Abstract
A strategy in drug design is to consider enhancing the affinity of lead molecules with structural modifications that displace water molecules from a protein binding site. Because success of the approach is uncertain, clarification of the associated energetics was sought in cases where similar structural modifications yield qualitatively different outcomes. Specifically, free energy perturbation calculations were carried out in the context of Monte Carlo statistical mechanics simulations to investigate ligand series that feature displacement of ordered water molecules in the binding sites of scytalone dehydratase, p38-αMAP kinase, and EGFR kinase. The change in affinity for a ligand modification is found to correlate with the ease of displacement of the ordered water molecule. However, as in the EGFR example, the binding affinity may diminish if the free energy increase due to the removal of the bound water molecule is not more than compensated by the additional interactions of the water-displacing moiety. For accurate computation of the effects of ligand modifications, a complete thermodynamic analysis is shown to be needed. It requires identification of the location of water molecules in the protein-ligand interface and evaluation of the free energy changes associated with their removal and with the introduction of the ligand modification. Direct modification of the ligand in free-energy calculations is likely to trap the ordered molecule and provide misleading guidance for lead optimization.
Introduction
Accurate computation of free energies of binding remains a key challenge for computer-aided drug design.1-3 Notably, hydration requires careful consideration, since interfacial water molecules contribute to protein stability, and release of water molecules from a binding site upon ligand binding is thought to provide a sizable contribution to the binding affinity.4 A popular approach to model bulk desolvation is to use implicit solvent theories,5-11 although they do not account well for ordered water molecules mediating protein-ligand interactions.12 The importance of these interactions is underscored by lead optimization strategies aimed at displacing ordered water molecules to gain entropy and improve affinity.13,14 In principle, water mediated protein-ligand interactions can be investigated by Monte Carlo (MC) or molecular dynamics (MD) simulations with the solvent explicitly represented.15 However, as the exchange of water molecules between a binding site and the bulk can be slow, specialized methodologies may be required to estimate accurately the locations and energetics of ordered water molecules.16,17
Li and Lazaridis have used inhomogeneous fluid solvation theory (IFST) to compute the thermodynamic properties of water molecules in binding sites.18 IFST allows the extraction of binding enthalpies and entropies as well as their components from a MD simulation and has proven useful to investigate water molecules in HIV-protease, concanavalin A, and cyclophilin A.19 The methodology has been expanded recently to locate all water molecules in a protein binding site and evaluate the favorability of their displacement.20 However, the binding free energies of water molecules computed by IFST are not directly comparable to absolute binding free energies, which are often computed by double-decoupling methods that compare the cost of annihilating the water molecule in bulk and in the binding site.21 As noted previously, the IFST results do not consider the relaxation of the protein and ligand after removal of a water molecule.22 Additionally, the approach assumes that a correct water distribution has been equilibrated by the MD simulation, which is not guaranteed for buried water molecules.17 Consequently, the resultant starting point has some ambiguity for typical free energy perturbation (FEP) or thermodynamic integration calculations, which could be used to predict the effects of ligand modifications that displace water molecules.
As an illustration of the complexities, Helms and Wade computed the absolute binding free energy of camphor bound to cytochrome P450 by simultaneously switching the ligand off and switching on six water molecules that had been shown previously to occupy the apo binding site by protein crystallography. Though the results were in good agreement with experiment,23 the approach required prior knowledge of the location and number of water molecules to switch on. Deng and Roux recently proposed an alternative protocol that combines grand canonical Monte Carlo and FEP techniques to sample dynamically the hydration level of binding sites during a perturbation so that prior detection of hydration sites is not necessary.24 However, simulations in the grand canonical ensemble require definition of a constant chemical potential. The value of this property should be chosen to match the usual experimental constant-NPT conditions, which typically requires simulations in large solvent boxes.25 In addition, information about the binding affinity of water molecules, useful during lead optimization efforts, is not readily obtained. Barillari et al. have computed absolute binding free energies for several water molecules in different binding sites and have shown that water molecules displaced by known ligands tend to have weaker binding affinities than water molecules that were never displaced by known ligands.26 Earlier results supporting this notion were provided by Hamelberg and McCammon.27 Yu and Rick have also recently reported MD results for the free energetics of introducing water molecules at the interface of DNA gyrase – inhibitor complexes that clarify effects of modifications of both the inhibitors and protein on binding affinities.28
This report focuses on the diversity of energetic effects incurred by displacement of water molecules from binding sites and their proper computational treatment. The approach combines an advanced algorithm for determining the location and likelihood of water molecules in binding sites, JAWS,17 and MC/FEP calculations. Detailed investigations are necessary to strengthen understanding of protein-ligand binding and to advance protocols for FEP-guided optimization of lead molecules.29-36 Protein-ligand systems were selected from the literature for which sufficient experimental data was available to assess reliably the energetic consequences of water displacement. Specifically, Chen et al. reported that conversion of the benzotriazine substructure in 1 to the3-cyano-cinnoline in 2 causes displacement of a bound water molecule in the binding site of scytalone dehydratase and a 30-fold improvement in Ki value (Figure 1A).37 Liu et al. followed a similar strategy, wherein the triazine core in 4, which is hydrogen-bonded to a water molecule in the binding site of p38α MAP kinase, was converted to the5-cyanopyrimidine unit in 5 (Figure 1B). Crystallographic evidence supported the notion that the cyano group had displaced a bound water molecule, leading to a 60-fold enhancement in Ki.38 Inspired by these results, Wissner et al. replaced the quinazoline in 7 with the 3-cyano-quinoline fragment in 8 intending to improve inhibition of the epidermal growth factor kinase domain (EGFR kinase). Surprisingly, this modification led to a 3-fold decrease in activity (Figure 1C).39 Given the similarities in the modifications for these three cases, elucidation of the different outcomes was pursued.
