Abstract
Background
Association analysis identified the homeobox transcription factor, ENGRAILED 2 (EN2), as a possible Autism Spectrum Disorder (ASD) susceptibility gene (ASD [MIM 608636]; EN2 [MIM 131310]). The common alleles (underlined) of two intronic SNPs, rs1861972 (A/G) and rs1861973 (C/T), are over-transmitted to affected individuals both singly and as a haplotype in three separate datasets (518 families total, haplotype P=0.00000035). Methods: Further support that EN2 is a possible ASD susceptibility gene requires the identification of a risk allele, a DNA variant that is consistently associated with ASD but is also functional. To identify possible risk alleles, additional association analysis and LD mapping were performed. Candidate polymorphisms were then tested for functional differences by luciferase (luc) reporter transfections and Electrophoretic Mobility Shift Assays (EMSAs). Results: Association analysis of additional EN2 polymorphisms and LD mapping with Hapmap SNPs identified the rs1861972-rs1861973 haplotype as the most appropriate candidate to test for functional differences. Luc reporters for the two common rs1861972-rs1861973 haplotypes (A-C and G-T) were then transfected into human and rat cell lines as well as primary mouse neuronal cultures. In all cases the A-C haplotype resulted in a significant increase in luc levels (P<.005). EMSAs were then performed and nuclear factors bound specifically to the A and C alleles of both SNPs. Conclusions: These data indicate the AC haplotype is functional and together with the association and LD mapping results support EN2 as a likely ASD susceptibility gene and the A-C haplotype as a possible risk allele.
Keywords: Autism, ENGRAILED 2, risk allele
INTRODUCTION
Autism Spectrum Disorder (ASD) is a polygenic disorder affecting CNS development 1,2. Individuals with ASD display deficits in language, emotional reciprocity as well as increased repetitive behaviors and movements. Although strong evidence for a genetic contribution to ASD exists, few causative genetic defects have been implicated in the etiology of the disorder 3-9.
Our previous analysis has focused on EN2, an important regulator of CNS development10-13. EN2 maps to the distal portion of chromosome 7 (7q36.3) and is encoded by two exons and a single 3.5kb intron spanning 8.1kb of genomic DNA. EN2 association was tested previously in nuclear pedigrees obtained from the Autism Genetic Resource Exchange (AGRE) and the NIMH. The pedigrees have at least two siblings diagnosed with ASD and can also include unaffected siblings.
Two intronic EN2 SNPs, rs1861972 and rs1861973, are significantly associated with ASD individually and as a haplotype under both a narrow (autism) and broad (autism, Asperger's syndrome or Pervasive Developmental Delay-Not Otherwise Specified) phenotypic definition. The common alleles for both SNPs (A-rs1861972; C-rs1861973) are over-transmitted to affected individuals and under-represented in unaffected siblings. These results were observed in an original dataset of 167 families (AGRE I, rs1861972-narrow: P=.026, broad: P=.016; rs1861973- narrow: P=.008, broad: P=.012; rs1861972-rs1861973 haplotype, narrow: P=.0009, broad: P=.0017) 14. Association was then replicated in two separate datasets (AGRE II, 222 families, haplotype, narrow: P=.0048, broad: P=.0016 and NIMH, 129 families, haplotype, narrow: P=.0463, broad: P=.0431). When all three datasets were combined strong evidence for association was observed (518 families, haplotype, narrow: P=.00000065; broad: P=.00000035)14,15. Rs1861972 and rs1861973 display strong inter-marker LD with each other in these three datasets (D'=.903, r2=.767). In the combined three datasets the frequencies of the common A and C alleles for rs1861972 and rs1861973 both individually and as a haplotype are ~72% (rs1861972 A allele- 73%, rs1861973 C allele- 72%, A-C haplotype- 71%). Four other groups have reported some association for EN2 with autism in datasets of different ethnicities: a Northern French population 16, one of largely Western-Northern European descent 17, and two Chinese datasets 18,19. However, polymorphic and allelic differences have been observed between these studies and our association data, suggesting that underlying causative genetic variant(s) may vary between datasets and ethnicities. Although many different rare and common variants are likely to contribute to ASD susceptibility, these data are consistent with EN2 being a likely ASD susceptibility gene. However further support for this possibility requires the isolation of a risk allele, an associated polymorphism that affects the expression or activity of EN2.
We expect candidate risk alleles to be in strong LD with rs1861972 and rs1861973 and to display at least as significant association with ASD as the A-C haplotype under both diagnoses. Our prior re-sequencing, association, and LD mapping data identified the rs1861972-rs1861973 A-C haplotype as a candidate for the EN2 risk allele. Previously 16 additional EN2 polymorphisms were typed in the AGRE I dataset. Only the intronic SNPs demonstrated high D' with rs1861972 and rs1861973, while one intronic SNP, rs2361688 (Minor Allele Frequency (MAF) = 27%), displayed high r2 (rs1861972 = .730; rs1861973 = .807). Re-sequencing of the intron identified one new SNP with a MAF of ~1%. Association analysis for all 16 EN2 polymorphisms demonstrated that none of them were as strongly associated as rs1861972 or rs1861973 individually or as a haplotype. Rs2361688 and another intronic SNP (rs3824068) displayed minimal association but only under one diagnostic criterion (rs2361688: narrow P=.13, broad P=.04; rs3824068: narrow P=.04, broad P=.10)15. This analysis identified the rs1861972-rs1861973 haplotype as one possible candidate risk allele but it was unknown whether additional polymorphisms were in strong r2 with either associated SNP. We now address this possibility and provide additional genetic and molecular data implicating the rs1861972-rs1861973 haplotype as a functional variant that may increase ASD risk.
