI] [PubMed] [Google Scholar]
- 2.Tuupanen S, Turunen M, Lehtonen R, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009 doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 3.Murphy DJ, Junttila MR, Pouyet L, et al. Distinct thresholds govern Myc's biological output in vivo. Cancer Cell. 2008;14:447–57. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–64. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 5.Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–66. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng H, Ying H, Yan H, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–33. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Chung HJ, Levens D. c-myc expression: keep the noise down! Mol Cells. 2005;20:157–66. [PubMed] [Google Scholar]
- 9.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–69. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–61. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeller KI, Zhao X, Lee CW, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103:17834–9. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer. 2004;4:562–8. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez PC, Frank SR, Wang L, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–29. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kidder BL, Yang J, Palmer S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS ONE. 2008;3:e3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medeiros RB, Papenfuss KJ, Hoium B, et al. Novel sequential ChIP and simplified basic ChIP protocols for promoter co-occupancy and target gene identification in human embryonic stem cells. BMC Biotechnol. 2009;9:59. doi: 10.1186/1472-6750-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JW, Dang CV. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–30. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 17.Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–6. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 18.Orian A, van Steensel B, Delrow J, et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003;17:1101–14. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermeking H, Rago C, Schuhmacher M, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci U S A. 2000;97:2229–34. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–90. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenneth NS, White RJ. Regulation by c-Myc of ncRNA expression. Curr Opin Genet Dev. 2009;19:38–43. doi: 10.1016/j.gde.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 22.Kim J, Lee JH, Iyer VR. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS ONE. 2008;3:e1798. doi: 10.1371/journal.pone.0001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F, Wang Y, Zeller KI, et al. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–34. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrish F, Giedt C, Hockenbery D. c-MYC apoptotic function is mediated by NRF-1 target genes. Genes Dev. 2003;17:240–55. doi: 10.1101/gad.1032503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrish F, Neretti N, Sedivy JM, Hockenbery DM. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle. 2008;7:1054–66. doi: 10.4161/cc.7.8.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato GJ, Barrett J, Villa-Garcia M, Dang CV. An amino-terminal c-myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990;10:5914–20. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blackwood EM, Lugo TG, Kretzner L, et al. Functional analysis of the AUG- and CUG-initiated forms of the c-Myc protein. Mol Biol Cell. 1994;5:597–609. doi: 10.1091/mbc.5.5.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blackwell TK, Kretzner L, Blackwood EM, Eisenman RN, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–51. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 29.Dominguez-Sola D, Ying CY, Grandori C, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–51. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 30.Lewis BC, Shim H, Li Q, et al. Identification of putative c-Myc-responsive genes: characterization of rcl, a novel growth-related gene. Mol Cell Biol. 1997;17:4967–78. doi: 10.1128/mcb.17.9.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shim H, Dolde C, Lewis BC, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94:6658–63. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–93. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim JW, Zeller KI, Wang Y, et al. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol Cell Biol. 2004;24:5923–36. doi: 10.1128/MCB.24.13.5923-5936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osthus RC, Shim H, Kim S, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 35.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–85. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–97. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Bonnet S, Archer SL, Allalunis-Turner J, et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11:37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 38.Gao P, Tchernyshyov I, Chang TC, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–5. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wise DR, DeBerardinis RJ, Mancuso A, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–7. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeBerardinis RJ, Mancuso A, Daikhin E, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–50. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem. 1979;254:2669–76. [PubMed] [Google Scholar]
- 43.Moreadith RW, Lehninger AL. The pathways of glutamate and glutamine oxidation by tumor cell mitochondria. Role of mitochondrial NAD(P)+-dependent malic enzyme. J Biol Chem. 1984;259:6215–21. [PubMed] [Google Scholar]
- 44.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu YC, Li F, Handler J, et al. Global regulation of nucleotide biosynthetic genes by c-Myc. PLoS ONE. 2008;3:e2722. doi: 10.1371/journal.pone.0002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mannava S, Grachtchouk V, Wheeler LJ, et al. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle. 2008;7:2392–400. doi: 10.4161/cc.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 48.Pickering MT, Stadler BM, Kowalik TF. miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate cell cycle progression. Oncogene. 2009;28:140–5. doi: 10.1038/onc.2008.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathupala SP, Ko YH, Pedersen PL. Hexokinase-2 bound to mitochondria: cancer's stygian link to the “Warburg Effect” and a pivotal target for effective therapy. Semin Cancer Biol. 2009;19:17–24. doi: 10.1016/j.semcancer.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pereira da Silva AP, El-Bacha T, Kyaw N, et al. Inhibition of energy-producing pathways of HepG2 cells by 3-bromopyruvate. Biochem J. 2009;417:717–26. doi: 10.1042/BJ20080805. [DOI] [PubMed] [Google Scholar]
- 51.Ralser M, Wamelink MM, Struys EA, et al. A catabolic block does not sufficiently explain how 2-deoxy-D-glucose inhibits cell growth. Proc Natl Acad Sci U S A. 2008;105:17807–11. doi: 10.1073/pnas.0803090105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–3. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 53.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–6. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 54.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–34. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 56.Xie H, Valera VA, Merino MJ, et al. LDH-A inhibition, a therapeutic strategy for treatment of hereditary leiomyomatosis and renal cell cancer. Mol Cancer Ther. 2009;8:626–35. doi: 10.1158/1535-7163.MCT-08-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deck LM, Royer RE, Chamblee BB, et al. Selective inhibitors of human lactate dehydrogenases and lactate dehydrogenase from the malarial parasite Plasmodium falciparum. J Med Chem. 1998;41:3879–87. doi: 10.1021/jm980334n. [DOI] [PubMed] [Google Scholar]
- 58.Alonso FJ, Segura JA, Lora J, et al. Sensitisation of Ehrlich ascitic tumour cells to methotrexate by inhibiting glutaminase. Anticancer Res. 2005;25:3315–20. [PubMed] [Google Scholar]
- 59.Lobo C, Ruiz-Bellido MA, Aledo JC, Marquez J, Nunez De Castro I, Alonso FJ. Inhibition of glutaminase expression by antisense mRNA decreases growth and tumourigenicity of tumour cells. Biochem J. 2000;348(Pt 2):257–61. [PMC free article] [PubMed] [Google Scholar]
- 60.Ollenschlager G, Roth E, Linkesch W, Jansen S, Simmel A, Modder B. Asparaginase-induced derangements of glutamine metabolism: the pathogenetic basis for some drug-related side-effects. Eur J Clin Invest. 1988;18:512–6. doi: 10.1111/j.1365-2362.1988.tb01049.x. [DOI] [PubMed] [Google Scholar]
- 61.Hidalgo M, Rodriguez G, Kuhn JG, et al. A Phase I and pharmacological study of the glutamine antagonist acivicin with the amino acid solution aminosyn in patients with advanced solid malignancies. Clin Cancer Res. 1998;4:2763–70. [PubMed] [Google Scholar]
- 62.Robinson MM, McBryant SJ, Tsukamoto T, et al. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES) Biochem J. 2007;406:407–14. doi: 10.1042/BJ20070039. [DOI] [PMC free article] [PubMed] [Google Scholar]