Figure 1.

Structure-based drug design by targeting water molecules in binding sites. (A) Scytalone dehydratase, (B) p38α MAP kinase, (C) EGFR kinase. The figures are estimated relative binding free energies (kcal/mol) based on Ki data for A and B and on IC50 data for inhibition of kinase activity for C.37-39 Question marks indicate a lack of experimental data. For EGFR kinase the inhibitory activity of the unsubstituted quinoline analogue 9 was not reported, but it can be estimated from the same modification on a pair of closely related analogs lacking the two methoxy groups.40
Methods
Protein Setup
The crystal structure of scytalone dehydratase in complex with 2 (PDB 3std),37 the crystal structure of a erlotinib (a quinazoline inhibitor related to 4-6) in complex with EGFR kinase (PDB 1m17),41 and the crystal structure of p38α MAP kinase in complex with a quinazoline inhibitor (PDB 1di9),42 were selected for this study. The protein preparation wizard of the program Maestro was used to add missing hydrogen atoms and to relax the protein structures to alleviate steric clashes.43 Protein-ligand Z-matrices were then prepared using the programs chop and pepz.44 Protein residues with any atom within ca. 15 Å of a ligand atom were retained. The degrees of freedom of side chains of the protein residues within 10 Å of a ligand atom were sampled during the Monte Carlo simulations with the exception of bond lengths, which were kept frozen along with backbone degrees of freedom. All degrees of freedom for the ligands were sampled. The net charge of the systems was set to zero by neutralizing protein residues distant from the ligands. The OPLS-AA force field was used for the proteins.45
Ligand Setup
In addition to the N to C-CN replacements in Figure 1, the CH analogs 3, 6, 9 were also considered. Though experimental binding data is only available for 3, simulations of the CH derivatives allowed comparison of the computed free energy changes between the N and C-CN derivatives from multiple pathways. Models of 1-3 complexed to scytalone dehydratase were created from the crystal structure of 2 in complex with scytalone dehydratase.37 Models of 7-9 were similarly constructed from the crystal structure of EGFR kinase in complex with a 4-anilinoquinazoline inhibitor.41 For p38α MAP kinase, Liu et al. published activity data for compound 6 as well as results of crystallographic studies (Figure 2 in reference 38), but the coordinate data was not released. A structure of p38α MAP kinase in complex with a related inhibitor was selected,42 and a binding mode for 5 was generated by energy minimization of the best poses generated by the docking program Glide.46 The top-ranked pose from Glide agreed well with the binding pose depicted in Figure 2 of Liu et al.38 Binding poses for 4 and 6 were then generated from that for 5. To determine starting conformations of the unbound ligands for the free energy calculations, the BOSS program was used to provide the lowest-energy conformer from a conformational search including GB/SA continuum hydration.44,47,48 For the MC simulations, the ligands were treated as fully flexible and their energetics were represented with the OPLS/CM1A force field.49 The CM1A atomic charges are scaled by 1.14 for neutral molecules.50
Solvent Setup
The TIP4P water model was used.51 Each protein-ligand complex was solvated by ca. 800 water molecules in a ca. 22-Å radius water cap. A half-harmonic potential with a force constant of 1.5 kcal/mol/Å2 was applied to water molecules at distances greater than 22 Å from the center of the system to prevent evaporation. For each protein-ligand complex, the location of bound water molecules in the vicinity of the binding site was determined using the water-placement algorithm JAWS.17 The methodology has been presented in detail previously,17 and only a brief summary is provided here. A 3-D cubic grid with 1-Å spacing is positioned to cover the binding site. The spatial domain of the grid, i.e., where water molecules can be present, is constructed from overlapping spheres of 4-5 Å radius, centered on ligand atoms or dummy atoms positioned in the binding site. A series of MC simulations is then performed. Putative hydration sites are detected by letting water molecules sample the grid positions, while they simultaneously have their intermolecular interactions scaled between “on” or “off”. The absolute binding affinity of a water molecule at a given hydration site is then estimated from the ratio of probabilities that the water molecule is “on” or “off”. The methodology accounts for entropic and enthalpic contributions to water binding affinities, requires only modest computational expense, and yields results comparable to standard free energy calculations.17 The water content of the grids was determined using 5 million (5M) MC configurations with sampling of just the water molecules, followed by 15M configurations that sampled the water, protein and ligand degrees of freedom to determine possible hydration sites, and finally 60M configurations to estimate the occupancy of the sites. The procedure was applied to scytalone dehydratase bound to ligand 3, p38α MAP kinase bound to ligand 4, and EGFR kinase bound to ligands 7-9. For the other ligands bound to scytalone dehydratase or p38α MAP kinase, possible substituent effects on the water distribution were examined using a smaller grid focused on the part of the ligand that is modified during the free-energy simulations. Because the focused grids are smaller, shorter simulations were conducted with 5M water-only MC configurations, 5M configurations to determine possible hydration sites, and 20M configurations to estimate the occupancy of these sites.
Free Energy Calculations
Relative binding free energies for the ligands were computed from a thermodynamic cycle that requires evaluation of a free energy change in solution and in complex with the protein.44,52 Free-energy changes for 1-3, 4-6, and 7-9 were computed with the MCPRO 2.1 program44 using Metropolis Monte Carlo simulations to sample configurations of the reference state,53 a single-topology technique to perturb the reference state,54 and 11 windows of simple overlap sampling to estimate the free-energy change between the reference and perturbed states.55,56 The initial solvent coordinates were provided by the JAWS calculations.17 Then, for the protein-ligand complexes, each FEP window involved 10M MC configurations of water-only sampling, followed by 10M configurations of full equilibration, and 30M configurations of averaging at 25 °C. For the ligands in water, a 22-Ǻ water cap was used containing ca. 1500 water molecules, and each FEP window consisted of 10M configurations of equilibration and 30M configurations of averaging. In all cases, evaluation of the potential energy employed 10-Å residue-based cutoffs.
