Abstract
Many synthetic and natural peptides are known to self-assemble to form various nanostructures such as nanofibers, hollow tubes, or ring-like structures. Some of the synthetic peptide molecules are specifically designed to produce well-defined nanostructures by controlling intermolecular interactions. Many environmental conditions such as salt concentration, pH, temperature, and surface characteristics influence intermolecular interactions, hence the process of the self-assembly. Here we studied self-assembly of a genetically engineered protein polymer composed of silk-like and elastin-like repeats on a mica surface. Silk-elastinlike protein polymers (SELPs) consist of tandem repeats of Gly-Ala-Gly-Ala-Gly-Ser from Bombyx mori (silkworm) and Gly-Val-Gly-Val-Pro from mammalian elastin. At a very low polymer concentration of 1 μg/ml, SELPs self-assembled into nanofibrous structures on a mica surface. Examination using atomic force microscopy (AFM) and dynamic light scattering techniques showed that SELPs self-assembled into nanofibers in the presence of the mica surface but not in the bulk state. Ionic strength had a significant influence on nanofiber growth, indicating the importance of electrostatic interactions between the polymer and the mica surface. At low ionic strength, the kinetics of nanofiber growth indicates that the mica surface effectively removed a lag phase by providing nucleating sites, facilitating nanofiber self-assembly of SELPs. Further examination of self-assembly on various surfaces such as silicon, positively charged surface, and hydrophobic surface revealed that negatively charged hydrophilic surface provides optimal surface to facilitate self-assembly of SELPs.
Keywords: Silk-elastinlike protein polymer, Self-assembly, Fibrillization, Nanofiber, Amyloid fiber, Ionic strength, Growth kinetics, Atomic force microscopy, Dynamic light scattering
Introduction
Many synthetic peptides are known to self-assemble to form various nanostructures such as nanofibers, hollow tubes, and ring-like structures.1 These synthetic peptide molecules are specifically designed to produce well-defined nanostructures by controlling intermolecular interactions.2,3 It is also well known that naturally occurring proteins such as amyloid beta, alpha-synuclein, or prion protein form self-assembled nanofibrillar structures.4–7 Self-assembly of peptide molecules into fibrillar structures under certain physiological conditions seems to be a general property of peptides in nature.8 Several self-assembled nanofibrous structures which mimic natural extracellular matrix have been applied to biomedical applications such as tissue engineering with varying degrees of success.9,10
The appearance of nanofibrils in general occurs during the transition of amorphous structure or protofibril structure into beta-sheet aggregates.11 The major driving forces for self-assembled structures are intermolecular interactions such as hydrogen bonding, hydrophobic interactions and electrostatic interactions.12 Self-assembled nanofibrous structures are usually formed in the bulk state but in certain cases the fibrillization is observed on surfaces.13 Fiber growth on surfaces has physiological relevance since some amyloidogenic fibers grow on the walls of blood vessels and basement membranes.14–17 The presence of surfaces and interfaces has been reported to affect nanofiber growth kinetics.18,19 Moreover, the final self-assembled morphologies are sensitive to the physicochemical properties of the substrate surface such as its hydrophobicity and surface charge.17,20,21
Here we studied self-assembly of a genetically engineered protein polymer composed of silk-like and elastin-like repeats on a mica surface. Silk-elastinlike protein polymers (SELPs) consist of tandem repeats of Gly-Ala-Gly-Ala-Gly-Ser from Bombyx mori (silkworm) and Gly-Val-Gly-Val-Pro from mammalian elastin.22 Using DNA recombinant techniques the sequence and length of SELPs can be controlled to modulate elasticity, gelation, biodegradation, interaction with cellular receptors, and sensitivity to environmental stimuli such as pH, temperature and ionic strength.23,24 The precise control over linear polymer structure provided by recombinant techniques promises the design and development of biomimetic SELPs with specific biorecognition function, tunable mechanical properties and biodegradability. Previous work described the self-assembly into nanoscale “whiskers” of an engineered protein polymer composed of repeats from silk and the cell adhesion domain of human fibronectin.25 Soluble fractions of spider dragline silk and exon 30 of human tropoelastin were also observed to form amyloid-like nanofibers but the effects of the substrates on the nanofiber self-assembly of these peptides were not probed in detail yet.26,27 In this paper we reports the role of the substrates on the self-assembly of silk-elastin peptide polymers. The effects of concentrations of SELP, ionic strengths, and physical properties of the substrates on nanofiber morphology and growth kinetics were investigated.
Material and Methods
Protein polymer
The silk-elastinlike protein polymer studied in this work, SELP-415K, was synthesized by recombinant DNA techniques and characterized as described previously.28 The repeating monomer of SELP-415K is composed of four silk-like units (GAGAGS), fifteen elastin-like units (GVGVP) and one lysine (K) modified elastin-like unit (GKGVP, single amino acid abbreviations are used). The complete amino acid sequence of the polymer with molecular weight of 71.5 kDa, including head and tail sequences is as follows:
MDPVVLQRRDWENPGVTQLVRLAAHPPFASDPMGAGSGAGS
[(GVGVP)4GKGVP(GVGVP)11(GAGAGS)4]7(GVGVP)4GKGVP(GVGVP)11(GAGAGS)2
GAGAMDPGRYQDLRSHHHHHH.
