Abstract
Background
HIV/HCV co-infected patients are known to have lower sustained viral response (SVR) rates than HCV mono-infected patients. However, the role of CD4+ T-cell counts on viral kinetics and outcome is not fully understood.
Methods
HCV-RNA kinetics (bDNA v3, LD=615 IU/ml) was analyzed in 32 HIV/HCV co-infected persons treated with Pegylated interferon-α2b (1.5 μg/kg weekly) and ribavirin (1–1.2 g daily) for 48 weeks and compared with results obtained from 12 HCV monoinfected patients treated with the same regimen.
Results
Baseline CD4+ T cell counts ≥450cells/mm3 were significantly (P<0.002) associated with SVR in co-infected genotype 1 patients. First phase decline was significantly lower among patients with low as compared to high CD4 counts (P<0.03) and among co-infected compared to mono-infected patients (P<0.002). Second phase decline slope showed a similar trend for co-infected patients.
Conclusions
Low baseline CD4+ T cell count is associated with slower HCV viral kinetics and worse response to treatment among HIV co-infected patients, suggesting HCV treatment response depends on immune status. HCV genotype 1 co-infected patients have slower first phase viral kinetics than HCV mono-infected patients. First-phase viral decline (>1.0 log) and second-phase viral decline slope (>0.3 log/week) are excellent predictors of SVR for co-infected patients.
Keywords (3-10): HCV, HIV, viral kinetics, sustained viral response
Introduction
Hepatitis C virus (HCV) infection is highly prevalent among human immunodeficiency virus (HIV) infected individuals, with prevalence directly related to risk categories including injection drug use and hemophilia 1. HIV/HCV co-infected patients have rapid progression of liver fibrosis when compared to individuals infected with HCV alone 2. Current standard of care (pegylated interferon and ribavirin for 48 weeks) therapy yields only modest cure rates among HIV/HCV co-infected individuals when compared to cure rates seen with HCV mono-infected individuals 3–8. Better understanding of underlying mechanisms responsible for lower response in co-infected patients is needed. Specifically, the impact of baseline CD4+ T-cell count on viral kinetics and response to treatment has not been studied in detail.
The biphasic viral kinetics of decline in HCV mono-infected patients 9 treated with combination therapy has been successfully used to predict sustained viral response (SVR) 10. Two studies of 10 and 12 HIV/HCV co-infected individuals each found a slower viral decline in co-infected subjects 11, 12. However, another study of 12 HIV co-infected patients did not show significant differences in key viral kinetics parameters 13. Additionally, several pharmacodynamic parameters were found to correlate with non-response to PEG-IFN among co-infected individuals, although IFN concentrations and pharmacokinetic parameters did not differ between persons with SVR and non-response (NR) 14.
Treatment with interferon and ribavirin carries the risk of serious adverse events such as cytopenias, depression and irritability. Thus, the identification of markers for SVR, which equate to cure rates, would help optimize anti-HCV therapy and avoid administering toxic and costly therapy to patients with little chance of successful outcome. Presently, lack of early viral response (EVR), defined as less than or equal to a 2 log reduction in HCV viral load at week 12, has been associated with more than 95% negative predictive value for SVR and is used to advise patients to discontinue therapy 3, 7, 8. Development of clinically validated predictors of SVR prior to week 12 will help to further optimize anti-HCV therapy among HIV/HCV co-infected individuals.
Our study, involving a larger group of HIV/HCV co-infected individuals, evaluated the influence of baseline CD4+ T-cell count on HCV viral kinetics and response to pegylated-interferon and ribavirin treatment and the early predictive value of CD4+ T cell counts and viral kinetics for SVR.. In addition, these results are compared with those obtained with 12 HCV monoinfected patients treated with the same therapeutic schedule.
Methods
Study Design
This was a pilot, prospective, open-label trial performed at the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) at Bethesda, MD from 2001 to 2004. Thirty-two HIV-infected subjects were treated with peginterferon alpha-2b at 1.5 μg/kg subcutaneously every week (Peg-Intron; Schering-Plough, Kenilworth, New Jersey, USA) and ribavirin daily (Rebetol, Schering-Plough, at 400mg Qam and 600mg Qpm for < 75kg, 600mg twice per day for > 75kg) for 48 weeks and followed up for 24 weeks after the termination of treatment. All patients signed informed consent approved by the NIAID institutional review board (IRB) prior to enrollment in the study. Twelve HCV-monoinfected patients were treated with the same therapeutic regimen at the Hospital General Universitario Valle Hebron at Barcelona (Spain) during the same period of time. Ten HCV mono-infected patients were treated with Peg interferon-alpha 2b 1ug/Kg with ribavirin at the University of Vienna were also studied.