For the ligands, the perturbations converted the cyano group C-CN to C-H and then to N, as in 2 → 3 → 1. To complete the thermodynamic cycles, the absolute binding free energies of water molecules displaced from the binding sites were computed using the double-decoupling formalism.21 The water molecule of interest was treated as a solute in MCPRO, and the perturbations were carried by first turning off the atomic partial charges and then removing the Lennard-Jones interactions yielding ΔGcoul and ΔGLJ. Each FEP calculation used 11 windows of simple overlap sampling.55,56 Each window consisted of 1M configurations of water-only MC sampling, followed by 4M configurations of full equilibration and 15M configurations of averaging. The reliable decoupling of a water molecule requires the use of a constraint to localize the decoupled water molecule.21,57 This was achieved using a spherical hard-wall potential with a of radius 2.8 Å, whose center was taken as the location of the hydration site for the water molecule, determined by the JAWS calculations. The chosen water molecule was forbidden to escape the hard-wall sphere. Furthermore, other water molecules were not permitted to diffuse into this excluded region, though solute and protein atoms could occupy the space. The absolute binding free energy of the water molecule was then determined using eq 1.
| (1) |
The first term is the free energy change for removing the intermolecular interactions of one water molecule in liquid water. It is the negative of the excess free energy of hydration of TIP4P water; a value of +6.4 kcal/mol was adopted as a consensus from the literature.26,58 The second and third terms are computed from the simulations, as described above. The fourth term accounts for the free energy cost to constrain an ideal particle occupying a volume V0 (1 molecule per 30 Å3, corresponding to a bulk concentration of water 55.55 mol/L) into a space of volume V defined by the hard-wall. With a hard-wall radius of 2.8 Å, the accessible volume is 91.9 Å3 and at room temperature and the correction term is 0.67 kcal/mol.
With MCPRO 2.1,44 free energy changes are computed using both simple overlap sampling (SOS) and double-wide sampling (DW).56 Averages are computed such that, over the course of a SOS run, forward and backward free energy changes are also obtained with a spacing that is effectively twice as large as the spacing between reference and perturbed states in the SOS calculations. This allows computation of the double-ended (DE) result with the larger spacing, Though this is expected to have significantly poorer precision than the SOS result,56 the hysteresis between the forward and backward free energy changes is useful in identifying problematic perturbations. Finally, over the course of this study, a number of different protocols were tested to take into consideration the effects of a water displacement in FEP studies; resultant recommendations are summarized in the Supporting Information.
Results and Discussion
Scytalone Dehydratase
The water distributions in the binding site for ligands 1, 2 and 3 were predicted with JAWS before conducting the free energy simulations. Figure 2 illustrates the water molecules in the vicinity of the ligands. For 2, the predicted hydration sites can be compared to the location of water molecules observed in the crystal structure.37 It is apparent that the four hydration sites observed within 5 Å of the ligand in the crystal structure have been found by the water equilibration algorithm (Figure 2C). When 1 is bound, one additional water molecule is predicted to occupy the space between the hydroxyl groups of Tyr30 and Tyr50 (Figure 2A). The predicted water locations are fully consistent with one water molecule being displaced by the cyano group of 2. For ligand 3, the JAWS analysis predicts two hydration sites of weak affinity, ca. -1 kcal/mol, in the vicinity of the two tyrosines. The first site is where the water displaced by 2 binds; the second site is sandwiched between the cinnoline ring and the nearest benzyl ring of 3 (Figure 2B). To assert with more confidence whether the water molecule bound with 1 had been displaced upon change to 3, absolute binding free energies were computed for the water molecules positioned at both sites. The results indicate that the free energy of the protein-ligand complex increases by 0.4 ± 0.2 kcal/mol for addition of the first water molecule and 0.3 ± 0.2 kcal/mol for addition of the second water molecule. Therefore, the optimum number of water molecules near the Tyr30 and Tyr50 hydroxyl groups is zero when 3 is bound, though partial occupancy cannot be ruled out. Ligand 3 is thus also predicted to displace the critical water molecule that is present when 1 is bound. This prediction is not obvious as there is space to accommodate a water molecule in this region; however, loss of hydrogen bonding with the ligand weakens significantly the binding affinity of a water molecule at this site such that it becomes energetically favorable to transfer it to the bulk solvent.
Figure 2.

Location of selected water molecules in the binding site of scytalone dehydratase in complex with, (A) ligand 1, (B) ligand 3, (C) ligand 2. The orange spheres indicate the position of water molecules observed in the crystal structure of 2 complexed to scytalone dehydratase.37 The displaced water molecule is represented in sticks. For clarity, non polar hydrogens and parts of the binding site have been omitted. Figures created with VMD.59
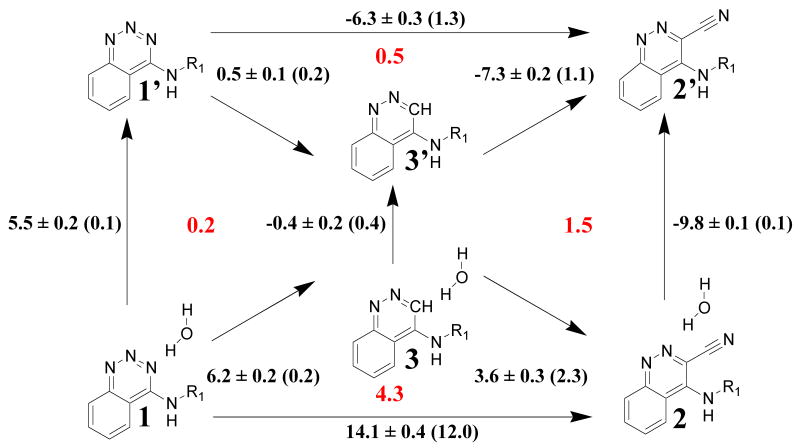
Figure 3 shows the relative binding affinities computed for ligands 1-3 with and without the displaceable water molecule. The bottom triangle represents the results with a water molecule initially present at the hydration site between the two hydroxyl groups of Tyr30 and Tyr50. This water molecule is tightly bound in the complex with 1, and it should be absent when 2 is bound. However, in this case as the cyano group is grown in, the water molecule is unable to escape from the binding site and it is trapped in an energetically unfavorable state. The result is a large overestimation of the relative binding free energy between 1 and 2 (+9.8 or +14.1 kcal/mol instead of -2.0 kcal/mol). Additionally, the closure of the thermodynamic cycle 1→2→3→1 is poor (+4.3 kcal/mol) indicating that the computed free energy changes are imprecise. The hystereses in the DE results also state that the results for 1 → 2 and 3 →2 are unreliable.
