Abstract
Purpose
Chronic lymphocytic leukemia (CLL) is a B-cell malignancy characterized by a variable clinical course. Several parameters have prognostic capabilities but are associated with altered response to therapy in only a small subset of patients.
Experimental Design
We used gene expression profiling methods to generate predictors of therapy response and prognosis. Genomic signatures that reflect progressive disease and responses to chemotherapy or chemo-immunotherapy were created using cancer cell lines and patient leukemia cell samples. We validated and applied these three signatures to independent clinical data from four cohorts representing a total of 301 CLL patients.
Results
A genomic signature of prognosis created from patient leukemic cell gene expression data coupled with clinical parameters significantly differentiated patients with stable disease from those with progressive disease in the training dataset. The progression signature was validated in two independent datasets, demonstrating a capacity to accurately identify patients at risk for progressive disease. In addition, genomic signatures that predict response to chlorambucil or pentostatin, cyclophosphamide, and rituximab were generated and could accurately distinguish responding and non-responding CLL patients.
Conclusions
Thus, microarray analysis of CLL lymphocytes can be used to refine prognosis and predict response to different therapies. These results have implications for standard and investigational therapeutics in CLL patients.
Introduction
The practice of oncology continually faces two major challenges – determining which patients are at risk for progression or recurrence of disease and identifying the most effective therapeutic regimen for the individual patient. Obstacles to address these challenges include the complexity of the disease processes, individual differences and comorbidities, and the paucity of markers to guide the use of available treatments. However, examples such as the use of trastuzumab to treat HER-2 positive breast cancer demonstrate that selecting therapies for patients based on tumor markers can improve overall response rates. Similarly, identifying predictors of sensitivity to cytotoxic agents that are able to select patients who will respond to these chemotherapeutic agents would directly impact current medical practice, where patients are often treated with one of several therapeutic regimens that, on a population basis, have equal efficacy.
Chronic Lymphocytic Leukemia (CLL) displays a wide spectrum of aggressiveness. Even among those patients with low or intermediate risk disease at diagnosis, accurate determination of which patients will progress and require therapy is inexact. For those patients requiring therapy, there are a variety of treatment options, varying in long-term efficacy and toxicity. Multiple factors, such as cytogenetic aberrations, immunoglobulin variable region heavy chain (IgVH) mutational status, and CD38 and ZAP-70 expression, are increasingly used to help refine prognosis and guide patient care in the previously untreated CLL patient. (1–5) However, at this time, only the interphase cytogenetic abnormality of 17p13 deletion has been consistently associated with poor response to purine-analogue based therapy. (6–8)
Recent advances using genomic technology, particularly the use of gene expression profiling, has provided an opportunity to further address these issues. Previous studies have described the development of gene-expression based profiles that correlate with clinical outcomes or surrogates of outcome. (9–21) These studies of gene expression differences include investigations of CLL and normal B-cells, CLL with specific cytogenetic anomalies, and mutated versus unmutated IgVH status. Here, we describe the generation of gene expression signatures with improved capacity to predict which low or intermediate risk patients are most likely to progress with CLL and, at the same time, can predict response to a variety of treatment approaches.
Materials and Methods
Patients and Leukemia Samples
Two CLL patient cohorts were used in this research: one from the Duke University and Durham VA Medical Centers (Duke/VA), and one from the Mayo Clinic and the Ohio State University (Mayo/Ohio State). The Duke/VA cohort was used to create the genomic signature of progressive disease, and the Mayo/Ohio State cohort was used to generate the genomic signature of response to the chemo-immunotherapy regimen of pentostatin, cyclophosphamide, and rituximab. Clinical data describing these cohorts were previously published. (22, 23)
Patients with a diagnosis of CLL and Rai stage 0 to 2 at diagnosis were recruited from the Duke University and Durham VA Medical Centers for participation in IRB approved protocols to donate blood for further study. Clinical data were determined according to the NCI Working Group criteria. (24) Blood was collected prior to therapy, CLL cells were purified by negative selection and frozen in pellets at −80°C. Using purified cells, we determined IgVH mutational status by sequencing genomic DNA amplified from the immunoglobulin heavy chain. We determined CD38 status by flow cytometry, and ZAP-70 expression by immunoblot and flow cytometry. We performed fluorescent in situ hybridization (FISH) to detect cytogenetic abnormalities of del(13q14), del(11q22.3), trisomy 12, and del(17p13.1). The methods used to perform these assays were previously described. (22) From samples containing ten to fifteen million CLL lymphocytes, total RNA was extracted using Qiashredder and RNeasy Mini Kits (Qiagen Inc, Valencia, CA) and quality was assessed by spectrophotometry and by Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
Patients enrolled in a phase II trial of pentostatin, cyclophosphamide, and rituximab at the Mayo Clinic and the Ohio State University had pretreatment CLL lymphocytes collected for a correlative study. All study subjects signed written informed consent to participate in this study in accordance with the Declaration of Helsinki and the Mayo Clinic Institutional Review Board. The description of patient characteristics and response to therapy was previously published. (23) Blood from patients enrolled in this trial was drawn into heparin tubes and peripheral blood mononuclear cells (PBMC) were isolated by Ficoll (Gallard-Schlesinger Industries, Inc.) density gradient centrifugation. CLL patient PBMCs that did not exceed >85% CD19+ cells were further purified using CD19 magnetic beads (Miltenyi Biotec, Auburn, CA). Cells to be purified were suspended with CD19 beads in PBS and 0.5% FCS and 2mM EDTA for 15 min at 4°C. Cells were washed, resuspended in the same buffer at a concentration of 5–10 million cells/ml and passed through the AutoMacs Magnetic Cell sorter (Miltenyi Biotec) to collect the CD19+ cells. Total RNA was isolated from cells using the Trizol reagent (Invitrogen, Carlsbad, CA). RNA was quantitated by reading the absorbance at 260/280 and the RNA quality was further tested using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Techniques used to determine prognostic markers (ZAP-70, IgVH mutation status, and CD38) were described previously. (23)
DNA Microarray Analysis
RNA samples were prepared and analyzed according to the manufacturer’s instructions and as described previously. (25) All analyses were performed in a MIAME (minimal information about a microarray experiment) compliant fashion. Raw data was normalized using the MAS 5.0 algorithm, using R. Genomic data is found in GEO (GSE # 10137, 10138, and 10139).
