Abstract
Iron-sulfur clusters are key iron cofactors in biological pathways ranging from nitrogen fixation to respiration. Due to the toxicity of ferrous iron and sulfide to the cell, in vivo Fe-S cluster assembly is carried out by multi-protein biosynthetic pathways. Fe-S cluster assembly proteins traffic iron and sulfide, assemble nascent Fe-S clusters, and correctly transfer Fe-S clusters to the appropriate target metalloproteins in vivo. The gram-negative bacterium E. coli contains a stress-responsive Fe-S cluster assembly system, the SufABCDSE pathway, that functions under iron starvation and oxidative stress conditions that compromise Fe-S homeostasis. Using a combination of protein-protein interaction and in vitro Fe-S cluster assembly assays, we have characterized the relative roles of the SufBCD complex and the SufA protein during Suf Fe-S cluster biosynthesis. These studies reveal that SufA interacts with SufBCD in order to accept Fe-S clusters formed de novo on the SufBCD complex. Our results represent the first biochemical evidence that the SufBCD complex within the Suf pathway functions as a novel Fe-S scaffold system to assemble nascent clusters and transfer them to the SufA Fe-S shuttle.
Protein-bound iron-sulfur (Fe-S) clusters are one of the most common enzyme prosthetic groups and play important roles in fundamental life processes such as electron transfer reactions, substrate binding and catalysis, transcriptional regulation, and sensing of reactive oxygen and nitrogen species (1, 2). In vivo formation of Fe-S clusters involves multiple components working in concert. Three primary Fe-S assembly pathways have been identified along with a large number of uncharacterized accessory proteins. The nif system is required for the formation of Fe-S clusters in the nitrogenase enzyme complex although nif homologues can be found in organisms that lack nitrogenase (3). The isc system works as a general pathway for the maturation of multiple Fe-S proteins in both bacteria and the mitochondria of eukaryotes (4-6). The third system, named suf, mediates Fe-S cluster assembly under oxidative stress and iron limitation conditions in E. coli (7-11), but is the sole cluster assembly system in other prokaryotes and in the chloroplast of some photosynthetic eukaryotes (11-13). All three systems utilize a cysteine desulfurase enzyme (NifS, IscS, SufS) to liberate sulfide from free cysteine during cluster assembly. In some bacterial phyla, the SufE protein acts in concert with SufS as a sulfur transfer partner for Fe-S cluster assembly. All three systems also contain members of the A-type carrier (ATC-II) family of Fe-S biosynthesis proteins (IscANif, IscA, and SufA) that contain three conserved cysteine residues involved in Fe-S cluster coordination (6, 14). Despite some early controversy concerning the role of ATC-II proteins, all recent biochemical and genetic analyses suggest that they bind Fe-S clusters in vivo and are able to transfer the clusters to target apoproteins (14-16).
The model organism E. coli carries the sufABCDSE operon that is required for stress-responsive Fe-S cluster assembly. Recently it was shown that E. coli SufA, co-expressed with the other Suf proteins, binds a [2Fe-2S]2+ cluster in vivo that can be transferred to target Fe-S apoproteins (15). However, recent studies have also shown that the SufB can assemble an iron-sulfur cluster in vitro (17). In vivo and in vitro, SufB forms a stable complex with SufC and SufD (referred to here as SufBCD) and all three proteins are necessary for in vivo Fe-S cluster assembly (8, 9, 18). Studies in our lab have shown that the SufBCD complex can also be reconstituted in vitro with an Fe-S cluster similar to SufB alone (A. Saini, manuscript in preparation). Since both SufA and the SufBCD complex can assemble Fe-S clusters, this raises the question of how they function in Suf-mediated Fe-S cluster assembly. Do SufA and SufBCD work together in a linear assembly pathway, where one protein functions as an Fe-S scaffold and the other as an Fe-S shuttle? Alternatively, do SufA and SufBCD work in parallel cluster assembly pathways, where each protein functions as a separate scaffold for particular cluster types or specific target apo-enzymes? In order to answer these questions we analyzed the protein-protein interactions among the Suf proteins and the ability of SufA and SufBCD to form Fe-S clusters in vitro. Our studies indicate that SufA is an Fe-S cluster shuttle protein that receives its cluster from the SufBCD scaffold complex prior to insertion of the cluster into target apo-enzymes.
Materials and Methods
Strains, plasmids, and growth conditions
SufA was amplified from MG1655 chromosomal DNA as template using the primers 5′-TAAACATATGGACATGCATTCAGGAACCTTTA-3′and 5′-ATAGGGATCCCTATACCCCAAAGCTTTCGCCACAG-3′. PCR products were digested with BamHI and NdeI and cloned into the corresponding sites of pET21a (Novagen), generating plasmid pET21a-SufA. The nucleotide sequences of the plasmid insert was confirmed by DNA sequencing. E. coli BL21(DE3) containing the pET21a-SufA expression vector was grown in LB at 30 °C. Isopropyl-1-thio-β-D-galactopyranoside was added at 500 μM final concentration for 6 h to induce SufA expression. The plasmid pGSO164 containing the entire suf operon under the control of arabinose-inducible promoter (18) was used to over-express SufABCDSE in the TOP10 strain of E. coli. The cells were grown in LB at 37 °C and L-arabinose was added to 0.2% final concentration by weight for 3 h to induce the expression of SufABCDSE. After induction, cells were harvested by centrifugation and cell pellets were frozen at -80 °C.
Protein purification
The SufBCD complex was purified as described previously (17), using Phenyl FF, Q-sepharose, and Superdex 200 chromatography resins in sequence. SufA was purified by freeze-thaw method as follows: Briefly, the cell pellet was thawed on ice and resuspended in buffer containing 25 mM Tris-Cl, pH 7.5, 100 mM NaCl, 10 mM βME, 2× EDTA-free protease inhibitor tablets. The pellet was refrozen at -80 °C for 1 h. The freeze-thaw cycle was repeated two more times. The freeze-thaw extract was centrifuged at 20,000×g for 20 min and the clear lysate was loaded onto a Q-sepharose anion exchange column. The protein was eluted with a linear gradient of 25 mM Tris-Cl, pH 7.5, 1 M NaCl, 10 mM βME. The fractions containing SufA were collected and concentrated to 3 mL and loaded onto HiLoad16/60 Superdex 75 gel filtration column equilibrated with 25 mM Tris-Cl, pH 7.5, 150 mM NaCl, 10 mM βME. Fractions containing SufA dimer were concentrated and frozen at -80 °C until further use.
Crossing-linking and Label Transfer
Purified SufA was labeled with a trifunctional cross-linker Mts-Atf-biotin (2-[N2-(4-azido-2,3,5,6-tetrafluorobenzoyl)-N6-(6-biotinamidocaproyl)-L-lysinyl]ethyl methane-thiosulfonate (Pierce). This crosslinker contains a sulfhydryl-specific methane-thiosulfonate (Mts) moiety that was used to attach Mts-Atf-biotin specifically to cysteine residues in SufA. It also contains a photo-activated tetrafluorophenyl azide moiety (Atf). The Atf moiety will insert into carbon-hydrogen bonds within 11.1 Å of the cross-linker upon exposure to UV light. SufA (50 μM) was mixed with 250 μM Mts-Atf-biotin in a total reaction volume of 300 μL in phosphate-buffered saline (0.1 M, pH 7.2). After 1 h incubation at room temperature, the unreacted Mts-Atf-biotin was removed by Zeba Desalting spin columns (Pierce) according to the manufacturer's protocol. Labeling reactions were carried out in the absence of ambient light to prevent premature activation of the Atf moiety. Addition of reductant to labeled SufA was able to remove the label, indicating that Mts-Atf-biotin binds SufA as expected via reducible disulfide bonds with cysteine residues.
Mts-Atf-biotin-labeled SufA (4 μM) was mixed with the other Suf proteins (2 μM) in 100 μL of phosphate-buffered saline. The reactions were incubated for 1 h at room temperature. Samples were irradiated with UV light for 5 min at a distance of 10 cm using a Spectroline Model BIB-150P UV lamp (312 nm) to initiate cross-linking with the Atf moiety. After UV light exposure, 4×LDS sample buffer (Invitrogen) with 1.2 M β-mercaptoethanol was added. The samples were separated by denaturing gel electrophoresis on 4–12% Bis-Tris gels and blotted to nitrocellulose membrane. Horseradish peroxidase-conjugated streptavidin (Pierce) was used to visualize proteins labeled with Mts-Atf-biotin. Where indicated, the relative intensity of labeled bands from the immunoblots was quantified using ImageJ software from NIH.