Patients were eligible for the study if they were > 18 years of age and had CD4+ T cell counts > 100 cells/mm3, absolute neutrophil counts > 1000 cells/mm3, HCV viral load > 2000 copies/mL, and had histologic evidence of chronic hepatitis C. Of the HIV/HCV co-infected patients, 85% were receiving ART and 68% had HIV-RNA<50 cp/ml at the initiation of therapy for HCV.
Laboratory Studies
Liver chemistry, immuno-phenotyping and safety laboratory tests were performed prior to treatment and during each study visit. HCV RNA was performed during all study visits (days 0, 1, 3, 5, 7, 10, week 2, week 3, week 4, week 6, week 8, and then every 4 weeks until week 48. HCV RNA concentration in plasma was measured by the VERSANT HCV RNA 3.0 Assay (Bayer Diagnostics, Puteaux, France). The assay has a quantitation range of 615 to 7.7 million HCV RNA IU/ml. HCV RNA from HCV mono-infected individuals were measured using commercial assays that had similar quantitation range at the respective institutions..
Statistical Analyses
First phase viral decline was calculated here as the log IU/ml decline at day 3 relative to baseline of HCV-RNA. It was in general equal to viral decline at day 1, but we chose to use the day 3 decline since a number of patients had a decline only after day 1. Second phase decline slope (log IU/ml/week) was calculated by log-linear regression of the viral decline at days 7, 14, 21 and 28, or until the first unquantifiable (<650 IU/ml) viral load. In 2 cases with undetectable viremia (<50 IU/ml) already at day 7 the second phase slope was assumed to be the fastest slope observed in the other patients (−0.89 log/week).
The non-parametric Mann-Whitney U-test was used to assess the significance of difference in distribution of continuous variables between groups of patients. The Fischer exact 2×2 test was used to compare frequencies between groups of patients. The Spearman non-parametric test was used to assess the significance of correlation between continuous variables. Significance level was assumed to be p=0.05.
Results
Relationship of SVR to of Baseline CD4+ T cell count and HCV Genotype
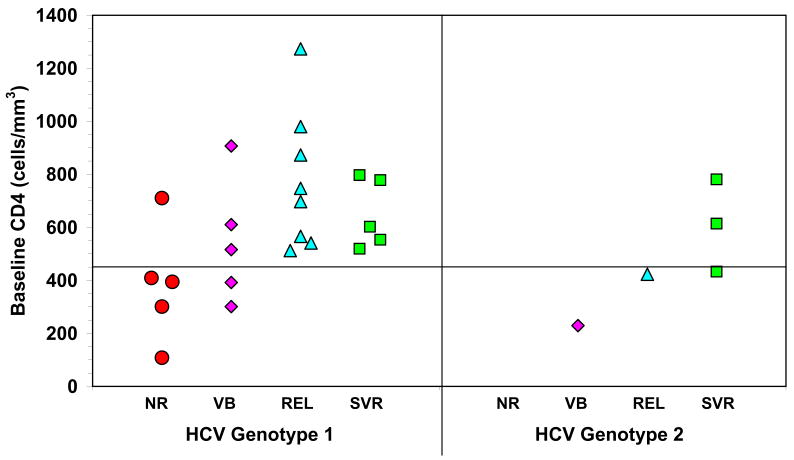
The baseline demographics of these patients are shown in Table 1 15. Among the 12 HCV monoinfected patients, 10 were males; mean age 44 Years (range 32–62 years). Four were infected by genotype 3, 5 by genotype 1, 2 by genotype 4 and 1 by genotype 2. Patients infected by genotype 1 and 4 were treated for 48 weeks and those infected by genotype 2 and 3 for 24 weeks. All patients completed therapy and a 24 weeks follow-up period. Of the 32 patients who participated in this study, 3 stopped treatment prior to week 2 due to social reasons and another discontinued due to the onset of psychosis. Of the remaining 28 patients, SVR was achieved in 5/23 (22%) of genotype 1 and 3/5 (60%) of genotype 2 patients. Among HCV genotype 1 patients with baseline CD4+ T cell counts < 450 cells/mm3, no one achieved end of treatment response (ETR) or SVR, while those with baseline CD4+ T cell counts ≥ 450 cells/mm3 show significantly (P<0.002) higher ETR of 76% and SVR of 29% (Fig. 1). The same trend was found among patients with HCV genotype 2, however, the sample size was too small to establish statistical significance. Note that 5 out of 6 patients with low CD4 counts were African-Americans as compared to 8 of 17 patients with higher CD4 count. While this difference was not statistically significant (P>0.1) we cannot rule out the influence of race on outcome since African Americans have been previously shown to have slower viral declines when compared to that seen with Caucasians in HCV-mono-infected patients treated with standard interferon alpha and ribavirin 16. Furthermore, the baseline HCV viral levels between HCV genotype 1-infected subjects with a CD4+ T cell counts less than 450cells/mm3 were not significantly different from those with a CD4+T cell counts more than 450 cells/mm3 (log 6,22 ± 0.21versus log 6.47 ± 0.08, respectively; p>0.5). Moreover, about half of the patients with high CD4 T cell counts are also African Americans, and nevertheless they respond better than those who have low CD4 T cell counts. There are no statistically significant differences in non-responders and SVR rates between Caucasians and African Americans, but there is nevertheless a significant (P=0.002, Mann-Whitney non-parametric test) difference in non-response rates between patients with low and high CD4 T cell count. Furthermore, logistic regression with race and CD4 T cell counts shows that CD4 T cell counts are better predictors of non response, but due to the small number of patients the results are not statistical significant.
