Abstract
Human serum paraoxonase-1 (HuPON1) has the capacity to hydrolyze aryl esters, lactones, oxidized phospholipids, and organophosphorus (OP) compounds. HuPON1 and bacterially expressed chimeric recombinant PON1s (G2E6 and G3C9) differ by multiple amino acids, none of which are in the putative enzyme active site. To address the importance of these amino acid differences, the abilities of HuPON1, G2E6, G3C9, and several variants to hydrolyze phenyl acetate, paraoxon, and V-type OP nerve agents were examined. HuPON1 and G2E6 have a ten-fold greater catalytic efficiency toward phenyl acetate than G3C9. In contrast, bacterial PON1s are better able to promote hydrolysis of paraoxon, whereas HuPON1 is considerably better at catalyzing the hydrolysis of the nerve agents VX and VR. These studies demonstrate that mutations distant from the active site of PON1 have large and unpredictable effects on the substrate specificities and possibly the hydrolytic mechanisms of HuPON1, G2E6, and G3C9. The replacement of residue H115 in the putative active site with tryptophan (H115W) has highly disparate effects on HuPON1 and G2E6. In HuPON1, variant H115W loses the ability to hydrolyze VR but has improved activity toward paraoxon and VX. The H115W variant of G2E6 has similar paraoxonase activity to wild type G2E6, modest activity with phenyl acetate and VR, and increased VX hydrolysis. VR inhibits H115W HuPON1 competitively when paraoxon is the substrate and non-competitively when VX is the substrate. We have identified the first variant of HuPON1, H115W, that displays significantly enhanced catalytic activity against an authentic V-type nerve agent.
Organophosphorus (OP) nerve agents are among the most toxic chemical substances identified (1). These compounds exert toxicity by readily binding to acetylcholinesterase (AChE) at the active site serine and inhibiting the ability of AChE to terminate cholinergic nerve transmissions (2). Existing pharmacologic treatments available to counteract the immediate effects of OP nerve agent intoxication, such as atropine, oximes, and diazepam, enhance survival but do not prevent performance deficits, behavioral incapacitation, loss of consciousness, or possible permanent brain damage (3). Current research has focused on the development of human butyrylcholinesterase as a stoichiometric bioscavenger to remove OP compounds from circulation before they can reach their physiological target (4-6).
In an effort to identify a human protein that can catalyze the hydrolysis of OP nerve agents, we have focused our attention on human serum paraoxonase-1 (HuPON1) (7). HuPON1 is an HDL-associated enzyme that can catalyze the hydrolysis of a diverse group of substrates including aryl esters, lactones, oxidized phospholipids, and OP compounds (8-12). Although the catalytic activity of HuPON1 toward OP nerve agents is too low to afford significant in vivo protection over butyrylcholinesterase, protein engineering could be used to increase the rate of nerve agent hydrolysis by the enzyme. Based on theoretical calculations, the catalytic efficiency of HuPON1 must be equal to or greater than 105 M−1 s−1 and the KM must be less than 10 μM for the enzyme to act in vivo as a highly efficient catalytic bioscavenger of OP nerve agents (13).
Functional HuPON1 has been notoriously difficult to express and purify in large quantities, despite attempts made in a variety of expression systems. For example, functional HuPON1 has been successfully produced from E. coli, but at low yields [(14), and Magliery, T.J. et al., unpublished data]. Using directed evolution via gene shuffling of human, rabbit, mouse, and rat PON1 genes, Aharoni et al. (15) expressed functional chimeric recombinant PON1 enzymes (G2E6 and G3C9, among others) in high yield in bacteria, thereby creating an opportunity to utilize high-throughput screening approaches to isolate variants with altered enzymatic function. Successful bacterial expression of the G2E6 enzyme enabled large-scale production of functional PON1 that was used in crystallization studies. As predicted by homology with DFPase (12), the X-ray structure of the G2E6 crystal revealed a six-fold β-propeller protein centrally arranged around two calcium ions (16, 17). Based on this structure, Tawfik and colleagues proposed the H115-H134 dyad within the putative active site as the catalytic machinery of the enzyme (16, 18, 19). Other studies suggest that this mechanism may only be correct for describing lactone hydrolysis, which may be the native substrate of PON1 (12, 17-20).
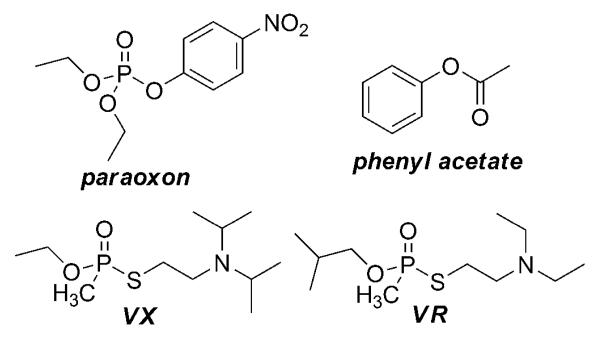
Because no crystal structures for HuPON1 or other variants have been reported, the structure of G2E6 has been used to approximate these enzymes (21). However, the amino acid composition of the PON1 clone G2E6 differs from that of the native HuPON1 in 59 amino acids, and from G3C9 in 20 amino acids; in each case the substitutions are distributed throughout the total 355 residues (Figure 1 and online supplemental Figure S1). It is important to consider what effects the amino acid differences between these enzymes may have on hydrolysis of the same substrates. The goal of this study was to examine the specificities of HuPON1, G2E6, G3C9, and several variants with respect to the hydrolysis of phenyl acetate and paraoxon, as well as of the nerve agents VX (ethyl-{[2-[di(propan-2-yl)-amino]ethylsulfanyl}-methylphosphinate) and VR (N,N-diethyl-2-(methyl-(2-methylpropoxy)phosphoryl)sulfanylethanamine; see Figure 2).
