Abstract
Blood is a tissue with a high cell turnover rate that is constantly being replenished by bone marrow hematopoietic stem cells (HSCs) seeded during fetal ontogeny from the liver. Here we show that the long-term (LT) reconstituting subset of cKit+Thy1.1(lo)Lin(-/lo)Sca1+Flk2- hematopoietic stem cells is CD150+. HSCs sourced from the fetal liver show long-term, multilineage engraftment from E14.5 onwards, and the CD150 cell surface molecule can readily substitute Thy1.1 as a positive marker of LT-HSCs in this tissue. From both fetal liver and adult bone marrow, cKit+Thy1.1(lo)Lin(-/lo)Sca1+Flk2-CD150+ cells exhibit robust LT competitive engraftment, self-renewal, multilineage differentiation capacity, and an accessible chromatin configuration consistent with high expression of erythroid/megakaryoid genes in purified cell subsets. Our data demonstrate that with appropriate combinations of cell surface markers, stem cells can be accurately isolated to high purity and characterized. This is important for the clarification of lineage relationships and the identification of bona fide regulators of stem cell self-renewal and differentiation, both in normal and neoplastic tissues.
Keywords: Hematopoietic stem cells, Fetal liver, CD150
Introduction
Hematopoiesis describes the production of all types of fully differentiated daughter blood cells from ancestral great-grandmother hematopoietic stem cells (HSCs) [1]. A single, rare, clonogenic, multipotent, self-renewing long-term HSC (LT-HSC) capable of replenishing the blood system of a lethally irradiated animal for life can be isolated from mouse bone marrow (BM) or fetal liver (FL) by fluorescence activated cell sorting (FACS) based on cell surface marker expression [2, 3]. Short-term HSCs (ST-HSCs) with limited self-renewal potential [4, 5] and a series of downstream transit-amplifying precursors with varying developmental spectra have also been isolated to high purity [6-11]. The outcome of these careful experiments has been a working hierarchy of early mammalian hematopoiesis which is constantly evolving to accommodate new laboratory findings.
Recent technical improvements for the enrichment and isolation of HSCs and progenitors have utilized genome-wide microarrays and advances in flow cytometric reagents to expand upon the well-established cKit+Thy1.1(lo)Lin(-/lo)Sca1+ phenotype. In particular, the SLAM family of cell-cell interaction molecules and signaling receptors that regulate leukocyte functions [12] are selectively expressed amongst stem and progenitor cells in both mouse BM [13, 14] and FL [15] and human mobilized peripheral and umbilical cord blood [16]. Here, we coalesce the recently identified LT-HSC SLAM marker CD150 with the more traditional KTLS phenotype. Following an examination of the reconstitution kinetics of differently-aged KTLS HSCs in FL and gene expression trends, we incorporate CD150 into our purification protocol, and compare the positive and negative subfractions according to their in vivo stem cell function. The chromatin configuration at the Gata1 erythroid-specific locus in these subfractions was of particular interest given the pattern of expression of lineage-affiliated genes in purified fractions of known HSCs and progenitors.
Materials and Methods
Mice
C57BL/6-Ka and -Thy1.1 strains were maintained at Stanford University's Research Animal Facility. Mice used were 8-12 weeks old. For FLs, the morning of vaginal plug observation was E0.5.
Flow Cytometry
Before sorting, stem/progenitor cells from FL/BM were prepared by lineage depletion with Dynabeads M-450 (Dynal, Oslo, Norway) or cKit-enrichment with streptavidin-conjugated magnetic beads (Miltenyi, Bergisch Gladbach, Germany). Unconjugated lineage mAbs were B220 (clone 6B2), CD3 (2C11), CD4 (GK1.5), CD5 (53-7.3/7.8), CD8 (53-6.7), Gr1 (8C5), Mac1 (M1/70), and TER119. Mac1 was only used in the Lin cocktail for BM [17] and IL7Rα (A7R34) included for myeloid progenitors. These were labeled with Tricolor- or PE Texas Red-conjugated goat anti-rat IgG (Caltag, Burlingame, CA) and stained with stem/progenitor cell markers: Sca1 (E13-161-7), cKit (2B8), Thy1.1 (19XE5), Flk2 (A2F10) (eBioscience, San Diego, CA), CD150 (TC15-12F12.2) (Biolegend, San Diego, CA), IL7Rα, CD34 (RAM34) (BD Pharmingen, San Diego, CA), and FcγR (CD16/32) (2.4G2) (93) (eBioscience). Integrin-specific antibodies were α1 (Ha31/8), α2 (Hmα2), α4 (R1-2), α5 (5H10-27), α6 (GoH3), and β1 (HMβ1-1) integrin (all BD). Unless otherwise indicated, all mAbs were prepared in I.L.W. Lab. Cells were analysed and sorted on an LSRII, FACSAria, or highly-modified FACSVantage cytometer (BD, Mountain View, CA). All cells were at least double-sorted. Dead cells were discriminated by high forward scatter and propidium iodide (PI) staining. FACS data was analyzed using FlowJo (Tree Star, Inc., Ashland, OR).
Adoptive Transfer Experiments
Competitive reconstitution assays were performed by intravenous or retro-orbital injection of freshly purified cells along with 3×105 unfractionated BM cells as competitor. Recipients were lethally irradiated (900 rad, single dose) by X-ray. Multilineage engraftment was monitored by FACS analysis of peripheral blood samples collected via tail vein into 500 μl EDTA (10 mM). Erythrocytes were pelleted by adding 500 μl 2% dextran and incubating at 37°C for at least 25 minutes. Donor-derived cells were distinguished from host by CD45.1 (A20.1.7) or CD45.2 (AL1-4A2) expression.
Gene Expression
Total RNA was isolated using TRIzol (InVitrogen, Carlsbad, CA) from equivalent cell numbers, digested with DNase I to remove DNA contamination, and used for reverse transcription (SuperScript II kit, InVitrogen). All reactions were performed in triplicate in an ABI-7000 (Applied Biosystems, Foster City, CA) using SYBR Green (Applied Biosystems) and cDNA equivalent of ~500 cells. Fold expression relative to whole BM was calculated following β-actin transcript normalization.
Bisulfite Sequencing
Genomic DNA isolation and bisulfite treatment were performed as described [18].
Statistics
Data were analyzed for significance between groups using a two-tailed Student's t test. Differences were considered significant at p < 0.05.
Results
Expression of Lineage-Affiliated Genes in Hematopoietic Stem and Progenitor Subsets
To gain an initial insight as to the genetic pre-programming of hematopoietic stem and progenitor cells, we first examined the expression of numerous lineage-affiliated genes in nine highly-purified cell subsets from mouse BM and compared the outcome to those of unfractionated whole BM cells (Table 1) [18]. The lymphoid-affiliated gene IL7Rα, which marks the common lymphoid progenitor (CLP) cell surface, was found to be most highly expressed on this same population (3.8-fold change relative to whole BM). Confirming previous datasets [19, 20] and emphasizing the unique genetic programs activated with the onset of lymphopoiesis from stem and multipotent progenitor cells, IL7Rα expression was practically undetectable in long- and short-term stem, multipotent, and bipotent myeloid progenitor cells. The B cell factor Pax5 showed a similar expression trend. By contrast, myeloid-, erythroid-, and megakaryoid-affiliated genes showed more ubiquitous expression in all nine cell subsets analyzed. As expected, myeloid genes C/EBPα (~35-fold increase relative to whole BM), MPO (144-fold increase), and GM-CSFRα (2.4-fold increase) were highest expressed in granulocyte/macrophage progenitors (GMPs). The erythroid genes EpoR (~14-fold increase) and Gata1 (~57-fold increase) [18] were highest expressed in megakaryocyte/erythrocyte progenitors (MEPs) and, unlike myeloid genes, also boasted robust expression in the two LT-HSC populations we purified, using either Thy1.1 or CD34 in combination with the well-characterized multipotent progenitor (MPP) marker Flk2 [4, 21] within the cKit+Lin(-/lo)Sca1+ (KLS) subfraction. An even greater stem cell bias was seen for the erythroid transcription factor Gata2 which was highest expressing in both LT-HSC populations (41- to 48-fold increase) followed next by MEPs (~31-fold increase). The megakaryocyte transcription factor NF-E2 showed a similar trend with highest expression in LT-HSCs (4- to 9-fold increase), while c-mpl/TpoR, known to have potent effects on HSCs [22], was expressed at very high levels in LT-HSCs (~4,000- to ~5,500-fold increase). For all erythroid- and megakaryoid-affiliated genes boasting robust LT-HSC expression, it was notable that their expression was considerably lower in the next differentiation subset immediately downstream of LT-HSCs having lost long-term self-renewal ability. These data collectively show that at the population level, LT-HSCs are most primed at the level of gene expression for the erythroid/megakaryoid lineages in preference of other cell fates.
