Abstract
The enzymes DdaG and DdaF, encoded in the Pantoea agglomerans dapdiamide antibiotic biosynthetic gene cluster, when expressed in E. coli,1 make the tandem amide bonds of the dapdiamide scaffold at the expense of ATP cleavage. DdaG uses fumarate, 2,3-diaminopropionate (DAP) and ATP to make fumaroyl-AMP transiently on the way to the Nβ-fumaroyl-DAP regioisomer. Then DdaF acts as a second ATP-dependent amide ligase, but this enzyme cleaves ATP to ADP and Pi during amide bond formation. However, DdaF will not accept Nβ-fumaroyl-DAP; the enzyme requires the fumaroyl moiety to be first converted to the fumaramoyl half amide in Nβ-fumaramoyl-DAP. DdaF adds Val, Ile, or Leu to the carboxylate of fumaramoyl-DAP to make the three dapdiamides A-C. Thus, to build the dapdiamide antibiotic scaffold, amidation must occur on the fumaroyl-DAP scaffold, after DdaG action but before DdaF catalysis. This is an unusual instance of two ligases acting sequentially in untemplated amide bond formations using attack of substrate carboxylates at Pα (AMP-forming) and then at Pγ (ADP-forming) of ATP cosubstrates.
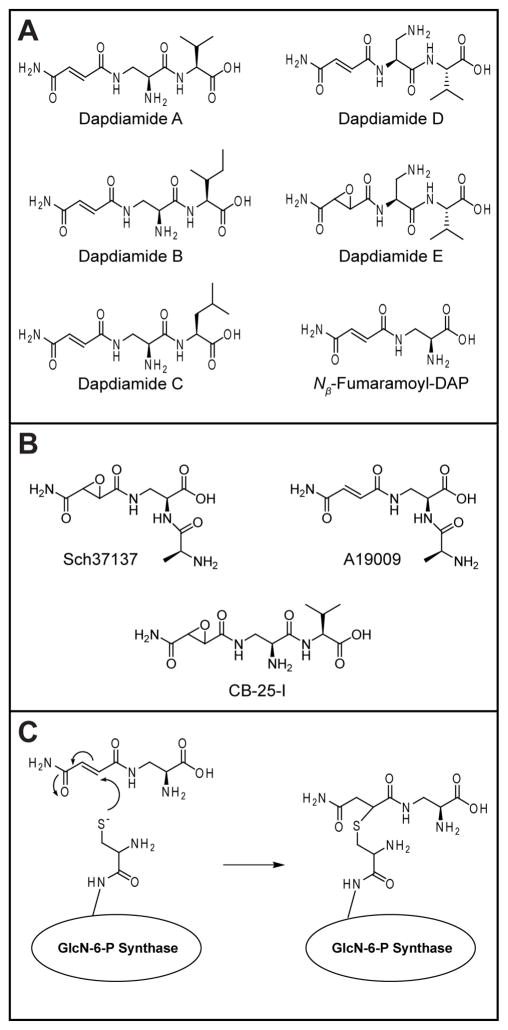
P. agglomerans strains produce a variety of antibiotics, as evidenced by the discoveries of the pantocins (1, 2), andrimid (3), and most recently dapdiamides A–E (4) (Figure 1A). This family of fumaramoyl- and epoxysuccinamoyl-dipeptides is named for the presence of DAP and the two amide linkages of the antibiotic scaffold. The compounds are similar in structure to synthetic methoxyfumaroyl-DAP-dipeptides which show broad spectrum activity by targeting bacterial and fungal glucosamine-6-phosphate (GlcN-6-P) synthase, the enzyme which catalyzes the rate-limiting step in the formation of the essential cell-wall building block UDP-N-acetyl-glucosamine (5, 6). An analogous mode of action has been proposed for three previously isolated antibiotics with similar structures to the dapdiamides, CB-25-I from Serratia plymuthica (7) and Sch37137 (8) and A19009 (9, 10) from actinomycete isolates (Figure 1B) (5). These compounds are likely cleaved intracellularly to generate acyl-DAP warheads which can inhibit GlcN-6-P synthase, as demonstrated for Nβ-fumaramoyl-DAP (11) (Figure 1A). The fumaramoyl-DAP warheads derived from A19009 and dapdiamides A-D would provide a conjugated eneamide system that is likely attacked by the thiolate side chain of the active site Cys in the glutaminase domain of GlcN-6-P synthase in analogy to the mechanism of inactivation observed for Nβ-methoxyfumaroyl-DAP (5) (Figure 1C). Alkylation of this residue blocks generation of the ammonia required to convert fructose-6-P into glucosamine-6-P. The epoxysuccinamoyl moiety in CB-25-I, Sch37137 and dapdiamide E could likewise function as an inactivating electrophile for the catalytic Cys in the target enzyme.
Figure 1.
A) The dapdiamide family of antibiotics and Nβ-fumaramoyl-DAP. B) Dapdiamide-related natural products. C) Proposed mechanism of GlcN-6-P synthase inhibition by Nβ-fumaramoyl-DAP.