Table 1.
Baseline Characteristics of Study Subjects.
Study subjects were predominantly male, and infected with HCV genotype 1, high ALT and varying levels of liver fibrosis.
| Age | Gender | Race | Risk | Base line CD4 (%) | log HIV VL | log HCV VL | HCV Genotype | ALT | GGT | Fibrosis (Ishak) | Outcome | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 43 | Male | AA | MSM/SN | 980 (31) | 3.42 | 6.38 | 1B | 55 | 80 | 2 | Relapser |
| 2 | 42 | Male | C | MSM | 747 (30) | 1.69 | 6.82 | 1A | 107 | 249 | 3 | Relapser |
| 3 | 48 | Male | AA | SN | 1273 (51) | 1.69 | 6.86 | 1 | 62 | 163 | 3 | Relapser |
| 4 | 39 | Male | C | MSM/SN | 873 (35) | 1.69 | 6.68 | 1 | 51 | 29 | 3 | Relapser |
| 5 | 38 | Male | C | MSM/SN | 697 (38) | 1.69 | 7.35 | 1A | 37 | 198 | 3 | Relapser |
| 6 | 51 | Female | AA | SN | 512 (22) | 1.69 | 5.87 | 1 | 74 | 125 | 4 | Relapser |
| 7 | 62 | Male | C | MSM | 541 (28) | 2.69 | 6.26 | 1 | 65 | 47 | 2 | Relapser |
| 8 | 52 | Male | AA | MSM | 547 (16) | 3.74 | 5.81 | 1A | 33 | 22 | 2 | Relapser |
| 9 | 50 | Male | AA | Heterosexual | 449 (16) | 1.69 | 7.12 | 2 | 17 | 32 | 1 | Relapser |
| 10 | 51 | Male | AA | MSM/SN | 710 (42) | 2.10 | 6.50 | 1A | 55 | 394 | 4 | Non responder |
| 11 | 44 | Female | AA | Heterosexual | 108 (6) | 1.69 | 6.55 | 1A | 25 | 112 | 2 | Non responder |
| 12 | 49 | Male | AA | SN | 392 (19) | 1.69 | 6.18 | 1B | 77 | 244 | 2 | Non responder |
| 13 | 48 | Male | C | SN | 610 (33) | 1.69 | 6.69 | 1 | 96 | 70 | 6 | Non responder |
| 14 | 49 | Male | AA | SN | 394 (22) | 1.69 | 6.66 | 1 | 34 | 179 | 2 | Non responder |
| 15 | 43 | Male | AA | MSM | 907 (31) | 1.69 | 5.92 | 1A | 29 | 29 | 3 | Non responder |
| 16 | 33 | Male | AA | MSM | 409 (22) | 1.69 | 6.72 | 1A | 81 | 93 | 3 | Non responder |
| 17 | 49 | Male | AA | SN | 516 (25) | 1.69 | 5.99 | 1 | 124 | 58 | 2 | Non responder |
| 18 | 35 | Male | C | MSM | 301 (20) | 2.84 | 6.30 | 1 | 226 | 57 | 2 | Non responder |
| 19 | 51 | Male | AA | SN | 301 (24) | 1.69 | 6.45 | 1 | 102 | 114 | 4 | Non responder |
| 20 | 55 | Male | AA | SN | 229 (16) | 1.69 | 6.59 | 2 | 64 | 29 | 3 | Non responder |
| 21 | 42 | Male | C | MSM | 797 (27) | 3.76 | 6.89 | 1 | 64 | 42 | 4 | SVR |
| 22 | 48 | Male | AA | MSM | 553 (34) | 3.17 | 6.21 | 1A | 70 | 60 | 1 | SVR |
| 23 | 46 | Male | C | MSM | 519 (36) | 1.85 | 3.32 | 1A | 33 | 31 | 1 | SVR |
| 24 | 38 | Male | C | MSM/SN | 602 (31) | 4.52 | 6.40 | 1A | 60 | 24 | 1 | SVR |
| 25 | 48 | Male | C | MSM | 778 (33) | 1.69 | 5.80 | 1A | 21 | 55 | 1 | SVR |
| 26 | 49 | Male | C | MSM | 614 (29) | 1.69 | 6.29 | 2B | 55 | 36 | 3 | SVR |
| 27 | 45 | Male | C | MSM | 481 (40) | 1.69 | 6.79 | 2B | 49 | 31 | 3 | SVR |
| 28 | 51 | Male | AA | MSM/SN | 781 (47) | 1.69 | 6.92 | 2 | 18 | 31 | 1 | SVR |
Figure 1. HCV response to treatment is associated with high baseline CD4+ T cell counts in HIV/HCV co-infected patients.