The series of silk-like (GAGAGS) and elastin-like (GVGVP) repeats are shown in bold. The isoelectric point of SELP-415K calculated based upon the amino acid composition is 10.4. (Protein Calculator v3.3; http://www.scripps.edu/~cdputnam/protcalc.html)
Sample preparation
Frozen SELP-415K stock solution of 1 mg/ml was thawed at 4°C and diluted with 1mM phosphate buffer (pH 7.2) with 5 mM NaCl to prepare various polymer concentrations (0.2 μg/ml, 1μg/ml, 5 μg/ml, and 10 μg/ml). 1 μg/ml SELP solutions with different ionic strengths were prepared by diluting the SELP-415K stock solution using the same phosphate buffer with various NaCl concentrations (5 mM, 10 mM, 50 mM and 100 mM). To prepare the samples for AFM imaging, 30 μl of SELP solutions were dropped on four different substrates; freshly cleaved 8×8 mm2 mica surfaces (Ted Pella, Inc., Redding, C A), positively modified mica substrate, silicon substrate (University Wafer, South Boston, MA), and highly ordered pyrolytic graphite (HOPG) (SPI supplies Inc., West Chester, PA). To prepare positively charged surface mica substrate was immersed in toluene with 0.1% 3-aminopropyl triethoxysilane (APTES, Gelest, Inc. Morrisville, PA) for 30 min. After the modification, APTES coated mica substrates were rinsed with toluene and dried with nitrogen gas. After adding SELP solution on the substrates, the samples were incubated in a humidity chamber at room temperature for 12 hours. To study nanofiber growth kinetics, the samples were incubated for 5, 15, 30, 60, 180, 360, and 540 minutes. SELP deposited substrates were then gently rinsed with 40 μL of phosphate buffer three times. After rinsing, the samples were kept at 4°C in a humidity chamber before the measurements. For pH experiments, concentrated HCl solution was added to 1 mM phosphate buffer (5 mM NaCl) until pH reaches 4. The solution of pH 9 was prepared by adding concentrated NaOH solution to the same buffer. The pH’s of the final solutions were checked using pH paper strips.
AFM imaging
AFM imaging was performed in aqueous state at room temperature using a Molecular Force Probe-3D instrument (MFP-3D, Asylum Research Inc., Santa Barbara, CA). The morphology of SELP on the mica surface was imaged in tapping mode at 0.5–1 Hz scan rate. Bio-lever (Asylum Research Inc., Santa Babara, CA) with 6 pN/nm spring constant was used for imaging. A target amplitude of ~350 mV and set point of ~320 mV were used to minimize disturbance to the SELP morphologies in the aqueous state.
Dynamic light scattering
Dynamic light scattering (DLS) experiments were performed using a Zetasizer ZS 90 (Malvern NANO series, Worcestershire, UK) with a 633 nm wavelength He-Ne laser at a fixed detector angle of 90 degrees. 10 mg/ml SELP-415K stock solution was diluted in 1 mM phosphate buffer (pH 7.2) with 5 mM NaCl to obtain a final SELP concentration of 10 μg/ml. The sample was transferred to a disposable microcuvette (Malvern Instruments, Worcestershire, UK) and total count rate was measured at 5, 15, 30, 60, 180, 360, and 720 minutes. The values reported are the average from 3 measurements at each time point.