Peptide antibiotics are typically formed by two biological strategies. The first involves ribosomal synthesis of protein precursors that undergo posttranslational proteolysis and/or modification and is exemplified by the Microcin B17, C7 and E492 molecules as well as the lantibiotics such as nisin, and the thiazolyl peptides such as thiostrepton and thiocillins (12). The second strategy uses nonribosomal peptide synthetase (NRPS) assembly lines to specify the order and composition of amino acid monomers incorporated and accounts for the biogenesis of penicillin, vancomycin, and daptomycin scaffolds among others (12). However, there is another way that peptides can be assembled, independent of ribosomal-based mRNA or NRPS machinery, via ATP–dependent coupling of amino acid monomers and elongating peptides by soluble enzymes. This route has precedents in glutathione biosynthesis (13) and bacterial cell wall pentapeptide elongation (14) where ATP is cleaved to ADP and intermediate acyl phosphates are formed as reactive species. A variant of this route occurs in the ATP-dependent elongation and cyclization of the siderophore ferrioxamine E (15) by cleavage of ATP to AMP and PPi and involves acyl-AMP intermediates. We have found that the acyl dipeptides of the dapdiamide antibiotic family are constructed by this third strategy, via tandem action of two soluble ligases, one (DdaG) that cleaves ATP to AMP and another (DdaF) that cleaves ATP to ADP.
While no biosynthetic gene information is available for CB-25-I, Sch37137 or A19009, a single gene cluster from P. agglomerans that makes all five dapdiamides has recently been cloned into E. coli (4) (Figure S1). This metabolic capacity from one gene cluster suggests both pathway enzyme promiscuity and the prospect of increased scaffold diversity from combinatorial biosynthesis once the catalysts have been identified. Inspection of the fumaramoyl/epoxysuccinamoyl-dipeptide scaffold of the dapdiamides and the encoding biosynthetic gene cluster has led to predictions about the possible functions of the encoded ORFs (4). Of particular importance here are DdaG and DdaF, which are predicted to be ATP-dependent amide ligases and thus are candidates for making the two peptide bonds. Intriguingly, DdaG has the signature elements of an adenylating ligase that cleaves ATP to AMP and PPi, while DdaF is predicted to be an ATP grasp family member (16) and instead cleave ATP to ADP and Pi in a phosphoryl transfer mechanism.
In this study we report heterologous expression, purification, and characterization of DdaG and DdaF and their amide ligase activities for making Nβ-fumaroyl-DAP and Nβ-fumaramoyl-DAP-Val/Ile/Leu, respectively.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, Materials, and Instrumentation
Oligonucleotide primers were synthesized by Integrated DNA Technologies (Coralville, IA). PCR was performed with Phusion High-Fidelity PCR Mastermix (New England Biolabs). Cloning was performed using the Gateway System (Invitrogen). One Shot Chemically Competent TOP10 E. coli (Invitrogen) and NovaBlue(DE3) (Novagen) were used for routine cloning and propagation of DNA vectors. Recombinant plasmid DNA was purified with a Qiaprep kit (Qiagen). DNA sequencing was performed at the Molecular Biology Core Facilities of the Dana Farber Cancer Institute (Boston, MA). Nickel-nitrilotriacetic acid-agarose (Ni-NTA) superflow resin and SDS-PAGE gels were purchased from Qiagen. Protein samples were concentrated using 30 kDa MWCO Amicon Ultra filters (Millipore). Protein concentrations were determined by Bradford assay with BSA as a standard. Chemicals were purchased from Sigma-Aldrich. NMR solvents were purchased from Cambridge Isotopes.
A pyruvate kinase/lactate dehydrogenase (PK/LDH) enzyme mix from rabbit muscle was purchased from Sigma as a buffered aqueous glycerol solution. Myokinase from chicken muscle was purchased from Sigma as a lyophilized powder and resuspended in 10 mM HEPES, pH 8. Synthetic dapdiamide A and the plasmid containing the dapdiamide gene cluster, pUC19 A10A, were provided by Jessica Dawlaty. Fumaramic acid was prepared from monomethyl fumarate as described previously (17) or from monoethyl fumarate via an analogous procedure.
1H-NMR spectra were recorded on Varian 400 or 600 MHz spectrometers. MS analysis was performed on an Agilent Technologies 6520 Accurate-Mass Q-TOF LC/MS, an Agilent Technologies 6210 Accurate-Mass TOF LC/MS, or by staff at the Harvard University Mass Spectrometry Laboratory (Cambridge, MA). HPLC data was collected on a Beckman Coulter System Gold (126 solvent module, 168 detector). An Alltech Alltima C18 (250 × 4.6 mm) column was used for routine analytical HPLC. A Chiralcel OD-RH (150 × 4.6 mm) chiral column was used for separation of FMOC-derivatized N-fumaroyl-DAP regioisomers. A Phenomenex Luna C18 (250 × 21.2 mm) column was used for preparative HPLC.