Surface Plasmon Resonance
Surface plasmon resonance experiments were performed on a Biacore 3000. SufA or SufE were covalently immobilized to the carboxylated dextran matrix on a CM5 sensor chip (Biacore) via primary amino groups using the amine-coupling protocol (Biacore). Levels of SufA or SufE immobilized to the chip were 1300-1600 RU (response units) for all experiments. Purified (His)6-SufS and/or SufE were injected at various concentrations. All experiments were performed at a flow rate of 20 μL/min in HBS-EP buffer (Biacore).
SufA iron donation to SufBCD
Apo-SufA (1 mM) was incubated with 5-fold excess of ferrous ammonium sulfate (FAS) in the presence of 5 mM DTT. After 1 h, Fe-SufA was purified by anion exchange chromatography using Hitrap Q FF 1-mL column. The purified, concentrated Fe-SufA contained 1.2 Fe/monomer and was added in excess to provide 5 equivalents Fe to a reaction buffer containing SufBCD (300 μM) and SufS-SufE/L-cysteine. The reaction mixture was incubated for 1.5-2 h and then separated by anaerobic gel filtration. As a comparison, SufBCD (300 μM) was reconstitiuted in the same way except that the Fe source was ferrous ammonium sulfate.
In vitro Fe-S reconstitution
The pure SufA and SufBCD proteins used in this study always had variable amounts of Fe (0.02 - 0.18 per monomer) in their as-purified forms and were converted to their fully apo forms following published procedures (19). Briefly, the apoproteins were obtained by incubating the proteins with EDTA and potassium ferricyanide (molar ratios 1:50:20) on ice for 5-10 min followed by desalting.
Both SufA and SufBCD (1 mM) were incubated separately in an anaerobic glove box (Coy) in reconstitution buffer containing 25 mM Tris, pH 7.5, 100 mM NaCl, 5 mM dithiothreitol (DTT), with 10-fold excess of L-cysteine and ferrous ammonium sulfate (FAS) and 4 μM SufS and SufE. After 1.5-2 h, the proteins were purified by anaerobic anion exchange chromatography using a Hitrap Q FF 1-mL column. The SufA or SufBCD fractions (colored due to cluster) were concentrated on Nanosep 10K or 30K centrifugal devices. In order to assess [Fe-S] cluster reconstitution on SufA and SufBCD in the SufABCDSE mixture, 500 μM each of SufA and SufBCD were incubated together in the reconstitution buffer with 2 μM SufS and SufE. After 1.5-2 h, proteins were separated anaerobically on a Superdex 200 gel filtration column. As controls, both SufA and SufBCD proteins were also reconstituted separately in the same manner.
Fe-S cluster transfer monitored by anaerobic chromatography
The concentrated holo-SufA and holo-SufBCD were used as Fe-S cluster donors for apo-SufBCD and apo-SufA respectively. Apo-SufA (300 μM) was incubated anaerobically in 200 μL of buffer (25 mM Tris, pH 7.5, 100 mM NaCl, 5 mM DTT), with 3-4 fold molar excess (relative to protein concentration) of holo-SufBCD containing 1.63 Fe/complex and 1.56 S/complex for 60 min. The proteins were separated anaerobically on a Superdex 200 gel filtration column. The same procedure was followed when apo-SufBCD was used as acceptor and holo-SufA (1.38 Fe/monomer, 1.30 S/monomer) as donor protein. For all assays, Fe content was determined colorimetrically using ferrozine (20). The acid labile sulfide was determined by a previously reported method (21).
Fe-S cluster transfer monitored by circular dichroism
ApoSufBCD (300 uM) was reconstituted anaerobically using SufSE (1.2 μM), L-cysteine (3 mM), and FAS (3 mM) in 25 mM Tris, pH 7.5, 100 mM NaCl, 5 mM DTT. After 2 h incubation, the sample was purified on a Hitrap Q FF 1-mL column. After purification the holoSufBCD sample was concentrated and split into two aliquots (280 μM each). An equimolar amount of apoSufA (280 μM) was added to one aliquot of holoSufBCD (300 uL total volume) and scans were taken at 10 min intervals for 1 h at 25°C in 0.2 cm path anaerobic cuvettes using a JASCO J-815 spectrometer. The same transfer was repeated (using the second aliquot of holoSufBCD) in the presence of EDTA (60 μM), which was added after the addition of apoSufA to holoSufBCD. The ratio of EDTA:protein was determined experimentally by monitoring stability of the Fe-S cluster on holoSufBCD and holoSufA (reconstituted separately) in the presence of increasing EDTA using UV-visible absorption spectroscopy (Fig. S3B). The highest ratio of EDTA:protein that does not significantly disrupt a preformed cluster on holoSufBCD or holoSufA was chosen for the CD experiments. For control experiments with holoSufBCD, the amount of Fe and S in the reconstituted holoSufBCD was determined (Fe = 1.7/complex, S=1.6/complex) and the same amount of Fe (470 μM) and sulfide (453 μM) was provided as FAS and Na2S in the 300 μL reaction for assembling cluster in apoSufA (280 μM). Scans of this reaction were taken at intervals of 10 min for 1 h at 25°C in 0.2 cm path anaerobic cuvettes using a JASCO J-815 spectrometer. The same control reaction was repeated in the presence of EDTA (60 μM), which was added after the addition of Na2S and FAS to apoSufA. For all CD experiments approximately 10 minutes passed from the initiation of the reaction until the first scan was taken due to transport of the anaerobically-sealed cuvette to the instrument. All spectra were processed using the Spectra Analysis program from JASCO.
Interaction of His6-SufA with apo- and holo-SufBCD
All steps were carried out anaerobically in a Coy chamber. The Ni-NTA 1-mL column was charged with 500 μg of His6-SufA protein. Next 200 μM apo-SufBCD or freshly prepared holo-SufBCD (reconstituted and purified as described above) were loaded on the column and allowed to incubate for 5 min prior to resuming flow. The column then was washed with 3 mL of binding buffer (20 mM sodium phosphate, pH 7.4, 0.5 M NaCl, and 5 mM imidazole). Then His6-SufA and any proteins bound to it were eluted with 20 mM sodium phosphate, pH 7.4, 0.5 M NaCl, and 500 mM imidazole. All wash and elution fractions were collected and concentrated separately using Nanosep centrifugal devices. Equal volumes of elution fractions (concentrated to the same final volume) were separated by SDS-PAGE. As controls, apo- and holo-SufBCD were loaded, washed, and eluted on a Ni-NTA column containing no His6-SufA protein. No non-specific SufBCD binding to the Ni-NTA column was observed for these controls (all SufBCD protein was found in the wash fractions).
Results
SufA interacts with SufB and SufC
To determine the stepwise interactions that occur between SufA and the other Suf proteins, we utilized the trifunctional cross-linker Mts-Atf-Biotin in a label transfer reaction as described previously (17). Briefly, we specifically labeled exposed cysteine residues in SufA with Mts-Atf-Biotin to generate the bait protein, and performed the label transfer reaction with all other Suf proteins (Fig. 1). Mts-Atf-Biotin specifically senses protein-protein interactions within 11 Å of a labeled cysteine residue. SufA contains three cysteines, all of which are highly conserved (13). Based on analysis of the SufA crystal structure, two of the three conserved cysteine residues of each SufA monomer (Cys114 and Cys116) are co-localized to the SufA dimer interface within 3 – 6 Å of each other while the third cysteine (Cys50) is nearby at a distance of approximately 8 - 9 Å from the other cysteines (22). Therefore, our label transfer assay will detect interactions that occur fairly close to this localized Fe-S cluster-binding site of SufA and may not indicate protein-protein interactions that involve more distant regions of SufA. However, given the importance of the conserved cysteines at the SufA dimer interface for in vivo function, protein-protein interactions that occur in the vicinity of that region also must be critical for Suf function.
Figure 1.
