Abstract
The use of indwelling medical devices (e.g. pacemakers, prosthetic joints, catheters, etc) continues to increase, yet these devices are all too often complicated by infections with biofilm-forming microbes with increased resistance to antimicrobial agents and host defense mechanisms. We investigated the ability of chitosan, a polymer isolated from crustacean exoskeletons, to damage biofilms formed by the pathogenic fungus Cryptococcus neoformans. Using 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium-hydroxide (XTT) reduction assay and CFU determinations, we showed that chitosan significantly reduced both the metabolic activity of the biofilms and cell viability, respectively. We further demonstrated that chitosan penetrated biofilms and damaged fungal cells using confocal and scanning electron microscopy. Notably, melanization, an important virulence determinant of C. neoformans, did not protect cryptococcal biofilms against chitosan. The chitosan concentrations used in this study to evaluate fungal biofilm susceptibility were not toxic to human endothelial cells. Our results indicate that cryptococcal biofilms are susceptible to treatment with chitosan, suggesting an option for the prevention or treatment of fungal biofilms on indwelling medical devices.
Keywords: Cryptococcus neoformans, fungi, biofilms, chitosan, planktonic, melanin
1. INTRODUCTION
Cryptococcus neoformans is an encapsulated opportunistic yeast-like fungus that is a relatively frequent cause of meningoencephalitis in immunocompromised patients, especially in individuals with AIDS or solid organ transplants, and also occasionally causes disease in apparently healthy individuals [1]. C. neoformans capsular polysaccharide is mainly composed of glucuronoxylomannan (GXM), which is a major contributor to its virulence since acapsularstrains are not pathogenic [2]. Copious amounts of GXM are released during cryptococcal infection, causing deleterious effects on the host immune response [2, 3]. Our laboratory previously reported that C. neoformans GXM release is necessary for adhesion to a solid support and subsequent biofilm formation [4], which facilitates the evasion of the yeast from host responses [5] and antifungal therapies [6].
Biofilms are communities of microorganisms attached to a solid surface enclosed in an exopolymeric matrix [7, 8]. A cryptococcal biofilm consists of a complex network of yeast cells enmeshed in a substantial amount of polysaccharide matrix [4, 6]. C. neoformans forms biofilms on polystyrene plates [4] and medical devices [9-12] after GXM shedding. For instance, Walsh et al. reported on C. neoformans biofilms in ventriculoatrial shunt catheters [9]. In addition, several reports of C. neoformans infection of polytetrafluoroethylene peritoneal dialysis fistula [11] and prosthetic cardiac valves [10] demonstrate the ability of this organism to adhere to medical devices. These observations highlight the importance of investigating the biofilm-forming properties of this organism.
Chitosan, a hydrophilic biopolymer industrially obtained by N-deacetylation of crustacean chitin, has antimicrobial activities [13]. This natural compound is inexpensive and nontoxic. Chitosan has been utilized in diverse applications, including as an antimicrobial compound in agriculture, as a potential elicitor of plant defense responses, as a flocculating agent in wastewater treatment, as an additive in the food industry, as a hydrating agent in cosmetics, and more recently as a pharmaceutical agent in biomedicine [13]. The antimicrobial activity of chitosan has been observed against a wide variety of microorganisms including fungi, algae, and bacteria [13]. Based on these applications and its antimicrobial activity, we hypothesized that chitosan could interfere with C. neoformans biofilm formation and, by penetrating mature biofilms, bind to yeast cells to deliver direct microbicidal activity.
Although considerable work on the effect of chitosan on bacterial biofilms has been done [14-16], no comparable studies have been done with fungal biofilms. In this study, we exploited the ability of C. neoformans to form biofilms in vitro on polystyrene microtiter plates to study the susceptibilities of cryptococcal biofilms to chitosan.
2. MATERIALS AND METHODS
2.1 C. neoformans
C. neoformans strain B3501 (serotype D) was acquired from the American Type Culture Collection (Rockville, MD) and grown in Sabouraud dextrose broth (Difco Laboratories, Detroit, MI) for 24 h at 30°C in a rotary shaker at 150 rpm (to early stationary phase). Stock cultures were maintained at -80 °C.
2.2 Biofilm formation
Fungal cells were collected by centrifugation, washed twice with phosphate-buffered saline (PBS), counted using a hemacytometer, and suspended at 107 cells/mL in a chemically defined minimal medium (20 mg/mL thiamine, 30 mM glucose, 26 mM glycine, 20 mM MgSO ·7H2O, and 58.8 mM KH2PO4). Then, 100 μl of the suspension was added to individual wells of polystyrene 96-well plates (Fisher, MA) and incubated at 37°C without shaking. Biofilms were allowed to form for 48 h and then the wells were washed three times with 0.05% Tween 20 in Tris-buffered saline (TBS) to remove non-adhered yeasts using a microtiter plate washer (Skan Washer 400; Molecular Devices, VA). Based on our prior work [4, 6], fungal cells that remained attached to the plastic surface were considered true biofilms. All assays were carried out in quadruplets unless otherwise specified and experiments were repeated at least twice on different days using different cultures.
2.3 Measurement of biofilm metabolic activity by XTT reduction assay
A semi-quantitative measurement of fungal biofilm formation was obtained from the 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium-hydroxide (XTT) reduction assay [17]. For fungal strains, 50 μl of XTT salt solution (1 mg/mL in PBS) and 4 μl of menadione solution (1 mM in acetone; Sigma) were added to each well. Microtiter plates were incubated at 37°C for 5 h. Fungal mitochondrial dehydrogenase activity reduces XTT tetrazolium salt to XTT formazan, resulting in colorimetric change that correlates with cell viability. The colorimetric change was measured using a microtiter reader (Labsystems Multiskan MS; Labsystems, Finland) at 492 nm. In all the experiments, microtiter wells containing heat-killed C. neoformans and minimal medium alone were included as negative controls.
2.4 C. neoformans planktonic cells
To determine the density of fungal planktonic cells used for comparison with the biofilms, we estimated the cell numbers from the XTT reduction signal using a dose-response curve. Briefly, yeasts were grown in minimal medium for 48 h 30°C in a rotary shaker at 150 rpm, collected by centrifugation, washed twice with PBS, counted with a hemacytometer, and suspended at various densities (5 × 106, 1 × 107, and 5 × 107 cells/mL) in minimal medium. Then, 100 μl of each suspension was added into individual wells of polystyrene 96-well plates to final densities of 5 × 105, 1 × 106, and 5 × 106 cells/mL. The viability was measured by determination of the amount of XTT reduction.
2.5 CFU killing assay
The toxicities of the chitosan for fungal biofilms and planktonic cells were compared by a CFU killing assay. After incubation with chitosan, biofilms were scraped from the bottom of the wells with a sterile 200-μl micropipette tip to dissociate yeast cells. A volume of 100 μl of suspension containing dissociated cells was aspirated from the wells, transferred to a microcentrifuge tube with 900 μl of PBS, and vortexed gently for 3 min. Then, serial dilutions were performed, and 100 μl of diluted suspension was plated on Sabouraud dextrose agar plates. The percentage of CFU survival was determined by comparing chitosan-exposed biofilms and planktonic cells to unexposed fungal cells.
2.6 Melanized fungal biofilms
Melanization was induced by growing the biofilms on defined minimal medium broth at 30°C with the addition of 1 mM L-dopa for 7 days. Nonmelanized controls were obtained by growing the yeast cells on defined minimal medium broth without L-dopa for 7 days.
2.7 Chitosan susceptibility of C. neoformans biofilms
2.7.1 Susceptibilities of fungal biofilms to chitosan
To evaluate the susceptibilities of fungal biofilms to chitosan, PBS containing different concentrations of chitosan (0, 0.625, 1.25, 2.5, and 5 mg/mL) in 200 μl was added to each well. Mature biofilms and chitosan were mixed for 1 min by use of a microtiter plate reader to ensure a uniform distribution and were incubated at 37°C for 0.5 and 1 h. After incubation, biofilm metabolic activity was quantified by the XTT reduction assay. The susceptibilities of the mature cryptococcal biofilms to chitosan were determined by comparing the metabolic activities of the biofilms co-incubated with chitosan with those of the biofilms grown in PBS.