Figure 3.
Relative binding free energies (kcal/mol) computed for ligands 1-3 bound to scytalone dehydratase in the presence or absence (prime superscript) of a bound water molecule. The binding free energies are quoted with one standard deviation and are obtained by SOS. Hystereses from the DE runs are in parentheses. Deviations from closure of thermodynamic cycles are shown in red.
For the perturbation 1→2, the hysteresis was plotted as a function of λ (Figure 4) and MC configurations were visualized. At the beginning of the perturbation, the position of the water molecule is almost invariant owing to its hydrogen bonding with the ligand and the hydroxyl groups of Tyr30 and Tyr50. Around λ = 0.5 the emerging cyano group begins to seriously interfere with the water molecule's hydrogen-bonding interactions. Towards the end of the perturbation the water molecule is pushed into a non-polar cavity, where it cannot form any hydrogen bonds with the protein. It is apparent in Figure 4 that the displacement of the water molecule near the end of the perturbation accounts for a large part of the total hysteresis of the perturbation. A smaller peak in the hysteresis occurs near the middle of the perturbation, when the water molecule loses its ability to form the three hydrogen bonds with the hydroxyl groups of Tyr30, Tyr50, and the benzotriazine ring. The inaccurate free energy estimates are associated with the simulation of the protein-2 complex in a state of higher free energy than the protein-ligand complex would reach upon release of the bound water molecule into the bulk. The forward and backward free-energy results become inconsistent for a window in such cases; typically, growth is more unfavorable than shrinking is favorable in an overly crowded environment.
Figure 4.

Average separation between the nitrogen/carbon atom of the ligand initially hydrogen-bonded to the oxygen atom of the bound water molecule during the perturbation of 1 → 2 (solid line). The dashed line shows the hysteresis from the DE calculations at each window. The error bars represent one standard deviation.
The top triangle in Figure 3 shows the relative binding affinities computed in the absence of the critical water molecule. The results differ markedly; conversion of 1′ into 2′ is predicted to improve the binding free energy by 6.3 kcal/mol, in large excess of the experimental estimate of 2.0 kcal/mol.37 3′ is predicted to be only 0.5 kcal/mol less potent than 1′, which contrasts with the 3.8 kcal/mol experimental difference (Figure 1). On the other hand, the strong preference for the cyano analogue 2′ over 3′ is well reproduced. The hystereses are lower than in the previous set of simulations and the closure of the thermodynamic cycle yields only a 0.5-kcal/mol discrepancy, which is well within statistical error. Hence, the results are better behaved when no water molecule is displaced during the perturbation. However, there are still severe discrepancies with experiment for the relative affinities of 1′ vs. 2′ and 3′. Clearly, the binding of 1′ in the absence of the critical water molecule is underestimated.
The results from eq 1 for removing the critical water molecule from the complexes are shown with the vertical arrows in Figure 3. The free energy increases by 5.5 kcal/mol for the perturbation 1→1′, indicating full water occupancy for this site. In contrast, when 3 is bound the free energy decreases by only 0.4 kcal/mol, and partial occupancy is predicted for this hydration site. For ligand 2, the free energy decreases by 9.8 kcal/mol, so the water molecule would not be present. The computed binding affinities of the water molecules can then be combined with the other FEP results in Figure 3. For conversion of 1 → 2′, the three pathways that avoid the non-physical state 2 yield differences of -0.8 ± 0.4, -1.3 ± 0.4, and -1.5 ± 0.4, which compare well with the estimate of -1.8 kcal/mol from the experimental data.37 Furthermore, the relative binding free energies between 1 and 3′ are 6.0 ± 0.2 and 5.8 ± 0.2 kcal/mol from the alternative two-step pathways. While the far greater inhibitory potency of 1 is reproduced, these figures overshoot the experimental estimate of 3.8 kcal/mol. Though the molecular change in going from 1 to 3 is small, the complex for 3 is ca. 6 kcal/mol less stable than for 1. The reduced affinity can be attributed largely to the lost hydrogen bonding with the water molecule; however, there also appear to be unfavorable steric contacts and electrostatic interactions due to the weak hydrogen-bond donor character of the aromatic hydrogen with the water molecule that is unable to reorient because of hydrogen bonding interactions Tyr30 and Tyr50.