Table 1.
Expression levels of lineage-affiliated genes in purified HSC and progenitor cell compartments as measured by quantitative real-time PCR (qRT-PCR)
| Population | IL7Rα | Pax5 | C/EBPα | MPO | GM-CSFRα | EpoR | Gata1 | Gata2 | c-mpl/TpoR | NF-E2 |
|---|---|---|---|---|---|---|---|---|---|---|
| LT-HSC (KLS Thy1.1(lo)Flk2-) | 0.0 | 0.0 | 1.6 | 0.1 | 0.1 | 1.7 | 1.3 | 48.0 | 5415 | 8.9 |
| LT-HSC (KLS CD34-Flk2-) | 0.3 | 0.2 | 0.8 | 0.0 | 0.0 | 1.6 | 3.5 | 40.8 | 4056 | 4.4 |
| ST-HSC (KLS CD34+Flk2-) | 0.0 | 0.0 | 2.3 | 1.4 | 0.1 | 0.5 | 1.7 | 22.6 | 1154 | 3.6 |
| MPP (KLS Thy1.1-Flk2+) | 0.2 | 0.0 | 5.0 | 0.3 | 0.0 | 0.0 | 0.1 | 3.4 | 470 | 3.1 |
| MPP (KLS CD34+Flk2+) | 0.1 | 0.0 | 5.3 | 0.8 | 0.1 | 0.0 | 0.1 | 5.5 | 415 | 3.0 |
| CLP | 3.8 | 0.5 | 1.1 | 1.1 | 0.0 | 0.1 | 0.0 | 0.1 | 19.7 | 0.9 |
| CMP | 0.0 | 0.0 | 10.2 | 20.4 | 0.3 | 1.5 | 7.2 | 20.9 | 207 | 3.1 |
| GMP | 0.0 | 0.0 | 34.6 | 144 | 2.4 | 0.1 | 0.4 | 2.9 | 7.5 | 3.1 |
| MEP | 0.2 | 0.0 | 0.7 | 0.2 | 0.1 | 13.6 | 57.2 | 30.6 | 39.0 | 3.0 |
Expression levels in all populations listed above are shown as mean fold change relative to mouse whole BM (expression set to 1.0). qRT-PCRs (with β-actin housekeeping control) were performed using the cDNA equivalent of ~500 purified cells with triplicate measurement of at least two independently double- or triple-sorted populations. All cells were sourced from adult mouse BM (8 weeks old). Progenitors were sorted as follows: CLP as Lin-Flk2+IL7Rα+cKit(lo)Sca1(lo) [48]; CMP as Lin-cKit+Sca1-FcγR(lo)CD34(lo); GMP as Lin-cKit+Sca1-FcγR(hi)CD34(lo); and MEP as Lin-cKit+Sca1-FcγR-CD34- [8]. For the HSC, MPP, and CLP populations, the Lin cocktail contained the following mature cell markers: B220, CD3, CD4, CD5, CD8, Gr1, Mac1, and TER119. The Lin cocktail for myeloid progenitors contained all the aforementioned markers and IL7Rα.
HSC Profiles Through Development
We next sought to monitor in vivo LT-HSC reconstitution kinetics during mouse ontogeny, directly comparing BM to differently-aged mouse FL. Using C57BL/6-Thy1.1 mice, we first examined normal steady-state FACS profiles of wild-type FL samples from mice at the first age when a robust tissue sample could be sourced (E12.5) and thereafter daily until just prior to birth (E18.5) (Fig. 1). Here, LT-HSCs were identified as the Thy1.1(lo)Flk2- fraction of the KLS population. Thy1.1 was solely used to positively identify LT-HSCs within the KLS population of FL since expression of CD34, a distinguishing cell cycle-related marker between LT- and ST-HSCs [5], only becomes downregulated on these highly cycling cells after around seven weeks of age [23]. Since LT-HSCs express low levels of Mac1 in FL [17], this antibody was excluded from the lineage cocktail for this tissue only.
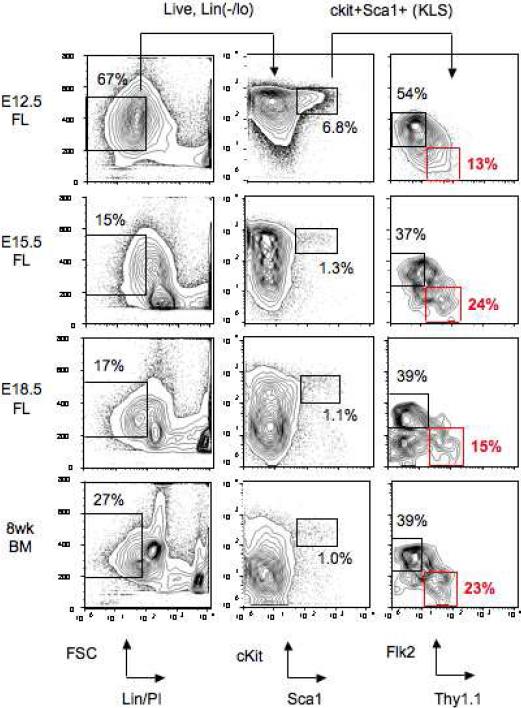
Figure 1.
FACS gating strategy used to purify mouse wild-type LT-HSCs for transplantation. Mouse LT-HSCs were purified with the phenotype cKit+Thy1.1(lo)Lin(-/lo)Sca1+Flk2- (KTLS cells) (red box). Multipotent progenitors (MPPs) are also highlighted as the cKit+Thy1.1(lo)Lin(-/lo)Sca1+Flk2+ population (black box in the far right KLS plot). E12.5, E15.5, and E18.5 fetal liver (FL) samples and 8 week bone marrow (BM) are shown. The percentage of total white blood cells (WBCs) within the gates is shown, with the exception of the KLS plots (far right) which show percentage of the KLS population. PI, propidium iodide.
As indicative of the nascent state of the hematopoietic system at this time during embryogenesis, and unlike other timepoints, the vast majority (at least 60%) of E12.5 FL cells were Lin(-/lo), with the greater proportion of these cKit+ (Fig. 1). More mature cells having downregulated cKit expression began to appear after E15.5 such that by E17.5, the FACS profiles of FL were comparable in appearance and cellular frequency to BM. At all developmental timepoints examined, a clear population of cKit+Thy1.1(lo)Lin-Sca1+Flk2- (KTLS) LT-HSCs was evident comprising around 15-25% of KLS cells, coupled with 2- to 3-fold more MPPs.