Cloning, Overexpression, and Purification of DdaG and DdaF
ddaG and ddaF were PCR amplified from pUC19 A10A. For ddaG, the forward primer was 5′-CACCAATAAGGGAGAAAACATGAATCTGGAAATG-3′ and the reverse primer was 5′-TTAATAGCGGCGTTTACTGACGTTGCCG-3′. For ddaF, the forward primer was 5′-CACCTCGATTTTGAACAATAAAGAAGTCATCGTAATC-3′ and the reverse primer was 5′-TTATTCATCAATTAGGCCCAAGTGATAAAAATCCG-3′. Unpurified PCR products were mixed with the pENTR/D-TOPO vector and the TOPO construct was then transformed into TOP10 E. coli. Purified vector was analyzed by PCR to confirm the presence of insert. The insert was recombined into pDEST17, and the product of this reaction was transformed into TOP10 E. coli. Purified vector was sequenced to confirm the insert. BL21 (DE3) E. coli were transformed with pDEST17-ddaF or pDEST17-ddaG. 25 mL cultures were grown overnight at 37 °C in Luria-Bertani (LB) medium supplemented with 50 μg/mL carbenicillin. 2 L LB with 50 μg/mL carbenicillin was inoculated with 10 mL of this overnight culture. The cultures were incubated at 37°C for 3.5–5 hours, then 15 °C for 0.5–1 hour. Cultures were induced with 0.5 mM IPTG. The cultures were incubated at 15 °C for 20 hours, then the cells were harvested by centrifugation at 4200 rpm for 12 minutes. Cell pellets were resuspended in 10 mL lysis buffer (10 mM Tris·HCl, pH 8, 200 mM NaCl or 10 mM HEPES, pH 8, 200 mM NaCl) and lysed at 5,000–10,000 psi in an Avestin EmulsiFlex-C5 high-pressure homogenizer. The cell debris was removed by ultracentrifugation at 35,000 rpm for 35 minutes. Supernatants were applied to a low pressure chromatography column containing Ni-NTA resin pre-equilibrated in lysis buffer. The resin was washed with lysis buffer containing 20 mM imidazole. The protein was eluted with lysis buffer containing 80 mM and 200 mM imidazole. Protein was concentrated and imidazole removed using 30 kDa MWCO tubes. In some cases additional purification was achieved by gel filtration chromatography on an Amersham Pharmacia Biotech AKTA FPLC using a Superdex 75 26/60 Hiload column. Aliquots of the proteins were flash-frozen in liquid nitrogen and stored at −80 °C.
DdaG and DdaF HPLC activity assays
50 μL reactions were incubated at room temperature and contained 9.5 μM enzyme, 2.5 or 5 mM amine substrate, 2.5 or 5 mM carboxylate substrate, 6 mM ATP, 10 mM MgCl2, and 100 mM HEPES, pH 8. Protein was removed by centrifugation in 5 kDa MWCO filter tubes at 12,000 × g for 15 minutes, 100 μL water was added, then filter tubes were spun again at 12,000 × g for 15 minutes. 80 μL of filtered reaction mixture was combined with 80 μL CH3CN, 50 μL water, 40 μL 10 mM FMOC-Cl (freshly prepared solution in CH3CN) and incubated at room temperature for at least 5 minutes. 80 μL 100 mM aminoadamantane (in 1:1 CH3CN-H2O) was added and reactions incubated at room temperature for at least 5 minutes before HPLC analysis using a gradient of 20–100% solvent B (0.1% TFA in CH3CN) in solvent A (0.1% TFA in water) over 25 minutes. For resolution of N-fumaroyl-DAP regioisomers, FMOC-derivatized samples were analyzed by chiral HPLC using a gradient of 20–70% solvent B (0.1% TFA in CH3CN) in solvent A (0.1% TFA in water) over 35 minutes. The absorbance was monitored at 263 nm.
Coupled Spectrophotometric Assays for AMP or ADP production
For all experiments, 200 μL reactions were incubated at room temperature and contained 10 mM ATP, 12 mM MgCl2, 200 μM NADH, 500 μM phosphoenolpyruvate, and 100 mM HEPES, pH 8. For characterization of DdaG, reactions additionally contained 43 units/mL LDH, 30 units/mL PK, 21 units/mL myokinase, 10 mM DAP, and 1.9 nM DdaG for fumarate assays or 9.5 nM DdaG for succinate assays. For comparison of Nα- and Nβ-fumar(am)oyl-DAP substrates for DdaF, reactions additionally contained 30 units/mL LDH, 15 units/mL PK, 2 mM Val, and 240 nM DdaF. For determination of the DdaF reaction velocity with varying concentrations of Nβ-fumaramoyl-DAP, reactions additionally contained 60 units/mL LDH, 42 units/mL PK, 240 nM DdaF, and 5 mM Val. For characterization of DdaF with respect to different amino acids, reactions additionally contained 60 units/mL LDH, 42 units/mL PK, 125 μM Nβ-fumaramoyl-DAP, and 240 nM DdaF. The consumption of NADH was monitored continuously in a Molecular Devices SPECTRAmax PLUS 96-well plate reader by measuring the absorbance at 340 nm every 20 seconds. Kinetic constants were derived from velocity vs. substrate concentration data using a non-linear, least squares fitting method with GraphPad Prism software.