HCV genotype 1 patients co-infected with HIV that have baseline CD4+ T cell counts < 450 cells/mm3 show 0% SVR and ETR, while those with baseline CD4+ T cell counts ≥ 450 cells/mm3 show significantly (P=0.002) higher ETR (76%) and SVR (29%). The same trend is found for HCV genotype 2 patients. NR-non response, VB-viral breakthrough, REL- end-of-treatment response with relapse, SVR-sustained viral response.
Relationship of Viral Kinetics to HCV genotype and baseline CD4+ T cell count
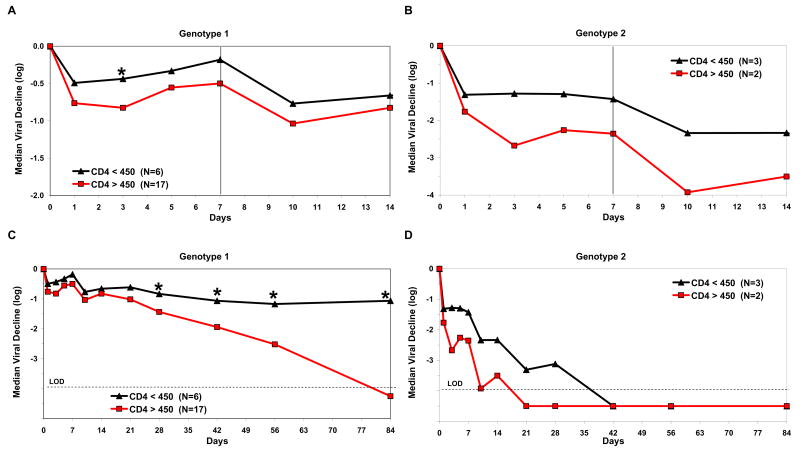
HIV co-infected subjects with HCV genotype 2 had significantly faster viral decline at days 1–84 than did HCV genotype 1 co-infected subjects(p<0.001) (Fig. 2). HIV co-infected subjects with HCV genotype 1 showed a significantly slower first-phase decline (at day 3) than did subjects with HCV genotype 2 (mean 0.75 ±0.44 log/wk vs. 2.0 ±0.72 log/wk, p<0.001) and a significantly slower second-phase slope (0.4 ±0.28 log/wk vs. 0.7 ±0.18 log/wk, p=0.03) (Fig 3). Transient rebounds (mean 0.4 ±0.3 log) were observed at the end of weeks 1 and 2, when viral load was measured during the week, before the next weekly Peg-IFN injection for both genotypes. Among subjects with HCV genotype 2, HCV viral load suppression <615 IU/mL was achieved in 3/5 (60%) patients by week 4. Only 2 of 23 (9%) of patients co-infected with HIV and HCV genotype 1 reached unquantifiable levels of viremia (<615 IU/mL) at week 4.
Figure 2. HCV kinetics as function of baseline CD4+ T cell counts and of HCV genotype in patients co-infected with HIV.
Co-infected patients with HCV genotype 1 show significantly (P<0.001) slower decline than do genotype 2 patients. For both genotypes, patients with baseline CD4+ T cell counts < 450 cells/mm3 show slower decline than do patients with higher baseline CD4+ T cell counts (* for genotype 1 patients P<0.03 in days 3, 28–84). Dashed horizontal line represents LOD-limit of detection (<615 IU/mL).