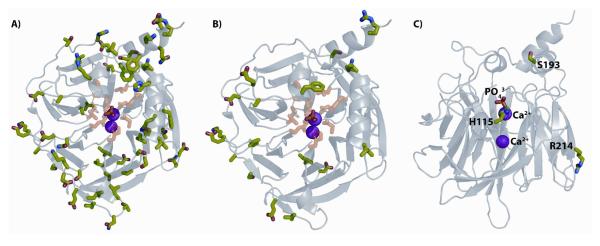
Figure 1. Structure of G2E6 PON1.
A) Residues that differ between human and G2E6 PON1 are rendered as sticks in green. The 59 amino acid differences are scattered throughout the sequence, mainly on the surface of the enzyme and not in the presumed active site. Residues with atoms within 5 Å of the phosphate ion in the crystal are rendered as sticks in pink. The purple spheres represent the calcium (Ca2+) ions. B) Residues that differ between G3C9 and G2E6 PON1 are rendered as sticks in green, where residues with atoms within 5 Å of the phosphate ion in the crystal are rendered as sticks in pink, as in 1A. C) Residue H115 is proximal to the ‘catalytic’ Ca2+ and phosphate ion found in the crystal. S193 lies at the top of the ‘lid’ that is proposed to be an HDL binding site. R214 is on the surface. Images are rendered with PyMOL (DeLano Scientific) from PDB entry 1V04.
Figure 2.
Structures of ester and OP substrates.
In this paper, we show that human and bacterially expressed forms of PON1 have very different enzymatic activities with respect to phenyl acetate, paraoxon, and V-type nerve agents. While G2E6 and G3C9 have improved hydrolysis of paraoxon compared to HuPON1, HuPON1 exhibits significantly better turnover of the nerve agents VX and VR. Of the five variants examined, only the H115W variant of either HuPON1 or G2E6 enhanced catalytic efficiency for the hydrolysis of paraoxon and VX.
EXPERIMENTAL PROCEDURES
Expression of HuPON1
cDNAs encoding wild type and the H115W, S193A, S193G, R214Q and S193A/R214Q variant HuPON1 proteins (with the Q192 allele and a C-terminal six histidine (6x-His) tag) were cloned into pcDNA3.1 and transiently transfected into human embryonic kidney 293T cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Media were harvested two days post-transfection, filtered and stored at 4 °C. Expression of HuPON1 and variants was confirmed by Western analysis using a mouse monoclonal anti-HuPON1 antibody kindly provided by Dr. Richard James (University Hospital of Geneva, Switzerland). HuPON1 protein concentrations were determined from quantitative Western blot analyses using densitometry (Un-Scan-It Version 5.1, Silk Scientific Corp., Orem, UT) with a HuPON1 standard of known concentration (Randox Life Sciences, Antrim, UK).
Expression and Purification of Bacterially Expressed PON1s
G3C9, G2E6 and the H115W, S193A, and R214Q variants of G2E6 were expressed and purified essentially as described (15, 16). G3C9 was expressed with a C-terminal 6x-His tag, whereas G2E6 and variants were expressed as thioredoxin fusion proteins with N-terminal 6x-His tags. The enzymes were expressed in Origami B (DE3) cells (Novagen, Madison, WI). When bacterial cultures reached an A600 of 0.8, they were induced with 0.1 mM IPTG for 3 h. Harvested cells were resuspended in lysis buffer (50 mM Tris-HCl, 50 mM NaCl, 1 mM CaCl2, 0.1 mM dithiothreitol, pH 8.0) and extruded through a syringe needle. After sonication, the lysate was incubated with 0.1% Tergitol NP-10 (Sigma-Aldrich, St. Louis, MO) with shaking at 4 °C for 150 min. Ni-NTA resin (Qiagen, Valencia, CA) was added to the lysate and shaken for 4 h. The resin was washed with activity buffer (50 mM Tris-HCl pH 8, 50 mM NaCl, 1 mM CaCl2, 0.1% Tergitol NP-10) including 10 mM imidazole followed by a wash with activity buffer supplemented with 25 mM imidazole. The fusion protein was eluted using activity buffer with 150 mM imidazole. After ten days at room temperature, there was no detectable spontaneous scission of G2E6 from the fused thioredoxin, in contrast to a previous report (16). Protein concentration was determined using the Bradford assay (Bio-Rad Laboratories, Hercules, CA).