2.7.2 Comparison of biofilm and planktonic cryptococcal cell susceptibility to chitosan
C. neoformans biofilms were incubated with 200 μl of PBS containing 0.625 mg/mL of chitosan. Wells containing cryptococcal biofilms treated with PBS alone were used as a control. C. neoformans planktonic cells were suspended at a density of 5 × 106 cells per mL in PBS alone or in the presence of chitosan. Either C. neoformans biofilms or planktonic cells were mixed with chitosan using a microtiter plate reader to ensure a uniform distribution of the biopolymer and were incubated at 37°C for 0.5 and 1 h. XTT reduction and CFU killing assays were used to determine the metabolic activity and fungal mass, respectively.
2.7.3 Comparison of melanized and nonmelanized fungal biofilm susceptibilities to chitosan
C. neoformans biofilms were incubated with 200 μl of PBS containing various concentrations of chitosan (0.625, 1.25, 2.5, and 5 mg/mL). Wells containing melanized and nonmelanized biofilms treated with PBS alone were used as a control. Melanized or nonmelanized biofilms and chitosan were mixed to ensure a uniform distribution of the biopolymer and were incubated at 37°C for 2 h. The XTT reduction assay was used to determine viability.
2.8 Capsule size measurements by India ink stain
An aliquot of 10 μL of yeast was mixed with India ink and visualized with an Olympus AX70 microscope. Pictures were taken using a QImaging Retiga 1300 Digital camera and QCapture Suite V2.46 software (QImaging, Burnaby BC, Canada). The capsule size of 100 cells was measured in these images using ImageJ 1.39u software (National Institutes of Health, USA). Capsule size was defined as the difference between the diameter of the total cell (capsule included) and the cell body diameter, defined by the cell wall.
2.9 ELISA spot
C. neoformans B3501 were collected by centrifugation, washed twice with PBS, counted using a hemacytometer, and suspended at 104 cells per mL in minimal medium. For each strain, 100 μl of the suspension was added into individual wells of polystyrene 96-well plates and incubated at 37°C. C. neoformans cells were allowed to adhere to the bottom of the wells over a series of time intervals (0.5, 1, and 2h). Following the adhesion stage, the wells containing C. neoformans biofilms were washed three times with 0.05% Tween 20 in TBS to remove non-adhered cryptococcal cells using a microtiter plate washer. All assays were carried out in five wells per strain. Between every step, the wells were washed with 0.05% Tween 20 in TBS. All incubations were done at 37°C for 1h or 4°C overnight. The wells were blocked for nonspecific binding by adding 200 μl of 1% BSA in PBS. Next, 2 μg/mL of GXM binding MAb 18B7 [18] in buffer (PBS with 1% BSA) was added followed by incubation with 1 μg/mL of biotin-labeled goat anti-mouse (GAM) IgG1. Next, a 50-μl volume of 1 mg of bromo-4-chloro-3-indolyl phosphate (BCIP; Amresco, Solon, OH) per mL diluted in AMP buffer (95.8 mL of 2-amino-2-methyl-1-propanol, 0.1 mL of Triton X-405, 0.2 g of MgCl2 · 6H2O in 800 mL of double-distilled water [pH 8.6]) (Sigma Chemical Co.) was added. After 1 h the wells were washed five times with distilled water and air dried.
2.10 Capture ELISA
Released C. neoformans capsular GXM was measured by capture ELISA as described. Briefly, microtiter polystyrene plates were coated with GAM IgM (1 μg/mL) and blocked with 1% bovine serum albumin in phosphate-buffered saline. Next, the IgM GXM binding MAb 2D10 (2 μg/mL) was added as a capture antibody [19], and the plate was incubated for 1 h. Culture supernatants were serially diluted on the plate and incubated for 1 h. The ELISA was completed by adding, in successive steps, MAb 18B7 (2 μg/mL) in buffer (PBS with 1% BSA), 1 μg of alkaline phosphatase-labeled GAM IgG1/mL in buffer, and 50 μl of p-nitrophenyl phosphate (5 mg/mL) in substrate buffer (0.001 M MgCl2 and 0.05 M Na2CO3, in 1 L [pH 9.8]). Between each step, the wells were washed with 0.05% Tween 20 in Tris-buffered saline. All incubations were done at 37°C for 1 h or 4°C overnight.
2.11 Isolation of exo-polysaccharide from culture supernatants by filtration
Polysaccharide was isolated by filtration and ultrafiltration as described. Briefly, C. neoformans B3501 strain cells were separated from culture supernatants by centrifugation and the resulting supernatant was concentrated approximately 20-fold using an Amicon (Millipore, Danvers, MA) ultrafiltration cell (cutoff = 100 kDa, total capacity of 200 mL) with stirring and Biomax polyethersulfone ultrafiltration discs (76 mm Millipore, Danvers, MA). After formation of a viscous film over the filtering disc, the fluid phase was discarded and the remaining jellified material was recovered with a cell scrapper. The final polysaccharide solution was lyophilized and the dry polysaccharide mass determined.
2.12 Release of capsular components by dimethyl sulfoxide
Capsular polysaccharide was isolated from yeast cells by DMSO as described [20]. The cells were suspended in 15 mL of DMSO and incubated for 30 minutes. This process was repeated twice and the polysaccharide-containing fractions were combined. Cells were removed by centrifugation and the supernatant was then dialyzed against water for 12 h, with replacements by fresh water at 2 h intervals. The polysaccharide fractions were again dialyzed against water for 3 days. The final polysaccharide solution was lyophilized and the dry polysaccharide mass determined.
2.13 Polysaccharide particle sizes
The effective diameter and the polydispersity of polysaccharide preparations were measured by quasy elastic light scattering (QE LS) in a 90Plus/BI-MAS Multi Angle Particle Sizing analyzer (Brookhaven Instruments Corp., Holtsville, NY). Polysaccharide solutions were prepared as described above. The fluctuating signal, originating from the random motion of particles in a liquid phase and the associated alterations in the intensity of the scattered light over time, were processed by the autocorrelation function C(t), C(t)=Ae2Γt+B. In this equation, t is the time delay, A is an optical constant determined by the instrument design, and Γ is related to the relaxation of the fluctuations by Γ=Dq2. The value of q is calculated from the scattering angle θ, the wavelength of the laser light λ0, and the index of refraction n of the suspended liquid, according to the equation q=(2πn/λ0)2Sin(θ/2). Particle size is related to the translational diffusion coefficient (D) as a function of molecular shape. Given that scanning transmission micrographs have shown polysaccharide molecules with disordered branching pattern and the observation that polysaccharide can have the same mass yet differ in radius of gyration consistent with branching and/or significant secondary structure, we used the equation D=(KBT)/(3πη(t)d) for a branched polymer form, where KB is Boltzmann's constant (1.38054 × 10−16 erg/K), T is the temperature in K (30°C), η(t) is the viscosity of the liquid in which the particles are moving, and d is the particle diameter. Polydispersity was defined as equal μ2/Γ2, where μ2 is proportional to the variance of the intensity weighted distribution. The multimodal distributions of particle size diameter were generated by a Non-Negatively constrained Least Squares algorithm (NNLS) based on the intensity of light scattered by each particle. All polysaccharide samples were analyzed under the same conditions.
2.14 Zeta Potential measurements
Zeta potential (ζ), particle mobility and shift frequency of polysaccharide samples were calculated in a Zeta potential analyzer (ZetaPlus, Brookhaven Instruments Corp., Holtsville, NY). ζ is a measurement of charge (in millivolts) defined as the potential gradient that develops across the interface between a boundary liquid in contact with a solid and the mobile diffuse layer in the body of the liquid. It is derived from the equation ζ = (4πηm)/D, where D is the dielectric constant of the medium, η is the viscosity, and m is the electrophoretic mobility of the particle.