In summary, once the binding free energy of the critical water molecule is taken into account, the computed results for ligand binding are in much better agreement with experiment. However, potential pitfalls with standard FEP calculations that do not include such a detailed analysis of changes in hydration are apparent.
p38α MAP Kinase
Figure 5 illustrates the location of key water molecules in the vicinity of the ligands identified from a JAWS analysis. As part of the binding site is solvent exposed, grids were positioned only in the pockets between the ligand and the protein where water molecules could provide bridging interactions. A water molecule is predicted to bridge between the backbone NH of Met109 and a triazine nitrogen with 4 (Figure 5a). However, it is displaced with 5 by the direct hydrogen bond between the Met109 NH and the cyano group (Figure 5c), in agreement with the crystallographic results of Liu et al.38 In addition, the JAWS analysis identified other likely hydration sites. In particular, the presence of the water molecule just above the displaceable one in Figure 5a is notable. It often participates in a hydrogen bond with the backbone C=O of His107 and sometimes with the proximal amino group of the ligand (Figure 5b) or with the displaceable water molecule (Figure 5a). When 6 is bound, there is also indication of partial occupancy for the displaceable water molecule near Met109, though it is shifted to be farther from the pyrimidine ring of the ligand. The JAWS analysis yielded a binding affinity of only 0-1 kcal/mol for this water molecule. This prediction is supported by double-decoupling results, which indicate an absolute binding free energy of 0.0 ± 0.1 kcal/mol.
Figure 5.

Location of key water molecules in the binding site of p38α MAP kinase in complex with, (A) 4, (B) 6, (C) 5. The displaced water molecule is represented in sticks. For clarity, non-polar hydrogens and parts of the binding site have been omitted.
Figure 6 summarizes the computed free-energy results for 4-6 bound to p38α MAP kinase. When the simulations are conducted with the displaceable water molecule initially present (lower triangle in Figure 6), 5 is predicted to be less well bound than 4 by 1.6 or 1.2 kcal/mol in disagreement with the experimental results in Figure 1.38 The SOS hystereses are smaller for this system, e.g., 0.5 kcal/mol for 4 → 5 → 6 → 4, and the DE hystereses are also significantly reduced with a maximum of 2.6 kcal/mol. The critical water molecule is in a less crowded environment than in the previous example, and it is displaced more smoothly during the perturbations. However, if the water molecule is removed from the binding site before computing the free energy changes (top triangle in Figure 6), the relative binding affinity of 5′ vs. 4′ becomes -6.8 kcal/mol, which is a significant overestimate of the experimental preference, -2.5 kcal/mol. The DE hystereses for all three perturbations are very small and the thermodynamic cycle closes well.
Figure 6.
Relative free energies of binding (kcal/mol) computed for ligands 4-6 bound to p38α MAP kinase in the presence or absence (prime superscript) of a bound water molecule. Other details are as in Figure 3.
The free energy changes for removal of the water molecule (vertical arrows in Figure 6) are 4.2, 0.0, and -5.1 kcal/mol for the complexes with 4, 6, and 5. This indicates full occupancy of this hydration site when 4 is bound, partial occupancy with 6, and full displacement of the water molecule by 5. Closure of the thermodynamic cycles, e.g., 4 → 6 → 6′ → 4′ → 4, is again satisfactory given that they involve 4 steps. Summing the free energy changes across different pathways yields relative binding free energies between 4 and 5′ of -2.6 ± 0.3, -2.1 ± 0.3, -2.9 ± 0.3, -3.5 ± 0.5 and -3.9 ± 0.3 kcal/mol. These figures are in acceptable accord with the experimental estimate of -2.5 kcal/mol (Figure1).38 Thus, for this case reasonable results can be obtained by removing the displaceable water molecule before or after the ligand perturbations. However, lack of consideration of the hydration details, as in just modeling 4 → 5 or 4′ → 5′, would lead to significant error.
EGFR Kinase
Figure 7 shows the location of water molecules in the binding site for ligands 7-9, as identified with JAWS. As for p38α MAP kinase, part of the binding site is solvent exposed, so the analysis was focused on the vicinity of the nitrile group for 8. A water molecule is predicted to bridge between one of the quinazoline nitrogen atoms of 7 and the hydroxyl group of Thr766 (Figure 7a), in agreement with the design in the experimental study.39 Unlike the other two systems, the water molecule is predicted to remain strongly stabilized in the binding site according to the JAWS analysis even after loss of the hydrogen bond with the heterocycle in progressing from 7 to 9 (Figure 7b). Nevertheless, it is displaced upon addition of the cyano group in 8 (Figure 7c). Interestingly, three other water molecules are predicted to occupy the cavity near the displaced water molecule. Display of individual MC configurations reveals that water molecules in the cavity can be connected to the bulk solvent through a narrow passage delimited by Asn747 and Cys751. The displaced water molecule receives a hydrogen bond from one of the additional water molecules in Figures 7a and 7b. This local water network was not obvious as only one water molecule is reported in the PDB structure used to initiate the calculations.41 However, these additional water sites are observed in other crystal structures of EGFR kinase. For instance in the crystal structure of the EGFR kinase domain in complex with the inhibitor lapatinib (PDB 1xkk), four hydration sites are located in this cavity, in good agreement with the predictions in Figure 7a.60
Figure 7.

Location of selected water molecules in the binding site of EGFR kinase in complex with, (A) 7, (B) 9, and (C) 8. In A the orange sphere indicates the position of a water molecule observed in the crystal structure of an analogue of 7 in complex with EGFR kinase.41 The green spheres indicate the positions of water molecules observed in the crystal structure of another analogue of 7 in complex with EGFR kinase.60 The displaced water molecule is represented in sticks. For clarity, non polar hydrogens and parts of the binding site have been omitted.
Figure 8 records the relative binding free energies computed for 7-9. If the displaceable water molecule is positioned initially in the binding site (lower triangle), it does not escape to bulk when the nitrile group is introduced. Thus, similar to the case in Figure 3, the binding affinity of 7 relative to 8 is far too favorable. In addition, large DE hystereses and poor closure of the thermodynamic cycle provide warnings of poor convergence. Though 9 was not reported by Wissner et al.,39 a relative affinity can be estimated from the IC50 values measured for a very similar pair of ligands, which differ from 7 and 9 only by the absence of the two methoxy groups.40 Given that these methoxy groups are solvent exposed and distant from the nitrogen involved in interactions with the displaceable water molecule, there is a reasonable basis for comparison.
