Abstract
We report on a mouse specific SuperMix immunoaffinity separation system for separating low abundance proteins from high and moderate abundance proteins in mouse plasma. When applied in tandem with a mouse IgY7 column that removes the seven most abundant proteins in plasma, the SuperMix column captures more than 100 additional moderate abundance proteins, thus allowing significant enrichment of low abundance proteins in the flow-through fraction. A side-by-side comparison of results obtained from 2D-LC-MS/MS analyses of flow-through samples from IgY7 and SuperMix columns revealed a nearly two-fold improvement in the overall proteome coverage. Detection of low abundance proteins was also enhanced, as evidenced by a more than two-fold increase in the coverage of cytokines, growth factors, and other low abundance proteins. Moreover, the tandem separations are automated, reproducible, and allow effective identification of protein abundance differences from LC-MS/MS analyses. Considering the overall reproducibility and increased sensitivity using the IgY7-SuperMix separation system, we anticipate broad applications of this strategy for biomarker discovery using mouse models.
Keywords: Immunoaffinity separation, LC-MS/MS, Proteomics, Mouse plasma, SuperMix
Introduction
The majority of disease biomarker discovery studies reported to date have been focused on analyzing blood plasma or serum, the primary clinical specimens for disease diagnosis and therapeutic monitoring. In spite of technological advances, the dynamic range of plasma protein concentrations, which spans more than 10 orders of magnitude, still presents significant challenges for analytical detection technologies, including mass spectrometry (MS)-based proteomics 1–3. To achieve in-depth coverage of the plasma proteome, many techniques have been developed for separating low abundance proteins from plasma and enhancing their detection by MS 3, 4. One of the most effective and reliable techniques is immunoaffinity chromatography using mixed antibodies for selected abundant proteins 5–8. While this technique has provided promising results, detection of low abundance plasma proteins is still limited by the number of proteins that the immunoaffinity columns can effectively capture.
Besides the proteome complexity, the substantial heterogeneity among biological subjects presents another major challenge for experiments using human samples. For this reason, animal models such as mice are commonly employed for studies of human biology and biomarker discovery. Because mouse can be generated from a single parental mating pair and bred with the same food and environment, there is significantly less genetic, biological, and environmental variability compared to human 9, 10. Mouse shares ~99% of its genome with human 11, and recapitulates the features of many human diseases, such as diabetes, cancer, and neurological disorders. The availability of mouse samples at early disease stages even makes it possible to find biomarkers for early diagnosis 12. Substantial biomarker discovery efforts using mouse plasma and proteomics tools have led to the identification of a number of proteins potentially associated with different human diseases, such as pancreatic tumor 12, 13, breast cancer 14, intestinal tumor 15, obesity and diabetes 16, and sepsis 17. Regardless, success remains limited by the ability to detect low abundance proteins in mouse plasma because of dynamic range and complexity issues similar to human plasma.
To facilitate more effective biological and biomarker discovery studies using mouse models, we describe a mouse-specific IgY7-SuperMix immunoaffinity separation strategy that enhances the detection of low abundance proteins within plasma. This strategy is based on a similar strategy we developed previously for analyzing human plasma samples in which IgY12 and SuperMix columns were used serially in tandem, which allowed for effective removal of the 12 most abundant proteins by IgY12 and ~50 moderate abundance proteins by the human SuperMix column18. By applying mouse-specific IgY7 and SuperMix columns in tandem, the SuperMix column can effectively capture more than 100 moderate abundance plasma proteins in addition to the 7 most abundant proteins captured by the IgY7 column. Moreover, the results from LC-MS/MS analyses of the SuperMix column flow-through sample reveal a >100% increase in proteome coverage compared to the results from IgY7 flow-through samples. The overall results illustrate the potential for enhanced detection of low abundance proteins and improved coverage of signaling pathways, as well as the reproducibility of the separation systems for identifying differential protein abundances.
Experimental Section
Generation of the Mouse SuperMix Column
Mouse plasma (CD-1 Mouse Plasma w/ EDTA-K3, male, 55.8 mg/mL) was obtained from Charles River Laboratories International, Inc. (Wilmington, MA). A custom SuperMix column was generated by GenWay Biotech (San Diego, CA) (now commercially available from Sigma-Aldrich, St. Louis, MO). A mouse plasma sample was initially depleted of its 7 most abundant proteins (serum albumin, IgG, fibrinogen, α1-antitrypsin, transferrin, haptoglobin and IgM) by using a Seppro™ mouse IgY7 LC10 column (Genway Biotech). Approximately 100 mg of the IgY7-depleted plasma were provided to GenWay Biotech as an antigen mixture for producing the custom mouse SuperMix column. The generation of the SuperMix LC5 column (12.7 × 39.5 mm) was similar to that previously described for producing the human SuperMix column 18.