2.15 Endothelial cell damage assay
HUVECs were grown in M199 (Sigma-Aldrich) supplemented with 10% FBS and 2 mM l-glutamine. For use in damage assays, HUVECs were grown on a collagen matrix in 96-well tissue culture plates (Fisher Scientific) to create confluent layers. All incubations were at 37°C in 5% CO2. LDH release from HUVEC monolayers was measured after treatment with chitosan (0.8, 2, 4, and 8 mg/mL). Growth medium alone was added to control wells with and without HUVEC layers. The plate was incubated at 37°C with 5% CO2 for 2 h. Maximal LDH release was obtained in a control set of wells by adding 15 μl of 0.9% Triton X-100 to each well and vigorously disrupting the HUVEC layers with a pipette tip 45 minutes before the end of the 8-hour incubation period; background LDH release was that from nondisrupted HUVEC layers. Samples (50 μl) were taken from each well, and LDH was assayed spectrophotometrically at 492 nm using the CytoTox96 kit (Promega) according to the manufacturer's instructions. The percentage lysis of HUVECs treated with chitosan was calculated as experimental LDH release-background/mean maximal LDH release-background.
2.16 Light (LM), immunofluorescence (IF), and confocal (CM) microscopy
Microscopic examinations of single cells or biofilms formed in microtiter plates were performed by LM with an Axiovert 200 M inverted microscope (Carl Zeiss MicroImaging, NY).
For immunofluoresence studies, slides were coated with poly-l-lysine (0.1 mg/mL; Sigma), and 106 yeast cells were allowed to air dry on slides so that organisms adhered. MAb 18B7 or melanin binding mAb 6D2 (IgM) [21] were added at 2 μg/mL in buffer (PBS with 1% BSA). FITC-labeled GAM-IgG1 (F-GAM-IgG1) or -IgM (F-GAM-IgM) (from Southern Biotechnology) were added at 2 μg/mL after application of unconjugated MAb. All incubations were done at 37°C for 30 min, and slides were washed three times with PBS between each application of reagents. Slides were washed again with PBS, 30 μl of mounting medium (0.1 M n-propyl gallate-50% glycerol in PBS) was added, and coverslips were placed. The slides were then viewed as described above and fluorescent images were recorded.
For CM, C. neoformans biofilms were incubated for 45 min in 75 μl of PBS containing the fluorescent stain FUN-1 (10 μM). Then, wells were blocked with PBS (1% BSA) for 1 h. Next, the IgG1 GXM binding MAb 18B7 (2 μg/mL) was added, and the plate was incubated for 1 h. F-GAM-IgG1 (1-μg/mL) in PBS (1% BSA) was applied for 1 h. Between steps, the wells were washed with 0.05% Tween 20 in TBS. All incubations were done at 37°C for 1h or 4°C overnight. FUN-1 (excitation wavelength, 470 nm; emission, 590 nm) is converted to orange-red cylindrical intravacuolar structures by metabolically active cells, while MAb 18B7-F-GAM-IgG1 (excitation wavelength, 488 nm; emission, 530 nm) binds to GXM and fluoresces green. Microscopic examinations of biofilms formed in microtiter plates were performed with CM. A 40x objective was used (numerical aperture of 0.6). Depth measurements were taken at regular intervals across the width of the device. To determine the structure of the biofilms, a series of horizontal (xy) optical sections with a thickness of 1.175 μm were taken throughout the full length of the biofilm. Confocal images of green (MAb 18B7-F-GAM-IgG1) and red (FUN-1) fluorescence were recorded simultaneously by CM using a multichannel mode. Z-stack imagesand measurements were corrected utilizing Axio Vision 4.4 software in deconvolution mode (Carl Zeiss MicroImaging, NY).
2.17 Scanning electron microscopy (SEM)
C. neoformans biofilms or planktonic cells were grown on glass coverslips in six-well microtiter plates with minimal medium at 37°C for 48 h. Then, coverslips with biofilms were washed three times with PBS and transferred to another six-well microtiter plate containing 2.5% glutaraldehyde and incubated for 1 h at room temperature. The samples were serially dehydrated in alcohol, fixed in a critical-point drier (Samdri-790; Tousimis, Rockville, M.D.), coated with gold-palladium (Desk-1; Denton Vacuum, Inc., Cherry Hill, N.J.), and viewed with a JEOL (Tokyo, Japan) JSM-6400 scanning electron microscope. Two separate sets of cultures were prepared.
2.18 Statistical analysis
Statistical analyses were done with GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA) or 90Plus/BI-MAS Software (Brookhaven Instruments Corp., Holtsville, NY). P values were calculated by analysis of variance and were adjusted by use of the Bonferroni correction. P values of <0.05 were considered significant.
3 RESULTS
3.1 Chitosan on C. neoformans biofilm formation
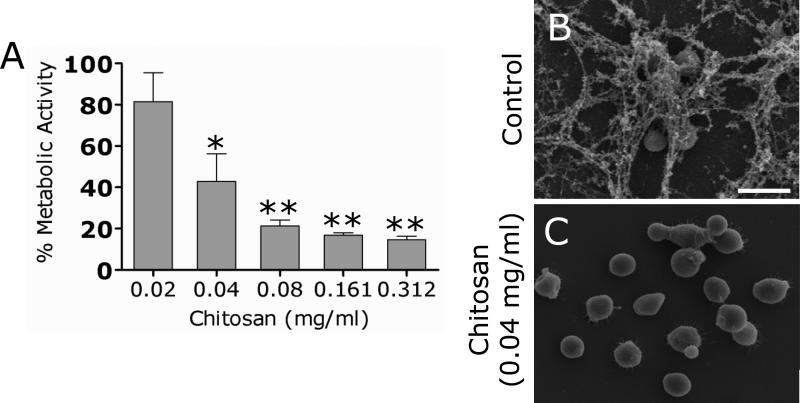
We investigated whether co-incubation of chitosan with yeast cells interfered with C. neoformans biofilm formation in vitro using the XTT reduction assay (Fig. 1A). Chitosan severely inhibited C. neoformans strain B3501 from forming biofilms at concentrations > 0.04 mg/ml.
Figure 1. Chitosan inhibits C. neoformans biofilm formation.
(A) Percent metabolic activity of untreated and chitosan-treated C. neoformans strain B3501 biofilms measured by the XTT reduction assay. Yeast cells were co-incubated with various concentrations (0.02, 0.04, 0.08, 0.16, and 0.31mg/mL) of chitosan for 48 h; and their biofilm production was compared to that of fungal cells incubated in PBS. Bars are the averages of four XTT measurements, and brackets denote standard deviations. *, P<0.05 and **, P<0.001 in comparing the untreated and chitosan-treating groups. This experiment was done twice, with similar results each time. (B) Scanning electron microscopy image of untreated C. neoformans B3501 biofilms formed on glass coverslips revealed that cryptococcal cells are internally connected by copious amounts of polysaccharide. (C) SEM image of C. neoformans B3501 grown with 0.04 mg/mL showed yeast cells with no exo- or capsular polysaccharide. Scale bar: 5 μm.
SEM examination was used to correlate the XTT reduction assay results with the visual effects on biofilm formation structure (Fig. 1B). C. neoformans yeast cells grown in the presence of PBS alone showed mature biofilms consisting of a dense group of cells enmeshed with large amount of polysaccharide. However, fungal cells treated with little as 0.04 mg/mL of chitosan appeared nearly acapsular (Fig. 1C). Notably, the minimum inhibitory concentration (MIC50) at which 50% of yeasts is inhibited, as determined by the CLSI M27-A method, was 0.625 mg/mL (data not shown).