Quantitative Reconstitution Kinetics and Lineage Output of Transplanted HSCs Through Development
Having observed KTLS cells at each developmental time examined, 25 of these cells with the CD45.2+ genotype were double-sorted by FACS and their in vivo engraftment potential was quantitatively and kinetically assayed in competitive reconstitution experiments. Peripheral blood analysis of donor-derived hematopoietic cells up to 16 weeks post transplantation revealed all KTLS populations were able to long-term multilineage reconstitute CD45.1+ lethally irradiated adult recipients at robust levels (Fig. 2). E12.5 FL KTLS cells engrafted very poorly in a competitive setting (only 1/5 mice long-term multilineage reconstituted) (Supplementary Table 1), although contribution by donor cells was at a high level (30%) when these LT-HSCs engrafted. The number of successfully engrafted animals from E13.5 KTLS donor cells increased (3/7 mice), although this was still clearly less than E14.5-18.5 FL KTLS cells which showed reconstitution from donor cells (at least 80% positive in all cases) at rates comparable to adult BM (8/10 mice) (Table S1). Engraftment was highest for KTLS cells sourced from older FLs with 40-50% donor-derived hematopoietic compartments from E16.5-18.5 KTLS cells compared to 20-30% from younger E13.5-15.5 cells (Fig. 2A). In agreement with prior studies [17, 24], overall LT-HSCs derived from FL gave more robust and rapid reconstitution of irradiated recipients relative to BM LT-HSCs.
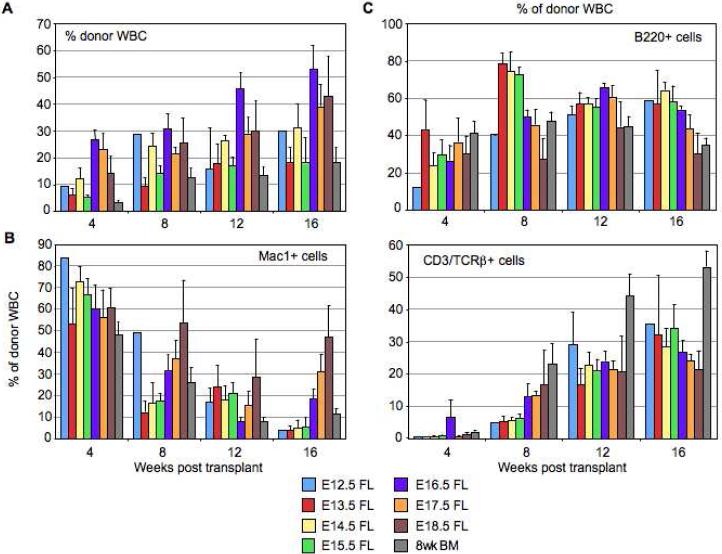
Figure 2.
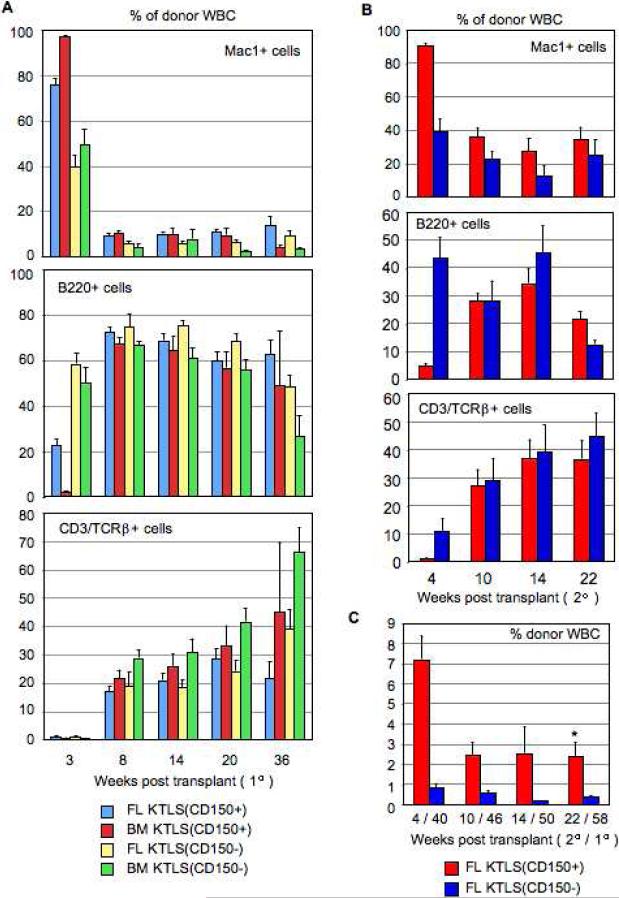
LT-HSCs differ in their engraftment levels and lineage potential throughout fetal development and early adulthood reminiscent of long-term stem cell aging reconstitution kinetics. (A): Quantitative (mean ± s.e.m.) and kinetic analysis of the percentage of donor-derived total WBC in the peripheral blood of lethally irradiated CD45.1+ mice transplanted with 25 KTLS cells from donor CD45.2+ animals (E12.5, E13.5, E14.5, E15.5, E16.5, E17.5 and E18.5 FL, and 8 week BM) and 3×105 recipient-type competitor adult whole BM (WBM). Donor cell reconstitution was assayed via readout in lysed, TER119- peripheral blood cells of recipients up to 16 weeks post transplantation. (B-C): Quantitative (mean ± s.e.m.) and kinetic analysis of the lineage potential of differently-aged KTLS cells showing the percentage of Mac1+ (myeloid, M lineage) (B), B220+ (B lineage) and CD3/TCRβ+ (T lineage) (C) cells within the donor-derived WBC compartment. The number of animals for which each of these statistics was calculated is shown in Table S1.
An analysis of donor-derived peripheral myeloid (Mac1+), B (B220+) and T (CD3/TCRβ+) cells showed expected temporal lineage reconstitution from all subsets of transplanted KTLS LT-HSCs (Fig. 2B, 2C). Short-lived myeloid cells were first produced following transplantation and comprised the bulk of the donor-derived hematopoietic compartment at 4 weeks, with long-lived lymphocytes appearing in greater proportions at later timepoints. The appearance of output myeloid cells before lymphoid following a HSC transplant is consistent with studies demonstrating LT-HSCs predominantly express myeloid over lymphoid genes [19, 20] and appear primed for myelopoiesis over lymphopoiesis in vivo [9, 20], and also reflects cellular emergence during ontogeny [25] and evolution [26]. A closer analysis of reconstitution frequencies at the longest timepoint examined (16 weeks) revealed a clear trend for reduced B/T lineage output as the transplanted LT-HSC aged across seven days of fetal development (from E12.5 to E18.5), with the mirror image seen for myeloid output with proportionally more donor-derived Mac1+ cells as the fetal LT-HSC aged.
CD150 as a Positive Marker of FL HSCs in Place of Thy1.1
Expression profiling and in vivo engraftment assays identified the SLAM family receptors as markers of HSCs/progenitors in both mouse BM [13, 14] and FL [15]. With CD34 unavailable for use in mouse FL to distinguish highly cycling LT-HSCs and Thy1.1 useful as a positive marker for LT-HSCs within the KLS fraction in only mouse strains expressing the Thy1.1 allele [2], CD150 represented the first marker to be positively expressed on LT-HSCs in a conserved manner across multiple mouse strains [13] including Trp53 null mice [27, 28].