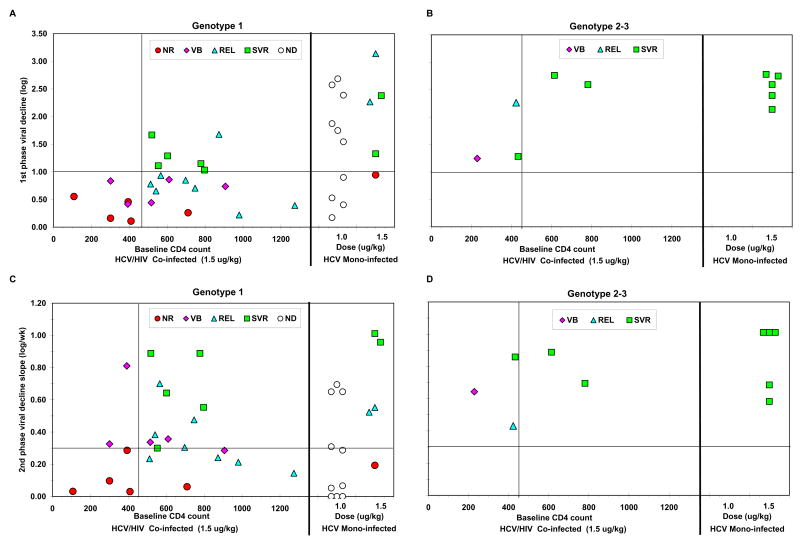
Figure 3. First phase decline and 2nd phase slope as function of CD4+ Tcell count and genotype in co-infected patients versus mono-infected patients.
Among co-infected patients, those with baseline CD4+ T cell counts < 450 cells/mm3 had significantly lower 1st phase decline (P<0.03) and a trend for slower 2nd phase decline slope than patients with higher CD4 counts. Moreover, mono-infected patients had significantly (P<0.02) faster 1st phase decline than co-infected patients, but non-significantly different 2nd phase decline slope (within the first 28 days). Co-infected genotype 2–3 patients had faster viral kinetics than genotype 1 patients, similar to genotype 2–3 mono-infected patients. ND: end-point not determined.
Among patients with HCV genotype 1, viral declines at both day 3 and days 28–84 were significantly faster in those with baseline CD4+ T cell counts ≥ 450 cells/mm3 than with those who had CD4+ T cell counts of <450 cells/mm3 (p<0.03). Indeed, first phase decline was significantly faster for genotype 1 patients with high baseline CD4+ T cell count (≥450) as compared to those with low baseline CD4 count (mean 0.87 ± 0.43 log vs. 0.42 ± 0.27, p<0.03). When comparing the first phase decline between co-infected and mono-infected patients (treated with either 1.0ug/Kg or 1.5ug/Kg QW) with HCV genotype 1, treated with same combination therapy (Fig 3A), co-infected patients show significantly slower declines at Day 3 (mean, 0.75 ± 0.44 log vs. 1.6 ± 0.9, p<0.002). We found that race is also a factor that affects 1st phase decline in HIV/HCV co-infected patients, as African Americans experienced a slower 1st phase decline when compared to Caucasians (p<0.01).
Second phase decline was also faster in patients with HCV genotype 1 with high baseline CD4+ T cell counts (≥450) compared to those with lower baseline CD4+ T cell counts (mean, 0.44 ± 0.35 log/week vs. 0.26 ± 0.33 log/week), however, this parameter showed a larger variation and the difference did not reach statistical significance (Fig 3C). In fact, there is also no significant difference in the second phase slope between co-infected patients and mono-infected patients treated with same regimen (Fig 3C). Genotype 1 patients with low CD4+ T cell counts continued to have a slower decline slope also during weeks 4–12 although it was not statistically significant (p=NS, Fig 2C).
The magnitude and frequency of the transient rebound at the end of the week is similar for mono-infected patients and co-infected patients with high or low CD4+ T cell counts (data not shown).
Also for co-infected patients with HCV genotype 2 there was a correlation between baseline CD4+ T cell counts and first phase decline (R=0.9, P=0.03), however, the difference between patients with lower baseline CD4+ T cell counts (< 450 cells/mm3) versus those with higher baseline CD4+ T cell counts, or between co-infected patients and mono-infected patients, did not reach statistical significance, possibly due to the small number of patients (Fig 3B). Second phase decline was similar among all patients with HCV genotype 2 (Fig 3D).