Arylesterase and Paraoxonase Activity Assays
To test for arylesterase activity, enzymes were incubated in reaction buffer (50 mM Tris-HCl, 10 mM CaCl2, pH 7.4) with increasing concentrations of phenyl acetate (Sigma-Aldrich) from 0.29 mM to 3.3 mM in a quartz cuvette. The rate of formation of phenol was measured at A270(ε=1,310 M−1 cm−1) using a UV/Vis SpectraMax Plus 384 spectrophotometer (Molecular Devices, Sunnyvale, CA) at room temperature for 1 min. Paraoxonase activity was measured using 60 to 750 μM paraoxon (Sigma-Aldrich) in reaction buffer. The formation of p-nitrophenol was followed at A412 (ε=17,000 M−1 cm−1) with a SpectraMax Plus 384 spectrophotometer for 20 min at room temperature in a 96-well microplate. Phenyl acetate and paraoxon were prepared as high concentration stocks in MeOH and diluted into reaction buffer on day of assay. Final MeOH concentration in assays was < 1%. Kinetic parameters (KM and kcat) with phenyl acetate and paraoxon were determined by Michaelis-Menten steady state kinetics. The data from 4 or more independent experiments were fit to the model using Prism 4.03 software (GraphPad, La Jolla, CA). R2 values for the non-linear regression were greater than 0.99.
V-Agent Hydrolysis Assays
VX and VR were obtained from the U.S. Army Edgewood Chemical Biological Center (ECBC, Aberdeen Proving Ground, MD) and diluted into reaction buffer. Using a modified Ellman-based colorimetric assay, enzyme samples were incubated with 0.75 mM 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB; Sigma-Aldrich) and a range of V-agent concentrations from 90 μM to 1.4 mM in a 96-well microplate. Turnover was followed at A412 (ε=13,600 M−1 cm−1) for 4 h at room temperature. Kinetic parameters (KM and kcat) with VX and VR were determined as described above.
VR Inhibition Assays
For H115W HuPON1 enzyme, assays with paraoxon (16 μM to 2 mM) plus a fixed concentration of VR (0, 62.5, 125, 250, or 300 μM) were conducted in a 96-well microplate. Hydrolysis of paraoxon was followed at A412 for 20 min at room temperature as described above. The data were fit using Michaelis-Menten steady state kinetics to derive the KM and Vmax of the enzyme at each concentration of VR. Based on inspection of the preliminary results, VR concentrations were plotted versus apparent KM values, and the linear plot was used to determine the competitive inhibitor constant. Separate assays with VX (75 μM to 1.2 mM) plus a fixed VR concentration (0, 29, 60, or 110 μM) were also performed with 0.75 mM DTNB in a 96-well microplate. VX hydrolysis was measured at A412 for 4 h at room temperature as described above. The data were fit using Michaelis-Menten steady state kinetics to derive the KM and Vmax of the enzyme at each VR concentration. Based on initial inspection of the results, VR concentrations were plotted versus Vmax values, and the linear plot was used to determine the non-competitive inhibitor constant.
RESULTS
Differential substrate specificities of HuPON1 versus G2E6 and G3C9 PON1s
The 293T-expressed HuPON1 and bacterially expressed chimeric recombinant PON1 G2E6 exhibited similar capacities to catalyze hydrolysis of phenyl acetate (with catalytic efficiencies of 1.5 × 106 and 1.4 × 106 M−1 s−1, respectively), while G3C9 was approximately ten-fold less efficient (0.15 × 106 M−1 s−1; see Table 1). G2E6 PON1 had a 13- and a six-fold greater catalytic efficiency for hydrolysis of paraoxon (the toxic metabolite of the pesticide parathion) versus HuPON1 and G3C9 respectively. This result suggested that bacterially expressed chimeric PON1s might be more efficient enzymes for catalyzing the hydrolysis of OP nerve agents (Table 1). These observations were unexpected since HuPON1 and the bacterial expressed PON1s do not differ in any positions in the presumed active site of the enzyme (Figures 1 and S1).
Table 1.
Kinetic constants of HuPON1, G2E6, G3C9, and variants with phenyl acetate and paraoxon
| Phenyl Acetate | Paraoxon | |||
|---|---|---|---|---|
| Enzyme | kcat (s−1) | KM (mM) | kcat (s−1) | KM (mM) |
| HuPON1 | 970 ± 70 | 0.66 ± 0.09 | 0.4 ± 0.07 | 0.57 ± 0.03 |
| H115W | --- | --- | 4.8 ± 0.9 | 0.19 ± 0.006 |
| S193A | 580 ± 120 | 0.50 ± 0.07 | 0.4 ± 0.07 | 1.00 ± 0.2 |
| S193G | 630 ± 80 | 0.23 ± 0.05 | 0.4 ± 0.05 | 0.60 ± 0.03 |
| R214Q | 600 ± 50 | 0.48 ± 0.06 | 0.3 ± 0.03 | 0.70 ± 0.2 |
| S193A/R214Q | 1130 ± 120 | 0.40 ± 0.1 | 0.5 ± 0.08 | 0.70 ± 0.3 |
| G2E6 | 1150 ± 70 | 0.80 ± 0.1 | 5.7 ± 0.2 | 0.60 ± 0.07 |
| H115W | 16 ± 1 | 1.70 ± 0.3 | 5.7 ± 0.1 | 0.30 ± 0.03 |
| S193A | 1000 ± 50 | 1.10 ± 0.2 | 4.0 ± 0.07 | 0.49 ± 0.03 |
| R214Q | 600 ± 33 | 0.44 ± 0.07 | 3.7 ± 0.08 | 0.55 ± 0.04 |
| G3C9 | 350 ± 120 | 2.30 ± 0.4 | 0.9 ± 0.07 | 0.60 ± 0.2 |
We also examined the activity of the enzymes with the nerve agent VX. Concentration limitations established for research with surety materials at USAMRICD prevented substrate saturation studies with VX at concentrations higher than 1.4 mM, and none of the enzymes analyzed reached saturation under these conditions. Since saturation kinetics could not be studied with VX, the assumption was made that binding affinity (KM) is much greater than the VX substrate concentration ([S]), and catalytic efficiency (kcat/KM) was estimated from the slope of the [S] versus rate plot (Table 2). Wild type HuPON1 hydrolyzed VX more effectively than did G2E6 or G3C9 PON1 with apparent kcat/KM values of 550 M−1 s−1 versus 22 M−1 s−1 or 10 M−1 s−1, respectively (Table 2).
