Abstract
Bone mineral density (BMD) is a heritable complex trait used in the clinical diagnosis of osteoporosis and the assessment of fracture risk. We performed meta-analysis of five genome-wide association studies of femoral neck and lumbar spine BMD in 19,195 subjects of Northern European descent. We identified 20 loci reaching genome-wide significance (GWS; P<5×10−8), of which 13 map to new regions including 1p31.3 (GPR177), 2p21 (SPTBN1), 3p22 (CTNNB1), 4q21.1 (MEPE), 5q14 (MEF2C), 7p14 (STARD3NL), 7q21.3 (FLJ42280), 11p11.2 (LRP4; ARHGAP1; F2), 11p14.1 (DCDC5), 11p15 (SOX6), 16q24 (FOXL1), 17q21 (HDAC5) and 17q12 (CRHR1). The metaanalysis also confirmed at GWS level, seven known BMD loci on 1p36 (ZBTB40), 6q25 (ESR1), 8q24 (TNFRSF11B), 11q13.4 (LRP5), 12q13 (SP7), 13q14 (TNFSF11), and 18q21 (TNFRSF11A). The numerous SNPs associated with BMD map to genes in signaling pathways with relevance to bone metabolism, and highlight the complex genetic architecture underlying osteoporosis and BMD variation.
Osteoporosis is a condition characterized by reduced bone mass and microarchitectural deterioration of bone tissue, leading to loss of bone strength and increased risk of fracture. Osteoporosis increases in incidence with age, and is a major health threat to hundreds of millions of elderly individuals worldwide. Linkage analysis in monogenic bone disorders like Osteoporosis-Pseudoglioma syndrome, sclerosteosis, high bone mass syndrome and Paget's disease, have yielded major advances in recent years and highlighted the importance of the Wnt signaling1 and the RANK/RANKL/Opg pathways in the regulation of bone mass and bone turnover2. Linkage studies for the common form of osteoporosis have had limited success however3. As with other complex diseases, most of the candidate gene association studies in osteoporosis have produced conflicting results with limited replication4 mostly due to small sample sizes5. However, large-studies of major candidate gene polymorphisms within the sufficiently powered setting of the GENOMOS consortium, have been successful in obtaining consistent evidence of association between some genetic variants, BMD and fracture6–10. Concurrently, genome-wide association studies (GWAS), which perform a hypothesis-free search for genetic determinants,11 have already been successful in identifying 10 loci associated at a genome-wide significance (GWS) level with BMD12,13. Four of these loci involve members of the Wnt and RANK/RANKL/Opg signaling pathways highlighting their role in monogenic forms as well as the common form of osteoporosis.
BMD is used in clinical practice for the diagnosis of osteoporosis and in the assessment of fracture risk14. BMD is usually measured at the hip (femoral neck) and lumbar spine, which are common sites of fracture. However, BMD measurements at different skeletal sites are predictive of fracture in general because of their high correlation (r2~0.60).15 From a genetic perspective, BMD at both spine and hip is a complex, but highly heritable trait (h2~0.60–0.80)16. As shown for human height17–19, dozens and possibly hundreds of loci with small effects can be expected to influence the variation in BMD. Detection and reliable documentation of these loci of weak effects requires studies with comprehensive coverage of the genome and very large sample sizes. Here, we report the findings of a large-scale meta-analysis of 19,195 adult individuals from five GWAS on BMD of the lumbar spine (LS) and the femoral neck (FN), within the setting of the GEnetic Factors of OSteoporosis (GEFOS) consortium.
RESULTS
Samples, genotyping and (meta) analysis of genome-wide scans
The five study populations included the Rotterdam Study (RS, n=4,987), Erasmus Rucphen Family Study (ERF, n=1,228), Twins UK Study (TUK, n=2,734), deCODE Genetics Study (dCG, n=6,743) and the Framingham Osteoporosis Study (FOS, n=3,503). The age of participants ranged from 18 to 96 years. All studies had a majority of women (range 57–88%) in their samples with TUK including women only. Additional characteristics of the study populations and subject exclusion criteria are presented in Supplementary Tables 1 and 2, respectively.
BMD loci identification
Association results (corrected by the genomic control method20) of all HapMap CEU imputed SNPs passing quality-control (QC) criteria in each study (Supplementary Table 3), were meta-analyzed using METAL21. We declared results genome-wide significant at α=5×10–8 after adjusting for all common variant tests in the human genome22,23. We investigated if there was an excess of significant associations by comparing the test statistics to those expected under the null distribution using inter quantile-quantile (QQ) plots (corrected for overall meta-analysis genomic control λLS-BMD=1.09 and λFN-BMD=1.08). As observed in Figure 1, strong (and not early) deviation of the observed statistics from the null distribution was observed for both BMD traits, corresponding to an excess of significant and likely true associations. Excluding all SNPs within 500 Kb of the SNPs associated at a GWS level and correction for overall meta-analysis genomic control, still left many SNPs associated with BMD more than expected by chance alone. This suggests that among many false positives appearing at less stringent statistical thresholds, additional truly associated BMD variants may exist.
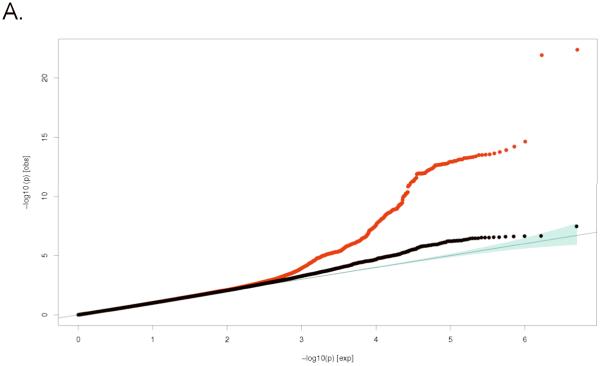
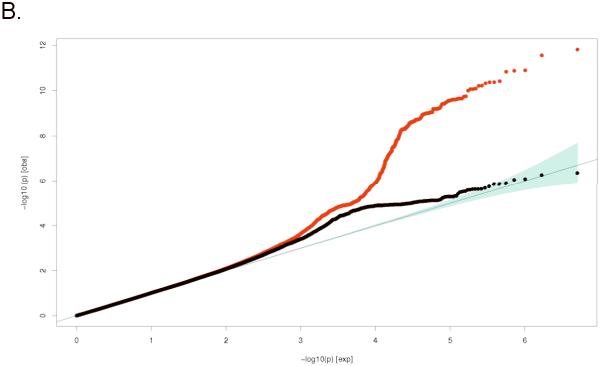
Figure 1.
Quantile-Quantile (Q-Q) plots for: A. Lumbar Spine BMD and B. Femoral Neck BMD; comparing additive model statistics to those expected under the null distribution using fixed-effects for all analyzed HapMap CEU imputed SNPs passing quality-control (QC) criteria in the studies (red lines), and after exclusion of all genome-wide significant and correlated (r2>0.1) SNPs (blue lines).
The meta-analysis identified 467 SNPs from 20 genomic loci exceeding the GWS threshold of association with the BMD traits (Figure 2). Of these, 15 loci associated with LS-BMD (Supplementary Tables 4A and 5A) and 10 with FN-BMD (Supplementary Tables 4B and 5B); five of these loci were associated with both skeletal sites. The effect sizes and significance of the top SNPs from the 20 regions containing markers associated with LS- and FN-BMD at GWS are presented in Table 1. Applying correction for overall-meta-analysis genomic control resulted in five of the 20 loci not to be GWS (with 7.1×10−7<P<5.0×10−8). For most markers heterogeneity was not very large or statistically significant.
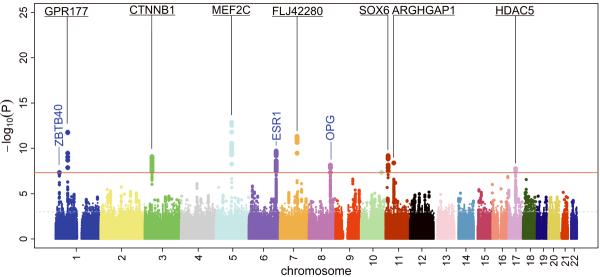
Figure 2.
Manhattan plots displaying novel and previously reported (known) loci associated at genome-wide significant level (GWS) with Lumbar Spine BMD (top) and Femoral Neck BMD (bottom) for all 2,543,686 HapMap CEU-imputed SNPs analyzed using fixed-effects. The 13 new GWS loci are in bold and underlined type. Already reported GWS loci are in blue type.
Table 1.