MATERIALS AND METHODS
Hapmap
CEU Hapmap genotypes for rs1861973 were obtained from the Hapmap consortium. All other genotypes were directly acquired from Hapmap (PhaseII, January 2007, NCBI: dbSNP b125). Haploview program (version 4.0) determined inter-marker LD relation between CEU Hapmap SNPs and rs1861973. For other datasets, LD data was directly downloaded from the Hapmap website.
Genotyping, association and LD analysis
Details concerning the genotyping, error checking, LD, and association analysis for eight EN2 3' polymorphisms typed as part of this analysis are available as Supplemental Information. The AGRE I, AGRE II, and NIMH datasets 15 were subdivided by ethnicity and rs1861972 and rs1861973 were analyzed for association in the White non-Hispanic subset (489 families, 2266 individuals, 790 individuals with narrow autism diagnosis, 938 individuals with broad ASD diagnosis). Three SNPs that displayed minimal association in the AGRE I dataset (rs2361688, rs3824068, and rs12533271) were also tested for association in the White non-Hispanic subset of AGRE I (154 families, 686 individuals, 241 individuals with autism narrow diagnosis, 298 individuals with broad ASD diagnosis).
Luciferase assays
Details concerning the generation of luc constructs are available as Supplemental Information. HEK293T cells were maintained in D-MEM supplemented with 10% FBS and 1% Penicillin/Streptomycin. PC12 cells were maintained as above except with an additional 5% horse serum. Granule cells were isolated from P6 C57BL6 mice by standard protocols and maintained at 35°C under 5% CO2 5μg of pGL3 constructs and 300ng of phRL-null vector (Promega) were transfected by Amaxa electroporation into 5 million granule and PC12 cells. HEK293T cells were transfected-null vector using the lipofectamine 2000 system. 24 hours following transfection, cells were collected and lysed using a 1X Promega passive lysis buffer. Luciferase activities were measured using the VeritasTMMicroplate Luminometer where 85μl of Promega luciferase substrate (LARII) and 100μl of Promega Renilla luciferase substrate (Stop & Glow) were consecutively added to 35μl of cell lysates.
Splicing RTPCR
HEK293T cells were transfected as described above with TATA-Luc-Intron A-C and G-T constructs. 24 hours after transfection, RNA was isolated and cDNA was generated. Primer sequences and RT-PCR conditions are available as Supplemental Information. The expected RT-PCR products are 1758bp and 342bp using the F1/R and F2/R primers respectively. Cerebellar post-mortem samples (lobule 6) were obtained from The Harvard Brain Tissue Resource Center. The rs1861972 and rs1861973 genotype was determined as described previously 15. Total RNA was isolated from two affected (1 A-C/G-T, 1 G-T/G-T) and two psychiatrically normal (1 A-C/G-T, 1 G-T/GT) individuals by standard RNA purification procedure using RNAlater-ICE (Ambion) and mirVana PARIS kit (Ambion). cDNA was generated and RT-PCR was performed (see Supplemental Information). The predicted size of the amplicon indicative of correct splicing is 134bp.
qRT-PCR
HEK293T cells were transfected with TATA-Luc, TATA-Luc-Intron A-C, and G-T constructs as described above. Primers for qPCR were designed using the Primer Express® software version 2.0 and available as Supplemental Information. qPCR was performed by adding 20μM of each primer, 12.5μl of 2x SYBR® Green and 2.5μl of cDNA using the ABI PRISM® 7000HT Sequence Detection System.
Electrophoretic mobility shift assays
Nuclear extracts prepared from P6 mouse granule neurons cultured for 24 hours were isolated using Panomics nuclear extraction kit (AY2002). Biotin-labeled, sense and anti-sense 21bp probes were designed such that 10bp of sequence both 5' and 3' flanked the polymorphic alleles of rs1861972 and rs1861973 (see Supplemental Information). Using the Panomics EMSA kit (AY1000),100ng of nuclear extracts was incubated with 1μg of Poly d(I-C) for 5 minutes at room temperature. 2μl of 5x binding buffer and 10ng of biotin labeled probes were then added to a final volume of 10μl and incubated for 30 minutes at 20°. For competition assays, 100 to 80 fold molar excess of competitors was added to the mixture prior to the 30 minutes incubation. The protein/DNA complex was separated on a non-denaturing 6% acrylamide gel in 0.5X Tris-borate-EDTA (TBE) buffer and wet-transferred onto a Biodyne Nylon membrane (PALL) which was exposed to a HyBlotCLTM Autoradiography film (Denville Scientific Inc) for chemiluminescence detection.
RESULTS
Association and LD mapping analysis
Candidate risk alleles responsible for rs1861972-rs1861973 ASD association are anticipated to fulfill the following three criteria: i) display high inter-marker r2 with rs1861972 and rs1861973, ii) exhibit at least as strong association as the rs1861972-rs1861973 haplotype under both narrow and broad diagnostic definitions, and iii) demonstrate a functional difference between alleles.
Because the region immediately 3' of EN2 was not densely analyzed in our previous study, eight additional polymorphisms were typed in AGRE I. None of these polymorphisms displayed pairwise r2 values exceeding 0.05 with rs1861972 or rs1861973 (Supplemental Table 1). In addition, one SNP (rs12533271) was marginally associated with ASD but only under the broad diagnosis (Supplemental Table 2).
To extend our LD map, we then examined publicly available Hapmap data, which was typed for rs1861973 but not rs1861972. To validate the applicability of the HapMap data to the AGRE and NIMH samples, the following was performed. First, r2 and D' values were first determined for 5 SNPs (rs1861973, rs6460013, rs3824067, rs3808331 and rs1861958) typed in both the Hapmap and our ASD datasets. Because 70.3% of the AGRE datasets tested for association were of Northern/Western European descent, the CEU inter-marker LD values were evaluated first. Very similar r2 and D' values were observed in both datasets (Supplemental Table 3). Second, the three ASD datasets tested previously for association (518 families) were then subdivided by ethnicity and 489 White non-Hispanic families were selected for analysis. Individual and haplotype association for rs1861972 and rs1861973 association was very similar between the White non-Hispanic subset and our previously reported results (Supplemental Tables 4 and 5). These studies validate using Hapmap CEU data to identify additional candidate risk alleles.