Results and Discussion
Self-assembly of SELPs to nanofibers on the mica substrates
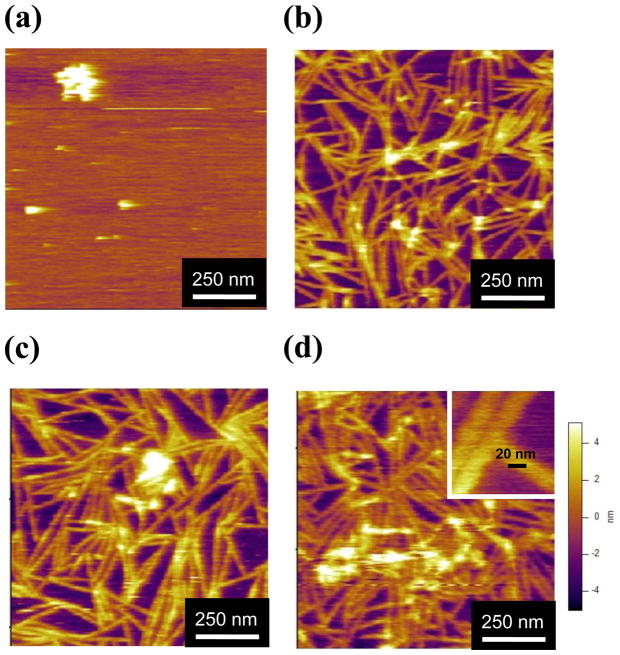
AFM imaging revealed that the SELP-415K polymer self-assembled into a nanofibrillar morphology in a concentration dependent manner (Figure 1). At 0.2 μg/ml (Figure 1a), sphere-like aggregates were observed with little or no nanofiber formation. Above 1 μg/ml polymer concentration (Figures 1b–d), nanofiber morphology was clearly evident. The height of the nanofibers was 3.90±0.6 nm (n=60), 3.64±1 nm (n=56), and 3.98±1 nm (n=45) at 1 μg/ml, 5 μg/ml, and 10μg/ml, respectively. The height was very similar to the height of the amyloid fibers measured using AFM.21,29 The widths of the nanofibers were 23.4±2.6 nm (n=64), 23.1±3.1 (n=56), and 24.5±3.4 nm (n=45) at 1 μg/ml, 5 μg/ml, and 10 μg/ml respectively. These results indicated that the height and width of nanofibers were not dependent on SELP concentrations once the nanofibers were formed whereas the morphologies of the SELP on mica surface drastically changed from sphere-like aggregates to nanofibers as the concentration increased. The height can be very accurately measured using AFM but the width can be different from its actual width due to the tip broadening effect.30 The width reported here is also very similar to the width of amyloid fiber measured using AFM29 but it is more than three times the width of amyloid beta fibers measured using electron microscopy.31,32 The high resolution image of SELP (Figure 1d, inset) did not show any modular or helical pattern which was observed in high resolution images of nanofibers formed from amyloid beta or alpha-synuclein.31,33 The growth direction of the SELP nanofiber was random, indicating that the crystallographic plane of the mica surface did not affect the direction of the fiber growth.21,34,35 The density of the nanofibers on the mica surface increased with increase in SELP concentration. This is probably due to the greater availability of polymer for participating in the self-assembling process at the higher concentrations. The length of the nanofiber defined as the distance between junction points in the network-like structure was 266.1±92.8 nm (n=181) for 1 μg/ml SELP-415K. Overall, the morphology of the SELP nanofibers on the mica surface appeared to be a network composed of many short fibers rather than long ones. This observation implies that the mica surface plays a significant role in nanofiber assembly, providing nucleating sites for fiber growth. In addition the junction points in the network-like morphology seemed to be formed by physical blocking of growing nanofibers upon contact with adjacent fibers. This implies that the growing nanofibers stop growing when they meet each other rather than continued to grow on top of each other.
Figure 1.
The morphology of SELP-415K at various polymer concentrations. The protein concentrations were (a) 0.2 μg/ml, (b) 1 μg/ml, (c) 5μg/ml, and (d) 10 μg/ml in 1 mM phosphate buffer (pH 7.2) with 5 mM NaCl. All samples were incubated on mica surface for 12 hours at room temperature. The images were taken in aqueous state in tapping mode AFM. The inset in (d) shows higher magnification (the scale bar is 20 nm).