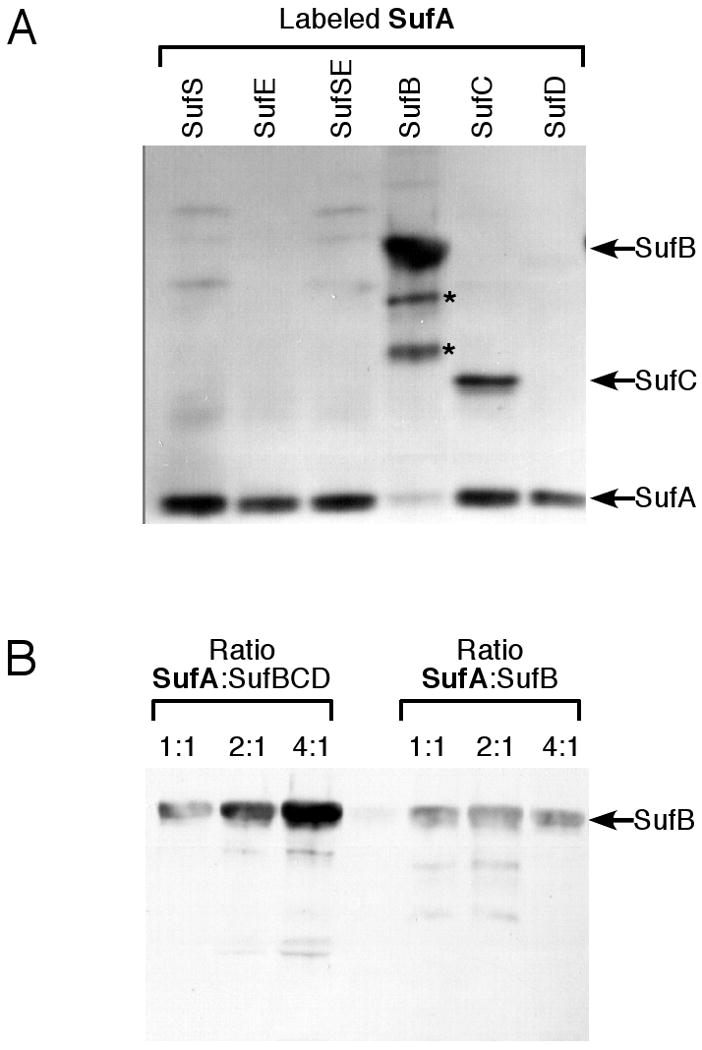
Label transfer analysis of SufA interactions with the other Suf proteins. (A) SufA (4 μM) pre-labeled with Mts-Atf-Biotin was incubated for 1 h with 2 μM of the other Suf proteins individually or in various combinations. Lower molecular weight bands below SufB (indicated by *) were confirmed by mass spectrometry to be proteolysis products of SufB. (B) Increasing amounts of SufA pre-labeled with Mts-Atf-Biotin were incubated for 1 h with 2 μM of SufB or the SufBCD complex. After UV-light induced cross-linking, samples from (A) and (B) were separated by reducing SDS-PAGE and the location of the biotin tag was determined by immunoblot using streptavidin conjugated to horseradish peroxidase.
After activation of protein cross-linking by Mts-Atf-Biotin with UV light, samples were reduced and biotin transfer from SufA to the other Suf proteins was detected via immunoblot (Fig. 1). SufA interacted with both SufB and SufC, resulting in detectable label transfer to those proteins (Fig. 1A). No strong interactions were observed between SufA and the other SufS, SufE, or SufD proteins. Next, increasing concentrations of labeled SufA were mixed with SufB alone or the SufBCD complex (Fig. 1B). SufA label transfer to SufB increased when SufB was bound as part of the SufBCD complex as compared to SufB alone. In contrast, the interaction of SufA with SufC seemed to diminish if SufC is present as part of the SufBCD complex (Fig. 1B). To further confirm this result, increasing concentrations of labeled SufA were mixed with SufC alone or the SufBCD complex. As initially observed, SufA interaction with SufC is diminished if SufC is part of the SufBCD complex (Fig. S1A). The results from these experiments suggest that the conformation of SufB in the SufBCD complex is altered (as compared to SufB alone) to enhance overall SufA binding or to bring the labeled SufA cysteines closer to SufB. Since there is currently no clearly defined functional role for SufC ATPase activity, we also tested if ATP affects the interaction between SufA and SufBCD. Increasing concentrations of ATP in the label transfer reaction showed no effect on the SufA-SufBCD interactions (Fig. S1B), indicating that the ATPase activity of SufC is not involved in the SufA and SufB interaction under our in vitro conditions.
SufS and SufSE reduce SufA label transfer to SufBCD
We previously demonstrated that SufE binds to SufB in the SufBCD complex in order to donate persulfide sulfur for Fe-S cluster assembly (17). To determine if the presence of the sulfur donation system SufS and SufE alters the interaction between SufA and SufBCD, we repeated the SufA-SufBCD label transfer reaction with unlabeled SufS and SufE added individually or together (Fig. 2). The label transfer reactions were conducted with a constant amount of labeled SufA and SufBCD but increasing concentrations of unlabeled SufS, SufE, or SufSE. The label transfer between SufA and SufBCD was slightly diminished if SufE was present at a 4-fold excess over SufA (Fig. 2A). However, SufS began to block SufA label transfer at equimolar protein ratios and further diminished the label transfer as its concentration increased (Fig. 2A). The SufSE complex also blocked label transfer between SufA and SufBCD in a manner similar to SufS alone although it was slightly less efficient than SufS alone based on quantification of the relative intensity of the labeled SufB band using ImageJ software (Fig. 2B).
Figure 2.
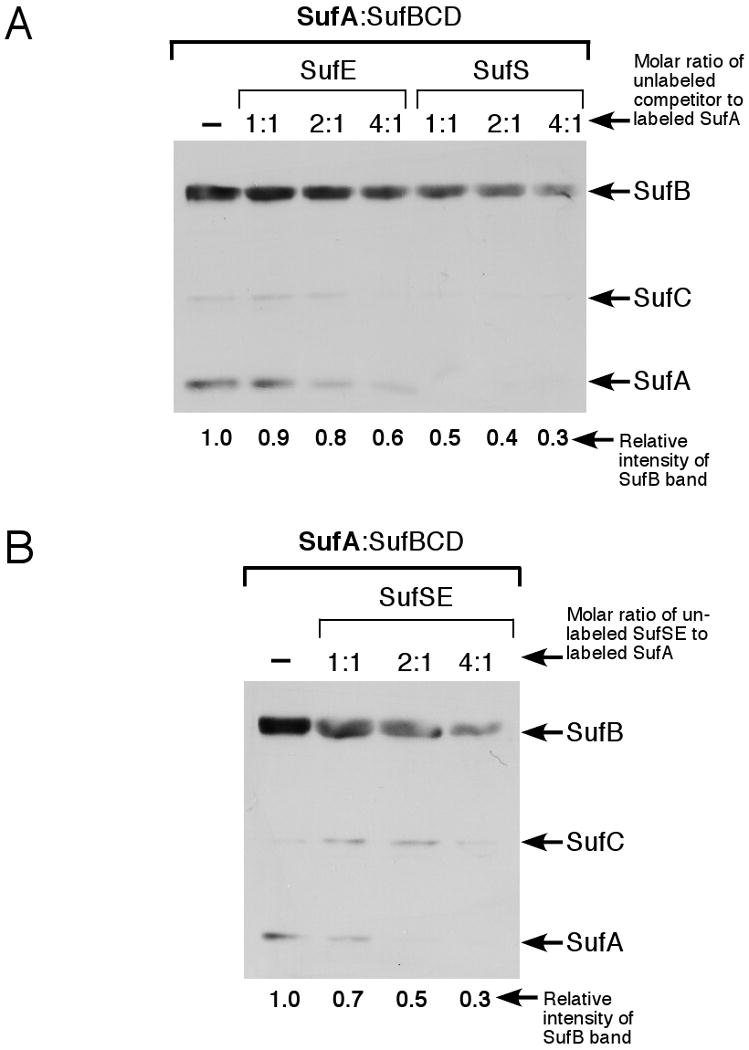
Label transfer analysis of SufA interactions with the SufBCD complex in the presence of SufS and SufE. (A) Increasing amounts of unlabeled SufE or SufS or SufSE complex (B) were added to a mixture of 4 μM pre-labeled SufA and 2 μM SufBCD complex. (B) Increasing amounts of unlabeled SufSE complex were added to a mixture of 4 μM pre-labeled SufA and 2 μM SufBCD complex. After UV-light induced cross-linking, samples were separated by reducing SDS-PAGE and the location of the biotin tag was determined by immunoblot using streptavidin conjugated to horseradish peroxidase. The relative intensity of the SufB band in each blot was quantified by ImageJ software and normalized to lane 1 of each blot.
There are two interpretations of these results. First, it is possible that both SufA and SufSE interact with SufBCD at a common binding site or at two binding sites that at least partially overlap. Such a common binding site would preclude simultaneous binding by both SufA and SufSE. Alternatively, SufS or SufSE may interact with SufA and block SufA binding to SufB. However, if SufS and SufA do interact, the site of interaction must be distant from the labeled SufA cysteines since we see no label transfer from SufA to SufS or SufSE (Fig. 1). To test these possibilities we further analyzed the interactions between SufA and SufS using surface plasmon resonance. SufA was covalently immobilized while SufS was added in solution. SufA and SufS did interact in this assay. The KD for the SufA-SufS interaction (1.4 μM) was calculated using observed kon and kof and was approximately three orders of magnitude higher than the KD for the strong SufE-SufS interaction (1.1 nM) measured under similar conditions. To determine if the SufSE complex interacts with SufA, we immobilized SufA while SufSE (pre-mixed prior to injection) were added in solution. We found that the weak interaction between SufA and SufS was completely abrogated as concentrations of SufE were increased (Fig. S2A). Even a 1:1 ratio of SufE:SufS was enough to block binding of SufS to immobilized SufA. SufE itself did not interact with immobilized SufA (data not shown), in good agreement with the label transfer results shown in Figure 1 and with our previous studies (17).