To generate genomic signatures, we performed supervised analyses using the Bayesian binary regression algorithm in Matlab (Mathworks, Natick, MA), as described previously. (26, 27) The signature of progressive disease was created with a training set comprised of all the samples from the Duke/VA cohort. The signature of response to chlorambucil was created using cancer cell lines, and the signature of response to the pentostatin, cyclophosphamide, and rituximab regimen was created with half the samples in the Mayo/Ohio State cohort. The number of probes in a signature was selected that conferred the best accuracy in the training set. Leave-one-out cross validation was performed to assess the accuracy of the signature and the supervised analysis method.
A genomic signature of chlorambucil sensitivity was created using the National Cancer Institute drug-screening panel (NCI-60 panel) of cancer cell lines and expression data (http://dtp.nci.nih.gov/mtargets/download.html) from Affymetrix U133A GeneChips from selected cell lines (Supplemental Table 2), with techniques described previously. (25) Cancer cell lines that represent the extremes of sensitivity and resistance to chlorambucil were selected based on in vitro 50% cytotoxic dose (LC50) and total growth inhibition (TGI) measurements.
A genomic signature of response to the PCR regimen was created using a similar process. From the total of 40 available pretreatment CLL lymphocyte samples, microarray data from ten long-term responders (time to progression greater than the median time to progression amongst non-progressors) and ten early progressors (time to progression less than the median time to progression amongst progressors) were used as a training set.
We validated the genomic signatures with external data sets using the same algorithm. Two independent data sets were used to validate the signature of progressive disease: one from the Spanish National Cancer Centre and the second from the National Cancer Institute. (17, 20) Both validation data sets had been created on cDNA microarrays. We applied the genomic signature of response to chlorambucil using microarray data from a subset of the Duke/VA cohort that were from CLL lymphocytes collected prior to treatment with chlorambucil. The genomic signature of response to the chemo-immunotherapy regimen of pentostatin, cyclophosphamide, and rituximab was tested using microarray data from the Mayo/Ohio State cohort that was independent of the data used in the training set.
We joined the training and validation sets for the signature of progressive disease using concordant gene symbols from the cDNA arrays and Affymetrix probes obtained from NetAffx1. cDNA probes that had null values were eliminated and only probes that represented shared genes between the two platforms were used, yielding combined datasets with fewer probes than either data set alone. Further processing included quantile normalization and standardization using ComBat. (28) When comparing training and validation data for the signatures of response to therapy from Affymetrix U133A and U133 Plus 2.0 GeneChips, we included only shared probe set IDs.
Statistical Analysis
We generated and validated genomic signatures using Matlab (Mathworks, Natick, MA). Kaplan-Meier analyses, Wilcoxon rank sum test, Fisher’s exact test, log-rank test, and Cox proportional hazards models were calculated using the statistical environment, R.
Results
We have made use of data from cancer cell lines and CLL patient leukemia samples to develop genomic signatures to guide the management of CLL. In particular, the goal was to create gene expression signatures to refine prognosis and predict therapeutic responses for CLL patients, thereby identifying patients who require therapy and determining the best treatment regimen for those patients.
Patient Characteristics
We studied low and intermediate risk CLL patients who had no immediate indication for therapy at the time of diagnosis in order to determine if a microarray based test could be created to help prognosticate need for subsequent therapy.
As patients often live for years to decades after a diagnosis of CLL, we used freedom from progression as our primary clinical outcome. We defined progressive disease as the need for therapy, and stable disease as requiring no therapy. In the Duke/VA, the 33 patients with stable disease were followed for 2.4 years to 26 years from diagnosis. Samples were collected from patients with stable disease 0 to 20.4 years after diagnosis. The 28 patients with progressive disease were followed for 2.3 years to 16.1 years from diagnosis, and were treated 0.05 to 11.9 years after diagnosis. Samples collected from patients with progressive disease were collected 0 to 10.6 years after diagnosis. Other characteristics of the patients and CLL cell samples are detailed in Table 1 and Supplemental Table 3. Notably, the IgVH mutation status was statistically different between the two groups.
Table 1.
Characteristics of Patients and CLL lymphocytes in the Genomic Signature of Progression Training Set
| Stable Disease (n = 33) |
Progressive Disease (n = 28) |
P-value * | |
|---|---|---|---|
| Age at diagnosis (years) | 0.06 | ||
| Median | 63 | 57.5 | |
| Range | 40 – 81 | 32 – 79 | |
| Sex | 0.40 | ||
| Male | 25 | 18 | |
| Female | 8 | 10 | |
| Race | 0.76 | ||
| White | 29 | 24 | |
| Black | 3 | 2 | |
| Other | 1 | 2 | |
| Stage at diagnosis | 0.48 | ||
| Rai 0 | 24 | 17 | |
| Rai 1 | 7 | 7 | |
| Rai 2 | 2 | 4 | |
| IgVH status † | 0.02 | ||
| Mutated | 24 | 11 | |
| Unmutated | 8 | 15 | |
| NA | 1 | 2 | |
| Cytogenetics † | 0.10 | ||
| Normal | 6 | 6 | |
| 13q deletion | 6 | 9 | |
| Trisomy 12 | 1 | 5 | |
| 11q deletion | 1 | 0 | |
| 17p deletion | 1 | 1 | |
| Complex | 4 | 3 | |
| NA | 14 | 4 | |
| CD38 † | 0.07 | ||
| Positive | 4 | 9 | |
| Negative | 28 | 17 | |
| NA | 1 | 2 | |
| ZAP-70 † | 1.0 | ||
| Positive | 21 | 19 | |
| Negative | 9 | 7 | |
| NA | 3 | 2 | |
P-value determined by Wilcoxon Rank Sum Test for continuous variables and Fisher’s Exact Test for categorical variables.