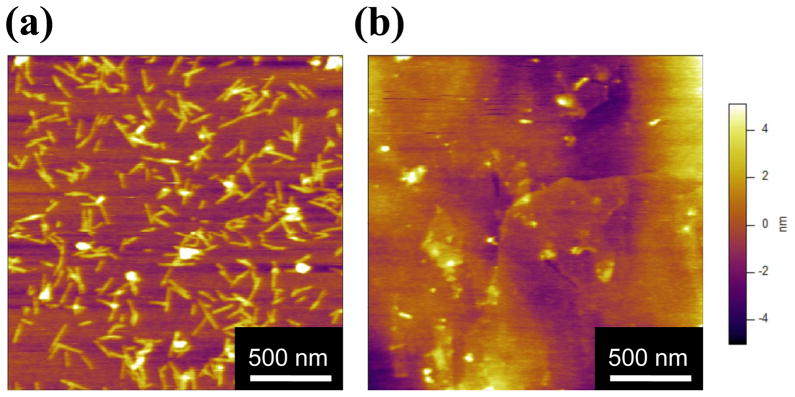
Surface facilitated self-assembly
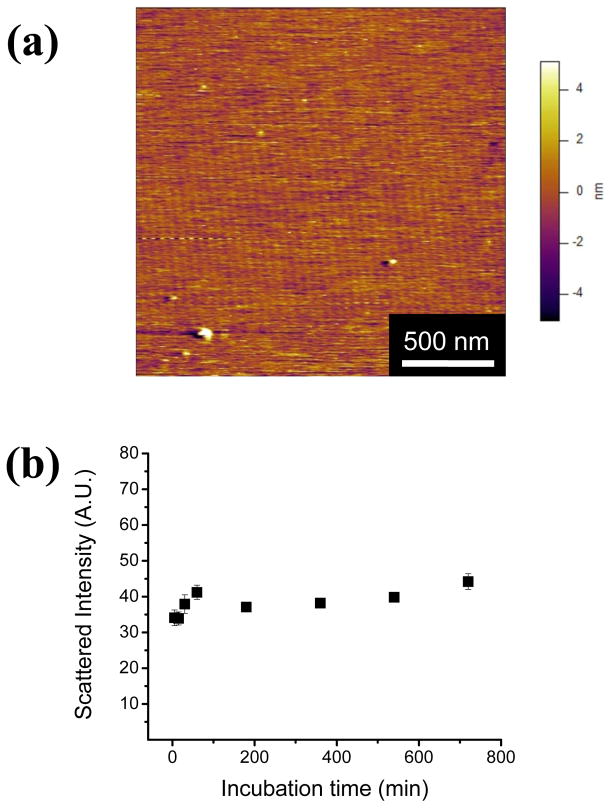
To further investigate the role of the mica surface in SELP-415K nanofiber assembly, 1 μg/ml polymer solution was incubated in the bulk state over 12 hours, and then deposited on a mica surface. The morphology of the nanofibers produced during the bulk incubation was analyzed by AFM. Some sphere-like aggregates but no nanofibrillar morphologies were present (Figure 2a) in sharp contrast to the nanofibrillar morphology observed after incubation of the polymer solution on the mica surface. These results highlight the importance of the mica surface for polymer self-assembly into nanofibers. To further probe whether SELP-415K can self-assemble in the bulk state, the light scattering intensity of the solution was measured at various time points. During the bulk incubation, the scattered intensity measured by DLS (Figure 2b) did not show any increase, which indicates that there was no significant change in the size of the particles during bulk incubation. The polymer concentration used in the DLS experiment was 10 μg/ml which showed a dense network of nanofibers on the mica surface (Figure 1d). These observations that no nanofibers were seen in the bulk incubated samples in the AFM image and that there were no changes in particle size in the DLS experiments, strongly indicate that in the range of concentrations tested SELP-415K self-assembled into nanofiber only in the presence of the mica surface.
Figure 2.
(a) AFM image of bulk incubated SELP-415K. 1 μg/ml SELP-415K with 5 mM NaCl in phosphate buffer was incubated in the bulk state for 12 hours at room temperature; (b) The scattered intensity was measured using dynamic light scattering. Polymer concentration was 10 μg/ml at 5 mM NaCl. The time points were 5, 15, 30, 60, 180, 360, and 720 minutes.
The effect of ionic strength on SELP self-assembly
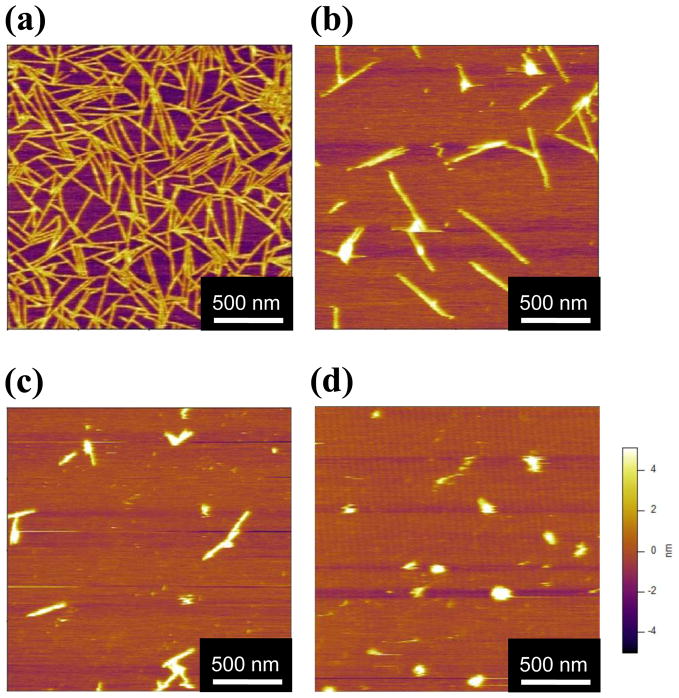
SELP-415K samples (1 μg/ml) in phosphate buffer with varying NaCl concentrations from 5 mM to 100 mM were incubated on mica surfaces for 12 hours. AFM images (Figure 3) show a morphological transition from fibrillar structure to sphere-like aggregates as ionic strength increased. At 5 mM NaCl concentration nanofiber network-like morphology was observed (Figure 3a) but at 10 mM salt concentration only isolated nanofibers with shorter length were observed (Figure 3b). At 50 mM NaCl concentration the length of the nanofibers became shorter and particles with sphere-like morphology, rather than fibrillar morphology started to appear (Figure 3c). At 100 mM salt concentration few nanofibrillar morphologies were observed and most of the adsorbed particles were sphere-like aggregates with sizes varying from 60 to 120 nm (Figure 3d). The width and height of the nanofibers were not dependent on salt concentration but length and shape dramatically changed as the ionic strength was increased. In addition, the density of SELP structures adsorbed on the surface decreased significantly with increasing salt concentration, which suggests that high ionic strength reduced overall adsorption of SELPs on the mica surface.