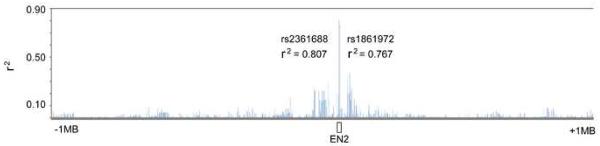
The Hapmap pairwise r2 values with rs1861973 were then ascertained in the CEU dataset for 3120 SNPs within 2 Mb of EN2 (~1 SNP/641bp; ~66% of validated Ensembl SNPs). This region was selected because it is likely to include most of the important cis-regulatory elements for EN2 expression 20-22. We found that all Hapmap SNPs within the 2 Mb region were in weak r2 with rs1861973 (<.370)(Fig 1). In addition, little difference in inter-marker r2 values with rs1861972 and rs1861973 was noted in the White non-Hispanic subset (Fig 1, Supplemental Fig 1) or the other Hapmap datasets (Supplemental Table 6).
Figure 1.
ENGRAILED 2 LD map. Inter-marker r2 values for rs1861973 are shown. The map includes 26 EN2 polymorphisms typed in the AGRE I dataset (167 families not subset on ethnicity) plus 3120 Hapmap SNPs within 2Mb of EN2 (+1Mb 5', -1Mb 3') typed in the CEU dataset. Only rs1861972 and rs2361688 display high r2 values (>.75) with rs1861973. However, rs2361688 is not consistently associated with ASD 15, identifying rs1861972 and rs1861973 as the most appropriate candidates to test for functional allelic differences.
Finally, the three other SNPs (rs2361688, rs3824068, and rs12533271) demonstrating minimal association in the AGRE I dataset were analyzed in the White non-Hispanic subset (n=154). Rs2361688 is not associated under either diagnostic definition while rs3824068 and rs12533271 display minimal association only under one diagnostic criterion (Supplemental Table 7).
Thus only rs1861972 and rs1861973 fulfill the first two criteria for an ASD risk allele responsible for our previously reported EN2 association. One, they are in high r2 with each other, and two both SNPs display consistent association with ASD under both diagnostic criteria. For these reasons we first decided to test the possible functionality of rs1861972 and rs1861973.
Luciferase assays
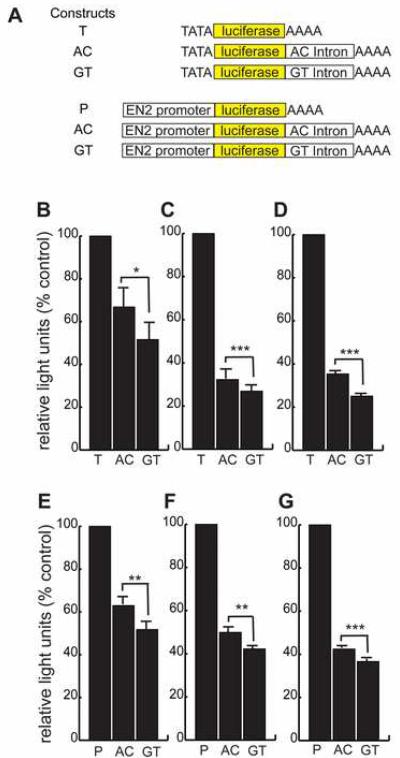
To investigate whether a functional difference could be observed between the two common rs1861972-rs1861973 haplotypes (A-C and G-T), luciferase (luc) assays were performed. Luc assays measure quanta of light and due to their reproducibility and sensitivity are commonly used to test functional activity of cis-regulatory sequences. Since the activity of cis-regulatory elements can be affected by position and functional variants associated with common disorders often have subtle effects on gene regulation 23-29, we designed the luc constructs to approximate the endogenous locus. The intron was cloned 3' of the promoter and the luc protein coding sequence but 5' of the SV40 poly-adenylation site so that the intron would be transcribed and spliced as the endogenous gene. Two promoters were used: the SV40 minimal promoter or the EN2 promoter (-1 to -5500) that is evolutionarily conserved from humans to rodents. These constructs were transiently transfected into three different cell types: a human nonneuronal cell line (HEK293T), a rat neuronal cell line (PC12) and primary cultures of mouse post-natal day 6 (P6) cerebellar granule neurons. Immunohistochemistry and in situ analysis have established that En2 is expressed abundantly in P6 post-mitotic granule neurons 13,30. Our RTPCR experiments demonstrated that En2 transcripts are detected in P6 primary granule cell cultures and HEK293T cells but not PC12 cells (Supplemental Fig 2). In all three cell types and for both promoters, the A-C haplotype resulted in a significant increase in luc levels compared to the G-T haplotype (Fig 2).
Figure 2.