ZAP-70, CD38, cytogenetics, and IgVH determination as described in Reference 22.
NA = not available
Forty patients of the total 64 patients in the Mayo/Ohio State cohort were included in the analysis to create a genomic signature of response to therapy. Patients were included if they gave a pretreatment blood sample and microarray analysis was successfully performed. The clinical and laboratory parameters and the response data for the entire cohort were described previously. (23) There was no significant difference between the group from which microarrays were performed compared to the remaining patients with regards to progression-free survival, CD38, ZAP-70, or IgVH mutational status (Fisher’s exact test, data not shown), however there were more patients that had microarrays performed that achieved complete or nodular partial responses (p = 0.007, Fisher’s exact test).
Genomic Signature of Progression
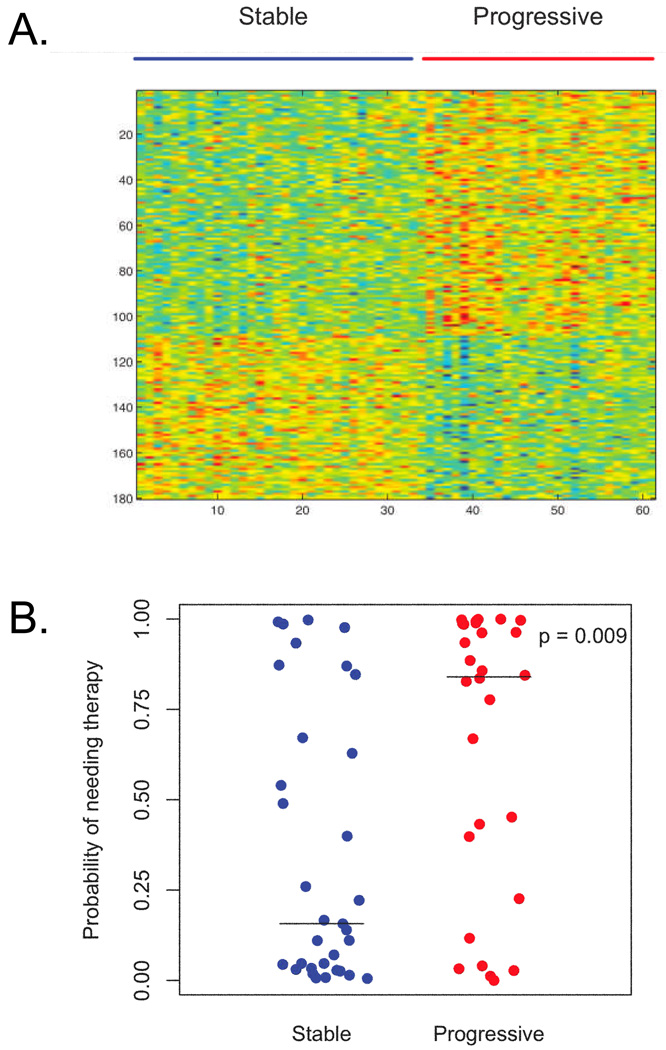
By applying the supervised analysis method of Bayesian binary regression to the gene expression data from the 33 samples from patients with stable disease and the 28 samples from patients with progressive disease, a gene expression signature comprised of 180 gene probes was generated (Figure 1A, Supplemental Table 1). Figure 1B demonstrates that leave-one-out cross validation distinguished stable and progressive disease states. Using a cut-off of 0.5, the sensitivity of this signature was 64%, the specificity was 67%, the positive predictive value was 62% and the negative predictive value was 69% (Supplemental Table 2). The signature’s association with the phenotype is independent of the prognostic markers IgVH mutational status, CD38 and ZAP-70 expression and cytogenetic aberrations.
Figure 1. Genomic Signature of Disease Progression.
Panel A: Heatmap of the 180-gene signature differentiating CLL samples from patients who did not require therapy (stable disease, n = 33 samples) from those who did require therapy (progressive disease, n = 28 samples). Red color represents upregulated genes and blue color represents downregulated genes.
Panel B: Leave-one-out cross validation score of fitting the 180-gene signature for the 33 samples in the stable group and the 28 samples in the progressive group. The median values for the stable and progressive groups are denoted by horizontal lines and were 0.157 and 0.840, respectively. P value was determined by Wilcoxon rank sum test.
We then compared the predictive ability of the genomic signature to that of IgVH mutational status, another parameter that is increasing used clinically. Prior evaluation of the entire Duke/VA cohort demonstrated that the sensitivity and specificity of unmutated IgVH status for predicting need for therapy was 48% and 77%, with a positive predictive value of 71% and a negative predictive value of 56%. (22) Thus, compared with the currently used parameter, IgVH mutation status, the genomic signature has a higher sensitivity but lower specificity.