The sum of the label transfer and surface plasmon resonance experiments indicate that SufS can weakly interact with SufA but that this interaction does not take place in the vicinity of the labeled cysteines in SufA (Fig. 1). The reduction of SufA label transfer to SufBCD in the presence of SufS (Fig. 2A) may result from direct SufA-SufS interactions or from competition between SufA and SufS for a common binding site on SufBCD. With the data in hand we cannot directly distinguish between those two possibilities. In contrast, both label transfer and surface plasmon resonance methods show that the SufSE complex does not interact with SufA (Fig. 1 and Fig. S2A). Therefore, the disruption of SufA label transfer to SufBCD in the presence of SufSE logically results from competition between SufA and SufSE for a common binding site on SufBCD (rather than from SufSE binding and sequestration of SufA). While a subtle point, this distinction has important implications for establishing the step-wise progression of the Suf Fe-S cluster assembly pathway. Our results indicate that SufA interacts with the SufBCD complex and not with the physiological SufSE sulfur transfer system. In fact, SufA and SufSE seem to compete for a common binding site on the SufBCD complex.
SufA does not donate iron to SufBCD for Fe-S cluster assembly
The observed protein-protein interactions between SufA and the other Suf proteins suggest that SufA works with the SufBCD complex downstream of the SufSE sulfur donation system. Previous studies have shown that the SufA homologue IscA can act in vitro as an iron donor for cluster assembly on the IscU scaffold (23-25). A role for SufA as an iron donor to the SufBCD complex is consistent with our protein-protein interaction data. We tested if SufA may act as potential iron donor for Fe-S cluster reconstitution on SufBCD. SufA protein was incubated with Fe to generate Fe-SufA protein (1.22 Fe/monomer). Anaerobic Fe-S cluster reconstitution on apo-SufBCD was carried out using Fe-SufA in 4-5 fold molar excess over SufBCD as the source of iron. In a parallel control, apo-SufBCD was reconstituted using the same fold excess of ferrous ammonium sulfate (FAS) as iron source. In both cases purified SufSE with L-cysteine were used for sulfur donation. Following reconstitution, the reaction components were separated anaerobically by gel filtration, which allows a complete separation of SufA dimer (∼30 kDa) and the SufBCD complex (∼160 kDa), and SufA and SufBCD fractions were analyzed separately for Fe-S cluster content. After reconstitution and purification, SufBCD incubated with Fe-SufA did not display any characteristic absorption bands in the 300-600 nm region (Fig. S2B). Analysis of Fe and acid-labile sulfide (S) content of SufBCD reconstituted with Fe-SufA showed little to no cluster formation (0.1 Fe/complex, 0.2 S/complex). In contrast, SufBCD reconstituted with FAS as iron source showed UV-visible features consistent with the presence of an Fe-S cluster (Fig. S2B) and contained 1.5 Fe/complex and 1.4 S/complex.
After reconstitution and separation, SufA showed two broad bands at 315 nm and 420 nm (Fig. S2B) suggesting an Fe-S cluster rather than binding of Fe only. SufA contained 1.1 Fe/monomer and 1.0 S/monomer) indicating that SufA retained most of its originally bound iron and may have been partially reconstituted with a cluster, perhaps due to the presence of sulfide generated by SufSE or to some other function performed by SufBCD (see below).
SufBCD enhances Fe-S cluster formation in SufA
Based on the UV-visible absorption spectrum of SufBCD reconstituted with FAS and SufSE plus L-cysteine, we propose that SufBCD contains an Fe-S cluster similar to SufB alone. In vivo and in vitro characterization of the SufBCD cluster is currently under investigation (A. Saini, manuscript in preparation). These results raise the question of whether SufA and SufBCD are in the same linear pathway or two parallel pathways for Fe-S cluster assembly. If SufA and SufBCD serve as parallel scaffolds for different target apoproteins, then they may compete for Fe and S such that the formation of cluster on one protein would be affected by the presence of the other.
To determine how the presence of apo-SufA and apo-SufBCD influence Fe-S cluster formation on each other, the following strategy was used: Apo forms of both SufA and SufBCD were incubated together in the presence of FAS and SufSE plus L-cysteine under anaerobic conditions. For comparison, the SufA and SufBCD were reconstituted separately under the same reaction conditions. After anaerobic gel filtration chromatography, fractions containing SufBCD and SufA were collected and concentrated for analysis. The SufA fractions were reddish in color, whereas those containing SufBCD were slightly yellowish brown. UV-visible absorption spectrum of SufA displayed absorption bands at 330, 420 and 460 nm that are characteristic of a [2Fe-2S]2+ cluster on SufA (Fig. 3A) (15, 16, 26). The UV-visible absorption spectrum of SufBCD also indicated the presence of an Fe-S cluster with absorption maxima at 414 and 460 nm and a broad shoulder at around 315 – 320 nm (Fig. 3B).
Figure 3.
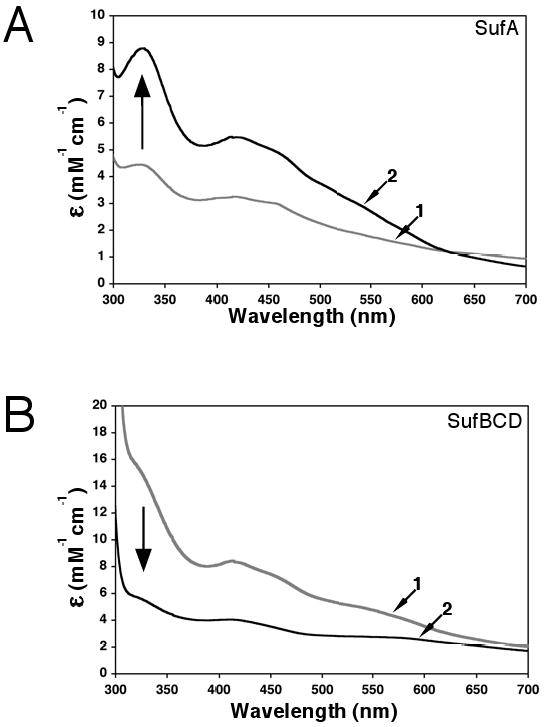
Fe-S cluster reconstitution using the entire Suf pathway. SufA and the SufBCD complex (500 μM each) were incubated anaerobically with 2 μM SufSE, L-cys, and FAS for 1.5 hours. SufA and SufBCD were separated by anaerobic gel filtration and analyzed for Fe-S cluster content. Control reconstitutions using only SufA or SufBCD were carried out under the same conditions. Arrows indicate direction of change for spectra of SufA or SufBCD reconstituted alone compared to spectra from samples reconstituted together. (A) UV-visible absorption spectra of SufA reconstituted alone (trace 1, ε456 =3 mM-1cm-1) or SufA reconstituted with SufBCD (trace 2, ε456 = 4.95 mM-1cm-1) or. (B) UV-visible absorption spectra of SufBCD reconstituted alone (trace 1, ε456 = 7.2 mM-1cm-1) or SufBCD reconstituted with SufA (trace 2, ε456 = 3.4 mM-1cm-1).
SufA reconstituted with SufBCD displayed a UV-visible absorption spectrum maxima at 330, 420 and 460 nm and these peaks were all more intense and better defined than the UV-visible features of SufA reconstituted alone (Fig. 3A). In the case of SufBCD reconstituted with SufA, the absorption maxima at 414 and 460 nm and the broad shoulder at 315 – 320 nm were decreased in intensity and less well-defined when compared to SufBCD reconstituted alone. The iron and acid-labile sulfur analyses of three independent reconstitution trials showed that the cluster content of SufA reconstituted with SufBCD (1.51 ± 0.05 Fe/monomer, 1.38 ± 0.06 S/monomer) was always higher than SufA reconstituted alone (1.03 ± 0.03 Fe/monomer, 1.30 ± 0.06 S/monomer). UV-visible absorption spectra of SufA from representative reconstitution assays are shown in Fig. 3A. In contrast, the iron and acid-labile sulfur analyses from the same trials showed that the cluster content of SufBCD reconstituted with SufA (0.78 ± 0.16 Fe/complex, 1.06 ± 0.09 S/complex) was always reduced as compared to that in SufBCD reconstituted alone (1.50 ± 0.19 Fe/complex, 1.35 ± 0.16 S/complex). UV-visible absorption spectra of SufBCD from representative reconstitution assays are shown in Fig. 3B.