ATP/[32P]-PPi Exchange Assays
1.2 mL reactions contained 1.9 μM DdaG, 5 mM substrate, 1 mM ATP, 1 mM MgCl2, 40 mM KCl, 1 mM DTT, 5 mM Na[32P]-PPi (3.3 × 104 cpm/mL), and 50 mM Tris·HCl, pH 8. Reactions were incubated at 37 °C for 30 minutes and then 350 μL aliquots were removed and quenched with 750 μL charcoal suspension (100 mM NaPPi, 350 mM HClO4, 16 g/L charcoal). The mixtures were vortexed, then centrifuged at 13,000 rpm for 3 minutes. The pellets were washed twice with 750 μL wash solution (100 mM NaPPi, 350 mM HClO4). Charcoal-bound radioactivity was measured on a Beckman LS 6500 scintillation counter.
RESULTS
Expression and Purification of DdaG and DdaF in E. coli
ddaG and ddaF were PCR amplified from pUC19 A10A, a plasmid containing the dapdiamide gene cluster, and cloned into an expression vector encoding an N-terminal His6 tag. The proteins were overexpressed in E. coli BL21 (DE3) and purified by Ni-NTA affinity chromatography (Figure S2). Yields ranged from 12–16 mg/L for DdaG and 6–11 mg/L for DdaF.
DdaG is a Regioselective ATP-dependent Fumaroyl-DAP Ligase
A small amount of Nβ-fumaramoyl-DAP was isolated with the dapdiamides from fermentation (4), suggesting that acylation of DAP precedes the formation of the DAP-amino acid amide bond. To determine which enzyme is responsible for this acylating activity, purified DdaG and DdaF were incubated with DAP and potential carboxylate substrates, and reaction mixtures were derivatized with FMOC and analyzed by HPLC.
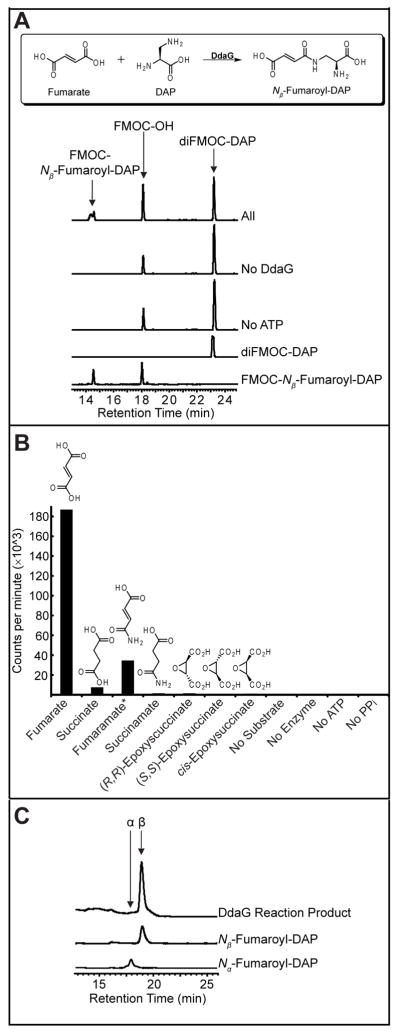
DdaG ligates fumarate and DAP to form N-fumaroyl-DAP in an ATP-dependent manner (Figure 2A and Table S1). The enzyme also forms N-succinoyl-DAP from succinate and DAP (Figure S3 and Table S1). In contrast, the ligase showed no activity in these assays with the corresponding half-amides, fumaramate and succinamate, or with the two enantiomers of trans-epoxysuccinate (Figure S4). A coupled spectrophotometric assay for AMP formation was used to kinetically characterize DdaG, and these experiments demonstrated that the enzyme is specific for fumarate over succinate, with a kcat/Km of 21 mM−1s−1 for fumarate and 0.19 mM−1s−1 for succinate (Table S2).
Figure 2.
Characterization of DdaG activity. A) HPLC analysis showing ATP-dependent ligation of fumarate and DAP by DdaG. B) ATP/[32P]-PPi exchange data. Graph shows counts per minute arising from [32P]-ATP that is adsorbed to charcoal after incubation of DdaG, [32P]-PPi, unlabeled ATP, and carboxylate substrate. *Activity with fumaramate results from 2–5% contamination with fumarate.2 C) Coelution of the FMOC-derivatized DdaG product with FMOC-Nβ-fumaroyl-DAP.