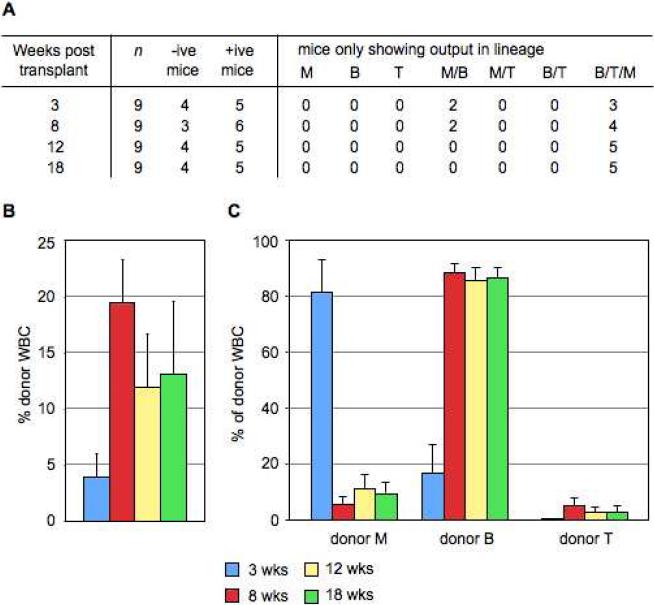
We sought to combine the use of CD150 as a positive FL LT-HSC marker with the established cKit+Lin-Sca1 (KLS) phenotype readily analyzed in the majority of mouse strains and so FACS sorted and competitively transplanted 10 CD45.2+ KLS(CD150+) E14.5 FL cells into lethally irradiated CD45.1+ animals. Peripheral blood analysis of donor-derived hematopoietic cells up to 18 weeks post transplant showed that KLS(CD150+) cells successfully engrafted adult recipients (5/9 mice long-term multilineage reconstituted) (Fig. 3A) at rates comparable to prior studies [15]. With 13% mean long-term donor engraftment levels (Fig. 3B), the hematopoietic lineage kinetics – primarily myeloid readout early and later B/T lymphocytes – from KLS(CD150+) cells was in agreement with E14.5 KTLS LT-HSCs (Fig. 3C). These data confirm KLS(CD150+) cells as bona fide LT-HSCs and that CD150 can substitute Thy1.1 as a positive marker of LT-HSCs in FL.
Figure 3.
KLS(CD150+) cells from E14.5 FL are long-term, multilineage engrafting HSCs. (A): 10 donor CD45.2+ E14.5 FL cells were sorted as cKit+Lin(-/lo)Sca1+CD150+ and transplanted into lethally irradiated CD45.1+ recipient animals along with recipient-type 2×105 competitor cells from adult WBM for radioprotection. Recipient animals were considered positive for engraftment if they boasted a robust population of large (SSC(med-hi)) myeloid (Mac1+) donor cells above background (>0.3%). (B): Quantitative (mean ± s.e.m.) and kinetic analysis of the percentage of donor-derived total WBC in the peripheral blood of positively-engrafted recipient mice transplanted with 10 KLS(CD150+) cells. (C): Quantitative (mean ± s.e.m.) and kinetic analysis of the lineage potential of KLS(CD150+) cells showing the percentage of Mac1+ (myeloid, M, lineage), B220+ (B lineage) and CD3/TCRβ+ (T lineage) cells within the donor-derived WBC compartment.
KTLS(CD150+) Cells are the Most Robustly-Engrafting, Long-Term Reconstituting, Self-Renewing HSCs
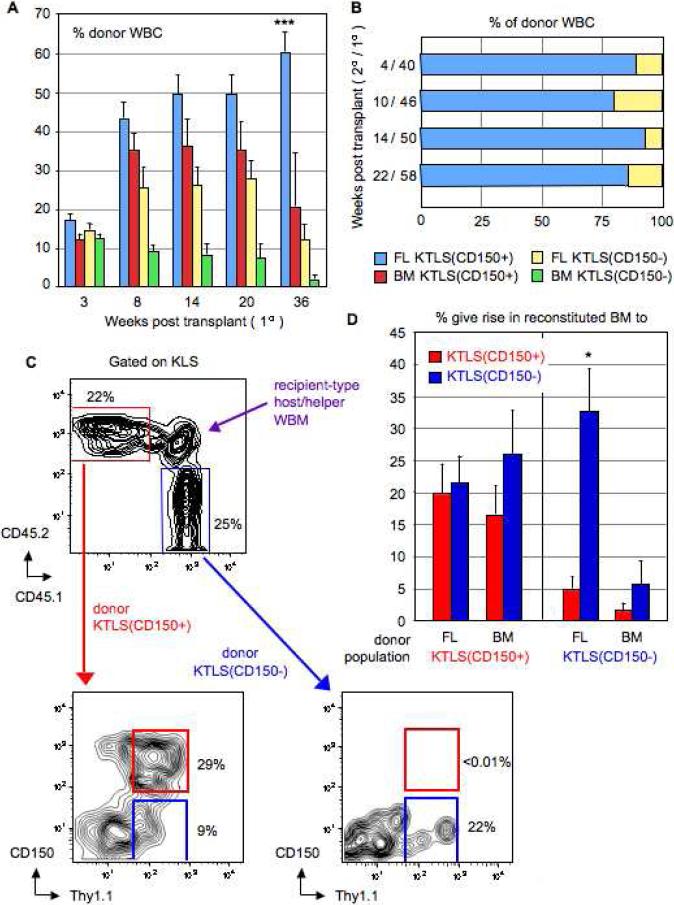
We next sought to gain insight as to which fraction, CD150+ or CD150−, of the cKit+Thy1.1(lo)Lin-Sca1+Flk2- (KTLS) population the long-term HSC potential predominantly resided within by directly comparing the function of these cells as LT-HSCs in vivo. A corresponding analysis we undertook using Ikaros point mutant mice [29] had found that homozygous mutant E14.5 FLs were selectively and significantly reduced in KTLS(CD150+) but not KTLS(CD150−) cells (P. Papathanasiou, unpublished observations). We sorted 100 donor CD45.2+CD45.1− KTLS(CD150+) and 100 donor CD45.2−CD45.1+ KTLS(CD150−) cells from either wild-type E15.5 FL or 8 week BM and competitively transplanted these into lethally irradiated CD45.2+CD45.1+ recipients. To safeguard against any engraftment advantages conferred by one CD45 allotype over the other, reciprocally-labelled transplants were also performed with a second cohort of purified KTLS(CD150+) and KTLS(CD150−) cells. In both cases, 300,000 recipient-type CD45.2+CD45.1+ whole BM cells were also transplanted for radioprotection. Peripheral blood analysis of donor-derived hematopoietic cells (see Supplementary Figure 1) up to 36 weeks post transplant showed KTLS(CD150+) cells engrafted at significantly higher levels compared to KTLS(CD150−) cells (Fig. 4A). At 36 weeks, KTLS(CD150+) consistently engrafted at 5-fold higher levels than KTLS(CD150−) cells, with peak donor reconstitution rates (~60%) from FL KTLS(CD150+) cells. Both KTLS(CD150+) and KTLS(CD150−) cells showed multilineage engraftment and an archetypal timecourse of mature blood cell production from precursor cells with primarily myeloid readout first followed later by lymphocytes (Fig. 5A).
Figure 4.