Functional difference between rs1861972-rs1861973 A-C and G-T haplotypes. (a) The functional difference between the A-C and G-T intronic haplotypes was investigated by generating the diagrammed luc reporter constructs: T- SV40 minimal promoter 5' of luc without EN2 intron, P- EN2 promoter (-1 to -5735) 5' of luc without EN2 intron, AC- EN2 intron with rs1861972-rs1861973 A-C haplotype cloned 3' of luciferase but 5' of the SV40 polyadenylation signal to approximate the endogenous locus, GT- EN2 intron with rs1861972-rs1861973 G-T haplotype cloned 3' of luciferase but 5' of the SV40 polyadenylation signal. (b-d) Relative light units of luciferase normalized to Renilla reniformis and expressed as percent of control, pgl3 promoter vector (T), is shown for the SV40 minimal promoter constructs transiently transfected into (b) P6 cerebellar granule neurons (n=6), (c) PC12 cells (n=6) and (d) HEK293T cells (n=6). (e-g) Normalized relative light units of luciferase for luc EN2 promoter constructs expressed as percent of control (P) is shown for (e) P6 cerebellar granule neurons (n=7), (f) PC12 cells (n=6) and (g) HEK293T cells (n=6). * P<.005, ** P<.001, *** P < .00001, two tailed paired Student's T test
We also transfected the SV40 minimal promoter intronic constructs into HEK-293T cells and measured luc mRNA levels by q-RTPCR. A similar difference in normalized luc RNA levels was observed between haplotypes (Supplementary Fig 3). These results demonstrate a consistent functional difference between the A-C and G-T haplotypes.
Splicing assays
Because the intron is transcribed, we also investigated whether the A-C haplotype affects splicing. For the above A-C and G-T constructs the intron also included the splice acceptor and donor sequences of each EN2 exon so that potential splicing effects of the haplotype could be investigated. The SV40 minimal promoter intronic constructs were then transfected into HEK-293T cells and RTPCR experiments with multiple primer sets to luc and the SV40 polyA sequence were performed (Supplemental Fig 4A). Appropriate cycling conditions were used to amplify the intron if it was present in the cDNA. Only amplicons of the correctly spliced transcripts were observed, indicating that neither haplotype resulted in cryptic splicing (Supplemental Fig 4B, C). This was confirmed by performing RTPCR for EN2 on cerebellar post-mortem samples with and without the risk allele (Supplemental Fig 4D).
Electrophoretic Mobility Shift Assay (EMSA) analysis
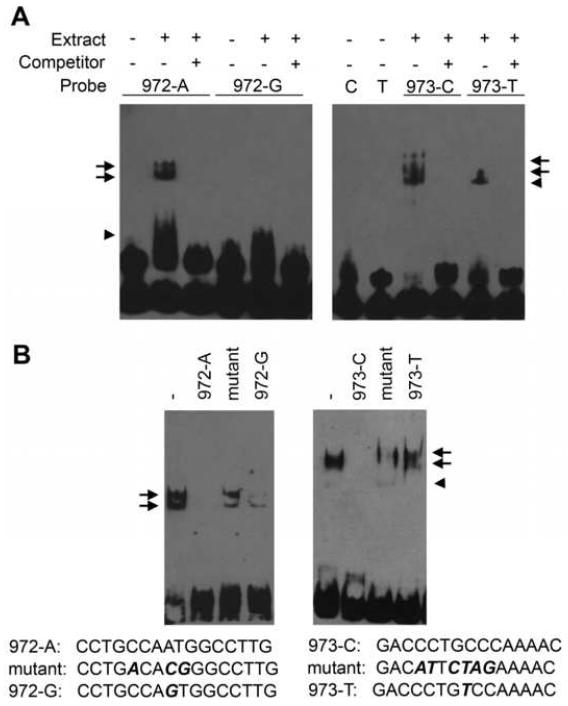
To investigate whether the associated SNPs affect the binding of DNA proteins, EMSAs were conducted. Nuclear extracts from P6 post-mitotic cerebellar granule cells were isolated and incubated with labeled oligonucleotides containing either allele of rs1861972 and rs1861973. For rs1861972, we detected two bands that specifically interacted with the common A allele but not the rare G allele (Fig 3A). These protein-DNA complexes were consistently observed in all nuclear extract preps (n=4). A third complex that interacted with both alleles was also detected but its presence was more variable between extracts (Fig 3A, B).
Figure 3.
Differential binding of nuclear proteins to rs1861972 and rs1861973 associated alleles. (A) To investigate whether the associated SNPs affect the binding of nuclear proteins, EMSAs were conducted with biotinylated 20-mer oligonucleotides and nuclear extract isolated from P6 mouse cerebellar granule cells. Extract was incubated with oligonucleotides specific to each allele, separated on a denaturing acrylamide gel, transferred to a membrane and detected by chemiluminescence. Several protein-DNA complexes were observed for both SNPs. Specificity was determined by competing with 100 molar excess of unlabelled oligonucleotide. Protein-DNA complexes specific to the associated rs1861972 A allele or rs1861973 C allele were observed (arrows) that were not detected for the corresponding rs1861972 G allele or rs1861973 T allele biotinylated oligonucleotides. In addition, protein-DNA complexes common to both alleles for rs1861972 or rs1861973 were observed (arrowheads). Abbreviations: 972-A: 20-mer oligonucleotide specific to the rs1861972 A allele, 972-G: 20-mer oligonucleotide specific to the rs1861972 G allele, 973-C and C: 20-mer oligonucleotide specific to the rs1861973 C allele, 973-T and T: 20-mer oligonucleotide specific to the rs1861973 T allele, + or -: presence or absence respectively of extract or 100 molar excess of unlabelled oligonucleotide. (B) To examine allele-specific binding of nuclear proteins to rs1861972 (left) and rs1861973 (right), additional competitions were performed. 80 molar excess of 3 different unlabelled oligonucleotides were each added individually to the probe and nuclear extract: oligonucleotide with the same sequence as the biotinylated probe (972-A, 973-C), mutant oligonucleotides predicted to disrupt NF1, NFY, C/EBP binding to the A allele of rs1861972 or Sp1 and Ets binding to the C allele of rs1861873, and oligonucleotides for the non-associated G (972-G) and T (973-T) alleles. The sequence for each oligonucleotide is shown. Abbreviation: - absence of competitor
Similar results were observed for rs1861973. Two specific DNA-protein complexes were consistently detected for the common C allele but not the rare T allele while one shifted band was observed for both alleles in some extracts (Fig 3A, B). All rs1861972 and rs1861973 DNA-protein complexes were competed with 100 molar excess of unlabelled oligonucleotide. These data demonstrate the specific binding of factors to the common alleles of both SNPs, which are over-transmitted to individuals with ASD.