3.2 Chitosan on capsular polysaccharide released during C. neoformans biofilm formation
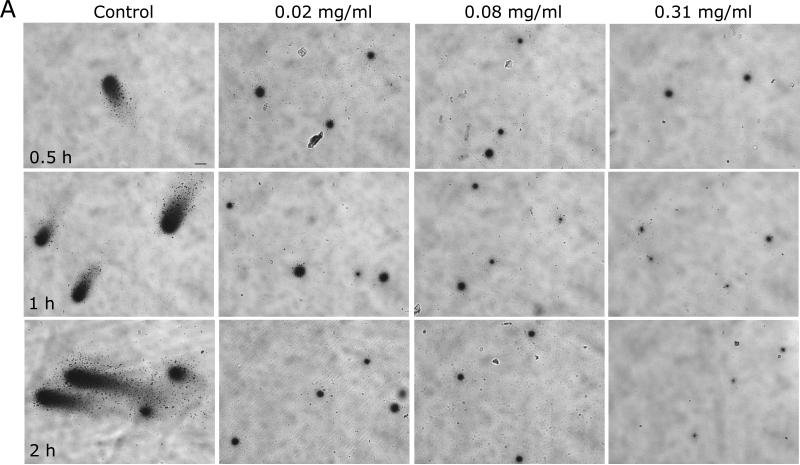
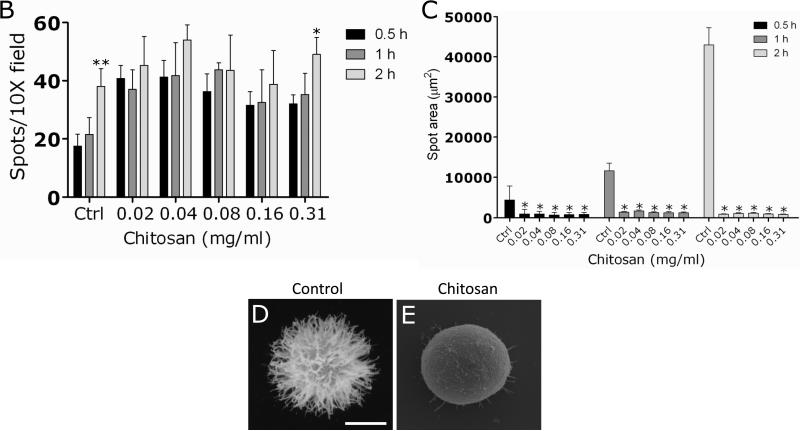
To determine the mechanism by which chitosan inhibits C. neoformans biofilm formation, the ELISA spot assay was utilized using sub-inhibitory concentrations of chitosan. This assay was used to examine local release of capsular polysaccharide by attached cryptococcal cells and the role of this polysaccharide in the creation of an exopolysaccharide matrix for C. neoformans biofilm formation. Light microscopy was used to quantify the number of cells that attached to plastic as measured by spot formation (Fig. 2A). Chitosan-treated C. neoformans B3501 cells produced significantly larger number of spots than control cells after a 1 h incubation period (Fig. 2B). However, the number of spots observed for control cryptococcal cells increased significantly after 2 h.
Figure 2. Chitosan interferes with GXM released during the adhesion stage of C. neoformans biofilm formation.
(A) Light microscopic images of spots formed by C. neoformans strain B3501 during the spot ELISA. Images were obtained after 0.5, 1, and 2 h of exposure of the fungal cells to various concentrations (0.02, 0.08, and 0.31 mg/mL) of chitosan; and the images were compared with those of yeast cells incubated in presence of PBS. The pictures were taken by using a x20 power field. Scale bars: 50 μm. The results are representative of two experiments. (B) Spot number in microtiter wells as a function of time after treatment with chitosan. Bars are the average numbers of spots in five power fields, and error bars denote standard deviations. *, P<0.01 and **, P<0.001 in comparing the untreated and chitosan-treating groups. (C) The release of C. neoformans strain B3501 GXM was visualized by the spot ELISA after exposure of the yeast cells for 0.5, 1, and 2 h to various concentrations (0.02, 0.04, 0.08, 0.16, and 0.31mg/mL) of chitosan. Fungal cells incubated in the presence of PBS were used as a control. Bars are the averages of the areas of 40 spots per power field, with the area being calculated by the equation πr2. Five power fields were observed for each time interval. Brackets denote standard deviations. *, P<0.01 in comparing the untreated and chitosan-treating groups. (D-E) SEM images demonstrated differences in polysaccharide released between (D) control and (E) chitosan-treated C. neoformans cells. The fibers of the capsule from an untreated cell (white arrow) were significantly more visible than those from a chitosan-treated cell, consistent with greater polysaccharide release to the plate at the site of cell-plastic contact. Scale bar: 2 μm.
In addition, the surface area of the spots was measured by tracing the circumference of the whole spot left by the organism at the equatorial plane (area = πr2) (Fig. 2C). This was done to determine the area involved in binding of GXM released by C. neoformans cells on the spot of attachment. Spots from control C. neoformans cells were significantly larger than those from chitosan-treated cells.
SEM examination was used to visualize differences in polysaccharide released between control and chitosan-treated C. neoformans cells. The fibers of the capsules on chitosan-treated cells were sparse, short and thin (Fig. 2E) compared to control yeast (Fig. 2D), consistent with greater and/or aberrant polysaccharide release.
3.3 Susceptibility of mature C. neoformans biofilms to chitosan
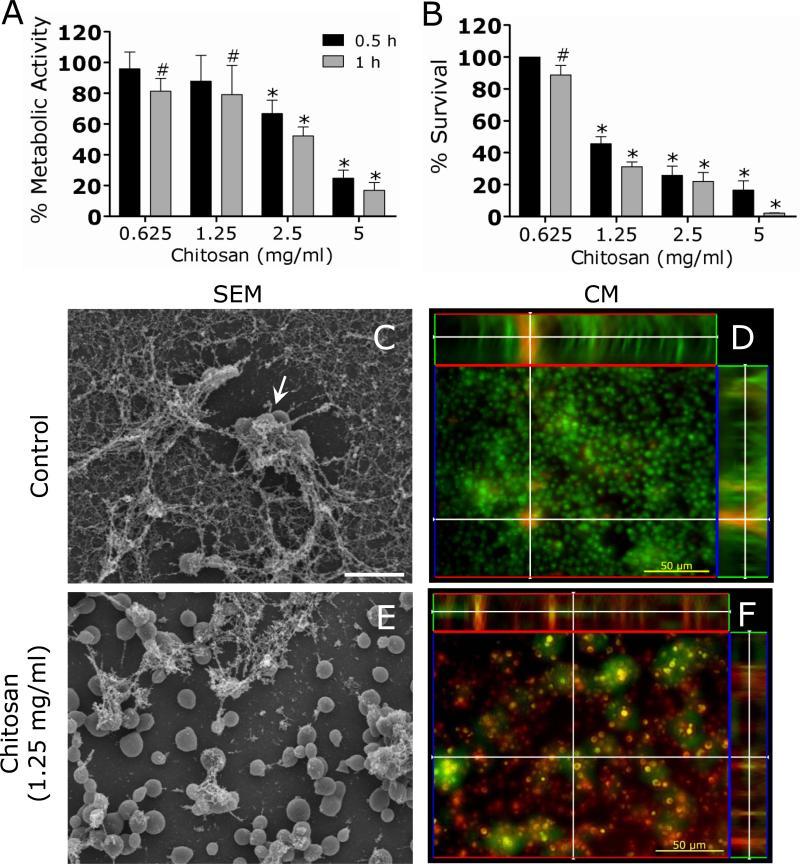
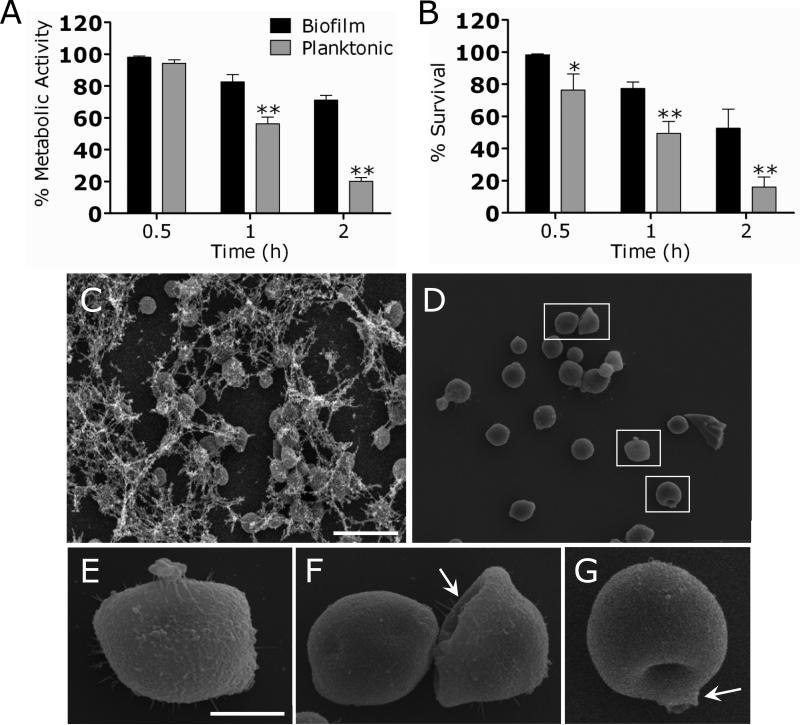
C. neoformans strain B3501 biofilms treated with chitosan showed a significant reduction in metabolic activity when viability was measured by the XTT reduction assay (Fig. 3A). For example, the metabolic activity of cryptococcal biofilms was reduced 40% after 0.5 h treatment with 2.5 mg/mL of chitosan.