ATP/[32P]-PPi exchange assays, which test for the reversible formation of adenylated intermediates, demonstrated that DdaG adenylates its carboxylate substrate as predicted based on its homology to AMP-forming enzymes (Figure 2B). As expected, activation of both fumarate and succinate was observed, whereas there was no activation of either the epoxysuccinates or succinamate. Exchange was seen with fumaramate as a result of contamination of this substrate with fumarate.2
In dapdiamides A–C, fumaramate and DAP are linked through the DAP β-amino group, while in dapdiamides D and E, it is the α-amino group that is acylated. This suggested that DdaG could act in a regiopromiscuous manner, accepting either the DAP α or β amine as a nucleophile. To test this hypothesis, both fumaroyl-DAP isomers were synthesized (Scheme S1 and S2) and their FMOC derivatives separated by chiral HPLC. The FMOC-derivatized product of DdaG catalysis coeluted exclusively with the β isomer, demonstrating that DdaG acts in a regioselective manner to form Nβ-fumaroyl-DAP exclusively (Figure 2C).
DdaF is an ATP-dependent Nβ-fumaramoyl-DAP-amino acid ligase
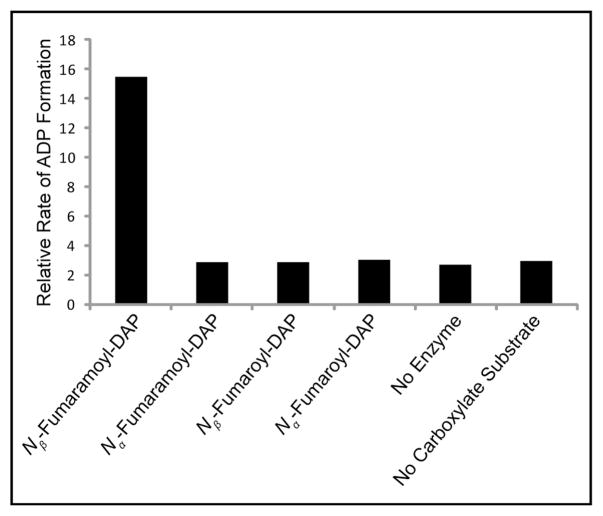
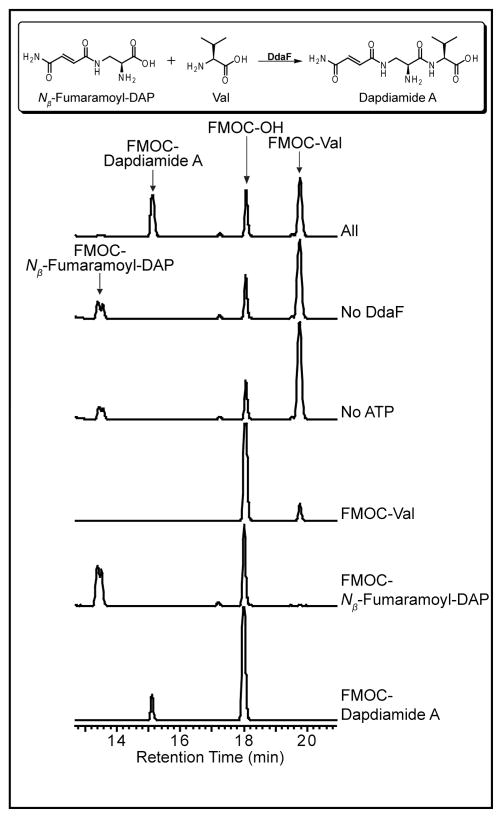
DdaF was characterized using both an ADP production assay and the same HPLC assay used to study DdaG. These experiments showed that DdaF catalyzes the second subunit condensation reaction in this pathway. The enzyme is not active with either the product of DdaG catalysis, Nβ-fumaroyl-DAP, or the corresponding α isomer (Figure 3 and S5). DdaF accepts only the corresponding amide Nβ-fumaramoyl-DAP, which it can ligate to Val, Ile, and Leu to form dapdiamides A-C (Figure 3, 4, S6, and Table S3). DdaF is specific for the Nβ-isomer of fumaroyl-DAP; it will not ligate Nα-fumaramoyl-DAP to Val (Figure 3 and S7).
Figure 3.
Relative rates of ADP formation on incubation of DdaF with potential carboxylate substrates and 2 mM Val.
Figure 4.
HPLC analysis showing ATP-dependent ligation of Nβ-fumaramoyl-DAP and Val by DdaF to form dapdiamide A.
Apparent substrate inhibition was observed at Nβ-fumaramoyl-DAP concentrations above 500 μM (Figure S8), so kinetics with respect to the three amino acid substrates were determined at a fixed Nβ-fumaramoyl-DAP concentration of 125 μM. While the apparent kcat values for each amino acid were similar, the apparent Km values were 260 μM for Val, 830 μM for Ile, and 7.82 mM for Leu (Table S4).