Predictive Ability of HCV Viral Kinetics and Therapeutic Response
A baseline CD4+ T cell count of <450 cells/mm3 shows a strong negative predictive value for SVR (no patient with SVR out of 6 patients, NPV=100%) among HCV genotype 1 co-infected patients, however, the positive predictive value for the CD4>450 cells/mm3 criteria is low (5 of 17, PPV=29%). In contrast, a first phase HCV decline faster than 1 log IU/mL, measurable at Day 3 of treatment, shows both a high negative predictive value (0 of 17, NPV=100%) and a high positive predictive value (5 patients with SVR out of 6, PPV=83%) (Fig 3A).
As with HCV mono-infected patients, co-infected patients with a slow second-phase viral decline slope (less than 0.3 log/week, or 0,1 ln/day) did not achieve SVR (NPV=100%), consistent with previous studies9, 10, 18. However, the positive predictive value for the second phase slope in co-infected patients was lower (PPV=38%). The number of co-infected patients with HCV genotype 2 is too small to draw any conclusions regarding the early prediction of SVR.
Discussion
The present study demonstrates a striking relationship between baseline CD4+ T cell counts and response to therapy with pegylated interferon-alpha-2b and ribavirin. Among HCV genotype 1 HIV co-infected patients, those with baseline CD4+ T cell count < 450 cells/mm3 did not achieve SVR or ETR (NPV=100%, PPV=30%), while patients with baseline CD4+ T cell counts ≥ 450 cells/mm3 achieved significantly higher ETR (76%) and SVR (29%) rates. Moreover, HCV genotype 1 co-infected patients with baseline CD4+ T cell counts <450 cells/mm3 had slower first phase kinetics in response to combination therapy than did either persons with higher baseline CD4+ T cell counts or persons with HCV mono-infection.
The effectiveness of interferon-alpha formulations are probably dependent on baseline immune status and hence there is a clear biological explanation for the relationship between baseline CD4 T cell counts and 1st phase HCV decline. Since race is also a major factor that would influence viral kinetics, larger clinical studies will be needed to answer the independent significance of race and CD4 T cell counts in influencing early viral kinetics and therefore SVR. However, our study
Previous studies in smaller number of patients have suggested that baseline CD4+ T cell count is independently associated with SVR 19–22; however, these studies are limited due to small sample sizes, use of non-pegylated interferon, and different dosing regimens and dosing intervals. Several larger studies have not shown low baseline CD4+ T cell count to be a predictor of SVR probably because these studies either did not analyze baseline CD4+ T cell count and SVR among isolated genotypes 4, 7, 8; were confounded by a disproportionately high percentage of genotype 2 and 3 patients 4, 8; did not present data in which CD4+ T cell count was used as a cut-off in analyses 3, 7; or used a CD4+ T cell cutoff of 500 cells/mm3, which may have missed the effect seen at lower CD4+ T cell levels 4, 8. The RIBAVIC study shows a trend among all patients in the pegylated interferon arm towards greater SVR among patients with CD4+ T cell counts > 500 (21% vs 33%) 8. In contrast, the present study focused on intense viral kinetics of all HCV genotypes, which were analyzed by various strata of CD4+ T cell count. Interestingly, a similar study of 21 HCV genotype 1 co-infected patients did not find similar correlation between CD4 count and SVR or rapid viral kinetics 14, possibly due to the small sample size of both studies. Both race and baseline HCV viral levels have also be shown to be independent determinants for SVR among HCV mono-infected individuals 16. In this study, the distribution of race and baseline HCV viral levels were statistically not different between those with high or low CD4 T cell counts. However, it is plausible that these factors could play a role in influencing HCV viral load declines. In this exploratory study, we chose to use a cut-off of CD4 T cell count of 450 cells/mm3 arbitrarily as our intention was to demonstrate the relationship between baseline characteristics and viral kinetics. Larger studies are required to determine whether race, baseline CD4 T cell counts or HCV viral levels plays a more significant role in virologic response to anti-HCV therapy.