Figure 3.
AFM images of SELP-415K (1 μg/ml) at NaCl concentrations of: (a) 5mM, (b) 10 mM, (c) 50 mM, and (d) 100 mM. All samples were incubated on the mica surface for 12 hours at room temperature.
These results show that ionic strength significantly affects the self-assembly of SELP-415K into nanofibers on a mica surface. Mica is known to have a negative surface charge density of −0.0025C/m2.36 Based only upon the amino acid sequence of SELP-415K (without considering the influence that the adjacent amino acids might have on pKa of individual residues in elastin-based polymer chains as reported by Urry and coworkers37), the isoelectric point of SELP polymer is calculated to be 10.4. At pH 7.4, the polymer has a net positive charge of 7.6. This suggests that there are electrostatically attractive interactions between the negatively charged mica surface and positively charged SELP-415K which could aid the surface facilitated nanofiber growth on mica surfaces at neutral pH. The formation of sphere-like aggregates of SELPs as ionic strength is increased is consistent with our observations in bulk solutions of other SELP analogs.28,38,39 Previously it was noticed that the transition temperature of glutamic acid-containing SELPs from soluble state to aggregated state decreased as ionic strength increased due to the shielding effect of counter-ions on charged side chains.38,39 We evaluated the influence of increased ionic strength on the degree of swelling of SELP-415K hydrogels and observed that as ionic strength increased their degree of swelling decreased.28 These previous observations clearly show that electrostatic interactions between SELP polymers play a significant role in phase separation behavior as well as swelling behavior of a hydrogel. At the nanoscale, we observe that higher ionic strength resulted in lower adsorbed density on the mica surface and increased aggregated morphology (Figure 3a–d), implying that increased ionic strength weakened the attractive interactions between the SELP polymer and the mica surface as well as reduced the repulsive interactions between the SELP polymers,
The kinetics of nanofiber growth
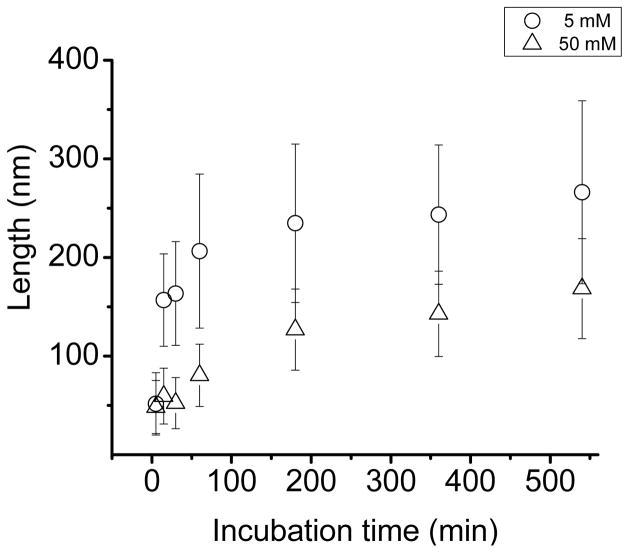
To study the effect of ionic strength on the growth rate of the nanofibers in a quantitative manner, the length of the nanofibers was measured at several time points under two different ionic strengths. Figure 4 shows that fiber length increased as incubation time increased and the rate of increase in length was much faster at 5 mM NaCl concentration compared to 50 mM. At lower ionic strength, the length of the nanofiber reached 60% of its maximum length in 5 minutes, whereas at higher ionic strength, nanofibers started to grow after 30 minutes, reaching a similar length after 9 hours. Nanofibrillar structure growth is typically characterized by three distinct phases; a lag phase, a burst phase, and a saturation (plateau) phase.40–42 The growth kinetics of SELP-415K nanofibers measured in 5 mM NaCl shows a burst phase in as early as 5 minutes during incubation without an apparent lag phase. The absence of a lag phase suggests that the mica surface plays a significant role in providing nucleating sites for nanofiber growth. In contrast, the growth kinetics at 50 mM NaCl concentration shows a lag phase during which the fiber had very limited growth for 30 minutes followed by a burst phase. These observations suggest that attractive electrostatic interaction between the polymer and the mica surface is a key factor in surface facilitated nanofiber growth.
Figure 4.
SELP-415K nanofiber growth kinetics measured in 5 mM (circles) and 50 mM (triangles) NaCl concentrations. Growth kinetics was assessed by measurement at various incubation time points (5, 15, 30, 60, 180, 360, and 540 minutes) using 1 μg/ml polymer solution incubated on the mica surface at room temperature for 12 hours for each salt concentration.