The decrease in cluster content of SufBCD in the presence of SufA could be consistent with SufA and SufBCD acting as separate scaffolds that compete with each other for iron and sulfur acquisition. However, the relative increase in Fe-S cluster content of SufA reconstituted with SufBCD as compared to that of SufA alone suggests a role for SufBCD in enhancing Fe-S cluster formation in SufA. Since both SufBCD and SufA form Fe-S clusters, SufBCD could be acting as a scaffold complex that assembles and transfers Fe-S clusters to SufA, thereby enhancing cluster formation. While either model is consistent with these results and with the recent finding that SufA co-expressed with SufBCDSE can be purified as a [2Fe-2S]2+ protein in vivo (15), the protein-protein interaction studies (Fig. 1-3) clearly show that SufA interacts with SufBCD but not with the SufSE sulfur donation system. The combined interaction and reconstitution data suggests that SufBCD interacts with SufA to transfer Fe-S cluster or otherwise enhance cluster assembly. Based on these results, we speculated that SufBCD may act as an Fe-S cluster scaffold that transfers its cluster to SufA thereby accounting for the observed enhancement of cluster formation on SufA if SufBCD is present.
SufBCD transfers an Fe-S cluster to SufA
To test if cluster transfer occurs between SufBCD and SufA, the pure SufA or SufBCD proteins were separately reconstituted to Fe-S holoproteins as described above. Each holoprotein was completely separated from SufSE, L-cysteine, and FAS using anaerobic anion exchange chromatography and then incubated with the corresponding apo form of the other protein. First, apo-SufA was incubated with 3-4 fold excess of holo-SufBCD (containing 1.63 Fe/complex, 1.56 S/complex). Following the transfer reaction, the proteins were separated by anaerobic gel filtration as above. The fractions containing SufA were reddish in color indicative of the presence of an Fe-S cluster (Fig. 4B). SufBCD fractions, on the other hand, were devoid of any visible color (Fig. 4A). The UV-visible absorption spectrum of SufA after Fe-S transfer displayed absorption bands at 330, 420 and 460 nm indicating the presence of a [2Fe-2S]2+ cluster (Fig. 4B). On the other hand, SufBCD fractions lost the UV-visible bands initially present before transfer (Fig. 4A). The disappearance of absorption bands in SufBCD fractions with subsequent appearance in SufA clearly showed the transfer of cluster from SufBCD to SufA. Furthermore, Fe and S content in SufA (1.1 Fe/monomer, 1.2 S/monomer) and the observed ε456= 3.1 mM-1cm-1 were consistent with 1 × [2Fe-2S]2+ cluster per SufA dimer. In contrast, the amount of Fe and S decreased to 0.63 Fe/complex to 0.44 S/complex in SufBCD. In order to ensure that the SufBCD cluster remains intact during the time course of transfer, an aliquot of holo-SufBCD alone was monitored by UV-visible spectroscopy for the same time period and under the same conditions as the transfer reaction (Fig. 4C). The UV-visible absorption spectrum of this control did not change over the time course of this experiment, indicating that the cluster on SufBCD does not degrade and further supporting direct cluster transfer to SufA (Fig. 4C).
Figure 4.
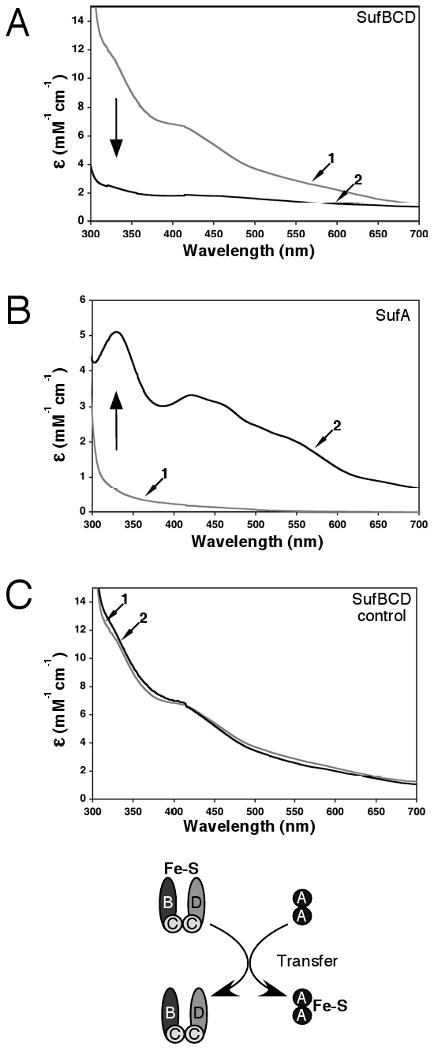
Fe-S cluster transfer from the SufBCD complex to SufA. Apo-SufA (300 μM) was incubated for 60 min with enough holo-SufBCD to provide a 3-4 fold molar excess of iron relative to the SufA concentration. SufA and SufBCD were then separated by anaerobic gel filtration and analyzed for Fe-S cluster content. Fe-S holo-SufBCD was prepared as described in Materials and Methods. Arrows indicate direction of change for spectra of holo-SufBCD or apo-SufA samples taken before transfer compared to spectra taken after transfer. (A) UV-visible absorption spectra of the SufBCD complex before (trace 1) and after (trace 2) the transfer reaction. (B) UV-visible absorption spectra of SufA before (trace 1) and after (trace 2) the transfer reaction. (C) Fe-S holo-SufBCD was incubated under the same conditions as the transfer reaction but without addition of apo-SufA. UV-visible absorption spectra of holo-SufBCD at time = 0 (trace 1) and time = 60 min (trace 2).
The transfer of cluster was also studied in the other direction, from holo-SufA to apo-SufBCD following the same procedure as above. In this case, apo-SufBCD was incubated anaerobically with 3-4 fold excess of holo-SufA (1.38 Fe/monomer, 1.30 S/monomer). The transfer reaction was then separated by anaerobic gel filtration and the fractions containing SufA and SufBCD were collected and concentrated. Interestingly, in this case, the SufA fractions were again reddish in color whereas those containing SufBCD remained colorless (Figure 6A). The UV-visible absorption spectrum of the SufA fraction was similar in intensity to that of initial holo-SufA protein but did show some sharpening of the peak at 330, 420, and 460 nm, possibly indicating subtle shifts in cluster redox state or coordination environment. In contrast, no UV-visible features were observed in the range of 300-600 nm for SufBCD after transfer (Figure 6B). Analyses of the Fe and S content after transfer showed essentially no significant change in the amount of cluster for SufA (1.27 Fe/monomer, 1.45 S/monomer, ε456=3.2 mM-1cm-1) or SufBCD (0.13 Fe/complex, 0.23 S/complex). These results indicate that in vitro transfer of Fe-S cluster from SufBCD to SufA occurs but that transfer from SufA to SufBCD is not observed under the same conditions.
Figure 6.
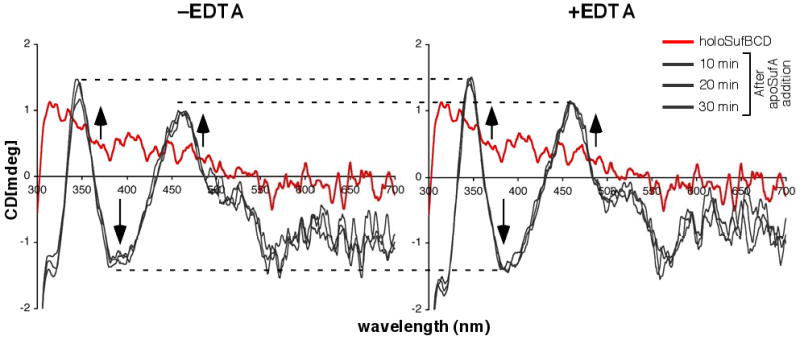
Monitoring Fe-S cluster transfer from holoSufBCD to SufA by CD spectroscopy. 280 μM holoSufBCD was mixed with 280 μM apoSufA in the absence (left panel) or presence (right panel) of 60 μM EDTA. Changes in CD spectra were monitored over time (grey traces). CD spectrum of holoSufBCD alone (red trace) is shown for reference. Dashed lines are shown to more easily compare relevant features between samples. Arrows indicate direction of change to CD spectra within each region over time upon apoSufA addition.