Combining Prognostic Markers
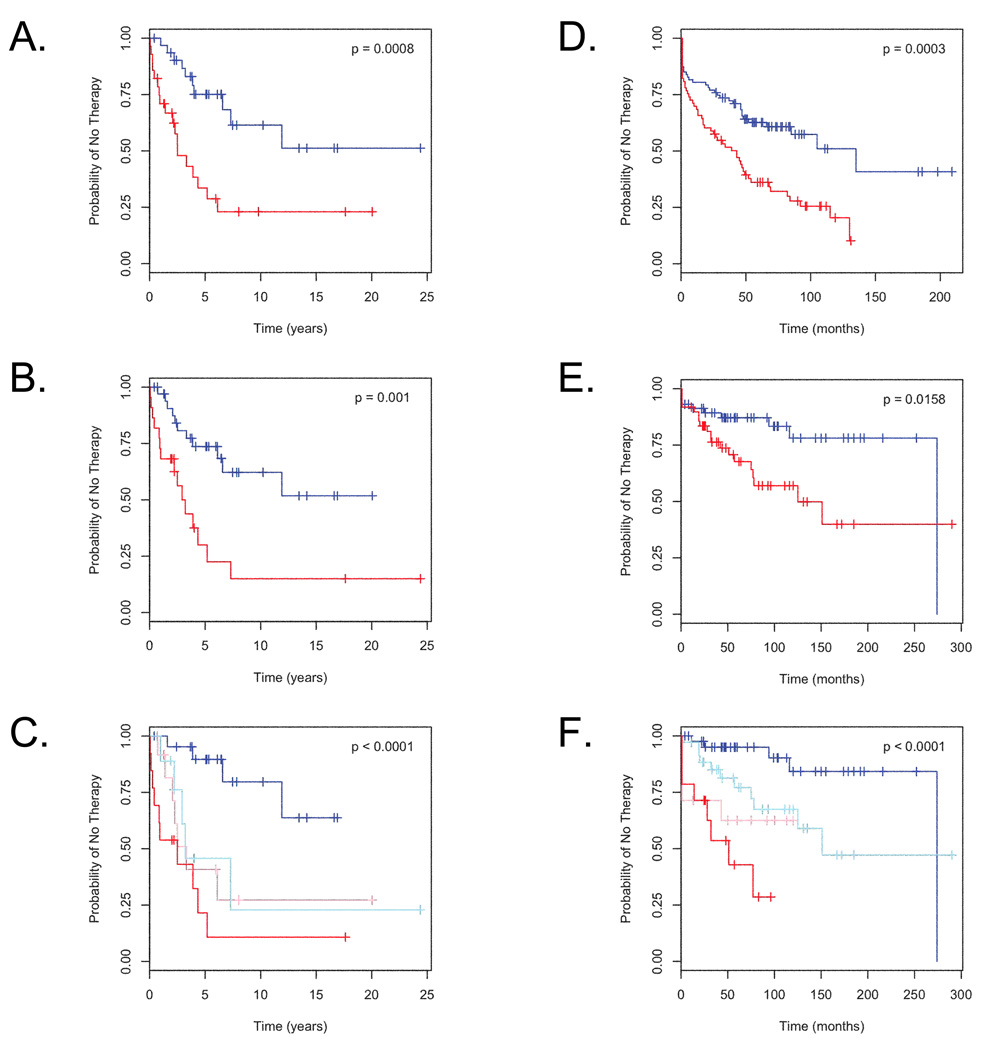
Using Kaplan-Meier analysis, two groups of patients defined by the genomic signature had significantly different times to treatment need (p = 0.0008, log-rank test, Figure 2A). IgVH mutational status could also define two groups with different times to progression of disease (p = 0.001, log-rank test, Figure 2B). We then evaluated the utility of combining the genomic signature with other markers associated with clinical outcomes using a multivariate Cox proportional hazards model. The inclusion of prognostic markers other than IgVH mutational status did not contribute significantly to the model. The genomic signature score and IgVH mutational status had no significant interaction in the multivariate model. As seen in Table 2, the multivariate model combining genomic signature score and IgVH mutational status outperformed each individual factor to identify a low-risk population. The hazard ratio of not needing therapy in the group of patients with high genomic signature score and mutated IgVH was 0.157, compared to hazard ratios of approximately 0.29 for either factor alone. In addition, the combination of a low progressive disease signature score and mutated IgVH status conferred the best prognosis and was greatest at selecting this low-risk group of patients (Figure 2C).
Figure 2. Refining Prognosis by Combining Genomic Signature Score with IgVH Mutation Status and Validating in Independent Datasets.
Panel A: Kaplan-Meier analysis of freedom from therapy in patients predicted to have stable disease by the genomic signature with a score < 0.5 (n = 39, blue color) compared to patients predicted to have progressive disease by the genomic signature with a score of ≥ 0.5 (n = 29, red color).
Panel B: Kaplan-Meier analysis of freedom from therapy in patients with mutated IgVH (n = 35, blue color) versus patients with unmutated IgVH (n = 23, red color).
Panel C: Kaplan-Meier analysis of freedom from therapy in patients subdivided based on a Cox proportional hazards model. High microarray score (≥ 0.5) combined with unmutated IgVH confers the worst prognosis (n = 14, red color), while low microarray score (< 0.5) combined with mutated IgVH confer the best prognosis (n = 22, blue color). Low microarray score with unmuated IgVH (n = 9, light blue color) or high microarray score with mutated IgVH (n = 13, pink color) confer intermediate prognoses. P values were calculated using the log-rank test.
Panel D: Kaplan-Meier analysis of freedom from therapy in patients from the Spanish National Cancer Centre predicted to have stable disease (prediction score < 0.5, n = 87, blue color) compared to patients predicted to have progressive disease (prediction score ≥ 0.5, n = 73, red color).
Panel E: Kaplan-Meier analysis of freedom from therapy in patients from the National Cancer Institute predicted to have stable disease (prediction score < 0.5, n = 58, blue color) compared to patients predicted to have progressive disease (prediction score ≥ 0.5, n = 49, red color).
Panel F: Combination of IgVH mutational status with genomic prediction of progressive disease reveals improved prognostic capabilities in the National Cancer Institute cohort. Low microarray score (< 0.5) combined with mutated IgVH confer the best prognosis (n = 44, blue color) conferred the best prognosis. High microarray score (≥ 0.5) combined with unmutated IgVH (n = 14, red color), low microarray score with unmuated IgVH (n = 14, pink color) and high microarray score with mutated IgVH (n = 35, light blue color) are also displayed. P values were calculated using the log-rank test.
Table 2.
Effect of combining prognostic markers to define need for therapy
| Duke/VA Cohort | |||
|---|---|---|---|
| Hazard Ratio | 95% CI Range | P-value* | |
| Univariate model with Microarray score ≤ 0.5 | 0.287 | 0.131 – 0.625 | 0.0017 |
| Univariate model with Mutated IgVH status | 0.291 | 0.132 – 0.638 | 0.002 |
| Multivariate model with Microarray score ≤ 0.5 and Mutated IgVH status |
0.157 | 0.054 – 0.461 | 0.0007 |
P-value determined by Wald test
Validation in External Datasets
Although the leave-one-out cross validation provides evidence for a model that can distinguish patients with progressive disease from those with stable disease, the utility of such a model requires validation in independent cohorts of patients. To address the utility of the model, we examined the signature and methods in two external data sets, one from the Spanish National Cancer Centre (17) (Array Express E-TABM-80, N = 160) and the other from the National Cancer Institute2 (20) (N = 107). Clinical characteristics of the CLL patients included in these cohorts were published previously. (17, 20)
Since these two validation data sets were generated using cDNA microarrays, we compared cDNA ratios to MAS5 calculated expression values. Because of the reduced number of probes in the merged data sets compared to the training set (3576 probes with the Spainish data set, and 6279 probes with the NCI data set), it was necessary to alter the parameters in the binary regression algorithm from those used initially (Supplemental Table 2). Thus, this analysis is a validation of the ability of the genomic data from the Duke/VA training group of patients to predict the outcomes of two separate independent cohorts, rather than testing the accuracy of a specific set of genes.