For HIV/HCV genotype 1 co-infected patients, a first phase viral decline of > 1.0 log/wk was associated with SVR with a NPV=100% and PPV=83%; a second phase decline of > 0.3 log/wk was associated with a NPV=100% and PPV=38%. Since our study is mainly intended to generate hypothesis, the NPV and PPV will need to be validated in larger treatment studies. Not surprisingly, the first and second-phase viral declines were much faster in genotype 2 than in genotype 1 co-infected patients, an observation previously made among HCV mono-infected individuals receiving pegylated interferon and ribavirin 23. A recent study examined data from 705 patients who were enrolled in the APRICOT study, came to similar conclusions that the ability to achieve SVR was significantly impaired in those who were infected with genotype 1 and had CD4+ T cell counts less than 350 cells/mm3.24
In conclusion, HIV/HCV co-infected patients with higher CD4+ T cell counts are more likely to achieve SVR when treated with pegylated interferon and ribavirin. However, although most of our patients were receiving HAART and had suppressed HIV viremia and it is not clear if our results can be extrapolated to patients with high CD4+ T cell counts before receiving HAART, it may be appropriate to treat for HCV while CD4+ T cell counts ≥ 450 cells/mm3, even before starting ART for HIV. Surely, along with our previous observation that the presence of advanced liver disease in genotype 1 HCV co-infected patients is associated with lower rates of SVR 15 these findings suggest that co-infected patients should be treated whether or not there is evidence of pathologic liver disease. Further validation of these concepts requires a larger, randomized study that is stratified by CD4+ T cell counts and race.
Acknowledgments
This research was supported in whole by the Intramural Research Program of the NIH [National Institute of Allergy and Infectious Diseases and National Institute of Digestive Diseases and Kidney].
Footnotes
Publisher's Disclaimer: The content of this publication does not necessarily reflect the views of policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organization imply endorsement by the U.S. Government.
Conflict of Interest Statement
None of the other authors have any conflicts of interest to report.
References
- 1.Sherman KE, Rouster SD, Chung RT, Rajicic N. Hepatitis C Virus prevalence among patients infected with Human Immunodeficiency Virus: a cross-sectional analysis of the US adult AIDS Clinical Trials Group. Clinical infectious diseases. 2002 Mar 15;34(6):831–837. doi: 10.1086/339042. [DOI] [PubMed] [Google Scholar]
- 2.Benhamou Y, Bochet M, Di Martino V, et al. Liver fibrosis progression in human immunodeficiency virus and hepatitis C virus coinfected patients. The Multivirc Group. Hepatology (Baltimore, Md) 1999 Oct;30(4):1054–1058. doi: 10.1002/hep.510300409. [DOI] [PubMed] [Google Scholar]
- 3.Torriani FJ, Rodriguez-Torres M, Rockstroh JK, et al. Peginterferon Alfa-2a plus ribavirin for chronic hepatitis C virus infection in HIV-infected patients. The New England journal of medicine. 2004 Jul 29;351(5):438–450. doi: 10.1056/NEJMoa040842. [DOI] [PubMed] [Google Scholar]
- 4.Perez-Olmeda M, Nunez M, Romero M, et al. Pegylated IFN-alpha2b plus ribavirin as therapy for chronic hepatitis C in HIV-infected patients. AIDS (London, England) 2003 May 2;17(7):1023–1028. doi: 10.1097/00002030-200305020-00011. [DOI] [PubMed] [Google Scholar]
- 5.Manns MP, McHutchison JG, Gordon SC, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001 Sep 22;358(9286):958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 6.Fried MW, Shiffman ML, Reddy KR, et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002 Sep 26;347(13):975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 7.Chung RT, Andersen J, Volberding P, et al. Peginterferon Alfa-2a plus ribavirin versus interferon alfa-2a plus ribavirin for chronic hepatitis C in HIV-coinfected persons. The New England journal of medicine. 2004 Jul 29;351(5):451–459. doi: 10.1056/NEJMoa032653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrat F, Bani-Sadr F, Pol S, et al. Pegylated interferon alfa-2b vs standard interferon alfa-2b, plus ribavirin, for chronic hepatitis C in HIV-infected patients: a randomized controlled trial. Jama. 2004 Dec 15;292(23):2839–2848. doi: 10.1001/jama.292.23.2839. [DOI] [PubMed] [Google Scholar]