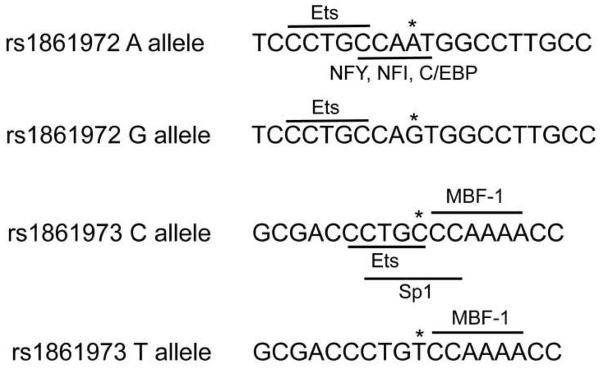
Bioinformatic analysis (Transcription Element Search Software-TESS)31 supports our EMSA results. The common A allele of rs1861972 (underlined) is situated in a canonical CCAAT binding site recognized by three transcription factor families (NF1, NFY and C/EBP) composed of multiple genes. The rare G allele (CCAGT) replaces one of the obligatory A nucleotides required for transcription factor recognition, which is predicted to completely disrupt binding of all three transcription factor families (Fig 4, Supplemental Table 8). For rs1861973, the sequence containing the common C allele is situated in overlapping consensus sites for the Sp1 and Ets family of transcription factors (Fig 4). Similar to rs1861972, the rare T allele of rs1861973 replaces a cytosine, which is required for the sequence-specific DNA binding of Sp1 and Ets family members. Transcription factors are also predicted to bind equally well to both alleles of rs1861972 and rs1861973, consistent with the common shifted complexes observed in some extract preps.
Figure 4.
Conservation of transcription factor binding sites for associated and non-associated alleles of rs1861972 and rs1861973. The 20 bp sequence encompassing rs1861982 and rs1861973 and used as probes in our EMSAs is depicted. Conserved transcription factor sites are underlined with the polymorphic allele for each SNP designated with an asterisk.
We further investigated the specificity of binding by performing additional competitions. Oligonucleotides mutated for either the CCAAT sequence for rs1861972 or the overlapping Sp1/Ets binding site for rs1861973 did not compete in our EMSAs (Fig 4B). Finally, oligonucleotides containing the rare alleles for rs1861972 (G allele) and rs1861973 (T allele) also did not compete as well as equimolar amounts of the associated alleles (Fig 4B). These studies are consistent with rs1861972 and rs1861973 affecting the binding of nuclear factors.
DISCUSSION
Our previous data demonstrated that the rs1861972-rs1861973 A-C haplotype is consistently associated with ASD in three separate datasets. LD mapping, association analysis and re-sequencing identified the rs1861972-rs1861973 haplotype as a possible risk allele. It was equally possible the associated SNPs were in strong LD with a risk variant mapping at a distance from EN2. In addition, no functional difference between the rs1816972-rs1861973 A-C and G-T haplotypes had yet been demonstrated 14,15. We have now extended the LD map and none of the new markers display high inter-marker r2 with rs1861973. These data are consistent with the shorter LD spans typically observed in telomeric positions 32 but it remains formally possible that rs1861972 and rs1861973 are in high r2 with other polymorphisms not typed in our analysis and these unidentified variants may also contribute to a functional difference. Nevertheless our LD mapping and association results identified the rs1861972-rs1861973 haplotype as the best candidate for functional experiments. Our luc assays demonstrate a consistent increase in levels for the A-C haplotype in three cell types using two different promoters. The specific binding of nuclear factors to the A and C alleles support this functional difference. In summary only rs1861972 and rs1861973 currently fulfill all three criteria of a risk allele responsible for our reported EN2 association: i) these SNPs are consistently associated with ASD under a narrow (autism) and broad (ASD) diagnostic criteria both individually and as a haplotype, ii) rs1861972 and rs1861973 are in high inter-marker r2 with each other, and iii) a functional difference between alleles has been observed. Together these data support EN2 as a likely ASD susceptibility gene and the A-C haplotype as a possible risk allele.
Rs2361688 is the only tested polymorphism, which is in high but not perfect r2 with both rs1861972 and rs1861973 and displays minimal association with ASD. These results could be explained in two ways. One, rs2361688 is a SNP that segregates frequently with rs1861972 and rs1861973 but individually is not functional. The difference in association for rs2361688 versus rs1861972 and rs1861973 is consistent with this possibility (rs2361688: narrow P=.128; broad P=.040; rs1861972: narrow P=.026, broad P=.016; rs1861973: narrow P=.008, broad P=.012). Alternatively, rs2361688 may function in concert with the A-C haplotype. However if this were the case, the common rs2361688-rs1861972-rs1861973 haplotype (G-A-C) would be expected to display more significant association than the A-C haplotype, which is not observed (G-A-C: narrow P=.009, broad P=.004; A-C: narrow P=.002, broad P=.004). Thus our current data suggests that rs2361688 is non-functional but segregates with the functional rs1861972-rs1861973 haplotype. Nevertheless to further investigate the possible involvement of rs2361688, additional association analysis in the AGRE II and NIMH datasets in the AGRE II and NIMH datasets are ongoing. If positive results are obtained, then functional experiments can be performed. Finally several other EN2 polymorphisms that are not in high r2 with rs1861972 or rs1861973 also exhibit minimal association in our study and other published reports. These data suggest the possible presence of additional EN2 risk alleles. Future association, LD mapping and functional experiments will test this possibility.