Scores reflecting the probability of progression in the two validation sets were calculated by applying the genomic signature created from the training set to gene expression data from the two independent cohorts. We used Kaplan-Meier analyses to demonstrate associations between the classifications of stable or progressive disease determined from the genomic signature and time to therapy (Figures 2D and 2E). This demonstrated the ability of the Duke/VA genomic data and signature to discriminate between stable and progressive disease in these cohorts in a statistically significant manner. The full predictive accuracy to predict need for therapy in these validation sets is shown in Supplemental Table 2.
A multivariate Cox proportional hazards model was created that combined the genomic signature prediction and the IgVH mutational status for the NCI data set (Figure 2F). As in the Duke/VA cohort, the combination of both the genomic signature and the IgVH mutation status was superior to either alone.
Genomic Signatures of Resistance to Chemotherapy
The selection of chemotherapy for a CLL patient depends in part on the patient’s performance status and comorbidities. Chlorambucil has been used, particularly in the elderly, because of its low toxicity profile. However, this agent rarely induces complete remissions, and it must be administered for as long as one year to control the disease. (29) To help identify patients who would respond to chlorambucil, we employed a genomic approach. (25)
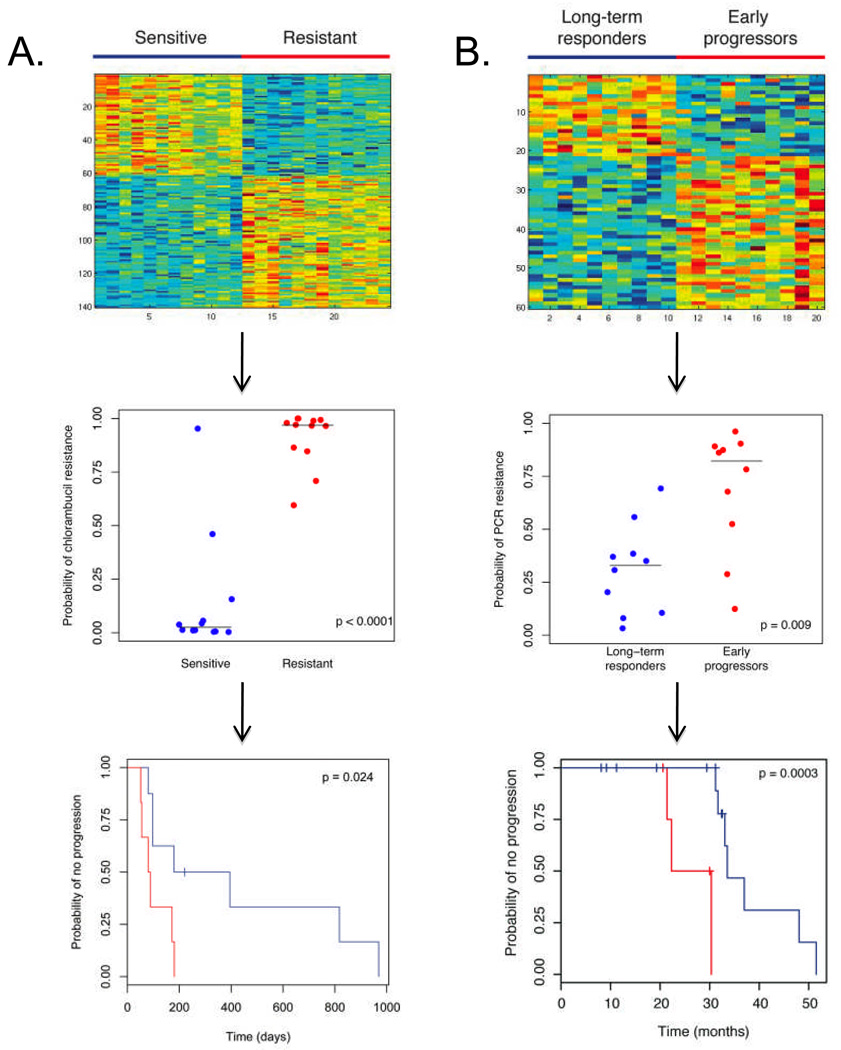
We used gene expression data coupled with drug sensitivity data from the NCI-60 data set to create a 140 probe genomic signature that reflects resistance to chlorambucil in cancer cell lines (Supplemental Table 1). As seen in Figure 3A, the leave-one-out cross validation predictions for the training set were statistically different, with an accuracy of 92% to predict chlorambucil resistance, sensitivity of 100%, and specificity of 92% (Supplemental Table 2). We next evaluated the signature’s applicability to patients’ clinical responses by evaluating a group of 14 Duke/VA patients treated with chlorambucil. These patients’ characteristics are described in Table 3. The genomic signature could define two groups of patients: those who had a meaningful response to chlorambucil and those who did not (Figure 3A).
Figure 3. Genomic Signatures of Chemotherapy Sensitivity Applied to Clinical Data.
Panel A: The top figure demonstrates the heatmap of the 140-gene signature of resistance to chlorambucil, created from gene expression profiling of cell lines sensitive and resistant to this agent. Red color represents upregulated genes while blue color represents downregulated genes. The middle figure displays the leave-one-out cross validation values for the training data set. The horizontal lines represent median predicted values for the sensitive and resistant groups, 0.025 and 0.969 respectively. As seen in the bottom figure, when applied to genomic data of CLL cells from patients subsequently treated with chlorambucil, this signature can discriminate patients based on clinical response to therapy, with blue color denoting a prediction of more durable response (prediction score < 0.5, n = 8) and red color denoting a prediction of less durable response (prediction score ≥ 0.5, n = 6). P-value determined by log-rank test.