The slower growth rate at higher ionic strength can also be caused by aggregation of SELP before it adsorbs to the surface. As observed previously, increased salt concentration can cause aggregation of elastin-based polymers presumably by the change of water of hydrophobic hydration surrounding apolar amino acid residues in SELP.28,38,39,43 Since elastin-like units (GVGVP) are the predominant component of SELP-415K, increasing salt concentration may have resulted in aggregation of the polymer as seen in Figure 3d. In addition, higher ionic strength screens the repulsive interactions between SELP polymers, further facilitating aggregation of the SELP. As SELP polymers aggregate, it is less likely that SELP would form well-defined nanofiber structure in the bulk state. However, in the presence of the mica, the negative charges on the substrate provide a charge neutralizing surface, electrostatically attracting positively charged SELP polymers. It is also possible that charge interactions may orient the repetitive sequences of SELP in a favorable position for self-assembly. At 50 mM salt concentration, SELP was still able to adsorb on the mica surface and grow into nanofibers although the adsorbed density was decreased significantly. This observation suggests that aggregation which may have occurred at higher salt concentration is not irreversible and SELP can still self-assemble into well-defined nanostructures in the presence of the mica substrate. The weakened electrostatic interaction between SELP and the mica surface as well as the increased hydrophobic interactions between SELP polymers at 50 mM NaCl concentration correlate with the observed lag phase and slow growth kinetics.
The effects of the surface properties on surface facilitated self-assembly
The three different substrates were further tested to examine surface induced SELP self-assembly. They are silicon wafer, highly ordered pyrolytic graphite (HOPG), and 3-aminopropyl triethoxysilane (APTES) modified mica. 10 μg/ml of SELP solution was used in the experiments for clear demonstration of the fibrillation of SELP-415K. A silicon substrate is known to be negatively charged and hydrophilic due to the oxide layer formed at the surface. HOPG provides atomically flat, chemically inert, and hydrophobic surface. When SELP was incubated on silicon substrate for 12 hours at room temperature, many short nanofibers were clearly observed whereas nanofibers were barely observed on HOPG surface.(figure 5) The nanofibers formed on silicon substrates were less than 200 nm, shorter than the ones formed previously on mica surface but the width and height were the same. This suggests that the nanofiber growth kinetics were slower on the silicon surface than on the mica surface. To further investigate the effect of the surface properties on the self-assembly, SELP was incubated on the APTES modified mica. APTES modified mica is positively charged at low pH but becomes negatively charged at high pH.44 At pH 4, no nanofibers were observed on both APTES modified mica (figure 6a) and bare mica surface (figure 6c). At pH 9, long nanofibrillar morphology was not noticed on APTES modified surface (figure 6b) whereas nanofibrillar network was observed on bare mica surface (figure 6d). The images obtained from three different substrates clearly show the importance of the surface properties in the surface facilitated nanofiber assembly. First, silicon surface has negatively charged and hydrophilic, which is very similar to mica surface. As expected, the nanofibers were readily formed on silicon substrate although the growth kinetics seemed to be slower on silicon than on the mica surface. The nanofibers were not formed on hydrophobic HOPG surface, indicating that hydrophobic interaction between the surface and SELP is not playing a major role in surface facilitated self-assembly. No nanofibers were observed on positively charged surface (APTES modified mica at pH 4) but as the surface charged becomes negative (APTES modified mica at pH 9), a lot of short fragments of nanofiber were observed. The growth of nanofiber on APTES modified mica was rather limited compared to the bare mica and silicon but it clearly suggests the critical role of the negative surface charge in nucleating nanofiber growth. It is speculated that more delicate interaction (such as degree of negative potential or hydrophilicity of the substrate) between SELP and the surface may influence on the growth rate of the nanofiber on the surface. No extensive nanofiber formation was observed on mica surface at pH 4. The surface potential of the mica is still negative at pH 4 but SELP undergoes drastic increase of the positive charges when pH decreased from 9 to 4 due to the protonation of the histidine tags.45 We suspect that strong electrostatic interaction between histidine tag and the mica surface may have prevented nanofiber formation on the mica surface at pH 4. At pH 9, SELP formed a nanofiber network structure on the mica surface which resembles the structure formed at pH 7.4. In summary, the surface facilitated SELP nanofiber tends to form on the negatively charged hydrophilic surface rather than hydrophobic or positively charged surface. Negatively charged surface will play a role in attracting positively charged SELP’s to the surface. Hydrophilic surface seems to aid the conformational change of SELP, accelerating the self-assembling process to form nanofibers.
Figure 5.
The effect of the surface characteristics on the fibrillation of SELP-415K. (a) silicon substrate, (b) HOPG surface. The images were obtained using the surfaces incubated with 10 μg/ml polymer solution for 12 hours at room temperature.
Conclusion
In this study we demonstrated that for SELP-415K, polymer concentration, surface characteristics, and environmental conditions such as ionic strength play important roles in nanofiber formation on a mica surface. Examination using AFM and dynamic light scattering techniques showed that SELP-415K formed nanofibers only on the mica surface but not in the bulk state. Ionic strength had a significant influence on nanofiber growth, implying the importance of electrostatic interactions between the polymer and the mica surface. The fiber growth kinetic data indicated that the mica surface aided the formation of nucleating sites for fiber growth, removing a lag phase during the fiber growing process. The investigation of nanofiber formation on the substrates with various surface properties showed that the surfaces with a negative potential and hydrophilic property facilitated surface induced nanofiber self-assembly of SELP.