Table 2.
Kinetic constants of HuPON1, G2E6, G3C9, and variants with VX and VR
| VX | VR | |||
|---|---|---|---|---|
| Enzyme | kcat/KM (M−1 s−1)* | kcat (s−1) | KM (mM) | kcat/KM (M−1 s−1) |
| HuPON1 | 550 ± 130 | 0.1 ± 0.04 | 0.20 ± 0.04 | 700 ± 180 |
| H115W | 2170 ± 320 | ---- | ---- | --- |
| S193A | 300 ± 30 | 0.09 ± 0.02 | 0.20 ± 0.04 | 470 ± 30 |
| S193G | 450 ± 40 | 0.1 ± 0.04 | 0.20 ± 0.01 | 520 ± 80 |
| R214Q | 280 ± 70 | 0.06 ± 0.01 | 0.20 ± 0.07 | 300 ± 70 |
| S193A/R214Q | 280 ± 20 | 0.08 ± 0.008 | 0.30 ± 0.07 | 280 ± 20 |
| G2E6 | 22 ± 0.3 | ND | ND | 18* |
| H115W | 85 ± 0.5 | ND | ND | 8.3* |
| S193A | 28 ± 0.7 | ND | ND | 23* |
| R214Q | 18 ± 0.03 | ND | ND | 13* |
| G3C9 | 10 ± 0.7 | ND | ND | 20 ± 2* |
Values are shown ± standard deviation. Where no deviation is indicated, the error was <1% of the value.
At the maximum regulated concentration of nerve agent, no evidence of saturation was detected indicating that KM>>[S] (1.4 mM for VX and VR). The Michaelis-Menten equation was thus reduced to v0 = kcat[E][S]/KM and used to calculate the values shown. ND = Not Determined
The enzymes were also tested for their capacities to hydrolyze VR, a structural isomer of VX. In this case, the affinity for VR was high enough that saturation could be achieved for HuPON1. Wild type HuPON1 catalyzed the hydrolysis of VR with a kcat/KM of 700 M−1 s−1 similar to what was observed with VX (Table 2). At 1.4 mM VR, saturation was not reached with G2E6 or G3C9 PON1, so apparent catalytic efficiencies were derived (vide supra). Both enzymes had a much lower capacity to hydrolyze VR than did HuPON1, with a ~35-fold decrease in kcat/KM. Therefore, the greater capacity of G2E6 and G3C9 PON1 to catalyze the hydrolysis of paraoxon did not correspond to a greater ability of the enzymes to hydrolyze the OP nerve agents VX and VR.
Differential effects of H115W substitution on substrate specificity
Residue H115 is located within the proposed active site of PON1 (Figure 1C). As previously reported (12), there was no detectable hydrolysis of phenyl acetate by the H115W active site variant of HuPON1. In contrast, the H115W substitution in G2E6 PON1 did retain activity against phenyl acetate, albeit with a marked decrease in kcat and a two-fold increase in KM, resulting in a 153-fold decrease in kcat/KM when compared to wild type G2E6 PON1 (Table 1). When the paraoxonase activity of the H115W variant of either G2E6 PON1 or HuPON1 was compared to the respective parent enzymes, kcat/KM improved only two-fold for H115W G2E6 PON1, whereas the same substitution on HuPON1 increased kcat/KM by 35-fold (Table 1). Interestingly, this improvement in catalytic efficiency was due primarily to an increase in kcat for H115W HuPON1 and a decrease in KM for H115W G2E6 PON1.
Since the H115W variants of both HuPON1 and G2E6 PON1 provided vastly different improvements in the turnover of paraoxon, we investigated whether these increases in activity would translate to an enhancement in hydrolysis of OP nerve agents. When the H115W substitution was examined in HuPON1, we observed a four-fold increase in apparent kcat/KM with VX when compared to the parent HuPON1 enzyme. Under these conditions it was not possible to determine if the improvement in turnover of H115W HuPON1 was due to an increase in kcat or a decrease in KM toward VX since saturation could not be achieved. A similar result was observed when the H115W variant was examined in G2E6 PON1 (Table 2). H115W G2E6 PON1 was able to catalyze the hydrolysis of VR, but less efficiently than did wild type G2E6 PON1. Surprisingly, H115W HuPON1 was unable to hydrolyze VR (Table 2).