Figure 3. C. neoformans biofilms are susceptible to chitosan.
(A) The percentage of metabolic activity of untreated and chitosan-treated C. neoformans strain B3501 biofilms was measured by the XTT reduction assay. (B) The percent survival of untreated and chitosan-treated C. neoformans strain B3501 biofilms was measured by determination of the numbers of CFU. For A and B, Biofilms were exposed to 1.25 mg/mL of chitosan for 0.5, and 1 h; and their metabolic activities were compared to those biofilms incubated in PBS. The bars are the averages of four measurements, and brackets denote standard deviations. #, P<0.05 and *, P<0.001 in comparing the untreated and chitosan-treating groups. These experiments were done twice, with similar results each time. (C and E) SEM of C. neoformans B3501 biofilms (C) untreated and (E) treated with chitosan. Biofilms grown in the presence of 1.25 mg/mL of chitosan showed fungal cells (white arrow) surrounded by large amounts of exopolymeric matrix. In contrast, biofilms co-incubated with chitosan displayed yeast cells lacking capsular and released polysaccharide. Scale bar for C and E: 10 μm. (D and F) CM of C. neoformans B3501 biofilms (D) untreated and (F) treated with chitosan. Orthogonal images of mature C. neoformans biofilms showed metabolically active (red, FUN-1-stained) cells embedded in the polysaccharide extracellular material (green, MAb 18B7-FITC-conjugated goat anti-mouse IgG1 stained), while the yellow-brownish areas represent metabolically inactive or nonviable cells. Images were obtained after 1 h of exposure of the fungal cells to 1.25 mg/mL of chitosan, and the images were compared with those of biofilms incubated in presence of PBS. The pictures were taken by using x40 power field. Scale bars: 50 μm. The results are representative of those of two experiments.
To confirm the results obtained by the XTT reduction assay, the percent survival of the cells in the biofilm was determined by CFU enumeration (Fig. 3B). C. neoformans biofilms were significantly susceptible to chitosan after 0.5 h treatment with 1.25 mg/mL of chitosan resulting in a reduction of approximately 50% when compared to untreated biofilms. Furthermore, treatment with ≥ 2.5 mg/mL of chitosan for 1 h resulted in ≥ 75% killing.
SEM examination was used to visualize architectural differences between untreated and chitosan treated C. neoformans biofilms (Fig. 3C and E). Untreated C. neoformans biofilms comprised a dense network of yeast cells surrounded with vast amounts of exopolymeric matrix (Fig. 3C). In contrast, biofilms co-incubated with chitosan displayed yeast cells lacking capsular and released polysaccharide (Fig. 3E). These biofilms showed scattered aggregates of polysaccharide after treatment with chitosan.
Confocal microscopic examination was used to correlate the XTT reduction and CFU killing assay results with the visual effects on biofilm metabolism and structure (Fig. 3D and F). Regions of red fluorescence (FUN-1) represent metabolically active cells, the green fluorescence (MAb 18B7-FITC-conjugated goat anti-mouse IgG1) indicates GXM, and yellow-brownish areas represent metabolically inactive or nonviable cells. C. neoformans biofilms grown in the presence of PBS alone showed regions of high metabolic activity (Fig. 3D). Biofilms treated with 1.25 mg/mL of chitosan manifested a decrease in the thickness of the exopolymeric matrix and metabolic activity (Fig. 3F).
The concentrations of chitosan used in this study to evaluate fungal biofilm susceptibility were not toxic to human endothelial cells (supplemental figure).
3.4 Susceptibility of C. neoformans planktonic cells to chitosan
The susceptibility of fungal biofilms to chitosan was investigated and compared with that of planktonic yeast cells. XTT reduction and CFU killing assays were utilized to quantify fungal metabolic activity and cellular mass, respectively.
C. neoformans biofilms were significantly more resistant to chitosan than their planktonic counterparts (Fig. 4A). For instance, the metabolic activities of cryptococcal biofilms were reduced approximately 20 and 30% after exposure to 0.625 mg/mL of chitosan for 1 and 2 h, respectively. In contrast, the metabolic activities of planktonic cells were significantly reduced 40 and 80% after treatment with similar concentration of chitosan for 1 and 2 h, respectively.
Figure 4. C. neoformans biofilms are more resistant to chitosan than planktonic cells.
(A) The percentage of metabolic activity of C. neoformans strain B3501 biofilms and planktonic cells was measured by the XTT reduction assay. Both phenotypes were exposed to 0.625 mg/mL of chitosan for 0.5, 1, and 2 h; and their metabolic activities were compared to those of fungal cells incubated in PBS. (B) The percent survival of C. neoformans strain B3501 biofilms and planktonic cells was measured by determination of the numbers of CFU. Both phenotypes were exposed to 0.625 mg/mL of chitosan for 0.5, 1, and 2 h; and their rates of survival were compared to those of fungal cells incubated in PBS. For panels A and B, the bars are the averages of four measurements, and brackets denote standard deviations. *, P<0.05 and **, P<0.01 in comparing biofilms and planktonic cells. This experiment was done twice, with similar results each time. (C-G) SEM of C. neoformans B3501 (C) biofilms and (D) planktonic cells treated with chitosan. C. neoformans biofilms grown in the presence of 0.625 mg/mL of chitosan showed fungal cells surrounded by large amounts of exopolymeric matrix. In contrast, planktonic cells treated with similar concentration resulted in significant decrease of capsular polysaccharide and (E-G; insets) cellular damage. White arrows denote cellular damage. Scale bar for C and D: 10 μm, and E-G: 2 μm.
To confirm the results obtained by the XTT reduction assay, the percent survival of the cells in the biofilm or the planktonic form was determined by counting the numbers of CFU in wells treated with chitosan and comparing these to the numbers of colonies obtained from untreated wells (Fig. 4B). C. neoformans planktonic cells were more susceptible to chitosan than biofilms after treatment for 0.5 h with 0.625 mg/mL. The survival of yeasts within biofilms was reduced approximately 50% after 2 h of treatment with 1.25 mg/mL of chitosan.
SEM examination was utilized to compare the effect of chitosan on C. neoformans biofilms and planktonic cells. C. neoformans biofilms grown in the presence of 0.625 mg/mL of chitosan showed fungal cells surrounded by large amounts of exopolymeric matrix (Fig. 4C). In contrast, planktonic cells treated with similar concentration resulted in significant decrease of capsular polysaccharide and cellular damage (Fig. 4D-G).