Although holoSufBCD appears to be stable under the strictly anaerobic conditions used for the cluster transfer reaction, there still exists the possibility that the SufBCD cluster disassembles into iron and sulfide that is then non-specifically bound by apoSufA. To address this possibility, we also analyzed holoSufBCD to apoSufA Fe-S cluster transfer over time by circular dichroism (CD) spectroscopy in the presence of the divalent metal chelator EDTA. CD analysis of holoSufBCD alone showed a broad positive (+) signal at around 315 – 320 nm but no other clearly defined features in the visible region (Fig. S3A). In contrast, the CD spectra of holoSufA alone has well-defined features in the visible region, including positive (+) maxima at 345 and 455 nm, negative (-) maximia at 317, 391, and 559 nm, and complex negative features from 500 – 700 nm (Fig. S3A). Therefore, CD spectroscopy can be used to specifically monitor the Fe-S cluster of SufA during the cluster transfer reaction without separating the mixture by anaerobic chromatography.
HoloSufBCD was reconstituted and purified as described above and mixed with equimolar apoSufA. The Fe-S cluster transfer reaction was monitored over time by CD spectroscopy (Fig. 6, left panel). A duplicate transfer reaction was carried out in the presence of EDTA (Fig. 6, right panel). We observed rapid transfer of Fe-S cluster from holoSufBCD to SufA such that the (+) maxima at 345 and 455 nm and the (-) maxima at 317 and 391nm reached their greatest intensity in the first 20-30 minutes and did not change at subsequent time points up to 60 minutes. The presence of EDTA during the transfer reaction did not alter the time course of cluster transfer or the overall maximum intensity of the SufA cluster features (Fig. 6, right panel). A similar experiment was performed using ferrous ammonium sulfate and sodium sulfide to provide iron and sulfide in amounts equivalent to that in holoSufBCD (Fig. 7). These iron and sulfide sources are roughly equivalent to what would be present in solution if the holoSufBCD cluster disassembled. In contrast to the holoSufBCD to SufA cluster transfer (Fig. 6), binding of the Fe-S cluster in apoSufA using FAS and Na2S was diminished by the presence of EDTA (Fig. 7). Taken together, the results in Fig. 6 and Fig. 7 support a model where Fe-S cluster transfer from holoSufBCD to apoSufA occurs without cluster disassembly and is resistant to the presence of EDTA.
Figure 7.
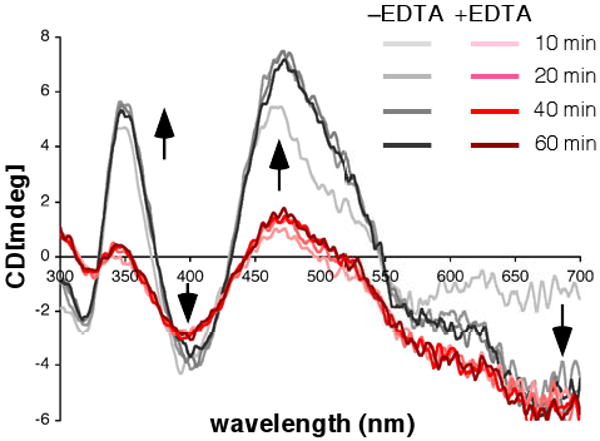
EDTA inhibits cluster acquisition by SufA. 280 μM apoSufA was incubated with 470 μM ferrous ammonium sulfate and 453 μM of sodium sulfide in the absence (grey traces) or presence (red traces) of 60 μM EDTA. Changes in CD spectra were monitored over time. Arrows indicate direction of change to CD spectra within each region over time after addition of iron and sulfide.
SufA interaction with SufB is enhanced for the SufBCD Fe-S holoprotein
Based on the enhancement of SufA Fe-S cluster content in the presence of SufBCD and the observed unidirectional transfer of Fe-S cluster from SufBCD to SufA, we tested if the presence of the Fe-S cluster on SufBCD alters SufA-SufB interactions. The protein-protein interaction studies carried out above (Fig. 1-3) were all performed with apoproteins. Due to the sensitivity of Fe-S clusters to oxidation, it is technically challenging to conduct label transfer or SPR analysis of Fe-S holoproteins. Therefore we utilized a slightly different approach to study such interactions.
His6-SufA (in the apo form) containing an N-terminal polyhistidine tag was bound to a Ni2+-NTA column under anaerobic conditions. Apo-SufBCD or reconstituted and purified Fe-S holo-SufBCD were then passed through the preloaded His6-SufA column under anaerobic conditions. After a brief incubation and subsequent washing step, His6-SufA and any proteins that interact with it were eluted from the column using a high imidazole buffer. The protein content of these elution fractions was analyzed by SDS-PAGE (Fig. 8). Separate controls were conducted to ensure that neither apo-SufBCD nor Fe-S holo-SufBCD bound non-specifically to the Ni2+-NTA column in the absence of His6-SufA (data not shown). The anaerobic interaction assay revealed that while His6-SufA does interact with apo-SufBCD (as observed using our label transfer assay), the His6-SufA-SufBCD interaction was greatly enhanced for Fe-S holo-SufBCD (Fig. 8). This interaction experiment was also repeated in the presence of ATP (in both the loaded holoSufBCD sample and running buffer) but ATP addition did not alter the amount of holoSufBCD that co-purified with His6-SufA (data not shown). These results are consistent with a linear model of Fe-S cluster assembly where cluster formation occurs first on the SufBCD complex followed by recruitment of apo-SufA and unidirectional Fe-S cluster transfer to the SufA Fe-S shuttle (Scheme 1).
Figure 8.
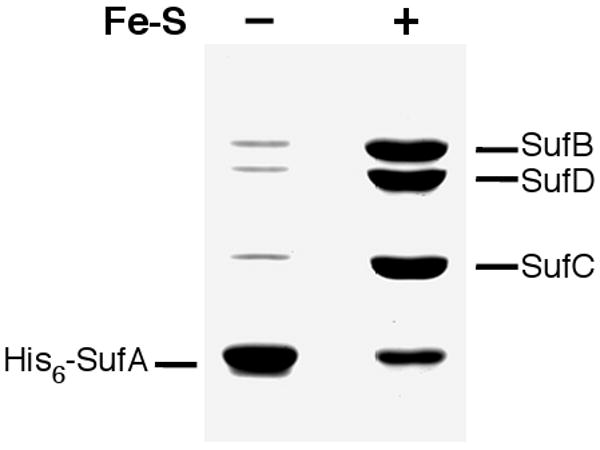
SufA interaction with apo and Fe-S holo forms of the SufBCD complex. His6-SufA was bound to a Ni2+-NTA column. Next, equal amounts of apo-SufBCD or Fe-S holo-SufBCD were passed over the His6-SufA-Ni2+-NTA column. After washing, all proteins bound to the column were eluted with high imidazole. All chromatography steps were carried out under strictly anaerobic conditions. Equal volumes of the elution fractions were then analyzed for protein content by SDS-PAGE.
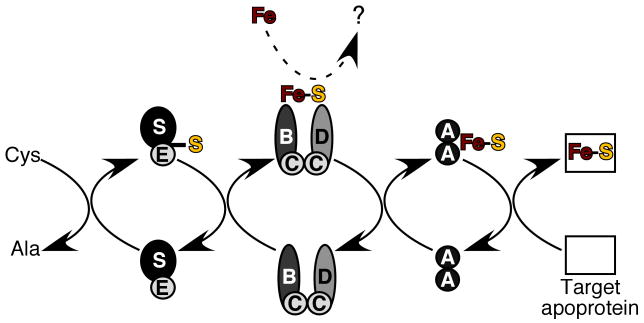
Scheme 1.
Current model of Suf-mediated Fe-S cluster assembly. Interactions and processes detailed in this work or previous studies are shown with bold arrows. The unknown process of iron donation is shown as a dashed arrow.
Discussion
Implications of protein-protein interactions for Suf-mediated Fe-S cluster assembly
Our label transfer results show that the labeled cysteines in the active site of SufA interact closely with SufBCD and SufB alone but do not interact with SufS, SufE or SufSE. While SufA can interact with SufB alone, the interaction is enhanced when SufB is present in the SufBCD complex (Fig. 1B). We previously reported that SufE interaction with SufB for sulfur transfer is also enhanced for the SufBCD complex compared to SufB alone, further confirming that the SufBCD complex is at the core of the Suf pathway (17). The SufSE complex reduces SufA binding to apo-SufBCD (Fig. 2B) but does not directly interact with SufA (Fig. 3), suggesting that SufSE and SufA share an overlapping binding site on SufBCD. The mutual exclusivity of SufSE and SufA interactions with SufBCD supports a model where SufA functions with the SufBCD complex to mediate a step downstream of the SufSE sulfur donation step during cluster assembly (Scheme 1). In this model SufA would not function as a scaffold and would carry out a function subsequent to de novo Fe-S cluster assembly. Such a model is consistent with previously published results showing that SufE and SufA do not interact (17) and that SufA does not enhance SufS or SufSE cysteine desulfurase activity (18).