The mechanism of inhibition of H115W HuPON1 by VR switches with different substrates
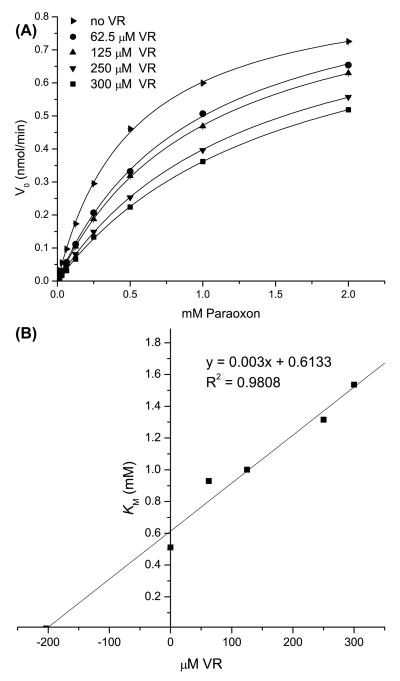
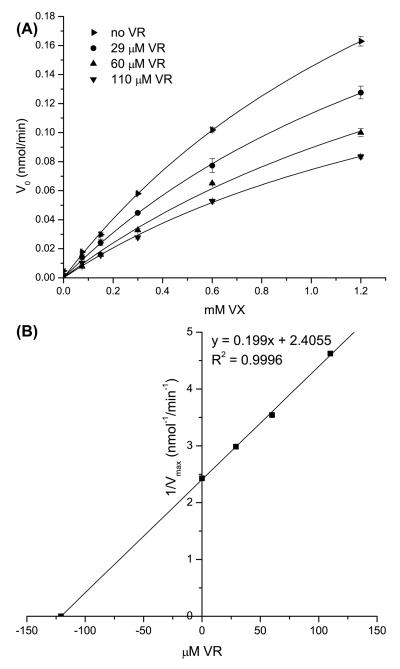
The absence of VR hydrolysis by H115W HuPON1 may result from the enzyme failing to bind the substrate or binding in a way that negatively affects substrate catalysis. To test these possibilities, we examined the capacity of H115W HuPON1 to hydrolyze increasing concentrations of paraoxon in the presence of fixed VR concentrations. The addition of VR reduced the rate of paraoxon hydrolysis (Figure 3A). The apparent KM increased with higher VR concentrations without producing any change in Vmax (data not shown). Based on these data, a competitive inhibition constant (Ki) of 204 μM was determined for the inhibition of paraoxon by VR (Figure 3B). The ability of VR to inhibit VX hydrolysis by H115W HuPON1 was also analyzed. In the presence of increasing concentrations of VR, the rate of VX hydrolysis by H115W was reduced (Figure 4A) as was observed when paraoxon hydrolysis was examined. However, Vmax decreased with higher VR concentrations without producing any change in KM (data not shown), indicating that VR acted as a non-competitive inhibitor of VX with a Ki of 121 μM (Figure 4B).
Figure 3. Inhibition of paraoxon hydrolysis by VR in H115W HuPON1.
A) Reaction velocity of paraoxon hydrolysis versus concentration of paraoxon (16 μM to 2 mM) in the presence of VR at 0 (▶), 62.5 (●), 125 (▲), 250 (▼), or 300 (■) μM. B) Plot of apparent KM values derived from A) versus concentration of VR. The x-intercept of this graph corresponds to (−Ki), calculated to be 204 μM.
Figure 4. Inhibition of VX hydrolysis by VR in H115W HuPON1.
A) Reaction velocity of VX hydrolysis versus concentration of VX (75 μM to 1.2 mM) in the presence of VR at 0 (▶), 29 (●), 60(▲), and 110 (▼) μM. B) Plot of 1/Vmax values derived from A) versus concentration of VR. The x-intercept of this graph corresponds to (−Ki), calculated to be 121 μM.
Mutations at two peripheral sites have little effect on specificity in either HuPON1 or G2E6
HuPON1 variants at sites on the periphery of the proposed active site of G2E6 PON1, (S193A/G), distal to the active site of G2E6 PON1 (R214Q), or a combination of both (S193A/R214; Figure 1C) were examined for their abilities to hydrolyze phenyl acetate, paraoxon, VX, and VR. Substitution at these sites had previously been found to slightly influence the capacity of HuPON1 to hydrolyze the OP nerve agent soman (GD; 2-(fluoromethyl-phosphoryl)oxy-3,3-dimethyl-butane) (22). No major differences in KM or kcat among these variants were observed when compared to wild type HuPON1. Variants S193A and R214Q were also studied in G2E6 PON1. As seen with HuPON1, no distinctions in KM or kcat were detected with these variants (Tables 1 and 2).