3.5 Impact of chitosan on cryptococcal capsular polysaccharide
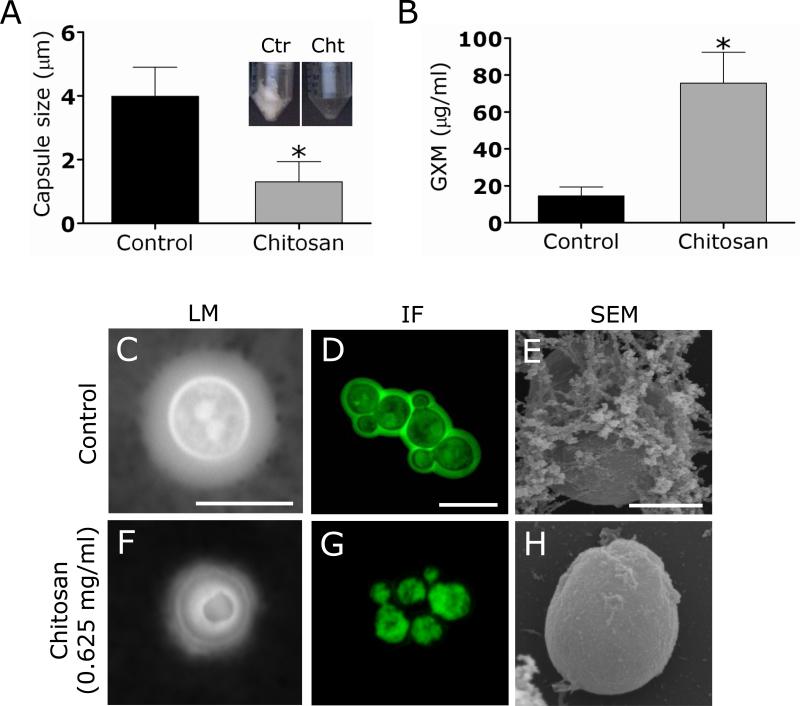
C. neoformans sheds large amounts of polysaccharide into culture media and infected tissues. The capsule size of C. neoformans B3501 cells was measured with India ink in cells grown in the presence or absence of chitosan to determine whether or not this biopolymer reduce capsule growth and to assess its effect in GXM released in culture. After 48 h, the capsule size of C. neoformans cells grown in the presence of chitosan was significantly reduced when compared with the control (Fig. 5A). Conversely, C. neoformans polysaccharide capsule release in the presence of chitosan was significantly greater than in the absence of the biopolymer (Fig. 5B). Using light (Fig. 5C and F), and immunofluorescence (Fig. 5D and G) microscopy, we demonstrated that chitosan significantly reduced the quantitiy of capsular polysaccharide, suggesting interference with capsular remodeling during growth. Similarly, SEM showed that treatment of cryptococcal biofilms with chitosan interfered with the formation of the exopolymeric matrix from biofilms (Fig. 5E and H) exposing yeasts within the biofilm to the antimicrobial activity of the biopolymer.
Figure 5. Chitosan trims the capsular polysaccharide from C. neoformans.
(A) Capsule size measurements of C. neoformans strain B3501 cells were done for cells grown in the presence and absence of 0.625 mg/mL chitosan. The insets show representative pictures of conical tubes containing the amount of polysaccharide recovered from culture supernatant of untreated control and chitosan-treated cryptococcal cells after co-incubation for 48 h. Capsule size of 100 cells was measured. (B) GXM concentration in the supernatant of C. neoformans cultures co-incubated in the absence and presence of chitosan. For A and B, average and standard deviations were determined. *, P<0.001 in comparing the untreated and chitosan-treated groups. (C and F) The effect of chitosan on capsule size of C. neoformans B3501 was measured with India ink. Untreated fungal cells exhibited smaller capsules than chitosan-treated C. neoformans cells. The pictures were taken by using x100 power field. Scale bar: 2 μm. (D and G) Chitosan shears capsular polysaccharide from cryptococcal cells. After co-incubation with chitosan, the cells were washed and incubated with MAb 18B7-FITC-conjugated goat anti-mouse IgG1 stained to label the capsular polysaccharide. Scale bar, 2 μm. (E and H) SEM showed that treatment of cryptococcal biofilms with chitosan clippers the exopolymeric matrix from biofilms. Scale bar for E and H: 2 μm.
3.6 Effect of chitosan on exo- and capsular-polysaccharide structure
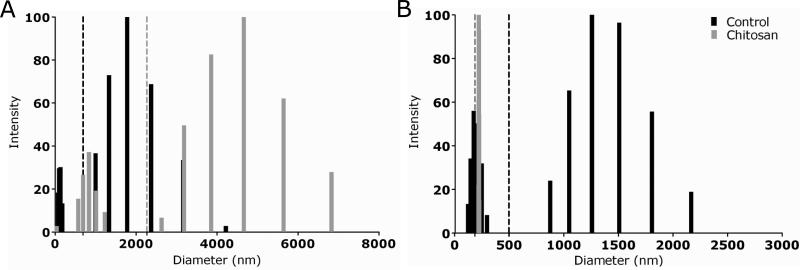
To gain additional insight into the structural alterations caused by chitosan on cryptococcal biofilms, we determined average effective diameters and size distributions of exo- and capsular polysaccharides from untreated and chitosan-treated samples using QE LS (Fig. 6). On average, the exo-polysaccharide molecules of chitosan-treated yeast cells (2,265 nm) exhibited larger diameters than untreated cells (665 nm) (Fig. 6A). Exo-polysaccharide from untreated and chitosan-treated cultures showed two different size populations. The size distribution range for each population of chitosan-treated cryptococcal cells were 40–1,225 and 2,630–6,828 nm, with the highest values at 837 and 4,662 nm, respectively. In contrast, the size distribution range for each population of untreated cells were 75–178 and 1,000–4,217 nm, with the highest values at 100 and 1,778 nm, respectively.
Figure 6. Multimodal size distribution analysis of polysaccharide fractions.
exopolysaccharide (A) and capsular-polysaccharide (B) obtained from untreated and chitosan-treated strain B3501. The x axis represents size distribution by particle diameter; y axis corresponds to the values of percentage intensity weighted sizes obtained from the NNLS algorithm.
We analyzed the size distribution of polysaccharide from C. neoformans B3501 strain capsule after treatment with chitosan (Fig. 6B). The capsular polysaccharide molecules of chitosan-treated yeast cells (179 nm) exhibited smaller diameter than untreated cells (494 nm). Treatment with chitosan yielded a homogeneous population of capsular polysaccharide on yeast cells. The size distribution range was 213-220 nm, with the highest value at 216 nm. However, two different populations in polysaccharide size were present for untreated cells. The range size for each population was 118–292 and 871–2,162 nm, with the highest values at 169 and 1,253 nm, respectively.
3.7 Chitosan treatment on C. neoformans zeta potential
The surface charge of untreated and chitosan-treated melanized and nonmelanized cryptococcal cells was measured (Table 1). Previous studies have shown that the polysaccharide capsule and melanin of C. neoformans are responsible for the high negative charge of the cells [22]. Chitosan-treated cells were significantly less negative (25.40±0.57) than untreated cells (-21.89±0.31) (P<0.001). Melanization significantly increased the negative charge (-34.75±0.57) of cryptococcal cells when compared to nonmelanized cells (-21.89±0.31) (P<0.001). However, treatment with chitosan imparted a high positive charge to melanized (26.15±0.38) and nonmelanized (25.40±0.57) C. neoformans cells.
Table 1.
Zeta potential of C. neoformans B3501 strain grown with (melanized) and without (nonmelanized) L-DOPA and untreated or treated with chitosan.
| Zeta Potential (mV) | |
|---|---|
| Control (PBS) | −21.89±0.31 |
| 0.312 mg/mL chitosan | 25.40±0.57a |
| L-DOPA | −34.75±0.57b |
| L-DOPA + 0.312 mg/mL chitosan | 26.15±0.38a |
Value significantly greater than the value for control (P < 0.001).
Value significantly less than the value for control (P < 0.001).