Top genome-wide significant markers of the 20 loci associated at GWS with lumbar spine and/or femoral neck BMD
| Marker Information |
Lumbar spine BMD |
Femoral neck BMD |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect estimate (in SD) |
Heterogeneity |
Effect estimate (in SD) |
Heterogeneity |
Site Specificity Ho:βLS=βFN |
|||||||||||||||
| Locus |
SNP |
A1*/A2 |
FREQ* |
Type |
Closest Gene |
Distance (kb) |
Beta (?LS) |
SE |
P-value |
OMA-GC P-value |
Q P-value |
I2 |
Beta |
SE |
P-value |
OMA-GC P-value |
Q P-value |
I2 |
P-value |
| Novel loci associated with BMD at GWS level | |||||||||||||||||||
| 1p31.3 | rs1430742 | T/C | 0.79 | I | GPR177 | 44.0 | 0.105 | 0.014 | 2.6E-13 | 2.5E-12 | 0.26 | 21 | 0.100 | 0.014 | 1.8E-12 | 1.2E-11 | 0.75 | 0 | 0.82 |
| rs2566755 | T/C | 0.79 | G | GPR177 | 44.3 | 0.104 | 0.014 | 3.3E-13 | 3.1E-12 | 0.27 | 20 | 0.100 | 0.014 | 1.7E-12 | 1.1E-11 | 0.76 | 0 | 0.83 | |
| 3p22 | rs87938 | A/G | 0.45 | G | CTNNB1 | 103.3 | −0.043 | 0.012 | 1.7E-04 | 3.1E-04 | 0.24 | 23 | −0.070 | 0.011 | 8.1E-10 | 3.4E-09 | 0.14 | 34.5 | 0.10 |
| 5q14 | rs1366594 | A/C | 0.55 | G | MEF2C | 197.0 | 0.005 | 0.012 | 0.65 | 0.66 | NC | NC | −0.085 | 0.011 | 1.3E-13 | 1.1E-12 | 0.62 | 0 | 3.2E-08 |
| 7p14 | rs1524058 | T/C | 0.40 | I | STARD3NL | 81.7 | −0.070 | 0.012 | 1.1E-09 | 5.2E-09 | 0.18 | 30 | −0.038 | 0.011 | 8.9E-04 | 1.4E-03 | 0.05 | 48 | 0.05 |
| 7q21.3 | rs4729260 | C/G | 0.68 | I | FLJ42280 | 14.9 | −0.081 | 0.013 | 1.7E-10 | 9.5E-10 | 0.14 | 35 | −0.085 | 0.012 | 9.4E-12 | 5.4E-11 | 0.77 | 0 | 0.82 |
| rs7781370 | T/C | 0.34 | I | FLJ42280 | 0.7 | −0.074 | 0.012 | 1.1E-09 | 5.5E-09 | 0.12 | 37 | −0.083 | 0.012 | 4.7E-12 | 2.9E-11 | 0.68 | 0 | 0.60 | |
| 11p14.1 | rs16921914 | A/G | 0.27 | G | DCDC5 | 61.6 | 0.077 | 0.013 | 2.3E-09 | 1.0E-08 | 0.52 | 0 | 0.038 | 0.013 | 0.003 | 0.005 | NC | NC | 0.03 |
| 11p15 | rs7117858 | A/G | 0.80 | G | SOX6 | 297.3 | −0.042 | 0.014 | 0.004 | 0.005 | NC | NC | 0.088 | 0.014 | 6.4E-10 | 2.7E-09 | 0.70 | 0 | 1.5E-10 |
| 16q24 | rs10048146 | A/G | 0.81 | G | FOXL1 | 95.4 | −0.093 | 0.016 | 1.7E-08 | 6.0E-08 | 0.21 | 28 | −0.085 | 0.016 | 1.7E-07 | 4.9E-07 | 0.96 | 0 | 0.73 |
| 17q12 | rs9303521 | T/G | 0.46 | G | CRHR1 | 56.5 | −0.068 | 0.012 | 1.4E-08 | 5.0E-08 | 0.05 | 49 | −0.055 | 0.012 | 3.6E-06 | 8.3E-06 | 0.07 | 45 | 0.46 |
| Suggestive loci now associated with BMD at GWS level | |||||||||||||||||||
| 2p21 | rs11898505 | A/G | 0.34 | G | SPTBN1 | 1.1 | 0.067 | 0.012 | 1.6E-08 | 6.3E-08 | 0.39 | 6 | 0.027 | 0.012 | 0.02 | 0.03 | NC | NC | 0.02 |
| 4q21.1 | rs1471403 | T/C | 0.34 | G | MEPE | 7.3 | 0.068 | 0.012 | 1.5E-08 | 5.7E-08 | 0.18 | 30 | 0.059 | 0.012 | 7.8E-07 | 2.0E-06 | 0.56 | 0 | 0.58 |
| 11p11.2 | rs7932354 | T/C | 0.29 | I | ARHGAP1 | 0.1 | 0.056 | 0.013 | 1.1E-05 | 2.4E-05 | 0.61 | 0 | 0.073 | 0.012 | 4.0E-09 | 1.5E-08 | 0.68 | 0 | 0.32 |
| 17q21 | rs228769 | C/G | 0.80 | I | HDAC5 | 7.8 | 0.067 | 0.014 | 4.0E-06 | 1.0E-05 | 0.87 | 0 | 0.081 | 0.014 | 1.7E-08 | 5.8E-08 | 0.94 | 0 | 0.49 |
| Known loci associated with BMD at GWS level | |||||||||||||||||||
| 1p36 | rs7524102 | A/G | 0.83 | G | ZBTB40 | 79.9 | 0.094 | 0.015 | 3.2E-10 | 1.7E-09 | 0.27 | 19.5 | 0.079 | 0.015 | 8.8E-08 | 2.6E-07 | 0.64 | 0 | 0.48 |
| rs6426749 | C/G | 0.17 | I | ZBTB40 | 66.9 | 0.107 | 0.017 | 7.6E-10 | 3.8E-09 | 0.74 | 0 | 0.082 | 0.015 | 4.8E-08 | 1.5E-07 | 0.64 | 0 | 0.26 | |
| 6q25 | rs2504063 | A/G | 0.40 | G | ESR1 | 38.0 | −0.078 | 0.012 | 6.1E-11 | 3.7E-10 | 0.03 | 52.7 | −0.066 | 0.012 | 3.0E-08 | 9.6E-08 | 0.84 | 0 | 0.45 |
| rs2941740 | A/G | 0.57 | I | C6orf97 | 67.3 | 0.070 | 0.012 | 2.0E-09 | 9.3E-09 | 0.01 | 59.0 | 0.073 | 0.012 | 2.0E-10 | 9.1E-10 | 0.02 | 57.2 | 0.84 | |
| 8q24 | rs2062377 | A/T | 0.56 | I | TNFRSF11B | 43.0 | 0.094 | 0.012 | 3.5E-16 | 5.7E-15 | 0.31 | 15.4 | 0.062 | 0.011 | 5.4E-08 | 1.7E-07 | 0.61 | 0 | 0.05 |
| rs11995824 | C/G | 0.45 | I | TNFRSF11B | 48.3 | −0.093 | 0.012 | 1.1E-15 | 1.6E-14 | 0.24 | 22.3 | −0.066 | 0.011 | 7.1E-09 | 2.6E-08 | 0.48 | 0 | 0.10 | |
| 11q13.4 | rs599083 | T/G | 0.69 | G | LRP5 | 24.4 | −0.067 | 0.012 | 4.7E-08 | 1.7E-07 | 0.50 | 0 | −0.047 | 0.012 | 9.7E-05 | 0.0002 | 0.76 | 0 | 0.25 |
| 12q13 | rs2016266 | A/G | 0.68 | G | SP7 | 1.6 | 0.070 | 0.012 | 1.3E-08 | 5.2E-08 | 0.93 | 0 | 0.046 | 0.012 | 1.9E-04 | 0.0003 | 0.81 | 0 | 0.16 |
| 13q14 | rs9533090 | T/C | 0.50 | I | AKAP11 | 54.0 | −0.120 | 0.012 | 5.4E-25 | 4.6E-23 | 0.02 | 57.1 | −0.041 | 0.011 | 3.9E-04 | 0.0006 | 0.39 | 5.7 | 1.1E-06 |
| 18q21 | rs884205 | A/C | 0.27 | I | TNFRSF11A | 1.4 | −0.078 | 0.0136 | 9.4E-09 | 3.8E-08 | 0.90 | 0 | −0.039 | 0.013 | 0.004 | 0.005 | NC | NC | 0.04 |
TypeSNP:G=Genotyped(atleastin1 study) I=Imputed
Distance: to coding region
Bold: P<5×10−8
OMA-GC: overall meta-analysis genomic control
NC: Not calculated P>0.001
Q P-value: Q-statistic P-value
Despite the correlation between LS- and FN-BMD measurements, site-specific effects were observed. Seven of the 20 loci showed evidence for skeletal site-specificity (P<0.05), three of which displayed strong evidence for site-specificity (P<1×10−6). This site-specificity is to be expected given the differences in heritability and that the genetic correlation (or fraction of “shared” heritability) between the measurements is considerably less than 1 (Supplementary Table 6).