DISCUSSION
The ability of plasma-derived HuPON1 to catalyze the hydrolysis of the chemical warfare agents sarin (GB; 2-(fluoro-methylphosphoryl)oxypropane) and soman (8, 23) suggests that this enzyme could be a promising scaffold for the development of a catalytic protein capable of providing in vivo protection against nerve agent exposure. Given the slow rate at which wild type HuPON1 hydrolyzes OP nerve agents, our research efforts were directed at identifying and substituting amino acid residues at and around the proposed active site of the enzyme to generate variants with enhanced catalytic activity (12, 20). Those efforts were hampered by the lack of a three-dimensional structure of HuPON1. The subsequent work of Harel et al. (16, 17), which provided crystals of a chimeric recombinant gene-shuffled mammalian form of PON1 (G2E6), has provided useful insights into the structure of HuPON1. The ability to express PON1 enzymes from a bacterial source has made it easier to rationally design PON1 mutants for testing with a variety of substrates (15). A concern associated with these advances is whether these recombinant PON1 proteins enzymatically perform like recombinant HuPON1 with the same substrates. There has been little effort to compare the activities of the two forms of PON1 with a wide variety of chemically related substrates. Despite presumably identical active sites, we report here substantial differences in the substrate specificities of HuPON1 versus bacterially expressed G2E6 and G3C9 PON1.
Substrate specificities of HuPON1 and bacterially expressed PON1s
HuPON1 and G2E6 had virtually identical catalytic efficiencies with phenyl acetate, while G3C9 was substantially less active against this substrate. The G2E6 PON1 variant is able to catalyze the hydrolysis of paraoxon about 13- and six-fold faster than HuPON1 and G3C9, respectively, despite having similar KM values. While these values are in close agreement with previously reported values for HuPON1 and G2E6, they differ from those reported for G3C9 (15); the basis for these differences are unclear. Although these results suggest the possibility of separate mechanisms of hydrolysis for phosphotriesters and aryl esters by PON1, a more likely explanation is that the requirements for docking of these two classes of substrates may differ for each enzyme.
When V-type nerve agents were presented as substrates for HuPON1 or the bacterial expressed PON1 enzymes, additional discrepancies became apparent. HuPON1 hydrolyzes VR about 35-fold more efficiently than does either G2E6 or G3C9 PON1. The human form also hydrolyzes VX about 25- and 55-fold more efficiently than does either G2E6 or G3C9 PON1, respectively. While we were unable to derive separate KM and kcat values for VX hydrolysis, we can estimate that the KM values for both enzymes were higher than 1.4 mM, which was the maximum VX concentration that was tested (due to safety regulations). From these data, we can approximate that the kcat value for wild type HuPON1 with VX is at least 0.8 s−1. Therefore, we conclude that wild type HuPON1 binds VX with lower affinity than it does VR, but once bound, VX is more quickly hydrolyzed than VR. VX and VR are structural isomers, with VX having a more bulky thiolate group and VR having a larger alkoxy group on the phosphorus atom (Figure 2). Significant differences in binding and turnover of these substrates with HuPON1 result from these fairly subtle changes.
Although G2E6 and G3C9 PON1 had greater capacities to hydrolyze paraoxon when compared to HuPON1, this improvement was lost when the bacterially expressed enzymes were tested for activity against the OP nerve agents VX and VR. Hence, we offer a word of caution about extrapolating changes in activities based on paraoxon as a substrate to activity of HuPON1 or bacterially expressed chimeric PON1s against other OP substrates (to include chemical warfare nerve agents).
Divergent effects of the H115W substitution
The H115W variant of HuPON1 was unable to hydrolyze phenyl acetate, as observed previously (12). Surprisingly, the H115W G2E6 PON1 variant retained activity with this substrate, although both KM and kcat for phenyl acetate were decreased relative to G2E6, resulting in a much lower kcat/KM. The H115W variant of G2E6 PON1 was two-fold more active against paraoxon than was the parent molecule; when H115W HuPON1 was studied using paraoxon as the substrate, the kcat/KM was improved 35-fold over wild type HuPON1. While the reason for this effect is not understood, it suggests that the differences in total amino acid composition between G2E6 PON1 and HuPON1, even with most of those differences distal from the active site, can alter the catalytic activity. The H115W substitution provided a four-fold increase in catalytic efficiency of HuPON1 with VX. But surprisingly, the same enzyme displayed no detectable turnover of VR. A similar trend was observed for H115W G2E6 PON1, although this variant retained some activity against VR.
Modes of inhibition for VR with H115W HuPON1
Given that the H115W HuPON1 variant did not catalyze the hydrolysis of VR, we expanded our studies to examine the effect of the presence of VR on either paraoxon or VX hydrolysis. Based on the structural differences between VR and VX (Figure 2), we felt that VR might be a useful probe to examine substrate orientation in the HuPON1 active site, particularly in light of the current results and previous reports that H115 is involved in substrate binding, but is not a part of the catalytic machinery (20, 22). When paraoxon was the substrate for H115W HuPON1, VR displayed classic competitive inhibition with a Ki of 204 μM. The capacity of VR to competitively inhibit hydrolysis of paraoxon suggests that paraoxon and VR bind the same locus in the active site of H115W HuPON1, where paraoxon can be hydrolyzed, but VR is in an orientation that precludes access of the active site machinery to the VR P-S bond. In contrast, when VX was the substrate, VR inhibition was non-competitive, with a Ki of 121 μM. The non-competitive inhibition of VX hydrolysis by VR could be explained if VX binds to H115W in both a non-productive orientation (or site) and in a separate orientation that results in VX hydrolysis. In such a model, the presence of VR blocks binding of VX in a productive orientation, but does not prevent binding of VX in a non-productive orientation. Such non-productive binding sites for VX in H115W HuPON1 have not been experimentally determined, but in silico studies have suggested that different classes of PON1 substrates interact with unique subsets of amino acid residues in the active site (21), and that VX may bind to PON1 in a variety of energetically favorable orientations, some of which are distal from the catalytic calcium (data not shown).