In vivo the Suf pathway must limit release of sulfide and/or oxidation of reactive sulfur species under oxidative stress. The in vivo sensitivity of the Fe-S cluster assembly process to oxidative stress necessitates tight protein-protein interactions to shield reactive sulfur species from the cellular milieu, a proposition supported by the crystal structures of SufS and SufE, in which their cysteine active sites are at least partially solvent excluded (27, 28). The label transfer assays conducted here show that neither SufS nor SufE come within 11 Å of the labeled cysteines on SufA (Fig. 1). This result does contradict other in vitro studies that seem to show direct sulfur transfer from SufSE to SufA (29). At present we have no explanation for this discrepancy, although it may reflect non-physiological sulfur transfer under in vitro conditions. Although we observed weak interaction between SufS and SufA using surface plasmon resonance, this interaction was abolished in the presence of SufE. Both SufS and SufE are co-expressed from the same polycistronic message and both are required in vivo for Suf function (9, 30). SufE also is necessary to elevate SufS cysteine desulfurase activity to levels comparable to other cysteine desulfurases (such as IscS) (18, 31). Therefore the SufSE complex is the physiological sulfur transfer pathway and it is unlikely that SufA interacts with SufS alone for sulfur transfer in vivo since SufE will also be present. Possibly weak SufS-SufA interactions are relevant in the context of stabilizing a larger macromolecular complex that includes SufBCD and SufE at some step of Fe-S cluster assembly. Resolving these mechanistic details will require co-structures of the Suf protein complexes.
We also found that SufA can interact with SufC alone but that SufA interaction with SufC is reduced in the SufBCD complex (Fig. 1). This may indicate a direct role for SufC in recruiting SufA to the SufBCD complex. Such an interaction must be short-lived since SufA only transfers label to SufB in the SufBCD complex (at least under the steady state conditions used in our assays). Possibly after initial binding to SufC, SufA quickly migrates to a more stable binding site that places its active site cysteines closer to SufB. Clearly the ATPase activity of SufC is not required and does not effect SufA protein-protein interactions with SufBCD. The exact role of SufC in mediating the SufA and SufB interaction remains to be clarified at the structural level.
Implications of in vitro Fe-S studies for Suf-mediated Fe-S cluster assembly
In vitro Fe-S reconstitution utilizing the entire Suf operon revealed that the presence of SufBCD enhances Fe-S cluster formation on SufA and diminishes Fe-S cluster formation on SufBCD (Fig. 4). Both SufA and SufBCD formed Fe-S clusters under these conditions (as well as when reconstituted separately), supporting the hypothesis that SufA and SufB function as Fe-S scaffolds or Fe-S transfer proteins rather than as iron donors or sulfur trafficking proteins. In fact, we found that SufA did not act as an in vitro iron donor for Fe-S cluster assembly on SufBCD (Fig. S2).
Reconstituted Fe-S holo-SufBCD was able to transfer cluster to apo-SufA without any observable cluster degradation (Fig. 5). In contrast there was no observable transfer in the reverse direction from holo-SufA to apo-SufBCD (Fig. 5). Apo-SufA showed a stronger interaction with Fe-S holo-SufBCD than with apo-SufBCD (Fig. 6). Together these results are consistent with de novo Fe-S cluster assembly on SufBCD followed by unidirectional Fe-S cluster transfer to SufA. Our in vitro results suggest a linear model of Fe-S cluster assembly on the SufBCD scaffold followed by cluster transfer to the SufA Fe-S shuttle protein (Scheme 1).
Figure 5.
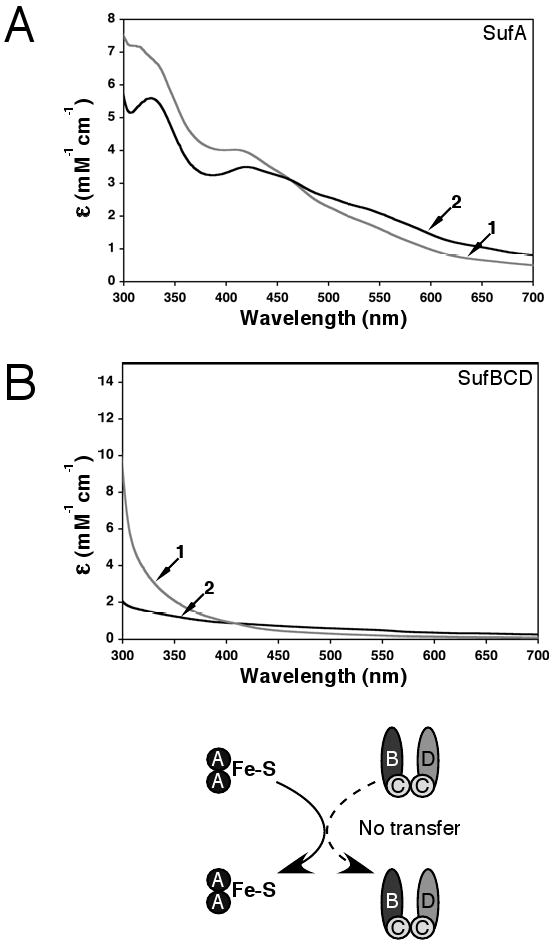
Fe-S cluster transfer from SufA to the SufBCD complex. Apo-SufBCD (300 μM) was incubated for 60 min with enough holo-SufA to provide a 3-4 fold molar excess of iron relative to the SufBCD concentration. SufA and SufBCD were then separated by anaerobic gel filtration and analyzed for Fe-S cluster content. Fe-S holo-SufA was prepared as described in Materials and Methods. (A) UV-visible absorption spectra of SufA before (trace 1) and after (trace 2) the transfer reaction. (B) UV-visible absorption spectrum of the SufBCD complex before (trace 1) and after (trace 2) the transfer reaction.
Elegant genetic studies in E. coli (14, 32) clearly show that the SufBCD complex can cross-talk with other Fe-S assembly systems in vivo. Genetic evidence suggests that SufBCD may be able to interact with IscA from the Isc basal Fe-S cluster assembly if SufA is absent (14). In addition to the Isc proteins, there are number of other proteins linked to in vivo Fe-S maturation that could interact with the Suf system, including ErpA, Grx4, and NfuA (33-36). Delineating the in vivo interactions among the various Fe-S cluster assembly and repair pathways should help provide a clear understanding of Fe-S metabolism.
Perhaps a question of greater importance for the Suf Fe-S cluster assembly pathway is the identity of the in vivo iron donor. Our in vitro assays utilize ferrous ammonium sulfate as an iron source but bioavailable iron is limited under the in vivo conditions where Suf is utilized (oxidative stress and iron starvation). The results presented here and other recent studies seem to rule out SufA as an iron donor for SufBCD (15). This suggests that SufC and/or SufD might play a role in iron acquisition in vivo. Current studies are underway to test these hypotheses.
This is the first study to present biochemical evidence that the SufBCD complex can function as an Fe-S cluster scaffold for the Suf pathway in E. coli. Our results confirm the central role of the SufBCD complex in Suf mediated Fe-S cluster assembly. Future studies will elaborate the biochemical mechanism of Fe-S cluster transfer from SufBCD to SufA and should provide insight into the roles of SufC and SufD for in vivo Fe-S cluster biosynthesis.
Supplementary Material
Acknowledgments
The authors thank C. E. Outten and M. Johnson for critical reading of the manuscript and helpful discussions.
Footnotes
Supporting Information Available. Supplemental Figures S1 – S3 are available on the World Wide Web at http://pubs.acs.org.