Genes in associated regions and their function
Of the 20 BMD loci identified in this genome scan at a GWS level, seven have been reported previously as GWS13,24, whereas the remaining 13 have not. Of these 13 loci not previously associated with BMD at a GWS level, four were suggestively associated in previous reports (12,13), whereas nine are novel loci. In Supplementary Table 7 we present a summary of relevant gene annotations including related pathways, monogenic syndromes, knockout mouse models, and additional functional details of the genes most likely to be underlying the associated signals in these 13 loci.
Novel loci associated with BMD at GWS level
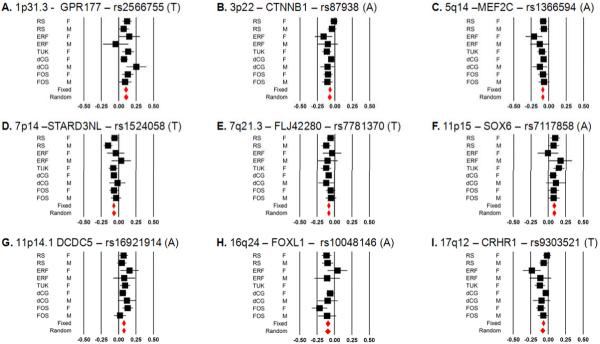
There are nine loci displaying novel associations with BMD for which we present Forest plots of effects (Figure 3) and regional association plots (Supplementary Figure 1) from the top SNPs.
Figure 3.
Forest Plots for the top SNPs for each of the 9 novel loci: A. 1p31.3, B. 3p22, C. 5q14, D. 7p14, E. 7q21.3, F. 11p15, G. 11p14.1, H. 16q24, I. 17q12. Black squares represent effect estimate and 95%CI for each study, and the red diamonds are summary effect. Measurements units are in BMD standard deviations (SD).
1p31.3 locus [GRP177]
Two common SNPs (MAF=0.21) in complete pair wise LD (rs1430742 and rs2566755) were associated at a GWS level with both FN- and LS-BMD. Both top SNPs are located within an intronic region of the G protein-coupled receptor 177 (GPR177, also named WNTLESS homologue) gene. GPR177 is part of the highly evolutionary conserved Wnt signaling pathway,25 involved in bone cell differentiation and development. The gene has been shown to be positive regulator of the I-kappaB kinase/NF-kappaB cascade, part of the RANK system. Cross-talk between the NF-kB and the mitogen-activated protein kinase (MAPK) pathway has been indicated by the identification of several overlapping genes expressed in both pathways, including several G protein-coupled receptors26.
3p22 locus [CTNNB1]
The rs87939 SNP (MAF=0.45) located 103 Kb upstream of the catenin (cadherin-associated protein), beta 1 (CTNNB1) gene on chromosome 3 was associated at a GWS level with FN-BMD. CTNNB1 is integral to the Wnt signaling pathway, and as such, is an excellent candidate for BMD regulation, considering that Wnt signaling controls the process of bone resorption by (negative) regulation of Opg (TNFRS11B) expression in osteoblasts27.
5q14 locus [MEF2C]
The rs1366594 top SNP (MAF=0.45) displayed skeletal site-specificity being associated with FN-BMD only, together with other 60 GWS markers at this locus. The SNP is located 197 Kb upstream of the MADS box transcription enhancer factor 2, polypeptide C (MEF2C) gene with no other known annotation within the large LD block. MEF2C is a transcription factor highly expressed in muscle which allows transcriptional cross-talk between the Ca2+/calmodulin-dependent kinase (CaMK) and mitogen-activated protein kinase (MAPK) signaling pathways by signal-dependent dissociation from histone deacetylases (HDACs). MEF2C interacts with HDAC4 and HDAC5 (see below 17q21 region), resulting in repression of the transcriptional activity of MEF2C. Potthoff et al. showed in mice that HDACs selectively degraded by the proteasome enables MEF2C to activate the slow myofiber gene program, resulting in enhanced endurance during physical exercise28.
7p14–p13 locus [STARD3NL]
The rs1524058 SNP (MAF=0.40) located on the short arm of chromosome 7 is 81 Kb upstream of the STARD3 n-terminal like (STARD3NL) gene, a cholesterol endosomal transporter, was associated at a GWS level with LS-BMD but less strongly with FN-BMD. Very recently, a study on Asian individuals29 identified a SNP in this 7p14 region consistently associated with BMD measurements of the radius, tibia and heel (lowest P=1.4×10−7). Yet, the rs1721400 SNP was not associated with either LS- or FN- BMD in our study (P>0.05). The rs1721400 marker (mapping close to SFRP4) is in very low LD with our top SNP rs1524058 in HapMap individuals of European descent (r2=0.015, D'=0.194), while LD between markers is considerably higher in Chinese or Japanese individuals (r2=0.230, D'=0.931). Thus, an underlying signal common to both Europeans and Asians may be still be captured by these SNPs considering the differences in LD across populations.
7q21.3 locus [FLJ42280]
Several SNPs are GWS in this region, with the two top SNPs being in moderate pair wise LD (r2=0.36; D'=0.84) and associated at GWS with both LS- and FN-BMD. The SNPs are located within (rs4729260) and just upstream (rs7781370) of FLJ42280, a hypothetical protein of unknown function. There are several genes within the ~480 kb LD stretch region, of which the split hand/foot malformation (ectrodactyly) type 1 (SHFM1) is the closest gene (87–185 kb away).
11p15 locus [SOX6]
The rs7117858 SNP (MAF=0.20) was associated at GWS level with FN- BMD only, displaying strong evidence for skeletal site-specificity. The SNP is located 297kb upstream from the SRY (sex determining region Y)-box (SOX6) gene, which is a transcription factor of the SOX gene family defined by a conserved high mobility group (HMG) DNA-binding domain. The gene is expressed in a wide variety of tissues, most abundantly in skeletal muscle. Sox6 knock-out mice exhibit early lethality due to cardiac insufficiency and present with mild skeletal abnormalities affecting size and mineralization of endochondral elements30. Other SOX-family genes regulate RUNX2-mediated differentiation of mesenchymal cells during endochondral ossification (skeletogenesis)31.
11p14.1 locus [DCDC5; DCDC1]
The rs16921914 SNP (MAF=0.27) is the only marker in the 11p14.1 region associated with LS-BMD at a GWS level. All other associated SNPs are in moderate LD with rs16921914 (r2<0.70) and display less strong associations (P>1×10−7). The SNP is located 62 kb downstream of the doublecortin domain containing 1 (DCDC1) and 73 kb upstream of the DCDC5 genes. Doublecortin domains are highly conserved elements which serve as protein-interaction platforms32. Mutations in members of this protein superfamily are linked to several neurogenetic diseases and to our knowledge are not expressed in bone.
16q24.3 locus [FOXC2; FOXL1]
The rs10048146 SNP (MAF=0.19) was associated with LS-BMD (−0.09 and is located on the subtelomeric region of chromosome 16, about 95Kb downstream from a cluster of small (1 Kb) genes of the “forkhead” (or winged helix) gene family. The genes are mainly expressed in the gastrointestinal mucosa (FOXL1) or are involved in adipocyte metabolism and early stage chondrogenic differentiation (FOXC2). FOXC2 stimulates osteoblast differentiation of mesenchymal cells through activation of canonical Wnt-beta-catenin signals33 while FOXC2 expression has been shown to occur via bone morphogenetic proteins34. Skeletal defects of the spine have been reported in FOXC2 mouse knockout models35 and recently, deletions and inactivating mutations affecting the FOX gene cluster have been identified as causing severe malformations of the VACTER type in humans, which include vertebral malformations.36
17q12–q22 [CRHR1]
The rs9303521 SNP (MAF=0.46) on chromosome 17 was associated with LS-BMD and is located 56 Kb from the corticotrophin-releasing factor receptor (CRHR1) gene. Among other genes in this LD region, MAP3K14 is another candidate potentially involved in bone-active pathways, particularly through the activation of NF-kappa-B.