Tandem Separations of Mouse Plasma by IgY7 and SuperMix Columns
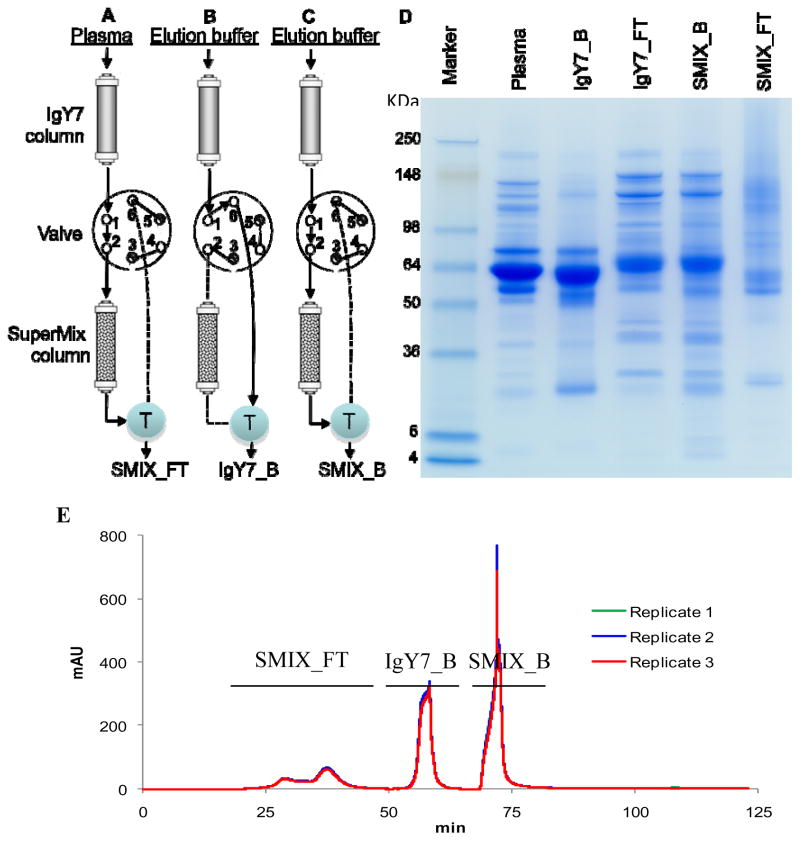
The HPLC setup18 used for plasma depletion was modified to allow the two separation steps to be applied in-series on-line to make the overall experiments automatic and less labor-intensive. Briefly, IgY7 LC10 and SuperMix LC5 columns were connected in tandem on an Agilent 1100 series HPLC system to allow sample flow through both columns following injection. A six-port valve (Agilent, Santa Clara, CA) was introduced between the two columns (Figure 1, A-C). Positions 3 and 5 were plugged to prohibit back-flush towards the waste or columns during valve switching. And a “T” connector was introduced to allow the liquid stream from IgY7 or IgY7-Supermix columns to be directed towards the UV detector and subsequently towards the fraction collector. This configuration allowed collection of the flow-through and bound portions in different fractions.
Figure 1.
The tandem IgY7-SuperMix separation strategy. (A) IgY7 and SuperMix columns are connected to ports 1 and 2 of a six-port valve. Port 1 is connected to port 2 when mouse plasma sample is injected into the columns. The flow-through is collected as SuperMix flow-through (SMIX_FT). (B) Port 1 is connected to port 6 following (A), and the elution buffer is injected into the IgY7 column. The eluted sample is collected as IgY7 bound (IgY7_B). (C) After IgY7_B collection, port 1 is again connected to port 2. Elution buffer is sent through the SuperMix column and the eluted SuperMix bound fraction (SMIX_B) is collected. (D) 15 μg proteins from each sample collected during steps (A) through (C) are separated on a 4–20% SDS-PAGE (Invitrogen) and stained with Coomassie Brilliant Blue G-250. Molecular markers (kDa) are labeled on the left. IgY7 flow-through (IgY7_FT) is collected when the plasma sample was injected into the IgY7 column and the valve was set as in (B). (E) LC chromatograms of 3 replicates of the tandem depletion experiment. The collection window of each fraction is marked on the peak.