KTLS(CD150+) cells, and not KTLS(CD150−) cells, are the more ancestral HSC population. (A): Quantitative and kinetic analysis of the percentage (mean ± s.e.m.) of donor-derived total WBC in peripheral blood of lethally irradiated recipients engrafted with KTLS(CD150+) and KTLS(CD150−) cells from E15.5 FL or 8 week BM on primary transplant (n = 7 per experimental group; n = 28 total). Statistically significant differences (p < 0.05) at only the long-term (36 week) timepoint are indicated by an asterisk (*); multiple asterisks indicate significant differences to more than one experimental group. (B): Quantitative and kinetic analysis of composition of the donor-derived WBC compartment from secondary adoptive transfer experiments comparing contribution of KTLS(CD150+) and KTLS(CD150−) E15.5 FL cells to hematopoietic recovery in lethally-irradiated recipients (n = 7 per experimental group; n = 14 total). Donor reconstitution was assayed via readout in lysed, TER119- peripheral blood cells of recipient mice at 4 (40), 10 (46), 14 (50), and 22 (58) weeks post secondary (primary) transplantation. (C): BM analysis at 28 weeks post transplantation of lethally irradiated CD45.2+CD45.1+ recipients reconstituted with 100 CD45.2+CD45.1− KTLS(CD150+) and 100 CD45.2−CD45.1+ KTLS(CD150−) donor cells from E15.5 FL or 8 week BM. (D): Quantitative analysis of percentage (mean ± s.e.m.) of donor experimental populations KTLS(CD150+) or KTLS(CD150−) from E15.5 FL or 8 week BM that in vivo give rise to either KTLS(CD150+) or KTLS(CD150−) in recipient BM (n = 4 per experimental group; n = 16 total) at 28 weeks post transplantation. Statistically significant differences (p < 0.05) between KTLS(CD150+) and KTLS(CD150−) groups are indicated by an asterisk (*).
Figure 5.
Lineage potential of KTLS(CD150+) and (CD150−) cells on primary and secondary transplant show initial myeloid and later lymphoid cell output. (A): Primary transplant data (n = 7 per experimental group; n = 28 total) shows the percentage (mean ± s.e.m.) of Mac1+ (myeloid, M, lineage), B220+ (B lineage) and CD3/TCRβ+ (T lineage) cells within the donor-derived white blood cell compartment. (B): Secondary transplant data (n = 7 per experimental group; n = 14 total) shows the percentage (mean ± s.e.m.) of M, B and T lineage cells within the donor-derived white blood cell compartment. (C): Quantitative and kinetic analysis of the percentage (mean ± s.e.m.) of donor-derived WBC reconstitution levels in the peripheral blood of irradiated recipient mice engrafted with 100 KTLS(CD150+) and 100 KTLS(CD150−) cells on secondary transplant.
After 9 months primary engraftment, donor E15.5 FL KTLS(CD150+) and KTLS(CD150−) cells (100 from each population) were re-harvested from BM of reconstituted recipients and secondary transplants were performed. As with primary transplants, CD45 reciprocally-labelled transplants were set up, and 300,000 recipient-type BM cells were transplanted for radioprotection. Peripheral blood analysis of donor-derived hematopoietic cells up to 22 weeks post secondary transplant showed KTLS(CD150+) cells had continued to engraft at 5-fold significantly higher levels compared to KTLS(CD150−) cells (p = 0.001) (Fig. 5C). At 58 weeks post first harvest and transplant, the contribution to the donor-derived hematopoietic compartment from KTLS(CD150+) cells was ~80% (Fig. 4B). As with primary adoptive transfer experiments, both transplanted populations multilineage engrafted and showed temporal lineage reconstitution as would be expected of multilineage stem/precursor cells (Fig. 5B).
We also examined the ontogeny relationship between KTLS(CD150+) and KTLS(CD150−) cells. At 28 weeks post primary transplant, we used the original HSC marker panel and CD45 allelic markers to analyze the BM compartment of recipients reconstituted with donor KTLS(CD150+) and KTLS(CD150−) cells. We first gated on KLS cells, second on either CD45.2+CD45.1− or CD45.2−CD45.1+ donor experimental cell populations, and third examined the Thy1.1 versus CD150 flow cytometric profile (Fig. 4C). We found KTLS(CD150+) cells from either FL or BM self-renewed and also equally gave rise to KTLS(CD150−) cells, whereas KTLS(CD150−) cells preferentially self-renewed (Fig. 4D). FL KTLS(CD150−) cells, in particular, were significantly skewed towards self-renewal (p = 0.002). Together, it was clear from these results that KTLS(CD150+), and not KTLS(CD150−), cells were the more ancestral HSC population by virtue of their superior self-renewal and long-term engraftment properties.
Reports of changes in expression of the integrin family of cell adhesion molecules affecting engraftment of LT-HSCs within the niche [30] prompted us to examine whether KTLS(CD150+) cells differed from KTLS(CD150−) cells in their integrin expression. A comparison of α1, α2, α4, α5, α6, and β1 integrin expression levels did not reveal any differences between the CD150+ and CD150− subunits (Supplementary Figure 2), thus suggesting altered cellular trafficking was not the underlying factor for the enhanced hematopoietic reconstitution by KTLS(CD150+) cells.
Bisulfite Sequencing of Gata1 Enhancer Reveals Heterogeneity Within KTLS HSCs Which Can be Separated with CD150
There is clear evidence of a functional link between gene expression, active histone modifications and CpG methylation states and, in turn, the maintenance of cellular identity, including at the stem cell level. To provide an epigenetic layer of molecular resolution beyond mRNA transcript levels as a representation of gene expression, we performed bisulfite genomic DNA sequencing of the Gata1 enhancer region in KTLS HSCs. We chose to focus on an erythroid-specific locus by virtue of data implicating the positioning of the HSC toward an erythroid fate (Table 1), as well as the corresponding lack of only KTLS(CD150+) LT-HSCs in anemic Ikaros mutants (P. Papathanasiou, unpublished observations).
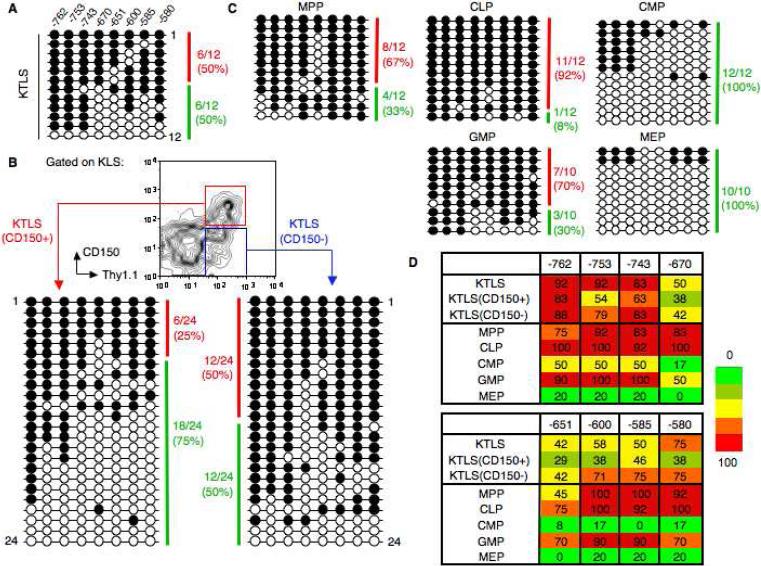
The Gata1 enhancer is located approximately 700bp upstream from the transcriptional start site and contains eight CpGs [31]. Unlike other lineage-specific loci we examined [18], Gata1 was notable in its bimodal epigenetic profile in purified HSCs: bisulfite analysis of purified wild-type KTLS cells from BM revealed a clear heterogeneity within a pool of sequenced templates with half the clones (6/12) appearing as heavily methylated (>80% methylated CpGs) and the other half (6/12) not heavily methylated (<80% methylation) (Fig. 6A). By contrast, all other bipotent progenitor populations examined showed a distinct skewing of methylation profile consistent with function: in CLPs and GMPs the Gata1 enhancer region was heavily methylated and reflected the dearth of erythroid potential from these progenitors, whereas in CMPs and MEPs it was unmethylated and thus accessible for gene transcription and resultant erythropoiesis (Fig. 6C). Having discerned from earlier experiments the KTLS population can be functionally separated according to CD150 expression, we re-purified the KTLS subset and repeated bisulfite sequencing on the CD150+ and CD150− cell fractions. The resultant 48 sequencing profiles showed the Gata1 enhancer was dramatically more accessible in KTLS(CD150+) cells (heavily methylated clones: 6/24, or 25%) compared to KTLS(CD150−) cells (12/24, or 50%) (Fig. 6B). Each individual CpG (8/8) showed consistently lower methylation levels in KTLS(CD150+) cells compared to KTLS(CD150−) cells and unfractionated KTLS cells (Fig. 6D). The -651 CpG site had the lowest overall methylation in KTLS(CD150+) (29%), KTLS(CD150−) (42%), and bulk KTLS (42%) cells. The methylation levels in CMPs (8%) and MEPs (0%) were also lowest at the -651 CpG.