3.8 Impact of melanin on biofilm resistance after chitosan treatment
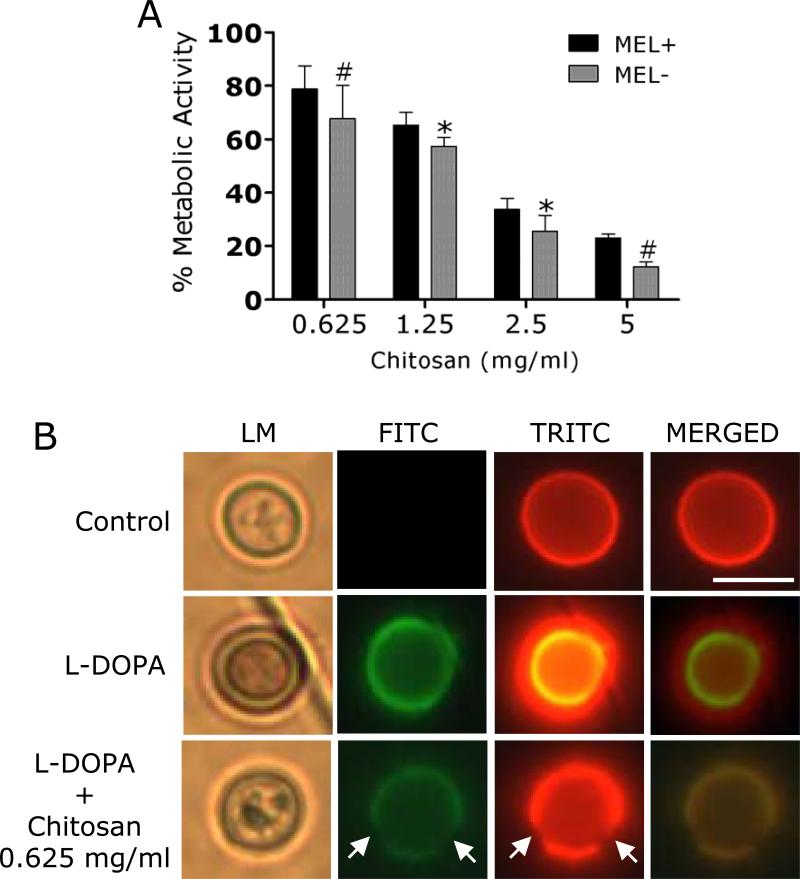
Melanized C. neoformans biofilms were more resistant to chitosan than their nonmelanized counterparts (Fig. 7A). For instance, the metabolic activities of melanized cryptococcal biofilms were reduced approximately 20% after exposure to 0.625 mg/mL of chitosan for 2h. In contrast, the metabolic activities of nonmelanized biofilms were significantly reduced 35% after treatment with similar concentration of chitosan. Although melanized biofilms showed less susceptibility to higher concentrations of chitosan than nonmelanized biofilms, melanin production did not provide significant resistance to chitosan's antifungal activity. Using IF, we determined that exposure to chitosan for 2 h effectively damages C. neoformans melanin enabling the biopolymer's antimicrobial activity (Fig. 7B).
Figure 7. Melanization does not reduce biofilm vulnerability chitosan.
(A) The percentage of metabolic activity of C. neoformans strain B3501 melanized and nonmelanized biofilms was measured by the XTT reduction assay. Both phenotypes were exposed to various concentrations of chitosan (0.625, 1.25, 2.5, and 5 mg/mL) for 2 h; and their metabolic activities were compared to those of fungal cells incubated in PBS. (B) Chitosan disrupts melanin from cryptococcal cells. After co-incubation with chitosan, the cells were washed and incubated with MAbs 6D2-FITC-conjugated goat anti-mouse IgM and 18B7-TRITC-conjugated goat anti-mouse IgG1 stained to label the melanin and capsular polysaccharide, respectively. Scale bar, 2 μm.
DISCUSSION
C. neoformans has substantial chitosan in its cell wall during vegetative growth and the polymer may be an essential factor for the proper maintenance of cell wall integrity [23]. Our results show that addition of exogenous chitosan to C. neoformans biofilms significantly reduces metabolic activity and prevents the adhesion of the yeast cells to the polystyrene surface. The cell wall is the structure that mediates the cell's interactions with the environment, and might be important in adhesion of fungi to solid surfaces such as indwelling medical devices. Bachmann et al. proposed the use of the cell wall as an attractive target for the development of strategies to combat biofilm-associated infections [24]. Chitosan has anti-adherent activity and prevents C. albicans biofilm development [25]. Other studies have proposed the treatment of medical devices with antifungal agents before they are implanted in patients [24, 26]. Chitosan may be a strong candidate for this endeavor, due to its anti-adherent and antifungal properties against fungal biofilms. Furthermore, we demonstrated that chitosan might be effective against C. neoformans, possibly because melanin production did not provide significant resistance to chitosan's antifungal activity. Using IF, we concluded that chitosan effectively damages melanin from yeast cells making C. neoformans cells more accessible to the chitosan's antimicrobial activity. Importantly, the concentrations used in these experiments were not toxic to human endothelial cells, which are the cells most readily exposed to chitosan if applied to a venous or arterial catheter.
Chitosan inhibits C. neoformans biofilm formation in polystyrene microtiter plates. A recent study showed that chitosan-coated surfaces have anti-biofilm properties in vitro against certain bacteria and fungi [14]. This phenomenon has been attributed to the ability of cationic chitosan to disrupt negatively charged cell membranes as microbes settle on the surface [13]. In this regard, our zeta potential analysis demonstrates that chitosan has a profound effect on the negative charge of the cryptococcal cellular membrane, which may translate into interference with surface colonization or adhesion and cell-cell interactions during biofilm formation [27]. For example, a net positive charge to the fungal surfaces may keep yeast cells in suspension, preventing biofilm formation [28].
The release of the cryptococcal polysaccharide capsule is important for adhesion of the yeast and biofilm development in C. neoformans [4]. An ELISA spot assay provided an alternate method to confirm the results obtained by XTT assay and zeta potential analysis in establishing the mechanism by which chitosan inhibits C. neoformans biofilm formation. The number and area of spots on microtiter plates were determined as a function of time using light microscopy. Biofilm formation by C. neoformans did not correlate with the number of yeast cells bound to the plastic support surface. For instance, C. neoformans biofilms grown in the presence of chitosan attached in larger quantities to the plastic surface of the microtiter plate but produced smaller spots than those formed by control biofilms. This result implies that cell adhesion was a necessary but not sufficient event for biofilm formation. In fact, we interpret the ELISA spot data as indicating that chitosan interferes with local release of capsular polysaccharide by attached cells to the plastic surface preventing the production of the exopolysaccharide matrix.
Chitosan decreased the metabolic activity and survival of C. neoformans biofilms, consistent with its fungicidal properties [13]. This phenomenon reflects physical stress on the biofilm structure due to permeabilization of the cellular membrane, which allowed increased penetration of chitosan and effective delivery of its antifungal activity [29]. Binding of chitosan with DNA and inhibition of mRNA synthesis occurs through chitosan penetration toward the nuclei of the microorganisms and interference with the synthesis of mRNA and proteins [29]. It is most likely that the interaction between positively charged chitosan molecules and negatively charged microbial cell membranes leads to the leakage of proteinaceous and other intracellular constituents causing cell death [30].
C. neoformans biofilms were significantly less susceptible than planktonic cells to chitosan. These results correlate with those presented in other reports that have suggested that the biofilm phenotype confers resistance to antifungal therapy [31, 32]. We found that chitosan mediated a significant reduction in the metabolic activity of C. neoformans cells. Baker et al. showed that chitosan helps to maintain cell integrity and normal capsule width and aids in bud separation [33]. Due to the absence of a protective exopolymeric matrix, there is a possibility that exogenous chitosan interacts more freely with the chitosan in the fungal cell wall making planktonic cells more susceptible to penetration and disruption of internal organelles. Therefore, our analysis indicates that exogenous chitosan might be an excellent antifungal therapeutic.
We assessed the effect of chitosan on C. neoformans capsular polysaccharide which is essential for biofilm formation. SEM and CM microscopy showed that chitosan removes most of the capsular polysaccharide from C. neoformans cells. The aberrant capsules on fungal cells might increase their susceptibility to chitosan. In this regard, structural studies using QE LS revealed that capsular polysaccharide on chitosan treated fungal cells was shorter than on their untreated counterparts. Moreover, we observed scattered aggregates of polysaccharide after fungal cell treatment with chitosan. This observation was further supported by QE LS that demonstrated that the exopolysaccharide fibers of chitosan treated cells were much larger than on untreated cells.
CONCLUSION
The findings of this study suggest that chitosan might offer a flexible, biocompatible platform for designing coatings to protect surfaces from infection. Our data suggests that chitosan can be potentially developed as an antimicrobial agent for prophylaxis against and/or the treatment of catheter or other medical device related fungal biofilm diseases.
Supplemental Figure. Chitosan is not toxic to human endothelial cells. Relative LDH activity measured in the tissue culture supernatant by HUVECs after 2 h co-culture in the absence or presence of various concentrations (0.8, 2, 4, and 8 mg/mL) of chitosan. Bars are the averages of the results for three measurements, and error bars denote standard deviations.