Figure 8.
Relative free energies of binding (kcal/mol) computed for ligands 7-9 bound to EGFR kinase in the presence or absence (prime superscript) of a bound water molecule. Other details are as in Figure 3.
The FEP calculations find that the binding affinity of ligand 9 is less favorable than of 7 by 3.9 kcal/mol, in good agreement with the experimental estimate of 3.2 kcal/mol.40 However, in the presence of the displaceable water molecule, the computed affinity of 9 is greater than for 8 by 2.2 kcal/mol, while the experimental data indicate the opposite preference, -2.6 kcal/mol. On the other hand, in the absence of this water molecule in the binding site, the preferences for 8′ over 9′ and 7′ are far too large, -6.7 and -5.8 kcal/mol, as compared to the experimental estimates of -2.6 and +0.6 kcal/mol. The results are precise and well behaved, as judged by the small DE hystereses and the low closure value of the thermodynamic cycle. Clarification again comes from the computed free energy changes for removal of the displaceable water molecule, 6.9, 4.3 to -3.1 kcal/mol for the complexes of 7, 9, and 8. Thus, the water molecule is displaced by the cyano group of 8, but its presence is strongly favored for both 7 and 9. In going from 7 to 9, the water molecule loses the hydrogen bond with the quinazoline nitrogen atom, but it can still form a π-type hydrogen bond with the 3-bromophenyl substituent (Figure 7b). This interaction likely contributes to the greater binding affinity for this water molecule in the complexes with 9 than for the corresponding complexes with 3 and 6.
The hydration analysis concludes that the correct forms for the complexes are represented by 7, 8′, and 9. Summing the free energy changes for the left-side pathway in Figure 8 then yields a greater binging affinity for 7 than 9 of 4.0 ± 0.3 or 3.9 ± 0.1 kcal/mol, in good agreement with the experimental estimate of 3.2 kcal/mol (Figure 1). For 9 and 8′, summing in the upper circuit, which has the smaller hystereses and avoids the non-physical 8, the computed affinity difference is -2.4 ± 0.2 or -2.9 ± 0.3 kcal/mol, in good agreement with the experimental estimate of -2.6 kcal/mol. For the pair 7 and 8′, the computed values favor the binding of 7 by 1.1 ± 0.3, 1.6 ± 0.3, 1.5 ± 0.3 for the three pathways that avoid 8. These values are also in reasonable accord with the observed greater inhibitory strength of 7, 0.6 kcal/mol.
Insight into why introduction of the cyano group in this case fails to provide enhanced binding emerges from comparison of the leftmost results in Figures 3, 6, and 8. The observed effects of introducing the cyano group for 1, 4, and 7 (-2.0, -2.5, and +0.6 kcal/mol in Figure 1) correlate with the costs of displacing the bound water molecule (5.5, 4.2, and 6.9 kcal/mol). By visualization, e.g., in Figures 2, 5, and 7, the order is not obvious as the displaced water molecule appears to participate in three hydrogen bonds in each case. Thus, displacement of water molecules in binding sites is not a viable, general strategy for lead optimization. The free energy increase due to the removal of a bound water molecule is not always offset by the additional interactions of the water-displacing moiety. Accurate computation of the free energy for replacement of specific water molecules is useful in helping to select the most promising ligand modifications.17-28 However, complete analyses as in Figures 3, 6, and 8 are necessary to gauge fully the effects of a ligand modification that includes potential displacement or addition of bound water molecules. This requires a combination of calculations to determine the locations of water molecules for the protein-ligand complexes17,20 and to evaluate the free energy changes for the ligand modifications and for any associated displacement or addition of water molecules.1-3 The present approach using JAWS, MC/FEP, and double-decoupling calculations provides an option to serve this purpose.
Conclusion
The three cases studied here provide clear examples of the importance and diversity of the contributions of specific water molecules to ligand binding. Though visualization of the binding sites or counts of hydrogen bonding interactions suggest that the displaced water molecules are well stabilized, detailed analyses with free energy calculations were necessary to estimate accurately the binding affinity of the water molecules and the ultimate impact of ligand modifications. Figures 3, 6, and 8 reflect both the complexity of achieving accurate computational estimates of relative binding affinities and a viable solution to the problem. In the absence of the key water molecule, conversion of an azine nitrogen to a C-CN unit in 1, 4, and 7 is computed to enhance binding by 6.3, 6.8, and 5.8 kcal/mol, while in the presence of the water molecule the modification is unfavorable by 14.1, 1.6, and 8.2 kcal/mol. In reality, the evidence from the JAWS and double-decoupling calculations is that the water molecule should be present in the complexes for the azine analogues, but not for the cyano-containing relatives. When this is taken into account in the free energy cycles, the computed changes in free energy of binding agree well with the observed activity trends including the diminished binding in the case of EGFR kinase. One could view the computational difficulties as reflecting sampling problems, i.e., the statistical mechanics calculations are not covering properly the configuration space. Grand canonical MC simulations warrant further exploration in this regard,24,25,61 since they can sample the insertion and deletion of water molecules. Simultaneous performance of well-converged FEP calculations is technically challenging.25 The present methodology provides a viable alternative that yields thorough characterization of the hydration of protein-ligand complexes and of the free energy changes associated with coupled modifications of a ligand and associated water molecules.
Supplementary Material
Acknowledgments
Gratitude is expressed to the National Institutes of Health (GM32136) and the National Foundation for Cancer Research for support of this research. J. M. also acknowledges support from a Marie Curie International Outgoing fellowship from the European Commission (FP7-PEOPLE-2008-4-1-IOF, 234796-PPIdesign).