Suggestive loci now associated with BMD at GWS level
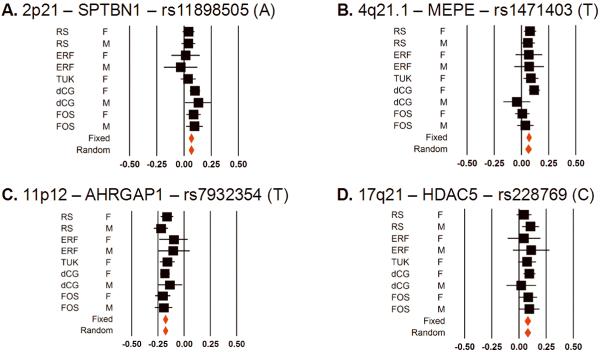
Four BMD loci which now reach for the first time a GWS level were “suggestively” associated with BMD in previous reports12,13. We present Forest plots of effects (Figure 4) and regional association plots (Supplementary Figure 2) from these SNPs.
Figure 4.
Forest Plots for the top SNPs for each of the 4 loci attaining GWS for the first time in this study: A. 2p21, B. 4q21.1, C. 11p11.2, D. 17q21. Black squares represent effect estimate and 95%CI for each study, and the red diamonds are summary effect estimates. Measurements units are in BMD standard deviations (SD).
2p21 locus [SPTBN1]
The rs11898505 (MAF=0.34) on chromosome 2 is intronic to the spectrin, beta, non-erythrocytic 1 (SPTBN1) gene which encodes a major cytoskeletal scaffolding protein, and was associated with LS-BMD. The same SNP was previously shown to be associated with BMD and fractures, even though it did not reach GWS level for BMD in the same study.13 In mice, disruption of beta-spectrin isoforms (Elf) leads to the disruption of TGF-beta signaling by Smad proteins37.
4q21.1 locus [MEPE]
The 4q21.1 region contains a cluster of structurally and phylogenetically related genes encoding matricellular phosphoglycoproteins with function in bone formation and growth38. The top associated rs1471403 SNP (MAF=0.34) is located 7 Kb 3′ to the matrix, extracellular, phosphoglycoprotein (MEPE) gene (also known as osteoblast/osteocyte factor 45), 42 Kb to the integrin-binding sialoprotein (IBSP) gene and 122 Kb 5′ to the secreted phosphoprotein 1 (SPP1) gene, also known as osteopontin. IBSP and SPP1 are highly expressed in osteoblasts, osteoclasts and hypertrophic chondrocytes. MEPE is predominantly expressed by osteocytes in human bone, playing an inhibitory role in bone formation. All three genes display diverse skeletal phenotypes in mice knock out (KO) models. MEPE (Of45) KO show increased bone mass and inhibition of age-related bone loss39, IBSP KO show high trabecular bone density with low bone turnover but respond to bone loss caused by disuse40 and the SPP1 KO have high trabecular bone mass and is resistant to bone loss41. Previously, a non-synonymous SNP in the IBSP gene (rs1054627, G195Q) in moderate LD with (r2=0.2, D'=0.8) was reported as suggestively associated with hip BMD.12
11p11.2 locus [ARHGAP1; LRP4]
At the 11p11.2 region a large LD block extends the region withholding several genes, including C11orf49, LRP4, CKAP5, F2, ZN408 and ARHGAP1 among others. Two fully correlated SNPs (r2=1;MAF=0.29) including rs7932354, which lies in the promoter region of the Rho GTPase activating protein 1 (ARHGAP1) gene; and rs2070852, located in intron 5 of the coagulation factor II (F2) gene were associated with FN-BMD at GWS level. Other correlated SNPs in the region (r2 between 0.2 and 0.8) were previously suggestively associated with hip BMD and attributed to the LRP4 gene13. ARHGAP1, a ubiquitous factor composing one of the GTPase activating proteins that represses RhoA, is another good candidate. RhoA is a small G-protein of the Rho family that regulates cell morphology via actin-cytoskeleton reorganization and which is thought to be a potential commitment switch in the differentiation of mesenchymal stem cells to osteoblasts42. In addition, data from ARHGAP1 KO mouse models show a strong skeletal phenotype, including a 3-fold reduction in BMD, decreased cortical thickness and bone fragility in older animals43.
17q21 locus [HDAC5;C17orf53]
The 17q21 region contains more than 30 genes in 1 Mb surrounding the top rs228769 SNP (MAF=0.20) which is located 8 Kb upstream of the histone deacetylase 5 (HDAC5) and 26 kb upstream of the C17orf53 genes. A non synonymous SNP in the C17orf53 gene, rs227584 (T126P), moderately correlated to rs228769 (r2=0.64) was found associated with hip BMD, albeit not at GWS level12. These SNPs likely represent the same signal. HDAC5 is a class histone deacetylase II (homologous to yeast Hda1), ubiquitously expressed and responsible for the deacetylation of lysine residues on the N-terminal part of the core histones (H2A, H2B, H3 and H4). Histone acetylation and deacetylation plays an important role in transcriptional regulation, cell cycle progression and developmental events, particularly for myocyte differentiation44. In undifferentiated myoblasts, HDAC5 is present in the nucleus where it binds to the myocyte enhancer MEF2C (see above) to repress transcription and detain muscle maturation. In bone, recruitment of class II histone deacytylases like HDAC5, is needed for TGF-β mediated osteoblast differentiation45, which occurs through inhibition of Run×2 function by Smad346.
Known loci associated with BMD at GWS level
We have replicated at GWS level 7 loci associated with BMD in previous GWA studies12,13,24. They include the 1p36 (ZBTB40), 6q25 (ESR1), 8q24 (TNFRSF11B;Opg), 11q13.4 (LRP5), 12q13 (SP7;Osterix), 13q14 (TNFSF11;RANKL), and 18q21 (TNFRSF11A;RANK) regions. Regional association plots for all seven regions are included in Supplementary Figure 3. In the 6q25 (ESR1) region at least two independent GWS signals are seen at the sides of a high recombination rate peak (Supplementary Figure 3B). This locus showed a complex pattern of association in a previous study indicating three independent signals at the locus13. In Supplementary Table 8 we report the associations observed in this study for all loci reported previously as attaining GWS or “suggestive” association12,13,24. SNPs from the 6p21.32 (MHC), 14q32 (MARK3) and 17q21 (SOST) regions described previously to be GWS13 were still significantly associated to BMD in our study, but not at a GWS level. It should be noted that in the previous study the 14q32 and the 17q21 regions were associated with total hip BMD, which differs from the femoral neck BMD phenotype used in the current study.
Gene expression (eQTL) associations
We tested the association of SNPs (or proxies) from the 13 newly GWS associated regions with cis-allelic expression of gene transcripts in primary human osteoblasts. All SNPs associated with gene expression at P<0.05 and located within the same LD block of the strongest associated variants (D'≥0.8) are presented in Supplemental Table 9. Associations were seen for transcripts of GPR177, MEF2C and FOXC2. Similarly, for variants in (or in LD with variants in) MEPE the most significant correlation with expression in osteoblasts was seen with the integrin bone sialoprotein (IBSP) gene, while MEPE seems not highly expressed in osteoblasts. Yet, the statistical evidence is not fully conclusive since only subtle overrepresentation of the associated loci was observed (10.5% vs 7% for non-associated control SNPs, Chi2=8.9 and P=0.003). The small overrepresentation of the associated loci suggests several of the associated genes may be expressed in cell & tissues other than osteoblast lineages.