Panel B: The top figure demonstrates the heatmap of the 60-gene signature of resistance to pentostatin, cyclophosphamide, and rituximab (PCR), created from genomic data from patients who progressed early or were long-term responders. Red color represents upregulated genes while blue color represents downregulated genes. The middle figure displays the leave-one-out cross validation values for the training data set. The horizontal lines represent median predicted values for the long-term responder and early progressor groups, 0.329 and 0.822 respectively. As seen in the bottom figure, when applied to the genomic data from an additional 20 patients treated with this regimen and using a cut-off of 0.5, this signature can separate patients based on response, with blue color denoting a prediction of long-term response (n = 15) and red color denoting a prediction of early progression (n = 5). P-value determined by log-rank test.
Table 3.
Characteristics of Patients and CLL lymphocytes in the Genomic Signatures of Treatment Response
| Chlorambucil Test Set (n = 14) |
PCR Training Set (n = 20) |
PCR Test Set (n = 20) |
|
|---|---|---|---|
| Sex | |||
| Male | 10 | 15 | 16 |
| Female | 4 | 5 | 4 |
| Race | |||
| White | 12 | 19 | 20 |
| Other | 2 | 0 | 0 |
| NA | 0 | 1 | 0 |
| Median Age at Treatment in years (range) |
64 (42 – 82) | 61 (40 – 78) | 65 (49 – 76) |
| Stage at Treatment | |||
| Rai 0 | 7 | 1 | 2 |
| Rai 1 | 4 | 0 | 4 |
| Rai 2 | 1 | 3 | 4 |
| Rai 3 | 1 | 11 | 4 |
| Rai 4 | 1 | 5 | 6 |
| Median Progression Free Survival in days (range) |
135 (51 – 969) | 822 (45 – 1803) | 923 (243 – 1546) |
| IgVH status * | |||
| Mutated | 9 | 8 | 7 |
| Unmutated | 5 | 12 | 13 |
| Cytogenetics * | |||
| Normal | 3 | 4 | 1 |
| 13q deletion | 5 | 6 | 8 |
| Trisomy 12 | 1 | 5 | 6 |
| 11q deletion | 0 | 3 | 5 |
| 17p deletion | 1 | 1 | 0 |
| Complex or Other | 1 | 1 | 0 |
| NA | 3 | 0 | 0 |
| CD38 * | |||
| Positive | 2 | 7 | 7 |
| Negative | 12 | 13 | 13 |
| ZAP-70 * | |||
| Positive | 10 | 7 | 8 |
| Negative | 4 | 10 | 11 |
| NA | 3 | 1 | |
To broaden the possibilities of using genomic signatures to predict patterns of response, we evaluated a larger cohort of progressive CLL patients who were treated with chemo-immunotherapy (CIT). Triple agent CIT is increasingly used in the treatment of CLL. (23, 30, 31) The recent phase II study of treatment with pentostatin, cyclophosphamide, and rituximab (the PCR regimen) in previously untreated patients demonstrated an overall response rate of 91% and a median progression free survival of 32.6 months. (23) Twenty of the total of 40 CLL patient samples collected prior to therapy were used to create a sixty-gene signature of sensitivity to this regimen that significantly differentiated the long-term responders from the early progressors (Figure 3B). There was no statistically significant difference in type of clinical response obtained by these two groups. Notably, the probes that comprise this signature are distinct from those that make up the signatures for progressive disease and chlorambucil resistance (Supplemental Table 1). We used leave-one-out cross validation to assess the accuracy of this signature, and found that the sensitivity and specificity were both 80% when a cut-off of 0.5 was used (Supplemental Table 2). As seen in Figure 3B, the prediction scores on leave-one-out cross validation in the long-term responders versus early progressors in this training set were statistically different (p = 0.0089 by Wilcoxon rank sum test), whereas there was no significant difference using IgVH, CD38, or ZAP-70.
We then applied the signature to the remaining 20 patients’ data to determine if classification based on the genomic signature was associated with clinical outcome. The clinical characteristics of the patients in the training and the test sets are shown in Table 3. Using Kaplan-Meier analysis, we demonstrated that the genomic signature defined two groups that differed based on clinical response of time to progression (p = 0.0003, log rank test, Figure 3B). Comparing the progressors to the non-progressors in this group, there was no statistically significant difference in pathologic response, IgVH mutation status, or CD38 or ZAP-70 expression. The genomic signatures of progressive disease and of chlorambucil sensitivity could not accurately classify patients by response (Supplemental Figure 1). These results indicate that unlike most current prognostic markers, a specific genomic signature appears capable of identifying CLL patients who will have rapid progression after treatment with pentostatin, cyclophosphamide, and rituximab, an effective chemo-immunotherapy regimen.
Discussion
Like many cancers, CLL is a heterogeneous malignancy in which there is uncertainty regarding optimal treatment selection and the identification of patients likely to have progressive disease. We utilized gene expression data from CLL patients from the Duke University and Durham VA Medical Centers, the Mayo Clinic and the Ohio State University, as well as published and publicly available data sets to demonstrate that genomic data from CLL patients can be used to address these critical issues.
We found that a genomic signature classifies patients into those likely to progress and those likely to have stable disease. Compared to IgVH mutational status in the entire Duke/VA cohort, the genomic signature of progressive disease was at least as good at predicting outcomes. There were insufficient numbers of patients with specific cytogenetic aberrations to evaluate a contribution from FISH analyses. The combination of the genomic score of progression and IgVH mutational status provided improved prognostic capability over either marker alone. Others have created models and nomograms that combine prognostic factors into one score. (22, 32) As genomic-based tests enter clinical use, understanding how to use this information in conjunction with other prognostic markers in a simple and efficient manner will be important.