Common functional variants reported to increase risk for other diseases typically affect the regulation of the associated gene 24-29,33. The significant increase in luc levels for the A-C haplotype is consistent with these published results and can be explained by two possible molecular mechanisms. One, since the intron is transcribed and spliced in our constructs, the functional difference could be due to the haplotypes affecting splicing efficiency or stability of nuclear pre-mRNA. This would reduce the amount of luc protein and be consistent with the functional effects of intronic SNPs for other common disorders23. Two, the rs1861972-rs1861973 haplotype could regulate transcription initiation. This possibility is supported by our EMSA data and the bioinformatics indicating that both associated alleles are situated in well-defined consensus transcription factor binding sequences. It is also well established that these transcription factors can function at a distance and in a position independent manner. Published reports for other intronic risk alleles are consistent with this idea 24,33,34. Finally, current bioinformatic data does not support another transcript or miRNA mapping to the EN2 intron and contributing to the functional difference between alleles (genome.ucsc.edu). Regardless of the molecular mechanism, our in vitro results indicate that the rs1861972-rs1861973 haplotype is functional and suggest the A-C haplotype will affect EN2 levels in vivo.
A large number of transcription factors are predicted to bind to the A and C alleles of rs1861972 and rs1861973. The A allele of rs1861972 is situated in a CCAAT box which is a consensus binding site for the C/EBP, NF1 and NFY transcription factor family of proteins. Each of these protein families is comprised of multiple genes (NFIA, B, C and X; C/EBPA, B, D, E, G and Z; NFYA, B and C). In addition each NFI and NFY gene also generates multiple protein isoforms through alternative splicing and processing 35. Approximately 40 different transcription factors could then bind to the rs1861972 A allele. For rs1861973, a similar large number of proteins are predicted to bind to the rs1861973 C allele, nine Sp1 members and ~25 Ets factors 36-38. Previous in situ studies have demonstrated that a large percentage of these genes are widely expressed in the developing and adult brain including neuronal cell types that transcribe EN2 such as post-mitotic granule cells 30. Microarray analysis has also determined that these putative transcription factors are expressed in HEK-293 and PC12 cells used in our transfection analysis (Gene Expression Omnibus). Interestingly, these transcription factor family members can function as either activators or repressors 35,39-41. Because EN2 is expressed in a variety of different developmental cell types, the magnitude and direction of the haplotype functional effect could vary between cells depending upon the expression of these various transcription factor isoforms. Alternatively, it is possible that other unidentified factors could be responsible for the observed protein-DNA complexes. Future experiments will be directed at identifying the nuclear proteins that bind to rs1861972 and rs1861973 using a variety of adult and developmental cell types, in which the haploype has been shown to be functional.
To investigate whether the rs1861972-rs1861973 haplotype affects EN2 levels in vivo, both post-mortem analysis and mouse models will be employed. Post-mortem cerebellar samples are currently being obtained to investigate whether affection status and/or haplotype are correlated with altered EN2 mRNA and protein levels. Transgenic mice have been created for both haplotypes where EN2 cis-regulatory sequences drive the expression of a fluorescent reporter. These mice will allow us to determine the potential regulatory effects of the haplotype in the developing and adult CNS. Knock-in mice are also being generated where the mouse locus is being replaced with either human haplotype. These knock-in mice will provide an important resource for determining potential phenotypic effects caused by altered EN2 levels. These ongoing in vivo studies will extend our current in vitro analysis and provide information regarding when, where, and how the haplotype is functional.
EN2 is a homeobox transcription factor that regulates gene expression during embryonic and post-natal CNS development and continues to be expressed in a subset of differentiated neurons in the adult. Mutational analysis using model organisms has demonstrated that En2 is necessary for the development of the cerebellum, ventral neurons of the serotonin, norepinephrine and dopamine neurotransmitter systems as well as the proper topographic mapping of retinal axons onto the tectum 10,13,42-46. Various anatomical, neurochemical and eye tracking studies have implicated these structures and neurotransmitter systems in the etiology of autism 1,47. Thus altered levels of EN2 may affect these or other developmental systems, which will be investigated in our rs1861972-rs1861973 knock-in mice.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by research grants from the National Institute of Mental Health (R01 MH076624), NARSAD, Autism Speaks and The New Jersey Governor's Council on Autism to J.H.M; from the National Institute of Mental Health (R01 MH70366 and R01 MH076435) and The New Jersey Governor's Council on Autism to L.M.B; Diversity Program in Neuroscience (T32MH018882) to R.B
We would like to thank Emanuel DiCicco-Bloom MD and Veronica Vieland PhD for many helpful discussions, David Altshuler MD PhD and Lincoln Stein MD PhD for CEU Hapmap data for rs1861973, Eileen White PhD for technical support and Max Tischfield for technical assistance. We would also like to thank the Harvard Brain Tissue Resource Center, which is supported in part by PHS grant number R24 MH068855.