Combined effect of the 20 GWS BMD loci and fracture risk
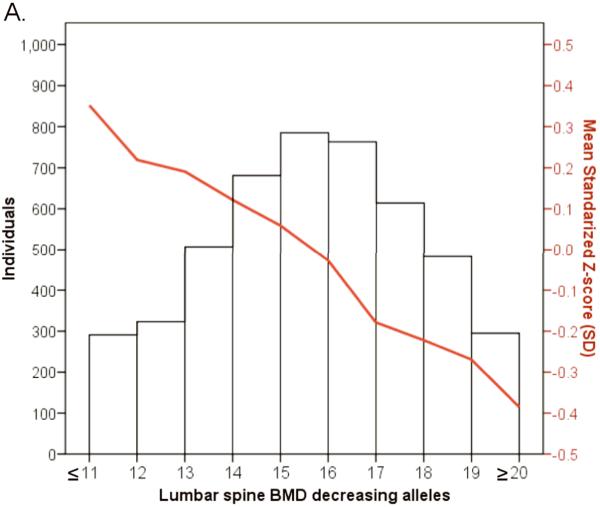
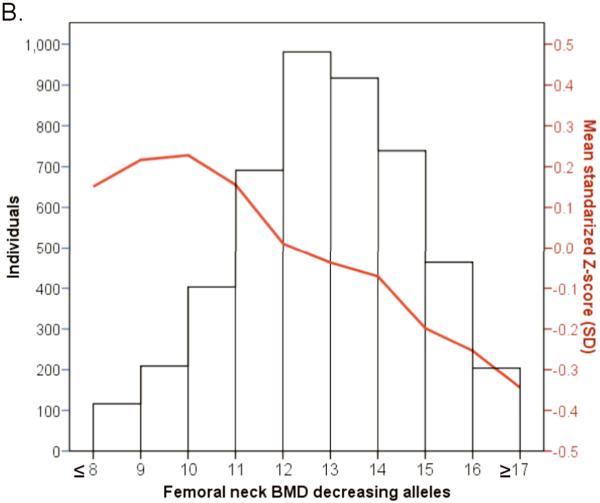
We examined the combined effect of the top SNPs arising from the 20 associated BMD loci in 4,983 individuals from the prospective population-based Rotterdam study. Risk allele counts were derived from the top associated SNPs from the 15 LS-BMD and 10 FN-BMD loci, all of which followed a normal distribution of their frequency in the study population (Figure 5). The 15 LS-SNPs combined explained ~2.9% of the variance in LS-BMD and the 10 FN-SNPs combined explained ~1.9% of the variance in FN- BMD. A highly significant linear decrease in the mean LS-BMD and FN- BMD of individuals was seen with increasing numbers of “low BMD” risk alleles, i.e. those carrying 20 or more alleles that associated with low LS-BMD (“low BMD” alleles) (n=300) had 0.65 SD (~0.12 g/cm2) lower BMD (P=3×10−8) compared to those that carried 11 “low BMD” alleles or fewer (n=360). A similar (yet less pronounced) trend was seen for FN-BMD. The association between the compound allelic scores and the risk of fracture were assessed in 2727 radiographically screened individuals (302 vertebral fracture cases) and in 4865 individuals followed-up 8.2 years on average (672 non-vertebral fractures). The compound LS-BMD allelic score was not significantly associated with the risk of vertebral fracture in the Rotterdam Study dataset (OR=1.018, 95%CI[0.966,1.073]; P=0.50), while it was borderline significant for association with the risk of incident non-vertebral fracture (HR=1.031, 95%CI[0.997,1.066]; P=0.07). In contrast, the compound FN- BMD allelic score was consistently associated with the risk of vertebral (OR=1.057, 95%CI[1.010,1.107]; P=0.02) and non-vertebral (HR=1.033, 95%CI[1.004,1.063]; P=0.03) fracture. Adjustment for FN-BMD showed that at least 46% of the genetic effect on vertebral fracture could be explained by FN-BMD (ORadjusted= 1.031, 95%CI[0.983,1.080]; P=0.21), and 54% of the genetic effect on incident non-vertebral fracture was through FN-BMD (HRadjusted= 1.015, 95%CI [0.986,1.044]; P=0.32). Power limitations are a very plausible explanation to the absence of significant association between the LS-BMD compound allelic score and the risk of vertebral fracture.
Figure 5.
Histogram and line plot modeling in the Rotterdam Study the combined allelic effect across all genome-wide significant associated loci for A. Lumbar Spine BMD and B. Femoral neck BMD. Subjects were classified according to the number of BMD decreasing (risk) alleles at the lumbar spine BMD (20 SNPs) and the femoral neck (15 SNPs). The mean for each risk allele-count group was determined and the extremes of the distribution counts were pooled into the nearest risk allele-count group of size >100 individuals.
DISCUSSION
The GEFOS consortium has been assembled to identify the genetic determinants of osteoporosis and fracture. This study represents the first step in a collaborative effort and expands the current knowledge on the underlying genetics of BMD -a clinical measurement used for the diagnosis of osteoporosis and the assessment of fracture risk. BMD measures of the lumbar spine and femoral neck were analyzed independently, because despite a relatively high phenotypic correlation, the genetic correlation is not perfect. This is illustrated by the site-specificity detected in some of the associations probably reflecting genuine biological mechanisms (i.e. differences in cortical vs trabecular bone content), but also intrinsic measurement differences (i.e. artifacts influencing BMD values like osteophytes of the lumbar spine or aortic calcifications).
Performing meta-analysis of GWAS has limitations. False positive associations generated from multiple hypothesis testing and population stratification are inherent possibilities. Nevertheless, we applied well-established methods to minimize the impact of multiple testing by applying stringent GWS thresholds for determining significance. Also, our findings are constrained by the power of our current sample suited to identify effect sizes explaining ~0.2% of the variance of the trait. This means we are not powered to detect real effects of the same (small) magnitude arising within specific sex and/or age groups. Similarly, due to power limitations we cannot address potential gene-gene and gene-environment interactions, the effects of rare alleles that are not captured by the haplotype tagging approach employed in GWAS nor determine the effect on fracture risk. Despite being underpowered to assess heterogeneity of effects, some markers displayed significant heterogeneity and were not GWS when analyzed under the random effects model. Further evaluation of such markers in larger populations is needed to determine the source(s) of heterogeneity across datasets, like inaccuracies in the imputations, subtle differences in phenotype ascertainment, differences in linkage to the culprit, differences in environmental modifying factors, or genuinely different genetic effects across populations.47 Achieving such sufficiently-powered setting is the target of the expanding GEFOS consortium. All our top associated markers displayed high quality imputation scores, high correlation after de-novo genotyping and/or at least one genotyped proxy in complete LD which was also associated at GWS level (Supplementary Table 10). Thus, we can exclude imputation inaccuracies as a source of heterogeneity and/or false-positive associations. In addition, we excluded individuals with non-European profiles strategy which confines our current findings to the context of populations of Northern European-descent. Similarly, test statistics corrected by the genomic inflation factors affecting each study, makes unlikely that population stratification or cryptic relatedness (like those observed in the ERF, TUK, and dCG populations) play an important role in our associations. In addition, we examined the effect of applying a second correction for overall meta-analysis genomic control in our results. Only one locus (LRP5) drifted away importantly from the GWS threshold after correction. This approach is likely to be over-conservative considering that the association of variants in LRP5 has been consistently replicated at GWS level in at least two previous efforts10,24.
In summary, we identified and/or confirmed at least 20 loci associated with lumbar spine and femoral neck BMD, highlighting the complex genetic architecture underlying the variation in BMD. Yet, these loci explain only a minor fraction of the variance in BMD, and hence, an even smaller fraction of the heritability for fracture risk. Nevertheless, these findings underscore molecules within novel and key-known biological pathways influencing BMD variation, particularly the Wnt and NF-kappa-B signaling pathways (including about half of the identified loci GPR177, CTNNB1, FOXC2, LRP5, SPTBN1, HDAC5, TNFSF11, TNFRSF11A and TNFRSF11B). None of the SNPs we have identified can be unequivocally designated as the underlying “true” variants driving the associations. Additional efforts to identify such variants are warranted to maximize the application of this genetic knowledge towards the prediction of risk in individuals and the translation into new pharmacological agents for the treatment of osteoporosis. Increasing further the sample size will aid the identification of additional loci associated not only with BMD, but more importantly will allow focusing on the risk of fracture, the ultimate consequence of osteoporosis.
OMIM48,49 accession numbers
GPR177[611514]; CTNNB1[116806]; MEF2C[600662]; STARD3NL[611759]; FLJ42280[None],SHFM1[183600]; SOX6[607257]; DCDC5[612321],DCDC1[608062]; FOXC2[602402],FOXL1[603252]; CRHR1[122561]; SPTBN1[182790]; MEPE[605912],IBSP[147563],SPP1[166490]; ARHGAP1[602732],LRP4[604270],F2[176930]; HDAC5[605315],C17orf53[None]; ZBTB40[612106]; ESR1[133430], C6orf97[None]; TNFRSF11B[602343]; LRP5[603506]; SP7[606633]; TNFSF11[602642],AKAP11[604696]; TNFRSF11A[603499].
METHODS
Study population
The GEnetic Factors of Osteoporosis (GEFOS) consortium is a coalition of teams of investigators working on the genetics of osteoporosis. The current meta-analysis incorporated 19,195 individuals of Northern European ancestry derived from five GWAS on BMD of the lumbar spine (LS-BMD) and the femoral neck (FN-BMD) including: the Rotterdam Study (RS, n=4,987), Erasmus Rucphen Family Study (ERF, n=1,228), Twins UK Study (TUK, n=2,734), deCODE Genetics Study (dCG, n=6,743) and the Framingham Osteoporosis Study (FOS, n=3503). All studies were approved by institutional ethics review committees at the relevant organizations and all participants provided written informed consent. The Rotterdam Study (RS) is a prospective population-based cohort study of chronic disabling conditions in Dutch elderly individuals age 55 years and over (http://www.epib.nl/ergo.htm).50,51 The Erasmus Rucphen Family (ERF) study is a family-based study of a genetic isolate in the South West Netherlands to identify genetic risk factors for complex disorders.52 The Twins UK (TUK) study is a population-based sample of twins from the UK studying the hereditary basis of a wide variety of age-related traits and diseases (http:/www.twinsUK.ac.uk).53 The Icelandic deCODE Genetics (dCG) study comprises a population-based sample to identify the genetic basis of complex diseases.13 The Framingham Osteoporosis Study (FOS) is embedded in the Framingham Heart Study (FHS), a community-based, longitudinal, prospective cohort comprising three generations of individuals in multigenerational pedigrees and additional unrelated individuals (http://www.framinghamheartstudy.org/). Individuals of “Generation 1” include those first examined in 194854, “Generation 2” includes those examined at the first cycle from 1971 to 197555, and “Generation 3” includes those examined at the first cycle beginning in 2002–2005.56 For these analyses, 812 members of the Generation 1 cohort (22.6% of the sample) and 2783 (77.4%) of the Generation 2 cohort who had BMD measured as part of the FOS were included.