Other substitutions
Variants at sites on the periphery of the proposed active site of G2E6 PON1 (S193A/G), distal from the active site of G2E6 PON1 (R214Q), or a combination of both (S193A/R214) were engineered in HuPON1 and examined for activity against phenyl acetate, paraoxon, VX, and VR. No significant changes were observed in either KM or kcat values when compared to wild type HuPON1 for any of these variants. Similar results were obtained when S193A and R214Q were examined on the G2E6 PON1 scaffold, suggesting that the substitutions in either HuPON1 or G2E6 PON1 at S193 or R214 do not affect hydrolysis of the substrates examined here. These results are interesting when compared with those of Davies et al. (23), who demonstrated that alterations at residue 192 of HuPON1 can alter activity with other OP compounds.
This study clearly demonstrates that both HuPON1 and the bacterially expressed PON1s are capable of catalyzing the hydrolysis of VX and VR, but that HuPON1 does so with substantially greater catalytic efficiency than do G2E6 or G3C9 PON1. The variation in enzymatic activity may be due to the >50 amino acid differences between HuPON1 and the bacterially expressed PON1s, the lack of glycosylation of bacterially expressed PON1, or a combination of these factors. The H115W substitution on either HuPON1 or G2E6 PON1 results in a four-fold improvement in catalytic efficiency with the nerve agent VX. This is the first variant of PON1 identified that improves hydrolysis of a bona fide nerve agent.
Supplementary Material
Abbreviations
- HuPON1
human serum paraoxonase-1
- OP
organophosphorus
- AChE
acetylcholinesterase
- DTNB
5,5′-dithiobis(2-nitrobenzoic acid).
Footnotes
The views of the authors do not purport to reflect the position or policies of the U.S. Army or the Department of Defense.
This work was supported by the NIH CounterACT Center of Excellence grant U54 NS058183 (to DEL (Center PI), DMC, and TJM) and by the Defense Threat Reduction Agency – Joint Science and Technology Office, Medical S & T Division (to DEL).
SUPPORTING INFORMATION
Additional supplemental material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Dacre JC. Toxicology of some anticholinesterases used as a chemical warfare agents - a review. In: Brzin M, Barnard EA, Sket D, editors. Cholinesterases, Fundamental and Applied Aspects. de Gruyter; Berlin, Germany: 1984. pp. 415–426. [Google Scholar]
- 2.Taylor P. Anticholinesterase agents. In: Hardman JG, Limbird LE, Gilman AG, editors. Goodman & Gillman's The Pharmacological Basis of Therapeutics. McGraw-Hill; New York: 2001. pp. 175–192. [Google Scholar]
- 3.Maynard RL, Beswick FW. Organophosphorus compounds as chemical warfare agents. In: Ballantyne B, Marrs TC, editors. Clinical and Experimental Toxicology of Organophosphates and Carbamates. Butterworth; Oxford, London: 1992. pp. 373–385. [Google Scholar]
- 4.Cerasoli DM, Griffiths EM, Doctor BP, Saxena A, Fedorko JM, Greig NH, Yu QS, Huang Y, Wilgus H, Karatzas CN, Koplovitz I, Lenz DE. In vitro and in vivo characterization of recombinant human butyrylcholinesterase (Protexia) as a potential nerve agent bioscavenger. Chem Biol Interact. 2005;157-158:363–365. doi: 10.1016/j.cbi.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 5.Huang YJ, Huang Y, Baldassarre H, Wang B, Lazaris A, Leduc M, Bilodeau AS, Bellemare A, Cote M, Herskovits P, Touati M, Turcotte C, Valeanu L, Lemee N, Wilgus H, Begin I, Bhatia B, Rao K, Neveu N, Brochu E, Pierson J, Hockley DK, Cerasoli DM, Lenz DE, Karatzas CN, Langermann S. Recombinant human butyrylcholinesterase from milk of transgenic animals to protect against organophosphate poisoning. Proc Natl Acad Sci USA. 2007;104:13603–13608. doi: 10.1073/pnas.0702756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lenz DE, Maxwell DM, Koplovitz I, Clark CR, Capacio BR, Cerasoli DM, Federko JM, Luo C, Saxena A, Doctor BP, Olson C. Protection against soman or VX poisoning by human butyrylcholinesterase in guinea pigs and cynomolgus monkeys. Chem Biol Interact. 2005;157-158:205–210. doi: 10.1016/j.cbi.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 7.Lenz DE, Broomfield CA, Yeung DT, Masson P, Maxwell DM, Cerasoli DM. Nerve agent bioscavengers: progress in development of a new mode of protection against organophosphorus exposure. In: Romano JA, Lukey B, Salem H, editors. Chemical Warfare Agents: Toxicity at Low Levels. CRC Press; Boca Raton: 2007. pp. 175–202. [Google Scholar]