Our goal with this work was to create and validate genomic signatures that might be used clinically in the future, irrespective of the genes that make up those signatures. However, analysis of the gene probes that constitute the genomic signature of progressive disease demonstrate several involved in the cytoskeleton (RDX, TNS3) or tumor necrosis factor (TNF) family members and genes involved in TNF cleavage (TNFSF13, ADAM17). Others have previously noted the prognostic significance of cytoskeletal genes and TNF in CLL. (33–35) Notably, probes for ZAP70 did not constitute this genomic signature, even though mean expression for ZAP70 probes in the samples from patients with progressive disease was higher than those from patients with stable disease. In part this may be due to different statistical methodology for supervised analysis of genomic data compared to other groups.
We found clinical heterogeneity and variable length of follow-up in our training and validation data sets, as well as methodological differences in sample collection and research techniques. Despite these substantial limitations, we could create a genomic signature that reflects prognosis, and this signature and methodology could be independently validated with other published cohorts. This points towards the potential of this tool to perform well in situations that are much better controlled than what was possible for this analysis.
The choice of optimal therapy in CLL is a balance between the efficacy of multi-drug regimens and the lower toxicity of less effective single agents. While no currently available biomarkers predict resistance to treatment, the deletions of chromosomes 17p13.1 or 11q22.3 are associated with incomplete and inferior responses to purine analogue based therapy. (6–8) Using gene expression profiling, we found that we could predict response of a patient’s leukemia to chlorambucil, an agent with less toxicity than other agents but lower long-term efficacy. The same genomic data could be tested against a distinct genomic signature of response to a more effective but more toxic regimen of chemo-immunotherapy with pentostatin, cyclophosphamide, and rituximab. Given the limited ability of available prognostic factors to predict responses to therapy, these findings suggest that it will be ultimately possible to make rational selections of therapies for CLL patients, moving towards the goal of “personalized medicine”.
In conclusion, gene expression profiling not only assists in refining prognosis of CLL, but may also be used in determining optimal therapies. Our results have potential applicability in guiding clinical and investigational medicine and may direct future care of CLL patients.
Statement of Translational Relevance
Chronic Lymphocytic Leukemia (CLL) is an incurable malignancy that exhibits clinical heterogeneity. Currently available prognostic markers only partially predict the aggressiveness of disease or response to treatment regimens. In this study, we have used gene expression microarray techniques and analyses to create and validate a genomic signature with prognostic capabilities that can be combined with a currently used prognostic marker, with improved performance over either alone. We also generated and validated genomic signatures that predict resistance to two chemotherapeutic regimens. These results demonstrate that gene expression profiling can be used to refine prognosis and chose optimal therapies for patients, indicating that this approach could be part of future personalized medical care of CLL patients.
Supplementary Material
Acknowledgements
We thank clinicians responsible for sample collection at the sites contributing to training data. We acknowledge Neil Kay, John Byrd, Diane Jelinek, Renee Tschumper, Jeanette Eckel-Passow, and Adrian Wiestner for data and input on data analysis. This research was supported in part by the American Society of Clinical Oncology Young Investigator Award (D.R. Friedman), the Leukemia and Lymphoma Society and the V.A. Research Service (J.B. Weinberg) and the National Institutes of Health grant 5U54 CA112952 (J.R. Nevins).
Footnotes
References
- 1.Crespo M, Bosch F, Villamor N, et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med. 2003;348:1764–1775. doi: 10.1056/NEJMoa023143. [DOI] [PubMed] [Google Scholar]
- 2.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- 3.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 4.Hallek M, Wanders L, Ostwald M, et al. Serum beta(2)-microglobulin and serum thymidine kinase are independent predictors of progression-free survival in chronic lymphocytic leukemia and immunocytoma. Leuk Lymphoma. 1996;22:439–447. doi: 10.3109/10428199609054782. [DOI] [PubMed] [Google Scholar]
- 5.Rassenti LZ, Huynh L, Toy TL, et al. ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med. 2004;351:893–901. doi: 10.1056/NEJMoa040857. [DOI] [PubMed] [Google Scholar]
- 6.Byrd JC, Gribben JG, Peterson BL, et al. Select high-risk genetic features predict earlier progression following chemoimmunotherapy with fludarabine and rituximab in chronic lymphocytic leukemia: justification for risk-adapted therapy. J Clin Oncol. 2006;24:437–443. doi: 10.1200/JCO.2005.03.1021. [DOI] [PubMed] [Google Scholar]
- 7.Dohner H, Fischer K, Bentz M, et al. p53 gene deletion predicts for poor survival and non-response to therapy with purine analogs in chronic B-cell leukemias. Blood. 1995;85:1580–1589. [PubMed] [Google Scholar]
- 8.Grever MR, Lucas DM, Dewald GW, et al. Comprehensive Assessment of Genetic and Molecular Features Predicting Outcome in Patients With Chronic Lymphocytic Leukemia: Results From the US Intergroup Phase III Trial E2997. J Clin Oncol. 2007;25:799–804. doi: 10.1200/JCO.2006.08.3089. [DOI] [PubMed] [Google Scholar]
- 9.Aalto Y, El-Rifa W, Vilpo L, et al. Distinct gene expression profiling in chronic lymphocytic leukemia with 11q23 deletion. Leukemia. 2001;15:1721–1728. doi: 10.1038/sj.leu.2402282. [DOI] [PubMed] [Google Scholar]
- 10.Dickinson JD, Smith LM, Sanger WG, et al. Unique gene expression and clinical characteristics are associated with the 11q23 deletion in chronic lymphocytic leukaemia. Br J Haematol. 2005;128:460–471. doi: 10.1111/j.1365-2141.2004.05344.x. [DOI] [PubMed] [Google Scholar]