Figure 6.
Bisulfite sequencing of the Gata1 enhancer reveals heterogeneity within the KTLS subset which can separated with CD150. (A): Bisulfite analysis of a 182-bp Gata1 enhancer in wild-type KTLS cells from 8 week BM. Each horizontal line represents an independently sequenced template with methylated (filled circles) and unmethylated CpGs (open circles). A heavily methylated clone (red) was classified as having >80% CpG dinucleotides methylated whereas a clone that was not heavily methylated (green) had <80% CpGs methylated. (B): Bisulfite analysis of Gata1 enhancer in KTLS cellular subsets purified according to expression of CD150 from BM. (C): Bisulfite analysis of Gata1 enhancer in five purified progenitor populations from BM. (D): Heatmap summary of the DNA methylation profile for the Gata1 enhancer in purified HSCs and progenitors. Methylation levels are represented in a gradation of colors from green (0-20%), olive (21-40%), yellow (41-60%), orange (61-80%), and red (81-100%). The number of methylated clones over total clones selected for sequencing (10-24 clones) is expressed as a percentage.
Discussion
Here, we have shown the long-term reconstituting subset of cKit+Thy.1.1(lo)Lin(-/lo)Sca1+Flk2- hematopoietic stem cells are CD150+. We defined and characterized this cell fraction according to: (a) six cell surface parameters; (b) long-term in vivo engraftment; (c) multilineage reconstitution in primary and secondary transplants; (d) self-renewal; and (e) chromatin accessibility. We also provided a snapshot of the kinetic variability of LT-HSCs across age and their gene expression pattern.
Mouse HSCs capable of long-term reconstitution develop at ~E10.5 (humans at E30-40) from the aorta-gonad-mesonephros (AGM) and have been prospectively isolated by flow cytometry [32]. These HSCs, and others from yolk sac [33], subsequently migrate to and colonize the FL around a day later when their cell surface phenotype changes to more resemble postnatal LT-HSCs. Although fetal HSCs have been largely characterized at ~E14.5, these cells can clearly transplant as LT-HSCs and differentiate to progenitors at a much earlier time during FL development. Although reconstitution rates were low in a competitive setting, overall levels were high from wild-type E12.5-13.5 donor KTLS cells, showing that these cells are capable of robust engraftment upon successful migration to the BM niche. An elimination of radiation-resistant host natural killer cell surveillance from residual embryonic MHC class I expression at low levels [34] through the use of alternate transplantation strategies such as Rag2-γc double knockout recipients could be one strategy by which to further improve engraftment by the youngest LT-HSCs.
Given the late fetal onset of hepatic growth factors [35], it was surprising that KTLS cells sourced from FLs older than E15.5 transplanted at levels higher than younger cells. However, the hematopoietic seeding of BM and spleen during late gestation is progressive rather than sudden [36], whilst many age-related changes of the hematopoietic system result directly from intrinsic changes at the HSC level [20, 37]. Our experiments with FL over individual developmental days in the embryo revealed clear trends in mature lineage output skewing with predominantly lymphoid potential from younger HSCs and primarily myeloid cells from older HSCs. This snapshot of short-term aging in the FL microenvironment was reminiscent of more dramatic differences in long-term aging previously seen across much larger expanses of time comparing young (1-2 month) and old (12-24 month) BM-derived HSCs [20]. This aging on a miniature scale in FL highlights the striking changes occurring in hematopoiesis over so short as a week of embryonic ontogeny that can also be seen in other developmental processes such as primitive erythroblast maturation [38]. Such rapid developments in blood cell production become further exaggerated following mutation of a critical genetic regulator in the cells supporting tissue generation, for example, by the rapid exhaustion of the KTLS(CD150+) pool and subsequent accumulation of all non-stem cell subsets within two developmental days in Ikaros point mutant homozygous mice (P. Papathanasiou, unpublished observations). This includes KTLS(CD150−) cells which only engraft for a limited time before system stress through serial transfer or genetic mutation. The finding of bona fide LT-HSC activity in CD150+ cells has also been demonstrated by numerous other studies [11, 13-15, 39], while another series of experiments has suggested CD150− cells combined with side population sorting parameters represent a functionally distinct population of LT-HSCs [40]. As with the gradual loss of developmental potential with cell maturation, we feel the very minor engraftment potential of KTLS(CD150−) cells is likely due to a progressive rather than sudden down-regulation of stem cell potential from KTLS(CD150+) cells, and is thus residual from these cells. The loss of CD150 expression from LT-HSCs may thereby represent one of the very first changes occurring with stem cell differentiation out of the self-renewing state.
Several mutant phenotypes provide the first evidence for a more intimate connection between LT-HSCs and non-lymphoid cells, in particular the erythrocyte/megakaryocyte lineage. PU.1 germline [41] and Scl/Tal-1 conditional [42] deletion HSCs were found to only give rise to MEPs, implying selective corruption of all but this cell fate. Second, the daily demands for the production of immense numbers of erythrocytes (~2 × 1011) [43] and platelets (~1011) [22] compared to other mature cell types and studies of asymmetric division and lineage commitment of HSC daughter cell pairs have called into question the number of cell divisions between HSCs and erythrocytes/megakaryocytes and the hitherto status of the CMP as precursor along this differentiation pathway. However, an alternate argument could equally be made that many intermediate, highly-proliferative progenitors are in fact needed to produce these mature cells which predominantly comprise blood in steady-state. Third, the isolation of cell subsets having diminished megakaryocyte/erythrocyte but retained lymphomyeloid potential by prospective purification [9, 11] is consistent with the erythrocyte/megakaryocyte fate as an early branchpoint from the HSC, although different experimental protocols have questioned this interpretation [44]. Perhaps both pathways are present, one acting in steady-state, the other in particular hematopoietic high-demand states; for example, single HSCs respond in serum-free medium to produce megakaryocytes but few other cell types [45]. Our qRT-PCR data here show that at the population level LT-HSCs are most primed for the erythrocyte/megakaryocyte lineages, and that this pattern of transcription also correlates with switching on of the CD150 gene (P. Papathanasiou, unpublished observations).