Footnotes
Supporting Information Available: Guidelines for the computation of relative binding free energies between pairs of ligands that involve the displacement of a water molecule, and complete citations for references 38 and 39. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Gohlke H, Klebe G. Angew Chem, Int Ed. 2002;41:2645–2676. doi: 10.1002/1521-3773(20020802)41:15<2644::AID-ANIE2644>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 2.Jorgensen WL. Science. 2004;303:1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 3.Jorgensen WL. Acc Chem Res. 2009;42:724–733. doi: 10.1021/ar800236t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Reichmann D, Phillip Y, Carmi A, Schreiber G. Biochemistry. 2008;47:1051–1060. doi: 10.1021/bi7019639. [DOI] [PubMed] [Google Scholar]; (b) Southall NT, Dill KA, Haymet ADJ. J Phys Chem B. 2002;106:521–533. [Google Scholar]
- 5.Guimaraes CRW, Cardozo M. J Chem Inf Model. 2008;48:958–970. doi: 10.1021/ci800004w. [DOI] [PubMed] [Google Scholar]
- 6.Su Y, Gallicchio E, Das K, Arnold E, Levy RM. J Chem Theory Comput. 2007;3:256–277. doi: 10.1021/ct600258e. [DOI] [PubMed] [Google Scholar]
- 7.Liu HY, Kuntz ID, Zou XQ. J Phys Chem B. 2004;108:5453–5462. [Google Scholar]
- 8.Kollman PA, Massova I, Reyes C, Kuhn B, Huo SH, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE. Acc Chem Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 9.Michel J, Taylor RD, Essex JW. J Chem Theory Comput. 2006;2:732–739. doi: 10.1021/ct600069r. [DOI] [PubMed] [Google Scholar]
- 10.Michel J, Verdonk ML, Essex JW. J Med Chem. 2006;49:7427–7439. doi: 10.1021/jm061021s. [DOI] [PubMed] [Google Scholar]
- 11.Michel J, Essex JW. J Med Chem. 2008;51:6654–6664. doi: 10.1021/jm800524s. [DOI] [PubMed] [Google Scholar]
- 12.Lu YP, Wang RX, Yang CY, Wang SM. J Chem Inf Model. 2007;47:668–675. doi: 10.1021/ci6003527. [DOI] [PubMed] [Google Scholar]
- 13.Lam PYS, Jadhav PK, Eyermann CJ, Hodge CN, Ru Y, Bacheler LT, Meek JL, Otto MJ, Rayner MM, Wong YN, Chang CH, Weber PC, Jackson DA, Sharpe TR, Erickson-Viitanen S. Science. 1994;263:380–384. doi: 10.1126/science.8278812. [DOI] [PubMed] [Google Scholar]
- 14.Ladbury JE. Chem Biol. 1996;3:973–980. doi: 10.1016/s1074-5521(96)90164-7. [DOI] [PubMed] [Google Scholar]
- 15.Samsonov S, Teyra J, Pisabarro MT. Proteins: Struct, Funct, Bioinf. 2008;73:515–525. doi: 10.1002/prot.22076. [DOI] [PubMed] [Google Scholar]
- 16.Monecke P, Borosch T, Brickmann R, Kast SM. Biophys J. 2006;90:841–850. doi: 10.1529/biophysj.105.065524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michel J, Tirado-Rives J, Jorgensen WL. J Phys Chem B. 2009;113:0000–0000. doi: 10.1021/jp9047456. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazaridis T. J Phys Chem B. 1998;102:3531–3541. [Google Scholar]
- 19.(a) Li Z, Lazaridis T. J Am Chem Soc. 2003;125:6636–6637. doi: 10.1021/ja0299203. [DOI] [PubMed] [Google Scholar]; (b) Li Z, Lazaridis T. J Phys Chem B. 2005;109:662–670. doi: 10.1021/jp0477912. [DOI] [PubMed] [Google Scholar]; (c) Li Z, Lazaridis T. J Phys Chem B. 2006;110:1464–1475. doi: 10.1021/jp056020a. [DOI] [PubMed] [Google Scholar]
- 20.(a) Young T, Abel R, Kim B, Berne BJ, Friesner RA. Proc Natl Acad Sci U S A. 2007;104:808–813. doi: 10.1073/pnas.0610202104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Abel R, Young T, Farid R, Berne BJ, Friesner RA. J Am Chem Soc. 2008;130:2817–2831. doi: 10.1021/ja0771033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilson MK, Given JA, Bush BL, McCammon JA. Biophys J. 1997;72:1047–1069. doi: 10.1016/S0006-3495(97)78756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Lazaridis T. Phys Chem Chem Phys. 2007;9:573–581. doi: 10.1039/b612449f. [DOI] [PubMed] [Google Scholar]
- 23.Helms V, Wade RC. J Am Chem Soc. 1998;120:2710–2713. [Google Scholar]
- 24.Deng YQ, Roux B. J Chem Phys. 2008;128:115103. doi: 10.1063/1.2842080. [DOI] [PubMed] [Google Scholar]
- 25.Speidel JA, Banfelder JR, Mezei M. J Chem Theory Comput. 2006;2:1429–1434. doi: 10.1021/ct0600363. [DOI] [PubMed] [Google Scholar]
- 26.Barillari C, Taylor J, Viner R, Essex JW. J Am Chem Soc. 2007;129:2577–2587. doi: 10.1021/ja066980q. [DOI] [PubMed] [Google Scholar]
- 27.Hamelberg D, McCammon JA. J Am Chem Soc. 2004;126:7683–7689. doi: 10.1021/ja0377908. [DOI] [PubMed] [Google Scholar]
- 28.Yu H, Rick SW. J Am Chem Soc. 2009;131:6608–6613. doi: 10.1021/ja809696s. [DOI] [PubMed] [Google Scholar]