Bone mineral density and anthropometric measurements
BMD was measured in all cohorts at the lumbar spine (either at L1–L4 or L2–L4) and femoral neck using dual-energy X-ray absorptiometry following standard manufacturer protocols (GE-Lunar Corporation, Madison, WI or Hologic Incorporated, Bedford, MA) see Supplementary Table 1 for details. All DXA and anthropometric measurements were performed in the RS at the baseline visit baseline between 1991–1992, in ERF between 2002–2003, in TUK at the latest follow-up, dCG at the baseline visit and in FOS Generation 1 between 1992–1997, and Generation 2 between 1996–2001.
Phenotype modeling
The overall strategy involved linear regression to adjust BMD measurements for effects of age, weight, sex and study using standardized residuals with mean 0 and standard deviation 1 in the genotype-phenotype association testing. Such residuals were obtained by regressing within each study the raw BMD measurements on age and weight (and principal components in FOS to adjust for population substructure using Eigenstrat57) in sex-specific models. Thus, in studies including both men and women the data for each gender are included as separate estimates in the meta-analysis.
Genotyping
The five GWAS were genotyped using the Illumina Infinium HumanHap550 Beadchip (RS), the Illumina Infinium HumanHap300 or HumanCNV370 Beadchip (ERF, TUK & dCG) or the Affymetrix Dual NspI/StyI GeneChip 2×250K with 50K gene-centered MIP set (FOS), all according to manufacturer's protocols and quality control standards. The exclusion/filtering criteria for individuals and SNPs are described in Supplementary Tables 2–3.
Genotype imputation
Imputation was used to evaluate associations for the same SNPs across study populations using scans from different genotyping platforms. Genotypes were imputed for all polymorphic SNPs oriented to the positive strand from phased autosomal chromosomes of the HapMap CEU Phase II panel (release 22, build 36)58. Hidden Markov Model-based algorithms were used to infer unobserved genotypes probabilistically as implemented in either MACH59 or IMPUTE60. Imputation quality control metrics from MACH and IMPUTE were used. Detailed descriptions of quality control and imputation procedures are summarized for all studies in Supplementary Table 3. We performed technical validation of the imputed genotypes in an independent set of 880 individuals of Icelandic origin using Centaurus (Nanogen)61 discrimination assays, for the top associated hits that reached GWS for the first time in this study (Supplementary Table 10).
Genotype-phenotype association testing62
Each study performed genome-wide association for BMD using sex-specific, age- and weight-adjusted standardized residuals analyzed under an additive (per allele) genetic model. Analysis of imputed genotype data accounted for uncertainty in each genotype prediction by utilizing either the dosage information from MACH or the genotype probabilities from IMPUTE. Studies used MACH2QTL59, which uses genotype dosage value (0 – 2, continuous) as a predictor in a linear regression framework, SNPTEST60, Merlin63 or the linear mixed effects model of the Kinship64 and ProABEL65 packages in R66 to account for relatedness (Supplementary Table 3). The genomic control method20 was used to correct the standard error by the square root of the genomic inflation factor (lambda): SEcorrected =SE , which is equivalent to the proposed correction of the Chi2-statistic by lambda. Genomic inflation factors for the studies are presented on Supplementary Table 3. Overall meta-analysis genomic control inflation factors were calculated as described previously.67 Genomic inflation factors scaled to a standard size (1000 individuals) to calibrate for the effect of sample size on λ67, showed residual genomic inflation was negligible (λLSBMD1000=1.005 and λFN-BMD1000=1.004).
Meta-analysis
The minor allele from HapMap CEU genotypes was used to define the coded allele in all analyses, regardless of frequency in individual cohorts. All meta-analysis calculations were done using the METAL21 software package applying inverse-variance methodology assuming fixed effects with Cochran's Q and I2 metrics used to quantify between-study heterogeneity. We also calculated the summary results by random effects using STATA software68 for those markers associated at GWS level with Cochran's Q P-value < 0.05 and/or I2 estimates > 50%, reflecting large heterogeneity beyond chance. Random effects models also incorporate in the calculations the between-study heterogeneity, and estimate the average genetic effect from the population of genetic effects that may differ in different studies. In the absence of between-study heterogeneity fixed and random effects calculations give identical results. We declared results genome-wide significant at α=5×10−8 after adjusting for all common variant tests in the human genome22,23. To test for BMD site specificity we estimated the effect difference (Δβ) as βfemoralneck − βlumbarspine, the SE of the mean difference (ΔSEM) as sqrt(SEfemoralneck2 + SElumbarspine2) and the Z-statistic as Δβ/ΔSEM from which the P-values were computed.
eQTL analysis in human osteoblast (HOb)
SNPs from new loci associated with BMD at the genome-wide significance (GWS) level were tested for association with cis-allelic expression of neighboring gene transcripts, in primary human osteoblasts (HOb) derived from 95 unrelated Swedish donors. Detailed cell culture methods have been described previously69. Expression profiling was performed using the Illumina HumRef-8 BeadChips according to the manufacturer protocol. Genotyping for genotype-expression association was performed using the Illumina HumanHap 550k Duo chip. Individuals with low genotyping rate and SNPs showing significant deviation from Hardy-Weinberg equilibrium (P < 0.05) were excluded. Similarly low frequency (MAF<0.05) SNPs and SNPs with high rates of missing data were excluded. The association of the expression levels was focused on cis-acting genetic variants, defined as being within 250kb window flanking the gene, using a linear regression model implemented in the PLINK70 software package with age and sex as covariates. SNPs included on the Illumina 550K chip were assessed for expression cis-associations directly. In addition, all genotyped SNPs included on the Illumina chip that were in strong LD (defined as D –≥0.8) and mapping ± 50kb from the GWS hit were included in the association study. To test for a significant enrichment of functional SNPs (i.e., SNPs associated with gene expression in HObs at p<0.05) among the candidate SNPs, a Chi2-statistic was obtained to test whether observed associations were different from expected associations in the expression data set beyond chance. Expected values (7.1%) were based on the proportion of SNPs with p<0.05 seen in the HOb gene expression data set and in a random selection of 1200 SNPs associated with both LS- and FN-BMD at P>0.90, MAF>0.20, and present in the Illumina HumanHap550 array (assessed as negative controls for association with HOb expression).
Combined effect of associated loci
Within the setting of the prospective population-based Rotterdam Study the combined effect of all 20 BMD loci was studied by classifying subjects according to the number of BMD decreasing (risk) alleles. This was based on 15 lumbar spine and 10 femoral neck BMD loci as follows:
Lumbar spine (15 SNPs) => rs7524102 [ZBTB40], rs1430742 [GPR177], rs11898505 [SPTBN1], rs1471403 [MEPE], rs2504063 [ESR1], rs1524058 [STARD3NL], rs4729260 [FLJ42280], rs2062377 [TNFRSF11B], rs16921914 [DCDC5], rs599083 [LRP5], rs2016266 [SP7], rs9533090 [TNFSF11], rs10048146 [FOXC2], rs9303521 [CRHR1] and rs884205 [TNFRSF11A].