The usefulness of bisulfite DNA sequencing in assaying cell purity is pertinent with regard both aforementioned topics discussing the stem cell identity of CD150 cells and the priming of the LT-HSC fraction for particular developmental lineages. Not only were these data from the Gata1 enhancer congruent with progentior cell surface phenotype, qRT-PCR expression data, and the distribution of histone modifications at the Gata1 enhancer in small numbers of purified primary HSCs and progenitors [18], but the lowest overall methylation levels were most notably observed at the −651 CpG dinucleotide. This particular CpG site is adjacent to a functionally critical Gata1 binding motif [31] which raises the possibility that this zinc finger transcription factor binds to this site and primes the LT-HSC to readily develop into erythroid cells. Given that Gata1 is essential for the formation of the β-globin active chromatin hub [46], chromosome conformation capture assays have found Ikaros plays an essential role in the formation of this complex [47], and the synchronous LT-HSC and anemia phenotype of Ikaros point mutant embryos (P. Papathanasiou, unpublished observations), alternate isoforms of Ikaros could act as binding partners participating in the lineage choice of the KTLS(CD150+) LT-HSC through simultaneously silencing or activating important, lineage-affiliated developmental loci.
The findings presented here demonstrate that in both fetal liver and adult bone marrow, the long-term reconstituting subset of cKit+Thy1.1(lo)Lin(-/lo)Sca1+Flk2- hematopoietic stem cells reside in the CD150+ subfraction. These cells boast long-term competitive engraftment on both primary and secondary transplant against CD150− cells, robust self-renewal, and multilineage differentiation kinetics consistent with the previously well-utilized KTLS fraction of LT-HSCs. The use of additional cell surface markers within the CD150+ subpopulation will be an important future area of research to further resolve precise lineage relationships within the hematopoietic hierarchy and facilitate the identification of genes that control LT-HSC self-renewal and differentiation.
Supplementary Material
Supplementary Table 1. Competitive reconstitution of lethally irradiated recipient mice with KTLS donor cells reveals differences in long-term, multi-lineage engraftment with age of donor HSCs
SUPPLEMENTARY FIGURE LEGENDS
Papathanasiou et al.
“Evaluation of the Long-Term Reconstituting Subset of Hematopoietic Stem Cells with CD150”
Supplementary Figure 1. FACS gating strategy used to analyze in vivo reconstitution and lineage distribution of donor CD45.2+CD45.1− KTLS(CD150+) and CD45.2− CD45.1+ KTLS(CD150−) cells in CD45.2+CD45.1+ recipient peripheral blood from 4 weeks post-transplant onwards. Cells were first gated according to Scatter, excluding FSC(small) (red cells) and FSC(large) (doublet) cells. Live (Propidium Iodide-negative), TER119- cells were then gated, to compare overall CD45.2 versus CD45.1 donor white blood cell (WBC) reconstitution. Each donor subset, whether CD45.2+CD45.1− or CD45.2−CD45.1+, was then analyzed according to the expression of B220+, CD3/TCRβ+ and Mac1+SSC(large) to assay the output of the B, T and M lineages, respectively. A similar gating strategy was utilized to assay the in vivo reconstitution and donor lineage distribution where only one CD45.2+ donor subset was transplanted into CD45.1+ recipients.
Supplementary Figure 2. Expression of α1 (Ha31/8), α2 (Hmα2), α4 (R1-2), α5 (5H10-27), α6 (GoH3), and β1 (HMβ1-1) integrins on the cell surface of KTLS(CD150+) (blue) and KTLS(CD150−) (red) bone marrow cells.
Acknowledgments
We thank D. Bryder, D.J. Rossi and E.C. Forsberg for stimulating discussions and technical guidance, S.J. Morrison for sharing unpublished results, L. Jerabek for excellent laboratory management, C. Richter for antibody preparations, and L. Hidalgo, J. Dollaga, and D. Escoto for animal care. This work was supported in part by NIH grants 5P01 DK53074 and R01 CA086065 (to I.L.W.) and R01 DK43726 (S.T.S.), and a National Health & Medical Research Council CJ Martin Fellowship (P.P.). S.T.S. is an Investigator of the Howard Hughes Medical Institute. I.L.W. has stock in Amgen and is a cofounder of Cellerant Inc. and Stem Cells Inc. The other authors have no financial interests to disclose.
Footnotes
Disclosure of potential conflicts of interest is found at the end of this article.
References
- 1.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 3.Osawa M, Hanada K, Hamada H, et al. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 4.Christensen JL, Weissman IL. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc Natl Acad Sci U S A. 2001;98:14541–14546. doi: 10.1073/pnas.261562798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang L, Bryder D, Adolfsson J, et al. Identification of Lin-Sca1+kit+CD34+flt3-short-term hematopoietic stem cells capable of reconstituting and rescuing myeloablated transplant recipients. Blood. 2005;105:2717–2723. doi: 10.1182/blood-2004-06-2159. [DOI] [PubMed] [Google Scholar]
- 6.Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 7.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 8.Akashi K, Traver D, Miyamoto T, et al. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 9.Adolfsson J, Månsson R, Buza-Vidas N, et al. Identification of flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential: a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Chen CC, Grimbaldeston MA, Tsai M, et al. Identification of mast cell progenitors in adult mice. Proc Natl Acad Sci U S A. 2005;102:11408–11413. doi: 10.1073/pnas.0504197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pronk CJ, Rossi DJ, Månsson R, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Calpe S, Wang N, Romero X, et al. The SLAM and SAP gene families control innate and adaptive immune responses. Adv Immunol. 2008;97:177–250. doi: 10.1016/S0065-2776(08)00004-7. [DOI] [PubMed] [Google Scholar]
- 13.Kiel MJ, Yilmaz ÖH, Iwashita T, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 14.Forsberg EC, Prohaska SS, Katzman S, et al. Differential expression of novel potential regulators in hematopoietic stem cells. PLoS Genetics. 2005;1:e28. doi: 10.1371/journal.pgen.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim I, He S, Yilmaz ÖH, et al. Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood. 2006;108:737–744. doi: 10.1182/blood-2005-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sintes J, Romero X, Marin P, et al. Differential expression of CD150 (SLAM) family receptors by human hematopoietic stem and progenitor cells. Exp Hematol. 2008;36:1199–1204. doi: 10.1016/j.exphem.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrison SJ, Hemmati HD, Wandycz AM, et al. The purification and characterization of fetal liver hematopoietic stem cells. Proc Natl Acad Sci U S A. 1995;92:10302–10306. doi: 10.1073/pnas.92.22.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Attema JL, Papathanasiou P, Forsberg EC, et al. Epigenetic characterization of hematopoietic stem cell differentiation using miniChIP and bisulfite sequencing analysis. Proc Natl Acad Sci U S A. 2007;104:12371–12376. doi: 10.1073/pnas.0704468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyamoto T, Iwasaki H, Reizis B, et al. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev Cell. 2002;3:137–147. doi: 10.1016/s1534-5807(02)00201-0. [DOI] [PubMed] [Google Scholar]
- 20.Rossi DJ, Bryder D, Zahn JM, et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci U S A. 2005;102:9194–9199. doi: 10.1073/pnas.0503280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adolfsson J, Borge OJ, Bryder D, et al. Upregulation of Flt3 expression within the bone marrow Lin-Sca1+c-kit+ stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15:659–669. doi: 10.1016/s1074-7613(01)00220-5. [DOI] [PubMed] [Google Scholar]