- 9.Neumann AU, Lam NP, Dahari H, et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998 Oct 2;282(5386):103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 10.Ferenci P. Predicting the therapeutic response in patients with chronic hepatitis C: the role of viral kinetic studies. The Journal of antimicrobial chemotherapy. 2004 Jan;53(1):15–18. doi: 10.1093/jac/dkh015. [DOI] [PubMed] [Google Scholar]
- 11.Torriani FJ, Ribeiro RM, Gilbert TL, et al. Hepatitis C virus (HCV) and human immunodeficiency virus (HIV) dynamics during HCV treatment in HCV/HIV coinfection. J Infect Dis. 2003 Nov 15;188(10):1498–1507. doi: 10.1086/379255. [DOI] [PubMed] [Google Scholar]
- 12.Talal AH, Shata MT, Markatou M, et al. Virus dynamics and immune responses during treatment in patients coinfected with hepatitis C and HIV. J Acquir Immune Defic Syndr. 2004 Feb 1;35(2):103–113. doi: 10.1097/00126334-200402010-00001. [DOI] [PubMed] [Google Scholar]
- 13.Sherman KE, Shire NJ, Rouster SD, et al. Viral kinetics in hepatitis C or hepatitis C/human immunodeficiency virus-infected patients. Gastroenterology. 2005 Feb;128(2):313–327. doi: 10.1053/j.gastro.2004.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talal AH, Ribeiro RM, Powers KA, et al. Pharmacodynamics of PEG-IFN alpha differentiate HIV/HCV coinfected sustained virological responders from nonresponders. Hepatology. 2006 May;43(5):943–953. doi: 10.1002/hep.21136. [DOI] [PubMed] [Google Scholar]
- 15.Wu L, Kottilil S, Lempicki R, et al. Hepatic histologic response (HR) to combination therapy among HCV/HIV-coinfected individuals: interferon induces HR independent of sustained virologic response (SVR) AIDS research and human retroviruses. 2006 Nov;22(11):1091–1098. doi: 10.1089/aid.2006.22.1091. [DOI] [PubMed] [Google Scholar]
- 16.Layden-Almer JE, Ribeiro RM, Wiley T, Perelson AS, Layden TJ. Viral dynamics and response differences in HCV-infected African American and white patients treated with IFN and ribavirin. Hepatology (Baltimore, Md) 2003 Jun;37(6):1343–1350. doi: 10.1053/jhep.2003.50217. [DOI] [PubMed] [Google Scholar]
- 17.Formann E, Jessner W, Bennett L, Ferenci P. Twice-weekly administration of peginterferon-alpha-2b improves viral kinetics in patients with chronic hepatitis C genotype 1. Journal of viral hepatitis. 2003 Jul;10(4):271–276. doi: 10.1046/j.1365-2893.2003.00446.x. [DOI] [PubMed] [Google Scholar]
- 18.Perelson AS, Herrmann E, Micol F, Zeuzem S. New kinetic models for the hepatitis C virus. Hepatology (Baltimore, Md) 2005 Oct;42(4):749–754. doi: 10.1002/hep.20882. [DOI] [PubMed] [Google Scholar]
- 19.Soriano V, Garcia-Samaniego J, Bravo R, et al. Interferon alpha for the treatment of chronic hepatitis C in patients infected with human immunodeficiency virus. Hepatitis-HIV Spanish Study Group. Clinical infectious diseases. 1996 Sep;23(3):585–591. doi: 10.1093/clinids/23.3.585. [DOI] [PubMed] [Google Scholar]
- 20.Soriano V, Bravo R, Garcia-Samaniego J, et al. A pilot study on the efficacy of escalating dosage of alpha-interferon for chronic hepatitis C in HIV-infected patients. The Hepatitis/HIV Spanish Study Group. The Journal of infection. 1997 Nov;35(3):225–230. doi: 10.1016/s0163-4453(97)92776-6. [DOI] [PubMed] [Google Scholar]
- 21.Mauss S, Klinker H, Ulmer A, et al. Response to treatment of chronic hepatitis C with interferon alpha in patients infected with HIV-1 is associated with higher CD4+ cell count. Infection. 1998 Jan–Feb;26(1):16–19. doi: 10.1007/BF02768746. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi K, Fukuda Y, Nakano I, et al. Poor response to interferon treatment for chronic hepatitis C in human immunodeficiency virus-infected haemophiliacs. Haemophilia. 2000 Nov;6(6):677–681. doi: 10.1046/j.1365-2516.2000.00444.x. [DOI] [PubMed] [Google Scholar]
- 23.Neumann AU, Lam NP, Dahari H, et al. Differences in viral dynamics between genotypes 1 and 2 of hepatitis C virus. The Journal of infectious diseases. 2000 Jul;182(1):28–35. doi: 10.1086/315661. [DOI] [PubMed] [Google Scholar]
- 24.Opravil M, Sasadeusz J, Cooper DA, et al. Effect of baseline CD4 cell count on the efficacy and safety of peginterferon Alfa-2a (40KD) plus ribavirin in patients with HIV/hepatitis C virus coinfection. Journal of acquired immune deficiency syndromes (1999) 2008 Jan 1;47(1):36–49. doi: 10.1097/QAI.0b013e31815ac47d. [DOI] [PubMed] [Google Scholar]