Acknowledgments
We acknowledge the support of the Maryland Nanocenter. This work was in part supported by a National Institutes of Health grant (R01-CA107621).
Abbreviations
- SELP
silk-elastinlike protein polymer
- G(Gly)
glycine
- A(Ala)
alanine
- S(Ser)
serine
- V(Val)
valine
- P(Pro)
proline
- L
leucine
- M
methionine
- D
aspartic acid
- Q
glutamine
- R
arginine
- W
tryptophan
- E
glutamic acid
- N
asparagines
- H
histidine
- P
phenylalanine
- K
lysine
- Y
tyrosine
- AFM
atomic force microscopy
- DLS
dynamic light scattering
- PBS
phosphate buffered saline
References
- 1.Zhang S. Nature biotechnology. 2003;21:1171–1178. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- 2.Ramachandran S, Tseng Y, Yu YB. Biomacromolecules. 2005;6:1316–1321. doi: 10.1021/bm049284w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang S, Holmes T, Lockshin C, Rich A. Proc Natl Acad Sci U S A. 1993;90:3334–3338. doi: 10.1073/pnas.90.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pastor MT, Esteras-Chopo A, Lopez de la Paz M. Curr Opin Struct Biol. 2005;15:57–63. doi: 10.1016/j.sbi.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Sipe JD, Cohen AS. J Struct Biol. 2000;130:88–98. doi: 10.1006/jsbi.2000.4221. [DOI] [PubMed] [Google Scholar]
- 6.Makin OS, Serpell LC. Febs J. 2005;272:5950–5961. doi: 10.1111/j.1742-4658.2005.05025.x. [DOI] [PubMed] [Google Scholar]
- 7.Ding TT, Lee SJ, Rochet JC, Lansbury PT., Jr Biochemistry. 2002;41:10209–10217. doi: 10.1021/bi020139h. [DOI] [PubMed] [Google Scholar]
- 8.Dobson CM. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 9.Venugopal J, Ramakrishna S. Tissue Engineering. 2005;11:847–854. doi: 10.1089/ten.2005.11.847. [DOI] [PubMed] [Google Scholar]
- 10.Haider M, Cappello J, Ghandehari H, Leong KW. Pharmaceutical Research. 2008;25:692–699. doi: 10.1007/s11095-007-9282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merlini G, Bellotti V. New England Journal of Medicine. 2003;349:583–596. doi: 10.1056/NEJMra023144. [DOI] [PubMed] [Google Scholar]
- 12.Knowles TP, Fitzpatrick AW, Meehan S, Mott HR, Vendruscolo M, Dobson CM, Welland ME. Science. 2007;318:1900–1903. doi: 10.1126/science.1150057. [DOI] [PubMed] [Google Scholar]
- 13.Zhu M, Souillac PO, Ionescu-Zanetti C, Carter SA, Fink AL. Journal of Biological Chemistry. 2002;277:50914–50922. doi: 10.1074/jbc.M207225200. [DOI] [PubMed] [Google Scholar]
- 14.Yoda M, Miura T, Takeuchi H. Biochemical and Biophysical Research Communications. 2008;376:56–59. doi: 10.1016/j.bbrc.2008.08.093. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Zhong J, Huang LX, Wang LJ, Hong YK, Sha YL. Journal of Physical Chemistry B. 2008;112:8950–8954. doi: 10.1021/jp802424h. [DOI] [PubMed] [Google Scholar]
- 16.Nayak A, Dutta AK, Belfort G. Biochemical and Biophysical Research Communications. 2008;369:303–307. doi: 10.1016/j.bbrc.2008.01.159. [DOI] [PubMed] [Google Scholar]
- 17.McMasters MJ, Hammer RP, McCarley RL. Langmuir. 2005;21:4464–4470. doi: 10.1021/la047044w. [DOI] [PubMed] [Google Scholar]
- 18.Yang H, Fung SY, Pritzker M, Chen P. Journal of the American Chemical Society. 2007;129:12200–12210. doi: 10.1021/ja073168u. [DOI] [PubMed] [Google Scholar]
- 19.Ku SH, Park CB. Langmuir. 2008;24:13822–13827. doi: 10.1021/la802931k. [DOI] [PubMed] [Google Scholar]
- 20.Kazlauskaite J, Sanghera N, Sylvester I, Venien-Bryan C, Pinheiro TJT. Biochemistry. 2003;42:3295–3304. doi: 10.1021/bi026872q. [DOI] [PubMed] [Google Scholar]
- 21.Kowalewski T, Holtzman DM. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:3688–3693. doi: 10.1073/pnas.96.7.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cappello J, Crissman J, Dorman M, Mikolajczak M, Textor G, Marquet M, Ferrari F. Biotechnol Prog. 1990;6:198–202. doi: 10.1021/bp00003a006. [DOI] [PubMed] [Google Scholar]