- 22.Kaushansky K. The molecular mechanisms that control thrombopoiesis. J Clin Invest. 2005;115:3339–3347. doi: 10.1172/JCI26674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito T, Tajima F, Ogawa M. Developmental changes of CD34 expression by murine hematopoietic stem cells. Exp Hematol. 2000;28:1269–1273. doi: 10.1016/s0301-472x(00)00535-x. [DOI] [PubMed] [Google Scholar]
- 24.Harrison DE, Zhong RK, Jordan CT, et al. Relative to adult marrow, fetal liver repopulates nearly five times more efficiently long-term than short-term. Exp Hematol. 1997;25:293–297. [PubMed] [Google Scholar]
- 25.Cumano A, Godin I. Ontogeny of the hematopoietic system. Annu Rev Immunol. 2007;25:745–785. doi: 10.1146/annurev.immunol.25.022106.141538. [DOI] [PubMed] [Google Scholar]
- 26.Hansen JD, Zapata AG. Lymphocyte development in fish and amphibians. Immunol Rev. 1998;166:199–220. doi: 10.1111/j.1600-065x.1998.tb01264.x. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Ellison FM, Keyvanfar K, et al. Enrichment of hematopoietic stem cells with SLAM and LSK markers for the detection of hematopoietic stem cell function in normal and Trp53 null mice. Exp. Hematol. 2008;36:1236–1243. doi: 10.1016/j.exphem.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Elf SE, Miyata Y, et al. p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell. 2009;4:37–48. doi: 10.1016/j.stem.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papathanasiou P, Perkins AC, Cobb BS, et al. Widespread failure of hematolymphoid differentiation caused by a recessive niche-filling allele of the Ikaros transcription factor. Immunity. 2003;19:131–144. doi: 10.1016/s1074-7613(03)00168-7. [DOI] [PubMed] [Google Scholar]
- 30.Wagers AJ, Allsopp RC, Weissman IL. Changes in integrin expression are associated with altered homing properties of Lin(-/lo)Thy1.1(lo)Sca-1(+)c-kit(+) hematopoietic stem cells following mobilization by cyclophosphamide/granulocyte colony-stimulating factor. Exp Hematol. 2002;30:176–185. doi: 10.1016/s0301-472x(01)00777-9. [DOI] [PubMed] [Google Scholar]
- 31.Nicolis S, Bertini C, Ronchi A, et al. An erythroid specific enhancer upstream to the gene encoding the cell-type specific transcription factor GATA-1. Nucleic Acids Res. 1991;19:5285–5291. doi: 10.1093/nar/19.19.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertrand JY, Giroux S, Golub R, et al. Characterization of purified intraembryonic hematopoietic stem cells as a tool to define their site of origin. Proc Natl Acad Sci U S A. 2005;102:134–139. doi: 10.1073/pnas.0402270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weissman IL, Papaioannou V, Gardner R. Fetal hematopoietic origins of the adult hematolymphoid system. In: Clarkson B, Marks PA, Till JE, editors. Differentiation of Normal and Neoplastic Hematopoietic Cells. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1978. pp. 33–47. [Google Scholar]
- 34.Ozato K, Wan YJ, Orrison BM. Mouse major histocompatibility class I gene expression begins at midsomite stage and is inducible in earlier-stage embryos by interferon. Proc Natl Acad Sci U S A. 1985;82:2427–2431. doi: 10.1073/pnas.82.8.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinoshita T, Sekiguchi T, Xu MJ, et al. Hepatic differentiation induced by oncostatin M attenuates fetal liver hematopoiesis. Proc Natl Acad Sci U S A. 1999;96:7265–7270. doi: 10.1073/pnas.96.13.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christensen JL, Wright DE, Wagers AJ, et al. Circulation and chemotaxis of fetal hematopoietic stem cells. PLoS Biol. 2004;2(3):e75. doi: 10.1371/journal.pbio.0020075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikuta K, Kina T, MacNeil I, et al. A developmental switch in thymic lymphocyte maturation potential occurs at the level of hematopoietic stem cells. Cell. 1990;62:863–874. doi: 10.1016/0092-8674(90)90262-d. [DOI] [PubMed] [Google Scholar]
- 38.Fraser ST, Isern J, Baron MH. Maturation and enucleation of primitive erythroblasts during mouse embryogenesis is accompanied by changes in cell-surface antigen expression. Blood. 2007;109:343–352. doi: 10.1182/blood-2006-03-006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiel MJ, Yilmaz OH, Morrison SJ. CD150- cells are transiently reconstituting multipotent progenitors with little or no stem cell activity. Blood. 2008;111:4413–4414. doi: 10.1182/blood-2007-12-129601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weksberg DC, Chambers SM, Boles NC, et al. CD150- side population cells represent a functionally distinct population of long-term hematopoietic stem cells. Blood. 2008;111:2444–2451. doi: 10.1182/blood-2007-09-115006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott EW, Fisher RC, Olson MC, et al. PU.1 functions in a cell-autonomous manner to control the differentiation of multipotential lymphoid-myeloid progenitors. Immunity. 1997;6:437–447. doi: 10.1016/s1074-7613(00)80287-3. [DOI] [PubMed] [Google Scholar]
- 42.Mikkola HK, Klintman J, Yang H, et al. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature. 2003;421:547–551. doi: 10.1038/nature01345. [DOI] [PubMed] [Google Scholar]
- 43.Giarratana M, Kobari L, Lapillonne H, et al. Ex vivo generation of fully mature human red blood cells from hematopoietic stem cells. Nat Biotech. 2005;23:69–74. doi: 10.1038/nbt1047. [DOI] [PubMed] [Google Scholar]
- 44.Forsberg EC, Serwold T, Kogan S, et al. New evidence supporting megakaryocyte-erythrocyte protential of Flk2/Flt3+ multipotent hematopoietic progenitors. Cell. 2006;126:415–426. doi: 10.1016/j.cell.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 45.Domen J, Weissman IL. Hematopoietic stem cells need two signals to prevent apoptosis; bcl-2 can provide one of these, kit1/c-kit signaling the other. J Exp Med. 2000;192:1707–1718. doi: 10.1084/jem.192.12.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vakoc CR, Letting DL, Gheldof N, et al. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 47.Keys JR, Tallack M, Crossley M, et al. Ikaros regulates human haemoglobin switching by facilitating developmental stage specific interactions between the individual globin gene promoters and a chromatin hub. Br J Haematol. 2008;141:398–406. doi: 10.1111/j.1365-2141.2008.07065.x. [DOI] [PubMed] [Google Scholar]
- 48.Karsunky H, Inlay MA, Serwold T, et al. Flk2+ common lymphoid progenitors possess equivalent differentiation potential for the B and T lineages. Blood. 2008;111:5562–5570. doi: 10.1182/blood-2007-11-126219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Competitive reconstitution of lethally irradiated recipient mice with KTLS donor cells reveals differences in long-term, multi-lineage engraftment with age of donor HSCs
SUPPLEMENTARY FIGURE LEGENDS
Papathanasiou et al.
“Evaluation of the Long-Term Reconstituting Subset of Hematopoietic Stem Cells with CD150”
Supplementary Figure 1. FACS gating strategy used to analyze in vivo reconstitution and lineage distribution of donor CD45.2+CD45.1− KTLS(CD150+) and CD45.2− CD45.1+ KTLS(CD150−) cells in CD45.2+CD45.1+ recipient peripheral blood from 4 weeks post-transplant onwards. Cells were first gated according to Scatter, excluding FSC(small) (red cells) and FSC(large) (doublet) cells. Live (Propidium Iodide-negative), TER119- cells were then gated, to compare overall CD45.2 versus CD45.1 donor white blood cell (WBC) reconstitution. Each donor subset, whether CD45.2+CD45.1− or CD45.2−CD45.1+, was then analyzed according to the expression of B220+, CD3/TCRβ+ and Mac1+SSC(large) to assay the output of the B, T and M lineages, respectively. A similar gating strategy was utilized to assay the in vivo reconstitution and donor lineage distribution where only one CD45.2+ donor subset was transplanted into CD45.1+ recipients.
Supplementary Figure 2. Expression of α1 (Ha31/8), α2 (Hmα2), α4 (R1-2), α5 (5H10-27), α6 (GoH3), and β1 (HMβ1-1) integrins on the cell surface of KTLS(CD150+) (blue) and KTLS(CD150−) (red) bone marrow cells.