Stability of the Nβ-acyl-DAP Compounds
The lack of DdaF activity with Nα-fumaramoyl-DAP makes the origin of the α-linked dapdiamides D and E mysterious. We suspected that these compounds could arise from nonenzymatic isomerization of the corresponding β-linked regioisomers, so we tested the stability of dapdiamide A in 500 mM NH4OH at room temperature overnight. However, we did not observe any isomerization (Figure S9). We also tested for possible isomerization of the pathway intermediates. There was no conversion of Nβ-fumaroyl-DAP to the α-regioisomer when it was incubated at pH 8 for three days (Figure S10). Preliminary results indicate that isomerization of the corresponding amide, Nβ-fumaramoyl-DAP, also does not happen readily at pH 9.5. On the other hand, some frozen samples of Nα-fumaramoyl-DAP (but not the Nβ isomer) decomposed but the process has not been examined thoroughly.
DISCUSSION
We observe that DdaG uses fumarate, DAP, and ATP to generate Nβ-fumaroyl-DAP as ATP is cleaved to AMP and PPi. DAP can in principle be used as a nucleophile either at the β-NH2 (on the way to dapdiamides A-C) or at the α-NH2 (on the way to dapdiamides D-E). In these studies only the β coupling regiochemistry was seen with purified DdaG. In turn DdaF cleaves ATP to ADP and Pi as Nβ-fumaramoyl-DAP is coupled to Val, Ile, or Leu. Most notably, while DdaG will not accept fumaramate in place of fumarate as coupling substrate, DdaF requires fumaramoyl-DAP rather than fumaroyl-DAP.
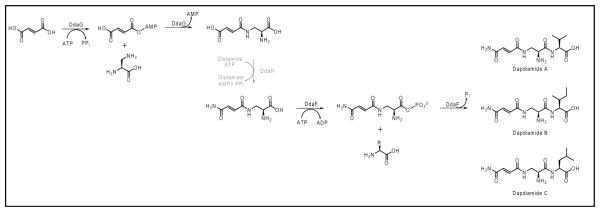
The characterization of DdaF and DdaG allows us to propose the timing of other catalytic events in this biosynthetic pathway. Bioinformatic analysis suggested that DdaH is an amidotransferase (4), and our results imply that this enzyme acts after DdaG, converting Nβ-fumaroyl-DAP to Nβ-fumaramoyl-DAP. DdaF can then ligate this compound to three branched chain amino acids (Scheme 1). DdaC is a putative Fe(II)/α-ketoglutarate-dependent dioxygenase and is likely responsible for the formation of the dapdiamide E epoxide. Because DdaG shows no activity with epoxysuccinate, it is probable that DdaC acts on a double bond of either fumar(am)oyl-DAP or fumaramoyl-DAP-Val. This epoxidation may occur while the substrate is tethered to the adenylation-thiolation enzyme DdaD, and the oxidized substrate could be released by the thioesterase homolog DdaE. The origin of the Nα-acyl-DAP linkage in dapdiamides D and E remains an open question as we have been unable to observe any Nα-acyl-DAP formation either from enzymatic or nonenzymatic conversion. These compounds could potentially be formed via the action of an enzyme that catalyzes an α/β isomerization or via the action of a DAP α-NH2-specific ligase, however these activities do not appear to be encoded in the dapdiamide gene cluster.
Scheme 1.
Biosynthetic route to dapdiamides A-C. The grayed-out step has not been biochemically validated.
The dapdiamides are the first example of a family of small molecules targeted against GlcN-6-P synthase that are produced by a single biosynthetic gene cluster. The production of a suite of compounds instead of a single antibiotic may allow the producing organism to target a broader range of microbes and/or blunt development of target resistance. It has been shown that the identity of the amino acid ligated to DAP in the methoxyfumaroyl-DAP-dipeptides influences the rate of target-cell uptake (6), and this is likely also true for the dapdiamides. The promiscuity of DdaF with respect to its amino acid substrate allows the formation of compounds with three different C-terminal amino acids, and each may interact differently with the nonspecific peptide permeases that import these types of compounds. Additionally, the two different electrophilic moieties (Michael acceptor or epoxide) may have different levels of reactivity with the target enzyme. It remains to be explored whether production of a family of structurally distinct compounds with the same target confers an advantage on the producer, but this may be one explanation for the evolution of substrate promiscuity in the dapdiamide pathway.
Here we focused on the two amide-bond forming enzymes that are required to generate the fumaramoyl-DAP-dipeptide scaffold of the dapdiamides. These catalysts are new members of an expanding class of ligases that assemble antibiotic peptides in a non-ribosomal and non-NRPS-dependent manner. Such enzymes typically use one of two modes carboxylate substrate activation: adenylation or phosphorylation. Like DdaG, the amide ligases CouL, NovL, and CloL use the first strategy to activate carboxylates for nucleophilic attack by donor amino groups to bring together monomers in the assembly of the aminocoumarin antibiotics coumermycin, novobiocin, and clorobiocin, respectively (18–20). In contrast, ATP grasp family members in the bacilysin (21, 22) and tabtoxin (23) pathways likely activate carboxylate substrates via acyl-phosphate formation, like DdaF. The dapdiamide pathway is an atypical example of the recruitment of two amide ligases with distinct ATP cleavage patterns to form tandem peptide bonds.