- 11.Durig J, Nuckel H, Huttmann A, et al. Expression of ribosomal and translation-associated genes is correlated with a favorable clinical course in chronic lymphocytic leukemia. Blood. 2003;101:2748–2755. doi: 10.1182/blood-2002-09-2683. [DOI] [PubMed] [Google Scholar]
- 12.Falt S, Merup M, Gahrton G, Lambert B, Wennborg A. Identification of progression markers in B-CLL by gene expression profiling. Exp Hematol. 2005;33:883–893. doi: 10.1016/j.exphem.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Ferrer A, Ollila J, Tobin G, et al. Different gene expression in immunoglobulin-mutated and immunoglobulin-unmutated forms of chronic lymphocytic leukemia. Cancer Genet Cytogenet. 2004;153:69–72. doi: 10.1016/j.cancergencyto.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 14.Haslinger C, Schweifer N, Stilgenbauer S. Microarray gene expression profiling of B-cell chronic lymphocytic leukemia subgroups defined by genomic aberrations and VH mutation status. J Clin Oncol. 2004;22:3937–3949. doi: 10.1200/JCO.2004.12.133. [DOI] [PubMed] [Google Scholar]
- 15.Huttmann A, Klein-Hitpass L, Thomale J, et al. Gene expression signatures separate B-cell chronic lymphocytic leukaemia prognostic subgroups defined by ZAP-70 and CD38 expression status. Leukemia. 2006 doi: 10.1038/sj.leu.2404363. [DOI] [PubMed] [Google Scholar]
- 16.Jelinek DF, Tschumper RC, Stolovitzky GA, et al. Identification of a global gene expression signature of B-chronic lymphocytic leukemia. Mol Cancer Res. 2003;1:346–361. [PubMed] [Google Scholar]
- 17.Rodriguez A, Villuendas R, Yanez L, et al. Molecular heterogeneity in chronic lymphocytic leukemia is dependent on BCR signaling: clinical correlation. Leukemia. 2007;21:1984–1991. doi: 10.1038/sj.leu.2404831. [DOI] [PubMed] [Google Scholar]
- 18.Rosenwald A, Alizadeh AA, Widhopf G, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194:1639–1647. doi: 10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stratowa C, Loffler G, Lichter P, et al. CDNA microarray gene expression analysis of B-cell chronic lymphocytic leukemia proposes potential new prognostic markers involved in lymphocyte trafficking. Int J Cancer. 2001;91:474–480. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1078>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 20.Wiestner A, Rosenwald A, Barry TS, et al. ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile. Blood. 2003;101:4944–4951. doi: 10.1182/blood-2002-10-3306. [DOI] [PubMed] [Google Scholar]
- 21.Zent CS, Zhan F, Schichman SA, et al. The distinct gene expression profiles of chronic lymphocytic leukemia and multiple myeloma suggest different anti-apoptotic mechanisms but predict only some differences in phenotype. Leuk Res. 2003;27:765–774. doi: 10.1016/s0145-2126(03)00015-8. [DOI] [PubMed] [Google Scholar]
- 22.Weinberg JB, Volkheimer AD, Chen Y, et al. Clinical and molecular predictors of disease severity and survival in chronic lymphocytic leukemia. Am J Hematol. 2007;82:1063–1070. doi: 10.1002/ajh.20987. [DOI] [PubMed] [Google Scholar]
- 23.Kay NE, Geyer SM, Call TG, et al. Combination chemoimmunotherapy with pentostatin, cyclophosphamide, and rituximab shows significant clinical activity with low accompanying toxicity in previously untreated B chronic lymphocytic leukemia. Blood. 2007;109:405–411. doi: 10.1182/blood-2006-07-033274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheson BD, Bennett JM, Grever M, et al. National Cancer Institute-sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood. 1996;87:4990–4997. [PubMed] [Google Scholar]
- 25.Potti A, Dressman HK, Bild A, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–1300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 26.Pittman J, Huang E, Dressman H, et al. Integrated modeling of clinical and gene expression information for personalized prediction of disease outcomes. Proc Natl Acad Sci U S A. 2004;101:8431–8436. doi: 10.1073/pnas.0401736101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potti A, Mukherjee S, Petersen R, et al. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med. 2006;355:570–580. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- 28.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 29.Rai KR, Peterson BL, Appelbaum FR, et al. Fludarabine compared with chlorambucil as primary therapy for chronic lymphocytic leukemia. N Engl J Med. 2000;343:1750–1757. doi: 10.1056/NEJM200012143432402. [DOI] [PubMed] [Google Scholar]
- 30.Keating MJ, O'Brien S, Albitar M, et al. Early results of a chemoimmunotherapy regimen of fludarabine, cyclophosphamide, and rituximab as initial therapy for chronic lymphocytic leukemia. J Clin Oncol. 2005;23:4079–4088. doi: 10.1200/JCO.2005.12.051. [DOI] [PubMed] [Google Scholar]
- 31.Wierda W, O'Brien S, Wen S, et al. Chemoimmunotherapy with fludarabine, cyclophosphamide, and rituximab for relapsed and refractory chronic lymphocytic leukemia. J Clin Oncol. 2005;23:4070–4078. doi: 10.1200/JCO.2005.12.516. [DOI] [PubMed] [Google Scholar]
- 32.Wierda WG, O'Brien S, Wang X, et al. Prognostic nomogram and index for overall survival in previously untreated patients with chronic lymphocytic leukemia. Blood. 2007;109:4679–4685. doi: 10.1182/blood-2005-12-051458. [DOI] [PubMed] [Google Scholar]
- 33.Bojarska-Junak A, Hus I, Szczepanek EW, Dmoszynska A, Rolinski J. Peripheral blood and bone marrow TNF and TNF receptors in early and advanced stages of B-CLL in correlation with ZAP-70 protein and CD38 antigen. Leuk Res. 2007 doi: 10.1016/j.leukres.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 34.Ferrajoli A, Keating MJ, Manshouri T, et al. The clinical significance of tumor necrosis factor-alpha plasma level in patients having chronic lymphocytic leukemia. Blood. 2002;100:1215–1219. [PubMed] [Google Scholar]
- 35.Nowakowski GS, Hoyer JD, Shanafelt TD, et al. Using smudge cells on routine blood smears to predict clinical outcome in chronic lymphocytic leukemia: a universally available prognostic test. Mayo Clinic proceedings. 2007;82:449–453. doi: 10.4065/82.4.449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.