References
- 1.Beinert H. Iron-sulfur proteins: ancient structures, still full of surprises. J Biol Inorg Chem. 2000;5:2–15. doi: 10.1007/s007750050002. [DOI] [PubMed] [Google Scholar]
- 2.Kiley PJ, Beinert H. The role of Fe-S proteins in sensing and regulation in bacteria. Curr Opin Microbiol. 2003;6:181–185. doi: 10.1016/s1369-5274(03)00039-0. [DOI] [PubMed] [Google Scholar]
- 3.Jacobson MR, Cash VL, Weiss MC, Laird NF, Newton WE, Dean DR. Biochemical and genetic analysis of the nifUSVWZM cluster from Azotobacter vinelandii. Mol Gen Genet. 1989;219:49–57. doi: 10.1007/BF00261156. [DOI] [PubMed] [Google Scholar]
- 4.Kispal G, Csere P, Prohl C, Lill R. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 1999;18:3981–3989. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schilke B, Voisine C, Beinert H, Craig E. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:10206–10211. doi: 10.1073/pnas.96.18.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng L, Cash VL, Flint DH, Dean DR. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 7.Nachin L, El Hassouni M, Loiseau L, Expert D, Barras F. SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase. Mol Microbiol. 2001;39:960–972. doi: 10.1046/j.1365-2958.2001.02288.x. [DOI] [PubMed] [Google Scholar]
- 8.Nachin L, Loiseau L, Expert D, Barras F. SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 2003;22:427–437. doi: 10.1093/emboj/cdg061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Outten FW, Djaman O, Storz G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol. 2004;52:861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 10.Patzer SI, Hantke K. SufS is a NifS-like protein, and SufD is necessary for stability of the [2Fe-2S] FhuF protein in Escherichia coli. J Bacteriol. 1999;181:3307–3309. doi: 10.1128/jb.181.10.3307-3309.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi Y, Tokumoto U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem. 2002;277:28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 12.Huet G, Daffe M, Saves I. Identification of the Mycobacterium tuberculosis SUF machinery as the exclusive mycobacterial system of [Fe-S] cluster assembly: evidence for its implication in the pathogen's survival. J Bacteriol. 2005;187:6137–6146. doi: 10.1128/JB.187.17.6137-6146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 14.Vinella D, Brochier-Armanet C, Loiseau L, Talla E, Barras F. Iron-Sulfur (Fe/S) Protein Biogenesis: Phylogenomic and Genetic Studies of A-Type Carriers. PLoS Genetics. 2009;5:e1000497. doi: 10.1371/journal.pgen.1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta V, Sendra M, Naik SG, Chahal HK, Huynh BH, Outten FW, Fontecave M, Ollagnier de Choudens S. Native Escherichia coli SufA, Coexpressed with SufBCDSE, Purifies as a [2Fe-2S] Protein and Acts as an Fe-S Transporter to Fe-S Target Enzymes. J Am Chem Soc. 2009;131:6149–6153. doi: 10.1021/ja807551e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ollagnier-de-Choudens S, Sanakis Y, Fontecave M. SufA/IscA: reactivity studies of a class of scaffold proteins involved in [Fe-S] cluster assembly. J Biol Inorg Chem. 2004;9:828–38. doi: 10.1007/s00775-004-0581-9. [DOI] [PubMed] [Google Scholar]
- 17.Layer G, Gaddam SA, Ayala-Castro CN, Ollagnier-de Choudens S, Lascoux D, Fontecave M, Outten FW. SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J Biol Chem. 2007;282:13342–13350. doi: 10.1074/jbc.M608555200. [DOI] [PubMed] [Google Scholar]
- 18.Outten FW, Wood MJ, Munoz FM, Storz G. The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli. J Biol Chem. 2003;278:45713–45719. doi: 10.1074/jbc.M308004200. [DOI] [PubMed] [Google Scholar]
- 19.Kennedy MC, Beinert H. The state of cluster SH and S2- of aconitase during cluster interconversions and removal. A convenient preparation of apoenzyme. J Biol Chem. 1988;263:8194–8198. [PubMed] [Google Scholar]
- 20.Riemer J, Hoepken HH, Czerwinska H, Robinson SR, Dringen R. Colorimetric ferrozine-based assay for the quantitation of iron in cultured cells. Anal Biochem. 2004;331:370–375. doi: 10.1016/j.ab.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 21.Beinert H. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal Biochem. 1983;131:373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- 22.Wada K, Hasegawa Y, Gong Z, Minami Y, Fukuyama K, Takahashi Y. Crystal structure of Escherichia coli SufA involved in biosynthesis of iron-sulfur clusters: implications for a functional dimer. FEBS Lett. 2005;579:6543–6548. doi: 10.1016/j.febslet.2005.10.046. [DOI] [PubMed] [Google Scholar]
- 23.Ding B, Smith ES, Ding H. Mobilization of the iron centre in IscA for the iron-sulphur cluster assembly in IscU. Biochem J. 2005;389:797–802. doi: 10.1042/BJ20050405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding H, Clark RJ, Ding B. IscA mediates iron delivery for assembly of iron-sulfur clusters in IscU under the limited accessible free iron conditions. J Biol Chem. 2004;279:37499–37504. doi: 10.1074/jbc.M404533200. [DOI] [PubMed] [Google Scholar]
- 25.Ding H, Yang J, Coleman LC, Yeung S. Distinct iron binding property of two putative iron donors for the iron-sulfur cluster assembly: IscA and the bacterial frataxin ortholog CyaY under physiological and oxidative stress conditions. J Biol Chem. 2007;282:7997–8004. doi: 10.1074/jbc.M609665200. [DOI] [PubMed] [Google Scholar]
- 26.Ollagnier-de Choudens S, Nachin L, Sanakis Y, Loiseau L, Barras F, Fontecave M. SufA from Erwinia chrysanthemi. Characterization of a scaffold protein required for iron-sulfur cluster assembly. J Biol Chem. 2003;278:17993–18001. doi: 10.1074/jbc.M300285200. [DOI] [PubMed] [Google Scholar]
- 27.Goldsmith-Fischman S, Kuzin A, Edstrom WC, Benach J, Shastry R, Xiao R, Acton TB, Honig B, Montelione GT, Hunt JF. The SufE sulfur-acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes. J Mol Biol. 2004;344:549–565. doi: 10.1016/j.jmb.2004.08.074. [DOI] [PubMed] [Google Scholar]
- 28.Lima CD. Analysis of the E. coli NifS CsdB protein at 2.0 A reveals the structural basis for perselenide and persulfide intermediate formation. J Mol Biol. 2002;315:1199–1208. doi: 10.1006/jmbi.2001.5308. [DOI] [PubMed] [Google Scholar]
- 29.Sendra M, Ollagnier de Choudens S, Lascoux D, Sanakis Y, Fontecave M. The SUF iron-sulfur cluster biosynthetic machinery: sulfur transfer from the SUFS-SUFE complex to SUFA. FEBS Lett. 2007;581:1362–1368. doi: 10.1016/j.febslet.2007.02.058. [DOI] [PubMed] [Google Scholar]
- 30.Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol. 2001;183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loiseau L, Ollagnier-de-Choudens S, Nachin L, Fontecave M, Barras F. Biogenesis of Fe-S cluster by the bacterial Suf system: SufS and SufE form a new type of cysteine desulfurase. J Biol Chem. 2003;278:38352–38359. doi: 10.1074/jbc.M305953200. [DOI] [PubMed] [Google Scholar]
- 32.Lu J, Yang J, Tan G, Ding H. Complementary roles of SufA and IscA in the biogenesis of iron-sulfur clusters in Escherichia coli. Biochem J. 2008;409:535–543. doi: 10.1042/BJ20071166. [DOI] [PubMed] [Google Scholar]
- 33.Angelini S, Gerez C, Ollagnier-de Choudens S, Sanakis Y, Fontecave M, Barras F, Py B. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J Biol Chem. 2008;283:14084–14091. doi: 10.1074/jbc.M709405200. [DOI] [PubMed] [Google Scholar]
- 34.Butland G, Babu M, Diaz-Mejia JJ, Bohdana F, Phanse S, Gold B, Yang W, Li J, Gagarinova AG, Pogoutse O, Mori H, Wanner BL, Lo H, Wasniewski J, Christopolous C, Ali M, Venn P, Safavi-Naini A, Sourour N, Caron S, Choi JY, Laigle L, Nazarians-Armavil A, Deshpande A, Joe S, Datsenko KA, Yamamoto N, Andrews BJ, Boone C, Ding H, Sheikh B, Moreno-Hagelseib G, Greenblatt JF, Emili A. eSGA: E. coli synthetic genetic array analysis. Nat Methods. 2008;5:789–795. doi: 10.1038/nmeth.1239. [DOI] [PubMed] [Google Scholar]
- 35.Loiseau L, Gerez C, Bekker M, Ollagnier-de Choudens S, Py B, Sanakis Y, Teixeira de Mattos J, Fontecave M, Barras F. ErpA, an iron sulfur (Fe S) protein of the A-type essential for respiratory metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:13626–13631. doi: 10.1073/pnas.0705829104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vilella F, Alves R, Rodriguez-Manzaneque MT, Belli G, Swaminathan S, Sunnerhagen P, Herrero E. Evolution and cellular function of monothiol glutaredoxins: involvement in iron-sulphur cluster assembly. Comp Funct Genomics. 2004;5:328–341. doi: 10.1002/cfg.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.