Femoral neck (10 SNPs) => (rs6426749 [ZBTB40], rs2566755 [GPR177], rs87938 [CTNNB1], rs1366594 [MEF2C], rs2941740 [ESR1], rs7781370 [FLJ42280], rs11995824 [TNFRSF11B], rs7117858 [SOX6], rs7932354 [ARHGAP1], and rs228769 [HDAC5]). The mean BMD for each risk allele-count group was determined, and at the extremes of the distribution counts were pooled into the nearest risk allele-count group of size >100 individuals. The approximate BMD difference in g/cm2 was obtained by multiplying in each group the mean Z-score LS-BMD by 0.18 g/cm2 and the mean Z-score FN-BMD by 0.13 g/cm2 (the SDs of BMD in the Rotterdam study). The allele score was obtained by dividing the number of “BMD decreasing alleles” by the total number of alleles. Also within the setting of the prospective population-based Rotterdam Study, we determined the risk for vertebral and non-vertebral fracture for the combined allelic scores constructed for all the top hits associated at a genome-wide significantly level with BMD. Risk ratio (RR) estimates were obtained from logistic regression (vertebral fractures) and Cox-proportional hazards (incident non-vertebral fractures) models adjusted for sex, age and weight. To determine the fraction of fracture risk explained by BMD, we applied the following formula: [lnRRunadjusted- lnRRBMDadjusted]/ lnRRunadjusted. Methods describing the fracture datasets have been published previously71,72. In summary, thoracolumbar radiographs of the spine were obtained in 3308 (genotyped) individuals who survived on average 6.4 ± 0.4 (SD) years and were scored for presence of vertebral fractures (n=329) using the McCloskey/Kanis method73. Record of the incident non-vertebral fractures (n=900) occurring between the baseline visit from 1990 through 1993 until January 1, 2002, was obtained from the computerized records of general practitioners and hospital registries for 5974 genotyped individuals followed on average 8.2 ± 2.7 (SD) years after the baseline visit.
Supplementary Material
ACKNOWLEDGEMENTS
Above all, we thank all study participants for making this work possible. This research and the GEnetic Factors of Osteoporosis (GEFOS) consortium (http://www.gefos.org) have been funded by the European Commission (HEALTH-F2-2008-201865-GEFOS). Rotterdam Study (RS): This study was funded by the Netherlands Organization of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012), the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) project nr. 050-060-810. We thank Pascal Arp, Mila Jhamai, Dr Michael Moorhouse, Marijn Verkerk, and Sander Bervoets for their help in creating the GWAS database. The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, Netherlands Organization for the Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The authors are very grateful to the staff from the Rotterdam Study, particularly Lydia Buist and J. Hannie van den Boogert and also with the participating general practitioners and pharmacists. Erasmus Rucphen Family (ERF): The study was supported by grants from The Netherlands Organization for Scientific Research (NWO), Erasmus MC and the Centre for Medical Systems Biology (CMSB). We are grateful to all general practitioners for their contributions, to Petra Veraart for her help in genealogy, Jeannette Vergeer for the supervision of the laboratory work and Peter Snijders for his help in data collection. Twins UK (TUK): The study was funded by the Wellcome Trust; the Arthritis Research Campaign; the Chronic Disease Research Foundation; the Canadian Institutes of Health Research (JBR); European Society for Clinical and Economic Aspects of Osteoporosis (JBR); the European Union FP-5 GenomEUtwin Project (QLG2-CT-2002-01254). The study also receives support from the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy's & St Thomas' NHS Foundation Trust in partnership with King's College London. We thank the staff from the TwinsUK, the DNA Collections and Genotyping Facilities at the Wellcome Trust Sanger Institute for sample preparation; Quality Control of the Twins UK cohort for genotyping (in particular Amy Chaney, Radhi Ravindrarajah, Douglas Simpkin, Cliff Hinds, and Thomas Dibling); Paul Martin and Simon Potter of the DNA and Genotyping Informatics teams for data handling; Le Centre National de Génotypage, France, led by Mark Lathrop, for genotyping; Duke University, North Carolina, USA, led by David Goldstein, for genotyping; and the Finnish Institute of Molecular Medicine, Finnish Genome Center, University of Helsinki, led by Aarno Palotie Icelandic deCODE Study (dCG): We thank the staff of the deCODE core facilities and recruitment centre for their important contributions to this work. Framingham Osteoporosis Study (FOS): The study was funded by grants from the National Institute for Arthritis, Musculoskeletal and Skin Diseases and the National Institute on Aging (R01 AR/AG 41398; DPK and R01 AR 050066; DK). The Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine were supported by the National Heart, Lung and Blood Institute's Framingham Heart Study (Contract No. N01-HC-25195) and its contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278). Analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. A portion of this research was conducted using the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. eQTL HOb Study: The study was supported by Genome Quebec, Genome Canada, and the Canadian Institutes of Health Research (CIHR). T.P. holds a Canada Research Chair. We thank Profs Olof Nilsson, Hans Mallmin and Östen Ljunggren at the Departments of Surgical and Medical Sciences, Uppsala University Hospital, Sweden for large-scale collection of primary bone samples.
Footnotes
AUTHOR CONTRIBUTIONS FR, KE, BVH, FKK, JPAI, ran meta-analysis; FR, KE, BVH, YHH, JBR, MCZ, NA, YAA, LAC, SD, NS, GT, YZ, ran statistical analysis in studies; FR, US, PD, JBvM, UT, AGU, coordinated GWA genotyping of studies; EG, TP, did expression studies; FR, US, MCZ, AH, BO, HAPP, GS, GT, FMKW, SGW, CMvD, TS, DPK, AGU, coordinated/collected phenotype information of studies; US, LAC, AH, AK, DK, BO, HAPP, UT, CMvD, TS, DPK, KS, AGU, designed studies; FR, US, JBvM, TS, UT, SHR, JPAI, AGU, established consortium and UT, SHR, JPAI, AGU obtained funding; all authors interpreted results; all authors read critically the manuscript; FR, wrote manuscript draft.
Note: Methods and Supplementary information is available on the Nature Genetics website
REFERENCES
- 1.Ellies DL, et al. Bone density ligand, Sclerostin, directly interacts with LRP5 but not LRP5G171V to modulate Wnt activity. J Bone Miner Res. 2006;21:1738–49. doi: 10.1359/jbmr.060810. [DOI] [PubMed] [Google Scholar]
- 2.Theoleyre S, et al. The molecular triad Opg/RANK/RANKL: involvement in the orchestration of pathophysiological bone remodeling. Cytokine Growth Factor Rev. 2004;15:457–75. doi: 10.1016/j.cytogfr.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Ioannidis JP, et al. Meta-analysis of genome-wide scans provides evidence for sex- and site-specific regulation of bone mass. J Bone Miner Res. 2007;22:173–83. doi: 10.1359/jbmr.060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ioannidis JP. Genetic associations: false or true? Trends Mol Med. 2003;9:135–8. doi: 10.1016/s1471-4914(03)00030-3. [DOI] [PubMed] [Google Scholar]
- 6.Ioannidis JP, et al. Differential genetic effects of ESR1 gene polymorphisms on osteoporosis outcomes. Jama. 2004;292:2105–14. doi: 10.1001/jama.292.17.2105. [DOI] [PubMed] [Google Scholar]
- 7.Langdahl BL, et al. Large-scale analysis of association between polymorphisms in the transforming growth factor beta 1 gene (TGFB1) and osteoporosis: the GENOMOS study. Bone. 2008;42:969–81. doi: 10.1016/j.bone.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Ralston SH, et al. Large-scale evidence for the effect of the COLIA1 Sp1 polymorphism on osteoporosis outcomes: the GENOMOS study. PLoS Med. 2006;3:e90. doi: 10.1371/journal.pmed.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uitterlinden AG, et al. The association between common vitamin D receptor gene variations and osteoporosis: a participant-level meta-analysis. Ann Intern Med. 2006;145:255–64. doi: 10.7326/0003-4819-145-4-200608150-00005. [DOI] [PubMed] [Google Scholar]
- 10.van Meurs JB, et al. Large-scale analysis of association between LRP5 and LRP6 variants and osteoporosis. Jama. 2008;299:1277–90. doi: 10.1001/jama.299.11.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 12.Styrkarsdottir U, et al. New sequence variants associated with bone mineral density. Nat Genet. 2009;41:15–7. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- 13.Styrkarsdottir U, et al. Multiple genetic loci for bone mineral density and fractures. N Engl J Med. 2008;358:2355–65. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 14.Kanis JA, Delmas P, Burckhardt P, Cooper C, Torgerson D. Guidelines for diagnosis and management of osteoporosis. The European Foundation for Osteoporosis and Bone Disease. Osteoporos Int. 1997;7:390–406. doi: 10.1007/BF01623782. [DOI] [PubMed] [Google Scholar]
- 15.Blake GM, Knapp KM, Spector TD, Fogelman I. Predicting the risk of fracture at any site in the skeleton: are all bone mineral density measurement sites equally effective? Calcif Tissue Int. 2006;78:9–17. doi: 10.1007/s00223-005-0127-3. [DOI] [PubMed] [Google Scholar]
- 16.Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of osteoporosis. Endocr Rev. 2002;23:303–26. doi: 10.1210/edrv.23.3.0464. [DOI] [PubMed] [Google Scholar]