- 8.Yeung DT, Smith JR, Sweeney RE, Lenz DE, Cerasoli DM. Direct detection of stereospecific soman hydrolysis by wild-type human serum paraoxonase. FEBS J. 2007;274:1183–1191. doi: 10.1111/j.1742-4658.2006.05650.x. [DOI] [PubMed] [Google Scholar]
- 9.Billecke S, Draganov D, Counsell R, Stetson P, Watson C, Hsu C, La Du BN. Human serum paraoxonase (PON1) isozymes Q and R hydrolyze lactones and cyclic carbonate esters. Drug Metab Dispos. 2000;28:1335–1342. [PubMed] [Google Scholar]
- 10.Gaidukov L, Rosenblat M, Aviram M, Tawfik DS. The 192R/Q polymorphs of serum paraoxonase PON1 differ in HDL binding, stimulation of lipolactonase, and macrophage cholesterol efflux. J Lipid Res. 2006;47:2492–2502. doi: 10.1194/jlr.M600297-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Rodrigo L, Mackness B, Durrington PN, Hernandez A, Mackness MI. Hydrolysis of platelet-activating factor by human serum paraoxonase. Biochem J. 2001;354:1–7. doi: 10.1042/0264-6021:3540001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeung DT, Josse D, Nicholson JD, Khanal A, McAndrew CW, Bahnson BJ, Lenz DE, Cerasoli DM. Structure/function analyses of human serum paraoxonase (HuPON1) mutants designed from a DFPase-like homology model. Biochim Biophys Acta. 2004;1702:67–77. doi: 10.1016/j.bbapap.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Masson P, Nachon F, Broomfield CA, Lenz DE, Verdier L, Schopfer LM, Lockridge O. A collaborative endeavor to design cholinesterase-based catalytic scavengers against toxic organophosphorus esters. Chem Biol Interact. 2008;175:273–280. doi: 10.1016/j.cbi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Stevens RC, Suzuki SM, Cole TB, Park SS, Richter RJ, Furlong CE. Engineered recombinant human paraoxonase 1 (rHuPON1) purified from Escherichia coli protects against organophosphate poisoning. Proc Natl Acad Sci USA. 2008;105:12780–12784. doi: 10.1073/pnas.0805865105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aharoni A, Gaidukov L, Yagur S, Toker L, Silman I, Tawfik DS. Directed evolution of mammalian paraoxonases PON1 and PON3 for bacterial expression and catalytic specialization. Proc Natl Acad Sci USA. 2004;101:482–487. doi: 10.1073/pnas.2536901100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harel M, Aharoni A, Gaidukov L, Brumshtein B, Khersonsky O, Meged R, Dvir H, Ravelli RB, McCarthy A, Toker L, Silman I, Sussman JL, Tawfik DS. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat Struct Mol Biol. 2004;11:412–419. doi: 10.1038/nsmb767. [DOI] [PubMed] [Google Scholar]
- 17.Harel M, Aharoni A, Gaidukov L, Brumshtein B, Khersonsky O, Meged R, Dvir H, Ravelli RB, McCarthy A, Toker L, Silman I, Sussman JL, Tawfik DS. Corrigendum: Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat Struct Mol Biol. 2004;11:1253. doi: 10.1038/nsmb767. [DOI] [PubMed] [Google Scholar]
- 18.Khersonsky O, Tawfik DS. The histidine 115-histidine 134 dyad mediates the lactonase activity of mammalian serum paraoxonases. J Biol Chem. 2006;281:7649–7656. doi: 10.1074/jbc.M512594200. [DOI] [PubMed] [Google Scholar]
- 19.Rosenblat M, Gaidukov L, Khersonsky O, Vaya J, Oren R, Tawfik DS, Aviram M. The catalytic histidine dyad of high density lipoprotein-associated serum paraoxonase-1 (PON1) is essential for PON1-mediated inhibition of low density lipoprotein oxidation and stimulation of macrophage cholesterol efflux. J Biol Chem. 2006;281:7657–7665. doi: 10.1074/jbc.M512595200. [DOI] [PubMed] [Google Scholar]
- 20.Yeung DT, Lenz DE, Cerasoli DM. Analysis of active-site amino-acid residues of human serum paraoxonase using competitive substrates. FEBS J. 2005;272:2225–2230. doi: 10.1111/j.1742-4658.2005.04646.x. [DOI] [PubMed] [Google Scholar]
- 21.Hu X, Jiang X, Lenz DE, Cerasoli DM, Wallqvist A. In silico analyses of substrate interactions with human serum paraoxonase 1. Proteins. 2009;75:486–498. doi: 10.1002/prot.22264. [DOI] [PubMed] [Google Scholar]
- 22.Yeung DT, Lenz DE, Cerasoli DM. Human paraoxonase 1: A potential bioscavenger of organophosphorus nerve agents. In: Mackness B, Mackness M, Aviram M, Paragh G, editors. The Paraoxonases: Their Role in Disease Development and Xenobiotic Metabolism. Springer; Dordrecht: 2008. pp. 151–170. [Google Scholar]
- 23.Davies HG, Richter RJ, Keifer M, Broomfield CA, Sowalla J, Furlong CE. The effect of the human serum paraoxonase polymorphism is reversed with diazoxon, soman, and sarin. Nat Genet. 1996;14:334–336. doi: 10.1038/ng1196-334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.