We gratefully acknowledge the resources provided by the Autism Genetic Resource Exchange (AGRE) Consortium* and the participating AGRE families. The Autism Genetic Resource Exchange is a program of Cure Autism Now and is supported, in part, by grant MH64547 from the National Institute of Mental Health to Daniel H. Geschwind (PI). We also thank Jay Tischfield and the Rutgers Cell and DNA Repository for providing the AGRE and NIMH samples. The NIMH samples were provided by the NIMH Center for Collaborative Genetic Studies on Mental Disorders grant MH068457 (to JT).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The AGRE Consortium:◻ ◻Dan Geschwind, M.D., Ph.D., UCLA, Los Angeles, CA;◻Maja Bucan, Ph.D., University of Pennsylvania, Philadelphia, PA;◻;W.Ted Brown, M.D., Ph.D., F.A.C.M.G., N.Y.S. Institute for Basic Research in Developmental Disabilities, Staten Island, NY;◻Rita M. Cantor, Ph.D., UCLA School of Medicine, Los Angeles, CA; St. Louis, MO; Herbert, M.D., Ph.D., Harvard Medical School, Boston, MA ◻Clara Lajonchere, Ph. D, Cure Autism Now, Los Angeles, CA;◻Davind H. Led better, Ph. D., Emory University, Atlanta, GA;◻Christa Lese-Martin, Ph.D., Emory University, Atlanta, GA;◻Janet Miller, J.D., Ph.D., Cure Autism Now, Los Angeles, CA;◻Stanley F. Nelson, M.D., UCLA School of Medicine, Los Angeles, CA;◻Gerard D. Schellenberg, Ph.D., University of Washington, Seattle, WA; ◻Carol A. Samango -Sprouse, Ed.D., George Washington University, Washington, ◻;D.C.;Sarah Spence, M.D., Ph.D., UCLA, Los Angeles, ◻CA;Matthew State, M.D., Ph.D., Yale University , New Haven, CT.◻Rudolph E. Tanzi, Ph.D., Massachusetts General Hospital, Boston, MA.
FINANCIAL DISCLOSURES The authors report no biomedical financial interests or potential conflicts of interest.
REFERENCES
- 1.DiCicco-Bloom E, Lord C, Zwaigenbaum L, Courchesne E, Dager SR, Schmitz C, Schultz RT, et al. The developmental neurobiology of autism spectrum disorder. J Neurosci. 2006;26:6897–906. doi: 10.1523/JNEUROSCI.1712-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta AR, State MW. Recent advances in the genetics of autism. Biol Psychiatry. 2007;61:429–37. doi: 10.1016/j.biopsych.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 3.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–88. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–97. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–9. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–75. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 7.Bakkaloglu B, O'Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Hum Genet. 2008;82:165–73. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–4. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–9. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baader SL, Sanlioglu S, Berrebi AS, Parker-Thornburg J, Oberdick J. Ectopic overexpression of engrailed-2 in cerebellar Purkinje cells causes restricted cell loss and retarded external germinal layer development at lobule junctions. J Neurosci. 1998;18:1763–73. doi: 10.1523/JNEUROSCI.18-05-01763.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baader SL, Vogel MW, Sanlioglu S, Zhang X, Oberdick J. Selective disruption of "late onset" sagittal banding patterns by ectopic expression of engrailed-2 in cerebellar Purkinje cells. J Neurosci. 1999;19:5370–9. doi: 10.1523/JNEUROSCI.19-13-05370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Millen KJ, Hui CC, Joyner AL. A role for En-2 and other murine homologues of Drosophila segment polarity genes in regulating positional information in the developing cerebellum. Development. 1995;121:3935–45. doi: 10.1242/dev.121.12.3935. [DOI] [PubMed] [Google Scholar]
- 13.Millen KJ, Wurst W, Herrup K, Joyner AL. Abnormal embryonic cerebellar development and patterning of postnatal foliation in two mouse Engrailed-2 mutants. Development. 1994;120:695–706. doi: 10.1242/dev.120.3.695. [DOI] [PubMed] [Google Scholar]
- 14.Gharani N, Benayed R, Mancuso V, Brzustowicz LM, Millonig JH. Association of the homeobox transcription factor, ENGRAILED 2, with autism spectrum disorder. Mol Psychiatry. 2004;9:474–84. doi: 10.1038/sj.mp.4001498. [DOI] [PubMed] [Google Scholar]
- 15.Benayed R, Gharani N, Rossman I, Mancuso V, Lazar G, Kamdar S, et al. Support for the homeobox transcription factor gene ENGRAILED 2 as an autism spectrum disorder susceptibility locus. Am J Hum Genet. 2005;77:851–68. doi: 10.1086/497705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petit E, Herault J, Martineau J, Perrot A, Barthelemy C, Hameury L, et al. Association study with two markers of a human homeogene in infantile autism. J Med Genet. 1995;32:269–74. doi: 10.1136/jmg.32.4.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brune CW, Korvatska E, Allen-Brady K, Cook EH, Jr., Dawson G, Devlin B, et al. Heterogeneous association between engrailed-2 and autism in the CPEA network. Am J Med Genet B Neuropsychiatr Genet. 2007;147B(2):187–93. doi: 10.1002/ajmg.b.30585. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Jia M, Yue W, Tang F, Qu M, Ruan Y, et al. Association of the ENGRAILED 2 (EN2) gene with autism in Chinese Han population. Am J Med Genet B Neuropsychiatr Genet. 2007;147B(24):434–8. doi: 10.1002/ajmg.b.30623. [DOI] [PubMed] [Google Scholar]
- 19.Yang P, Lung FW, Jong YJ, Hsieh HY, Liang CL, Juo SH. Association of the homeobox transcription factor gene ENGRAILED 2 with autistic disorder in Chinese children. Neuropsychobiology. 2008;57:3–8. doi: 10.1159/000123115. [DOI] [PubMed] [Google Scholar]
- 20.Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–25. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 21.Logan C, Khoo WK, Cado D, Joyner AL. Two enhancer regions in the mouse En-2 locus direct expression to the mid/hindbrain region and mandibular myoblasts. Development. 1993;117:905–16. doi: 10.1242/dev.117.3.905. [DOI] [PubMed] [Google Scholar]