Supplementary Material
ACKNOWLEDGEMENTS
L.R.M. is supported by a molecular pathogenesis training grant. J.D.N. is supported in part by NIH grant AI52733. J. M. F. is partially supported by DOD grant DAMD17-03-1-0127. A.C. and S.F. were supported by NIH Awards AI52733, AI033774, HL059842, and AI033142. A.J.F. is supported by Orlando Dermatology Aesthetic and Clinical Young Investigators and American Society for Dermatologic Surgery Cutting Edge Research Grants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Mitchell TG, Perfect JR. Cryptococcosis in the era of AIDS--100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vecchiarelli A. Immunoregulation by capsular components of Cryptococcus neoformans. Med Mycol. 2000;38:407–417. doi: 10.1080/mmy.38.6.407.417. [DOI] [PubMed] [Google Scholar]
- 3.Casadevall A, Perfect JR. Cryptococcus neoformans. ASM Press; Washington DC: 1998. [Google Scholar]
- 4.Martinez LR, Casadevall A. Specific antibody can prevent fungal biofilm formation and this effect correlates with protective efficacy. Infect Immun. 2005;73:6350–6362. doi: 10.1128/IAI.73.10.6350-6362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez LR, Casadevall A. Cryptococcus neoformans cells in biofilms are less susceptible than planktonic cells to antimicrobial molecules produced by the innate immune system. Infect Immun. 2006;74:6118–6123. doi: 10.1128/IAI.00995-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez LR, Casadevall A. Susceptibility of Cryptococcus neoformans biofilms to antifungal agents in vitro. Antimicrob Agents Chemother. 2006;50:1021–1033. doi: 10.1128/AAC.50.3.1021-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 9.Walsh TJ, Schlegel R, Moody MM, Costerton JW, Salcman M. Ventriculoatrial shunt infection due to Cryptococcus neoformans: an ultrastructural and quantitative microbiological study. Neurosurgery. 1986;18:373–375. doi: 10.1227/00006123-198603000-00025. [DOI] [PubMed] [Google Scholar]
- 10.Banerjee U, Gupta K, Venugopal P. A case of prosthetic valve endocarditis caused by Cryptococcus neoformans var. neoformans. J Med Vet Mycol. 1997;35:139–141. [PubMed] [Google Scholar]
- 11.Braun DK, Janssen DA, Marcus JR, Kauffman CA. Cryptococcal infection of a prosthetic dialysis fistula. Am J Kidney Dis. 1994;24:864–867. doi: 10.1016/s0272-6386(12)80683-4. [DOI] [PubMed] [Google Scholar]
- 12.Penk A, Pittrow L. Role of fluconazole in the long-term suppressive therapy of fungal infections in patients with artificial implants. Mycoses. 1999;42:91–96. doi: 10.1111/j.1439-0507.1999.tb00021.x. [DOI] [PubMed] [Google Scholar]
- 13.Rabea EI, Badawy ME, Stevens CV, Smagghe G, Steurbaut W. Chitosan as antimicrobial agent: applications and mode of action. Biomacromolecules. 2003;4:1457–1465. doi: 10.1021/bm034130m. [DOI] [PubMed] [Google Scholar]
- 14.Carlson RP, Taffs R, Davison WM, Stewart PS. Anti-biofilm properties of chitosan-coated surfaces. J Biomater Sci Polym Ed. 2008;19:1035–1046. doi: 10.1163/156856208784909372. [DOI] [PubMed] [Google Scholar]
- 15.Pasquantonio G, Greco C, Prenna M, Ripa C, Vitali LA, Petrelli D, et al. Antibacterial activity and anti-biofilm effect of chitosan against strains of Streptococcus mutans isolated in dental plaque. Int J Immunopathol Pharmacol. 2008;21:993–997. doi: 10.1177/039463200802100424. [DOI] [PubMed] [Google Scholar]
- 16.Busscher HJ, Engels E, Dijkstra RJ, van der Mei HC. Influence of a chitosan on oral bacterial adhesion and growth in vitro. Eur J Oral Sci. 2008;116:493–495. doi: 10.1111/j.1600-0722.2008.00568.x. [DOI] [PubMed] [Google Scholar]
- 17.Meshulam T, Levitz SM, Christin L, Diamond RD. A simplified new assay for assessment of fungal cell damage with the tetrazolium dye, (2,3)-bis-(2-methoxy-4-nitro-5-sulphenyl)-(2H)-tetrazolium-5-carboxanil ide (XTT). J Infect Dis. 1995;172:1153–1156. doi: 10.1093/infdis/172.4.1153. [DOI] [PubMed] [Google Scholar]
- 18.Casadevall A, Cleare W, Feldmesser M, Glatman-Freedman A, Goldman DL, Kozel TR, Lendvai N, Mukherjee J, Pirofski LA, Rivera J, Rosas AL, Scharff MD, Valadon P, Westin K, Zhong Z. Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob Agents Chemother. 1998;42:1437–1446. doi: 10.1128/aac.42.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukherjee S, Lee S, Mukherjee J, Scharff MD, Casadevall A. Monoclonal antibodies to Cryptococcus neoformans capsular polysaccharide modify the course of intravenous infection in mice. Infect Immun. 1994;62:1079–1088. doi: 10.1128/iai.62.3.1079-1088.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dadachova E, Nosanchuk JD, Shi L, Schweitzer AD, Frenkel A, Nosanchuk JS, Casadevall A. Dead cells in melanoma tumors provide abundant antigen for targeted delivery of ionizing radiation by a mAb to melanin. Proc Natl Acad Sci U S A. 2004;101:14865–1470. doi: 10.1073/pnas.0406180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryan RA, Zaragoza O, Zhang T, Ortiz G, Casadevall A, Dadachova E. Radiological studies reveal radial differences in the architecture of the polysaccharide capsule of Cryptococcus neoformans. Eukaryot Cell. 2005;4:465–475. doi: 10.1128/EC.4.2.465-475.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nosanchuk JD, Casadevall A. Cellular charge of Cryptococcus neoformans: contributions from the capsular polysaccharide, melanin, and monoclonal antibody binding. Infect Immun. 1997;65:1836–1841. doi: 10.1128/iai.65.5.1836-1841.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banks IR, Specht CA, Donlin MJ, Gerik KJ, Levitz SM, Lodge JK. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot Cell. 2005;4:1902–1912. doi: 10.1128/EC.4.11.1902-1912.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bachmann SP, VandeWalle K, Ramage G, Patterson TF, Wickes BL, Graybill JR, et al. In vitro activity of caspofungin against Candida albicans biofilms. Antimicrob Agents Chemother. 2002;46:3591–3596. doi: 10.1128/AAC.46.11.3591-3596.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soustre J, Rodier MH, Imbert-Bouyer S, Daniault G, Imbert C. Caspofungin modulates in vitro adherence of Candida albicans to plastic coated with extracellular matrix proteins. J Antimicrob Chemother. 2004;53:522–525. doi: 10.1093/jac/dkh099. [DOI] [PubMed] [Google Scholar]
- 26.Trampuz A, Zimmerli W. New strategies for the treatment of infections associated with prosthetic joints. Curr Opin Investig Drugs. 2005;6:185–190. [PubMed] [Google Scholar]
- 27.Miyake Y, Tsunoda T, Minagi S, Akagawa Y, Tsuru H, Suginaka H. Antifungal drugs affect adherence of Candida albicans to acrylic surfaces by changing the zeta-potential of fungal cells. FEMS Microbiol Lett. 1990;57:211–214. doi: 10.1016/0378-1097(90)90067-z. [DOI] [PubMed] [Google Scholar]
- 28.Savard T, Beaulieu C, Boucher I, Champagne CP. Antimicrobial action of hydrolyzed chitosan against spoilage yeasts and lactic acid bacteria of fermented vegetables. J Food Prot. 2002;65:828–833. doi: 10.4315/0362-028x-65.5.828. [DOI] [PubMed] [Google Scholar]
- 29.Sudarshan NR, Hoover DG, Knorr D. Antibacterial action of chitosan. Food Biotechnology. 1992;6:257–272. [Google Scholar]
- 30.Jung B, Kim C, Choi K, Lee YM, Kim J. Preparation of amphiphilic chitosan and their antimicrobial activities. J App Polym Sci. 1999;72:1713–1719. [Google Scholar]
- 31.Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 2001;183:5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hawser SP, Douglas LJ. Resistance of Candida albicans biofilms to antifungal agents in vitro. Antimicrob Agents Chemother. 1995;39:2128–2131. doi: 10.1128/aac.39.9.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker LG, Specht CA, Donlin MJ, Lodge JK. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot Cell. 2007;6:855–867. doi: 10.1128/EC.00399-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.