Recent studies of P. agglomerans antibiotics have led to the identification of several novel condensation catalysts (24) which are homologs of enzymes from primary metabolism, suggesting that nature’s repertoire of condensation catalysts for natural product biosynthesis may be broader than once suspected. A detailed understanding of the function of these catalysts may open the door to the study of new classes of compounds that are not produced by canonical biosynthetic enzymes and that may have structural features which set them apart from previously identified natural products.
Supplementary Material
Acknowledgments
We thank Jessica Dawlaty for providing a sample of synthetic dapdiamide A and the pUC19 A10A plasmid containing the dapdiamide gene cluster. We thank Elizabeth Sattely, Christopher Neumann, Emily Balskus, and Michael Fischbach for helpful discussions.
Footnotes
This work was supported in part by the National Institutes of Health GM 20011 (CTW), GM 086258 (JC), and Medical Scientist Training Program GM 07753 (MAH).
Abbreviations: P. agglomerans, Pantoea agglomerans; E. coli, Escherichia coli; ATP, adenosine-5′-triphosphate; DAP, 2,3-diaminopropionate; AMP, adenosine-5′-monophosphate; ADP, adenosine-5′-diphosphate; Pi, phosphate; GlcN-6-P, glucosamine-6-phosphate; UDP, uridine-5′-diphosphate; NRPS, nonribosomal peptide synthetase; PPi,, pyrophosphate; ORF, open reading frame; PCR, polymerase chain reaction; Ni-NTA, nickel-nitrilotriacetic acid-agarose; SDS-PAGE, sodium dodecyl sulfate – polyacrylamide gel electrophoresis; MWCO, molecular weight cut-off; BSA, bovine serum albumin; 1H-NMR, proton-nuclear magnetic resonance; PK, pyruvate kinase; LDH, lactate dehydrogenase; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; LC, liquid chromatography; MS, mass spectrometry; Q-TOF, quadrupole time-of-flight; HPLC, high-performance liquid chromatography; LB, Luria-Bertani medium; IPTG, isopropyl-β-D-galactopyranoside; FPLC, fast protein liquid chromatography; FMOC, 9-fluorenylmethoxycarbonyl; TFA, trifluoracetic acid; CH3CN, acetonitrile; DTT, dithiothreitol
Fumaramate used in this assay was prepared from commercial monoethyl fumarate that is contaminated with a small amount of fumarate. Kinetic analysis of the contaminated fumaramate is consistent with the hypothesis that DdaG activity with this substrate results from contamination with 2–5% fumarate.
SUPPORTING INFORMATION AVAILABLE
Figures S1–10, Tables S1–4, Schemes S1–4, and supplemental methods. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Brady SF, Wright SA, Lee JC, Suton AE, Zumoff CH, Wodzinski RS, Beer SV, Clardy J. Pantocin B, an antibiotic from Erwinia herbicola discovered by heterologous expression of cloned genes. J Am Chem Soc. 1999:11912–11913. [Google Scholar]
- 2.Jin M, Liu L, Wright SAI, Beer SV, Clardy J. Structural and functional analysis of Pantocin A: an antibiotic from Pantoea agglomerans discovered by heterologous expression of cloned genes. Angew Chem, Int Ed. 2003;42:2898–2901. doi: 10.1002/anie.200351053. [DOI] [PubMed] [Google Scholar]
- 3.Jin M, Fischbach MA, Clardy J. A biosynthetic gene cluster for the acetyl-CoA carboxylase inhibitor andrimid. J Am Chem Soc. 2006;128:10660–10661. doi: 10.1021/ja063194c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Dawlaty JA. PhD Thesis. Cornell University; Ithaca, NY: 2008. Bioactive natural products from Pantoea agglomerans, Janithinobacterium lividum, and Conocarpus erectus. [Google Scholar]; b Dawlaty J, Fischbach MA, Clardy J. Personal communications [Google Scholar]
- 5.Kucharczyk N, Denisot MA, Le Goffic F, Badet B. Glucosamine-6-phosphate synthase from Escherichia coli: determination of the mechanism of inactivation by N3-fumaroyl-L-2,3-diaminopropionic derivatives. Biochemistry (Mosc) 1990;29:3668–3676. doi: 10.1021/bi00467a012. [DOI] [PubMed] [Google Scholar]
- 6.Milewski S, Andruszkiewicz R, Kasprzak L, Mazerski J, Mignini F, Borowski E. Mechanism of action of anticandidal dipeptides containing inhibitors of glucosamine-6-phosphate synthase. Antimicrob Agents Chemother. 1991;35:36–43. doi: 10.1128/aac.35.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shoji J, Hinoo H, Sakazaki R, Kato T, Hattori T, Matsumoto K, Tawara K, Kikuchi J, Terui Y. Isolation of CB-25-I, an antifungal antibiotic, from Serratia plymuthica. J Antibiot. 1989;42:869–874. doi: 10.7164/antibiotics.42.869. [DOI] [PubMed] [Google Scholar]