- 22.Miyoshi G, Fishell G. Directing neuron-specific transgene expression in the mouse CNS. Curr Opin Neurobiol. 2006;16:577–84. doi: 10.1016/j.conb.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 23.Onouchi Y, Gunji T, Burns JC, Shimizu C, Newburger JW, Yashiro M, et al. ITPKC functional polymorphism associated with Kawasaki disease susceptibility and formation of coronary artery aneurysms. Nat Genet. 2008;40:35–42. doi: 10.1038/ng.2007.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tokuhiro S, Yamada R, Chang X, Suzuki A, Kochi Y, Sawada T, et al. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat Genet. 2003;35:41–8. doi: 10.1038/ng1267. [DOI] [PubMed] [Google Scholar]
- 25.Kakiuchi C, Iwamoto K, Ishiwata M, Bundo M, Kasahara T, Kusumi I, et al. Impaired feedback regulation of XBP1 as a genetic risk factor for bipolar disorder. Nat Genet. 2003;35:171–5. doi: 10.1038/ng1235. [DOI] [PubMed] [Google Scholar]
- 26.Ozaki K, Sato H, Iida A, Mizuno H, Nakamura T, Miyamoto Y, et al. A functional SNP in PSMA6 confers risk of myocardial infarction in the Japanese population. Nat Genet. 2006;38:921–5. doi: 10.1038/ng1846. [DOI] [PubMed] [Google Scholar]
- 27.Hata J, Matsuda K, Ninomiya T, Yonemoto K, Matsushita T, Ohnishi Y, et al. Functional SNP in an Sp1-binding site of AGTRL1 gene is associated with susceptibility to brain infarction. Hum Mol Genet. 2007;16:630–9. doi: 10.1093/hmg/ddm005. [DOI] [PubMed] [Google Scholar]
- 28.Sun T, Gao Y, Tan W, Ma S, Shi Y, Yao J, et al. A six-nucleotide insertion-deletion polymorphism in the CASP8 promoter is associated with susceptibility to multiple cancers. Nat Genet. 2007;39:605–13. doi: 10.1038/ng2030. [DOI] [PubMed] [Google Scholar]
- 29.Tuo J, Ning B, Bojanowski CM, Lin ZN, Ross RJ, Reed GF, et al. Synergic effect of polymorphisms in ERCC6 5' flanking region and complement factor H on age-related macular degeneration predisposition. Proc Natl Acad Sci U S A. 2006;103:9256–61. doi: 10.1073/pnas.0603485103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuller U, Kho AT, Zhao Q, Ma Q, Rowitch DH. Cerebellar 'transcriptome' reveals cell-type and stage-specific expression during postnatal development and tumorigenesis. Mol Cell Neurosci. 2006;33:247–59. doi: 10.1016/j.mcn.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 31.Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. Curr Protoc Bioinformatics. 2008;21:2.6.1–2.6.15. doi: 10.1002/0471250953.bi0206s21. [DOI] [PubMed] [Google Scholar]
- 32.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emison ES, McCallion AS, Kashuk CS, Bush RT, Grice E, Lin S, et al. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–63. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- 34.Wang GJ, Yang P, Xie HG. Gene variants in noncoding regions and their possible consequences. Pharmacogenomics. 2006;7:203–9. doi: 10.2217/14622416.7.2.203. [DOI] [PubMed] [Google Scholar]
- 35.Gronostajski RM. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249:31–45. doi: 10.1016/s0378-1119(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 36.Sharrocks AD, Brown AL, Ling Y, Yates PR. The ETS-domain transcription factor family. Int J Biochem Cell Biol. 1997;29:1371–87. doi: 10.1016/s1357-2725(97)00086-1. [DOI] [PubMed] [Google Scholar]
- 37.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–37. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 38.Zhao C, Meng A. Sp1-like transcription factors are regulators of embryonic development in vertebrates. Dev Growth Differ. 2005;47:201–11. doi: 10.1111/j.1440-169X.2005.00797.x. [DOI] [PubMed] [Google Scholar]
- 39.Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko VV, Nakatani Y, Wolffe AP, et al. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. Embo J. 1998;17:6300–15. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nerlov C. The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007;17:318–24. doi: 10.1016/j.tcb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Chen B, Li Y, Chen J, Lou G, Chen M, et al. Transcriptional regulation of the human PNRC promoter by NFY in HepG2 cells. J Biochem. 2008;143:675–83. doi: 10.1093/jb/mvn019. [DOI] [PubMed] [Google Scholar]
- 42.Sgaier SK, Lao Z, Villanueva MP, Berenshteyn F, Stephen D, Turnbull RK, et al. Genetic subdivision of the tectum and cerebellum into functionally related regions based on differential sensitivity to engrailed proteins. Development. 2007;134:2325–35. doi: 10.1242/dev.000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura H, Sugiyama S. Polarity and laminar formation of the optic tectum in relation to retinal projection. J Neurobiol. 2004;59:48–56. doi: 10.1002/neu.10339. [DOI] [PubMed] [Google Scholar]
- 44.Simon HH, Scholz C, O'Leary DD. Engrailed genes control developmental fate of serotonergic and noradrenergic neurons in mid- and hindbrain in a gene dose-dependent manner. Mol Cell Neurosci. 2005;28:96–105. doi: 10.1016/j.mcn.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 45.Brunet I, Weinl C, Piper M, Trembleau A, Volovitch M, Harris W, et al. The transcription factor Engrailed-2 guides retinal axons. Nature. 2005;438:94–8. doi: 10.1038/nature04110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alberi L, Sgado P, Simon HH. Engrailed genes are cell-autonomously required to prevent apoptosis in mesencephalic dopaminergic neurons. Development. 2004;131:3229–36. doi: 10.1242/dev.01128. [DOI] [PubMed] [Google Scholar]
- 47.McDougle CJ, Erickson CA, Stigler KA, Posey DJ. Neurochemistry in the pathophysiology of autism. J Clin Psychiatry. 2005;66(Suppl 10):9–18. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.