- 29.Jorgensen WL, Ruiz-Caro J, Tirado-Rives J, Basavapathruni A, Anderson KS, Hamilton AD. Bioorg Med Chem Lett. 2006;16:663–667. doi: 10.1016/j.bmcl.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 30.Kim JT, Hamilton AD, Bailey CM, Domaoal RA, Wang LG, Anderson KS, Jorgensen WL. J Am Chem Soc. 2006;128:15372–15373. doi: 10.1021/ja066472g. [DOI] [PubMed] [Google Scholar]
- 31.Ruiz-Caro J, Basavapathruni A, Kim JT, Bailey CM, Wang LG, Anderson KS, Hamilton AD, Jorgensen WL. Bioorg Med Chem Lett. 2006;16:668–671. doi: 10.1016/j.bmcl.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 32.Thakur VV, Kim JT, Hamilton AD, Bailey CM, Domaoal RA, Wang LG, Anderson KS, Jorgensen WL. Bioorg Med Chem Lett. 2006;16:5664–5667. doi: 10.1016/j.bmcl.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 33.Barreiro G, Guimaraes CRW, Tubert-Brohman I, Lyons TM, Tirado-Rives J, Jorgensen WL. J Chem Inf Model. 2007;47:2416–2428. doi: 10.1021/ci700271z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barreiro G, Kim JT, Guimaraes CRW, Bailey CM, Domaoal RA, Wang L, Anderson KS, Jorgensen WL. J Med Chem. 2007;50:5324–5329. doi: 10.1021/jm070683u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeevaart JG, Wang LG, Thakur VV, Leung CS, Tirado-Rives J, Bailey CM, Domaoal RA, Anderson KS, Jorgensen WL. J Am Chem Soc. 2008;130:9492–9499. doi: 10.1021/ja8019214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michel J, Harker EA, Tirado-Rives J, Jorgensen WL, Schepartz A. J Am Chem Soc. 2009;131:6356–6357. doi: 10.1021/ja901478e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen JM, Xu SL, Wawrzak Z, Basarab GS, Jordan DB. Biochemistry. 1998;37:17735–17744. doi: 10.1021/bi981848r. [DOI] [PubMed] [Google Scholar]
- 38.Liu, et al. J Med Chem. 2005;48:6261–6270. doi: 10.1021/jm0503594. [DOI] [PubMed] [Google Scholar]
- 39.Wissner, et al. J Med Chem. 2000;43:3244–3256. doi: 10.1021/jm000206a. [DOI] [PubMed] [Google Scholar]
- 40.Rewcastle GW, Denny WA, Bridges AJ, Zhou HR, Cody DR, McMichael A, Fry DW. J Med Chem. 1995;38:3482–3487. doi: 10.1021/jm00018a008. [DOI] [PubMed] [Google Scholar]
- 41.Stamos J, Sliwkowski MX, Eigenbrot C. J Biol Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 42.Shewchuk L, Hassell A, Wisely B, Rocque W, Holmes W, Veal J, Kuyper LF. J Med Chem. 2000;43:133–138. doi: 10.1021/jm990401t. [DOI] [PubMed] [Google Scholar]
- 43.Schrödinger LLC, New York, 2008.
- 44.Jorgensen WL, Tirado-Rives J. J Comput Chem. 2005;26:1689–1700. doi: 10.1002/jcc.20297. [DOI] [PubMed] [Google Scholar]
- 45.Jorgensen WL, Maxwell DS, Tirado-Rives J. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 46.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 47.Still WC, Tempczyk A, Hawley RC, Hendrickson T. J Am Chem Soc. 1990;112:6127–6129. [Google Scholar]
- 48.Jorgensen WL, Ulmschneider JP, Tirado-Rives J. J Phys Chem B. 2004;108:16264–16270. [Google Scholar]
- 49.Jorgensen WL, Tirado-Rives J. Proc Natl Acad Sci U S A. 2005;102:6665–6670. doi: 10.1073/pnas.0408037102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Udier-Blagovic M, De Tirado PM, Pearlman SA, Jorgensen WL. J Comput Chem. 2004;25:1322–1332. doi: 10.1002/jcc.20059. [DOI] [PubMed] [Google Scholar]
- 51.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 52.Kollman P. Chem Rev. 1993;93:2395–2417. [Google Scholar]
- 53.Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E. J Chem Phys. 1953;21:1087–1092. [Google Scholar]
- 54.Jorgensen WL, Ravimohan C. J Chem Phys. 1985;83:3050–3054. [Google Scholar]
- 55.Lu ND, Kofke DA, Woolf TB. J Comput Chem. 2004;25:28–39. doi: 10.1002/jcc.10369. [DOI] [PubMed] [Google Scholar]
- 56.Jorgensen WL, Thomas LL. J Chem Theory Comput. 2008;4:869–876. doi: 10.1021/ct800011m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Michel J, Verdonk ML, Essex JW. J Chem Theory Comput. 2007;3:1645–1655. doi: 10.1021/ct700081t. [DOI] [PubMed] [Google Scholar]
- 58.Jorgensen WL, Blake JF, Buckner JK. Chem Phys. 1989;129:193–200. [Google Scholar]
- 59.Humphrey W, Dalke A, Schulten K. J Mol Graph. 1996;14:33–45. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 60.Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, Ellis B, Pennisi C, Horne E, Lackey K, Alligood KJ, Rusnak DW, Gilmer TM, Shewchuk L. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 61.Pan CF, Mezei M, Mujtaba S, Muller M, Zeng L, Li JM, Wang ZY, Zhou MM. J Med Chem. 2007;50:2285–2288. doi: 10.1021/jm070014g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.