- 8.Cooper R, Horan AC, Gentile F, Gullo V, Loebenberg D, Marquez J, Patel M, Puar MS, Truumees I. Sch 37137, a novel antifungal compound produced by a Micromonospora sp. Taxonomy, fermentation, isolation, structure and biological properties. J Antibiot. 1988;41:13–19. doi: 10.7164/antibiotics.41.13. [DOI] [PubMed] [Google Scholar]
- 9.Molloy BB, Lively DH, Gale RM, Forman M, Boeck LD, Higgens CE, Kastner RE, Huckstep LL, Neuss N. A new dipeptide antibiotic from Streptomyces collinus, Lindenbein. J Antibiot. 1972;25:137–140. doi: 10.7164/antibiotics.25.137. [DOI] [PubMed] [Google Scholar]
- 10.van der Baan JL, Barnick JWFK, Bickelhaupt F. Antibiotic A 19009 Structural Investigation and Synthesis. J Antibiot. 1983;36:784–792. doi: 10.7164/antibiotics.36.784. [DOI] [PubMed] [Google Scholar]
- 11.Chmara H, Andruszkiewicz R, Borowski E. Inactivation of glucosamine-6-phosphate synthetase from Salmonella typhimurium LT2 by fumaroyl diaminopropanoic acid derivatives, a novel group of glutamine analogs. Biochim Biophys Acta. 1985;870:357–366. doi: 10.1016/0167-4838(86)90240-2. [DOI] [PubMed] [Google Scholar]
- 12.Nolan EM, Walsh CT. How Nature Morphs Peptide Scaffolds into Antibiotics. Chem Bio Chem. 2009;10:34–53. doi: 10.1002/cbic.200800438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mooz ED, Meister A. Tripeptide (glutathione) synthetase. Purification, properties, and mechanism of action. Biochemistry (Mosc) 1967;6:1722–1734. doi: 10.1021/bi00858a022. [DOI] [PubMed] [Google Scholar]
- 14.Walsh CT. Enzymes in the D-alanine branch of bacterial cell wall peptidoglycan assembly. J Biol Chem. 1989;264:2393–2396. [PubMed] [Google Scholar]
- 15.Kadi N, Oves-Costales D, Barona-Gomez F, Challis GL. A new family of ATP-dependent oligomerization-macrocyclization biocatalysts. Nat Chem Biol. 2007;3:652–656. doi: 10.1038/nchembio.2007.23. [DOI] [PubMed] [Google Scholar]
- 16.Galperin MI, Koonin EV. A diverse superfamily of enzymes with ATP-dependent carboxylate-amine/thiol ligase activity. Protein Sci. 1997;6:2639–2643. doi: 10.1002/pro.5560061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Talley EA, Fitzpatrick TJ, Porter WL. Formation of fumaramic acid from asparagine in phosphate buffer. J Am Chem Soc. 1959;81:174–175. [Google Scholar]
- 18.Schmutz E, Steffensky M, Schmidt J, Porzel A, Li SM, Heide L. An unusual amide synthetase (CouL) from the coumermycin A1 biosynthetic gene cluster from Streptomyces rishiriensis DSM 40489. Eur J Biochem. 2003;270:4413–4419. doi: 10.1046/j.1432-1033.2003.03830.x. [DOI] [PubMed] [Google Scholar]
- 19.Steffensky M, Li SM, Heide L. Cloning, overexpression, and purification of novobiocic acid synthetase from Streptomyces spheroides NCIMB 11891. J Biol Chem. 2000;275:21754–21760. doi: 10.1074/jbc.M003066200. [DOI] [PubMed] [Google Scholar]
- 20.Galm U, Dessoy MA, Schmidt J, Wessjohann LA, Heide L. In vitro and in vivo production of new aminocoumarins by a combined biochemical, genetic, and synthetic approach. Chem Biol. 2004;11:173–183. doi: 10.1016/j.chembiol.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 21.Steinborn G, Hajirezaei MR, Hofemeister J. bac genes for recombinant bacilysin and anticapsin production in Bacillus host strains. Arch Microbiol. 2005;183:71–79. doi: 10.1007/s00203-004-0743-8. [DOI] [PubMed] [Google Scholar]
- 22.Tabata K, Ikeda H, Hashimoto S-I. ywfE in Bacillus subtilis codes for a novel enzyme, L-amino acid ligase. J Bacteriol. 2005;187:5195–5202. doi: 10.1128/JB.187.15.5195-5202.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinscherf TG, Willis DK. The biosynthetic gene cluster for the beta-lactam antibiotic tabtoxin in Pseudomonas syringae. J Antibiot (Tokyo) 2005;58:817–821. doi: 10.1038/ja.2005.109. [DOI] [PubMed] [Google Scholar]
- 24.Fortin PD, Walsh CT, Magarvey NA. A transglutaminase homologue as a condensation catalyst in antibiotic assembly lines. Nature. 2007;448:824–827. doi: 10.1038/nature06068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.