- 23.Megeed Z, Cappello J, Ghandehari H. Advanced drug delivery reviews. 2002;54:1075–1091. doi: 10.1016/s0169-409x(02)00063-7. [DOI] [PubMed] [Google Scholar]
- 24.Dandu R, Ghandehari H. Prog Polym Sci. 2007;32:1008–1030. [Google Scholar]
- 25.Anderson JP, Cappello J, Martin DC. Biopolymers. 1994;34:1049–1058. doi: 10.1002/bip.360340808. [DOI] [PubMed] [Google Scholar]
- 26.Oroudjev E, Soares J, Arcdiacono S, Thompson JB, Fossey SA, Hansma HG. Proc Natl Acad Sci U S A. 2002;99(Suppl 2):6460–6465. doi: 10.1073/pnas.082526499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamburro AM, Pepe A, Bochicchio B, Quaglino D, Ronchetti IP. Journal of Biological Chemistry. 2005;280:2682–2690. doi: 10.1074/jbc.M411617200. [DOI] [PubMed] [Google Scholar]
- 28.Haider M, Leung V, Ferrari F, Crissman J, Powell J, Cappello J, Ghandehari H. Molecular pharmaceutics. 2005;2:139–150. doi: 10.1021/mp049906s. [DOI] [PubMed] [Google Scholar]
- 29.Blackley HKL, Sanders GHW, Davies MC, Roberts CJ, Tendler SJB, Wilkinson MJ. Journal of Molecular Biology. 2000;298:833–840. doi: 10.1006/jmbi.2000.3711. [DOI] [PubMed] [Google Scholar]
- 30.Fung SY, Keyes C, Duhamel J, Chen P. Biophysical Journal. 2003;85:537–548. doi: 10.1016/S0006-3495(03)74498-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malinchik SB, Inouye H, Szumowski KE, Kirschner DA. Biophys J. 1998;74:537–545. doi: 10.1016/S0006-3495(98)77812-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldsbury C, Kistler J, Aebi U, Arvinte T, Cooper GJS. Journal of Molecular Biology. 1999;285:33–39. doi: 10.1006/jmbi.1998.2299. [DOI] [PubMed] [Google Scholar]
- 33.Lee JH, Bhak G, Lee SG, Paik SR. Biophys J. 2008;95:L16–L18. doi: 10.1529/biophysj.108.135186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loo RW, Goh AC. Langmuir. 2008;24:13276–13278. doi: 10.1021/la803041v. [DOI] [PubMed] [Google Scholar]
- 35.Karsai A, Grama L, Murvai U, Soos K, Penke B, Kellermayer MSZ. Nanotechnology. 2007:18. [Google Scholar]
- 36.Israelachvili JN, Alcantar NA, Maeda N, Mates TE, Ruths M. Langmuir. 2004;20:3616–3622. doi: 10.1021/la0352974. [DOI] [PubMed] [Google Scholar]
- 37.Urry DW, Harris CM, Luan CX, Luan C-H, Gowda DC, Parker TM. Transductional protein-based polymers as new controlled release vehicles. American Chemical Society; Washington DC: 1997. [Google Scholar]
- 38.Nagarsekar A, Crissman J, Crissman M, Ferrari F, Cappello J, Ghandehari H. J Biomed Mater Res. 2002;62:195–203. doi: 10.1002/jbm.10272. [DOI] [PubMed] [Google Scholar]
- 39.Nagarsekar A, Crissman J, Crissman M, Ferrari F, Cappello J, Ghandehari H. Biomacromolecules. 2003;4:602–607. doi: 10.1021/bm0201082. [DOI] [PubMed] [Google Scholar]
- 40.Kad NM, Myers SL, Smith DP, Smith DA, Radford SE, Thomson NH. J Mol Biol. 2003;330:785–797. doi: 10.1016/s0022-2836(03)00583-7. [DOI] [PubMed] [Google Scholar]
- 41.Hortschansky P, Schroeckh V, Christopeit T, Zandomeneghi G, Fandrich M. Protein Sci. 2005;14:1753–1759. doi: 10.1110/ps.041266605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fezoui Y, Teplow DB. J Biol Chem. 2002;277:36948–36954. doi: 10.1074/jbc.M204168200. [DOI] [PubMed] [Google Scholar]
- 43.Reguera J, Urry DW, Parker TM, McPherson DT, Rodriguez-Cabello JC. Biomacromolecules. 2007;8:354–358. doi: 10.1021/bm060936l. [DOI] [PubMed] [Google Scholar]
- 44.Pham KN, Fullston D, Sagoe-Crentsil K. Aust J Chem. 2007;60:662–666. [Google Scholar]
- 45.Nishimura S, Tateyama H, Tsunematsu K, Jinnai K. J Colloid Interf Sci. 1992;152:359–367. [Google Scholar]