- 17.Weedon MN, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–83. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gudbjartsson DF, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–15. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 19.Visscher PM. Sizing up human height variation. Nat Genet. 2008;40:489–90. doi: 10.1038/ng0508-489. [DOI] [PubMed] [Google Scholar]
- 20.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 21.Abecasis GR, Willer C. Metal: Meta Analysis Helper. 1.0 14/4/2008 edn 2008. [Google Scholar]
- 22.Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–5. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 23.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards JB, et al. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet. 2008;371:1505–12. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banziger C, et al. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell. 2006;125:509–22. doi: 10.1016/j.cell.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 26.Matsuda A, et al. Large-scale identification and characterization of human genes that activate NF-kappaB and MAPK signaling pathways. Oncogene. 2003;22:3307–18. doi: 10.1038/sj.onc.1206406. [DOI] [PubMed] [Google Scholar]
- 27.Glass DA, 2nd, et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell. 2005;8:751–64. doi: 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 28.Potthoff MJ, et al. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J Clin Invest. 2007;117:2459–67. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho YS, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41:527–34. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 30.Smits P, et al. The Transcription Factors L-Sox5 and Sox6 Are Essential for Cartilage Formation. 2001;1:277–290. doi: 10.1016/s1534-5807(01)00003-x. [DOI] [PubMed] [Google Scholar]
- 31.Zhou G, et al. Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc Natl Acad Sci U S A. 2006;103:19004–9. doi: 10.1073/pnas.0605170103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reiner O, et al. The evolving doublecortin (DCX) superfamily. BMC Genomics. 2006;7:188. doi: 10.1186/1471-2164-7-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SH, et al. The forkhead transcription factor Foxc2 stimulates osteoblast differentiation. Biochem Biophys Res Commun. 2009 doi: 10.1016/j.bbrc.2009.06.071. [DOI] [PubMed] [Google Scholar]
- 34.Nifuji A, Miura N, Kato N, Kellermann O, Noda M. Bone morphogenetic protein regulation of forkhead/winged helix transcription factor Foxc2 (Mfh1) in a murine mesodermal cell line C1 and in skeletal precursor cells. J Bone Miner Res. 2001;16:1765–71. doi: 10.1359/jbmr.2001.16.10.1765. [DOI] [PubMed] [Google Scholar]
- 35.Winnier GE, Hargett L, Hogan BL. The winged helix transcription factor MFH1 is required for proliferation and patterning of paraxial mesoderm in the mouse embryo. Genes Dev. 1997;11:926–40. doi: 10.1101/gad.11.7.926. [DOI] [PubMed] [Google Scholar]
- 36.Stankiewicz P, et al. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am J Hum Genet. 2009;84:780–91. doi: 10.1016/j.ajhg.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang Y, et al. Disruption of transforming growth factor-beta signaling in ELF beta-spectrin-deficient mice. Science. 2003;299:574–7. doi: 10.1126/science.1075994. [DOI] [PubMed] [Google Scholar]
- 38.Alford AI, Hankenson KD. Matricellular proteins: Extracellular modulators of bone development, remodeling, and regeneration. Bone. 2006;38:749–57. doi: 10.1016/j.bone.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 39.Gowen LC, et al. Targeted disruption of the osteoblast/osteocyte factor 45 gene (OF45) results in increased bone formation and bone mass. J Biol Chem. 2003;278:1998–2007. doi: 10.1074/jbc.M203250200. [DOI] [PubMed] [Google Scholar]
- 40.Malaval L, et al. Bone sialoprotein plays a functional role in bone formation and osteoclastogenesis. J Exp Med. 2008;205:1145–53. doi: 10.1084/jem.20071294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshitake H, Rittling SR, Denhardt DT, Noda M. Osteopontin-deficient mice are resistant to ovariectomy-induced bone resorption. Proc Natl Acad Sci U S A. 1999;96:8156–60. doi: 10.1073/pnas.96.14.8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyers VE, Zayzafoon M, Douglas JT, McDonald JM. RhoA and cytoskeletal disruption mediate reduced osteoblastogenesis and enhanced adipogenesis of human mesenchymal stem cells in modeled microgravity. J Bone Miner Res. 2005;20:1858–66. doi: 10.1359/JBMR.050611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Yang L, Debidda M, Witte D, Zheng Y. Cdc42 GTPase-activating protein deficiency promotes genomic instability and premature aging-like phenotypes. Proc Natl Acad Sci U S A. 2007;104:1248–53. doi: 10.1073/pnas.0609149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–11. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schroeder TM, Westendorf JJ. Histone deacetylase inhibitors promote osteoblast maturation. J Bone Miner Res. 2005;20:2254–63. doi: 10.1359/JBMR.050813. [DOI] [PubMed] [Google Scholar]
- 46.Kang JS, Alliston T, Delston R, Derynck R. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. Embo J. 2005;24:2543–55. doi: 10.1038/sj.emboj.7600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ioannidis JP, Thomas G, Daly MJ. Validating, augmenting and refining genome-wide association signals. Nat Rev Genet. 2009;10:318–29. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Online Mendelian Inheritance in Man, OMIM (TM) McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University; Baltimore, MD: and National Center for Biotechnology Information, National Library of Medicine (Bethesda, MD), [Google Scholar]
- 49.MCKussic VA. Mendelian Inheritance in Man. A Catalog of Human Genes and Genetic Disorders. Johns Hopkins University Press; Baltimore: 1998. [Google Scholar]
- 50.Hofman A, et al. The Rotterdam Study: objectives and design update. Eur J Epidemiol. 2007;22:819–29. doi: 10.1007/s10654-007-9199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hofman A, Grobbee DE, de Jong PT, van den Ouweland FA. Determinants of disease and disability in the elderly: the Rotterdam Elderly Study. Eur J Epidemiol. 1991;7:403–22. doi: 10.1007/BF00145007. [DOI] [PubMed] [Google Scholar]
- 52.Aulchenko YS, et al. Linkage disequilibrium in young genetically isolated Dutch population. Eur J Hum Genet. 2004;12:527–34. doi: 10.1038/sj.ejhg.5201188. [DOI] [PubMed] [Google Scholar]
- 53.Arden NK, Baker J, Hogg C, Baan K, Spector TD. The heritability of bone mineral density, ultrasound of the calcaneus and hip axis length: a study of postmenopausal twins. J Bone Miner Res. 1996;11:530–4. doi: 10.1002/jbmr.5650110414. [DOI] [PubMed] [Google Scholar]
- 54.Dawber TR, Kannel WB, Lyell LP. An approach to longitudinal studies in a community: the Framingham Study. Ann N Y Acad Sci. 1963;107:539–56. doi: 10.1111/j.1749-6632.1963.tb13299.x. [DOI] [PubMed] [Google Scholar]
- 55.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol. 1979;110:281–90. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 56.Splansky GL, et al. The Third Generation Cohort of the National Heart, Lung, and Blood Institute's Framingham Heart Study: design, recruitment, and initial examination. Am J Epidemiol. 2007;165:1328–35. doi: 10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- 57.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 58.A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y, Abecasis GR. Mach 1.0: Rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet. 2006:2290. [Google Scholar]
- 60.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 61.Kutyavin IV, et al. A novel endonuclease IV post-PCR genotyping system. Nucleic Acids Res. 2006;34:e128. doi: 10.1093/nar/gkl679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Little J, et al. Strengthening the reporting of genetic association studies (STREGA): an extension of the STROBE statement. Eur J Epidemiol. 2009;24:37–55. doi: 10.1007/s10654-008-9302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 64.Therneau T. R package. version 1.1.0–19 edn 2008. Kinship: mixed effectss Cox models, sparce matrices, and modelling data from large pedigrees. [Google Scholar]
- 65.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–6. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 66.R Developmental Core Team . A language and environment for statistical computing. R Foundation for Statistical Computing; 2007. [Google Scholar]
- 67.de Bakker PI, et al. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–8. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.StataCorp . Stata Statistical Software. 10 edn. StataCorp LP; College Station, TX: 2007. Release. [Google Scholar]
- 69.Grundberg E, et al. Systematic assessment of the human osteoblast transcriptome in resting and induced primary cells. Physiol Genomics. 2008;33:301–11. doi: 10.1152/physiolgenomics.00028.2008. [DOI] [PubMed] [Google Scholar]
- 70.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rivadeneira F, et al. Estrogen receptor beta (ESR2) polymorphisms in interaction with estrogen receptor alpha (ESR1) and insulin-like growth factor I (IGF1) variants influence the risk of fracture in postmenopausal women. J Bone Miner Res. 2006;21:1443–56. doi: 10.1359/jbmr.060605. [DOI] [PubMed] [Google Scholar]
- 72.Schuit SC, et al. Fracture incidence and association with bone mineral density in elderly men and women: the Rotterdam Study. Bone. 2004;34:195–202. doi: 10.1016/j.bone.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 73.McCloskey EV, et al. The assessment of vertebral deformity: a method for use in population studies and clinical trials. Osteoporos Int. 1993;3:138–47. doi: 10.1007/BF01623275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.