For each injection, 80 μL of mouse plasma was diluted 12-fold in dilution buffer (10 mM Tris-HCl, 150 mM NaCl, pH 7.4), filtered through 0.45 μm cellulose acetate centrifuge tube filters (Sigma-Aldrich), and injected into the tandem IgY7 and SuperMix separation system to capture the high and moderate abundance proteins, respectively. The flow-through was collected as SuperMix flow-through. Elution buffer (100 mM glycine, pH 2.5) was used to collect the IgY7 bound fraction (valve connected to collecting plate, Figure 1B) and SuperMix bound fraction (valve connected to Supermix column, Figure 1C). The separation scheme consisted of sample loading and washing with dilution buffer (0.5 mL/min, 45 min), IgY7 eluting (2 mL/min, 15 min), SuperMix eluting (1 mL/min, 20 min), neutralization (100 mM Tris-HCl, pH 8.0) (1 ml/min, 20 min), and re-equilibration with dilution buffer (1 mL/min, 20 min), with a total cycle time of 120 min. The separation parameters for loading, washing, and elution were chosen based on manufacturer’s instructions on IgY-LC10 and LC5 columns. To collect the IgY7 flow-through portion, the valve between the two columns was switched from SuperMix column to collecting plate (Figure 1B) when plasma sample was injected, so that the sample will be collected without passing through the SuperMix column. All collected fractions were concentrated in Amicon Ultra-15 concentrators (3-kDa nominal molecular mass limit; Millipore, Billerica, MA) followed by buffer exchange to 50 mM NH4HCO3, pH 8.0. Protein concentration was determined using a BCA protein assay from Pierce (Rockford, IL).
To demonstrate the ability for measuring differential protein abundances, the following six non-mouse standard proteins were spiked into three 80 μL mouse plasma samples at 0.8, 4, and 20 μg/ml, respectively: 1) Bovine Carbonic Anhydrase 2 (BCA), 2) Bovine Beta-Lactoglobulin (BBL), 3) E. coli Beta-Galactosidase (EBG), 4) Bovine Alpha-Lactalbumin (BAL), 5) Equine Skeletal Muscle Myoglobin (ESMM), and 6) Chicken Ovalbumin (CO). The tandem separation was performed as described above. The SuperMix flow-through and bound fractions were collected separately for further analysis.
Protein Digestion
Protein samples were denatured using 50% 2,2,2-Trifluoroethanol (TFE) in 50 mM NH4HCO3 buffer (pH 8.0) at 60 °C for 2 h and reduced by 2 mM DTT at 37 °C for 1 h. The resulting protein mixture was diluted 6-fold with 50 mM NH4HCO3, and digested by sequencing grade modified porcine trypsin (Promega, Madison, WI) for 3 h at 37 °C in a trypsin:protein ratio of 1:50 (w/w). The final peptide concentration of the supernatant was determined using BCA protein assay. All samples were stored at −80 °C until further analysis.
Strong Cation Exchange (SCX) Fractionation
SCX fractionation of digested peptides was performed using an Agilent 1100 series HPLC system at a flow rate of 200 μL/min. 150 μg tryptic peptides from either the IgY7 flow-through or SuperMix flow-through fractions were resuspended in buffer A (25% acetonitrile, 10 mM ammonium formate, pH 3.0) and loaded onto a 2.1 × 200 mm (5 μm, 300 Å) Polysulfoethyl A LC column (PolyLC, Columbia, MD) preceded by a 2.1 × 10 mm guard column. The fractionation scheme consisted of 10 min at 100% buffer A, a 40 min linear gradient from 0 to 50% buffer B (25% acetonitrile, 500 mM ammonium formate, pH 6.8), a 10 min linear gradient from 50 to 100% buffer B, and 20 min at 100% buffer B. 25 fractions were collected for each sample via an automated fraction collector based on the UV trace. Each fraction was lyophilized prior to MS analysis.
Reverse-phase Capillary LC-MS/MS Analysis
Peptides were analyzed using a custom-built automated four-column high pressure capillary LC system coupled on-line to either an LTQ or LTQ-Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA) via a nanoelectrospray ionization interface manufactured in-house. The reverse-phase capillary column was prepared by slurry-packing 3-μm Jupiter C18 bonded particles (Phenomenex, Torrence, CA) into a 65-cm-long, 75-μm-inner diameter fused silica capillary (Polymicro Technologies, Phoenix, AZ). After loading 1 μg of peptides onto the column, the mobile phase was held at 100% A (0.1% formic acid) for 20 min, followed by a linear gradient from 0 to 70% buffer B (0.1% formic acid in 90% acetonitrile) over 85 min with a flow rate ~500 nL/min. Each full MS scan (m/z 400–2000) was followed by collision-induced MS/MS scans (normalized collision energy setting of 35%) for the 10 most abundant ions. The dynamic exclusion duration was set to 1 min, the heated capillary was maintained at 200 °C, and the ESI voltage was held at 2.2 kV.
Data Analysis
LC-MS/MS raw data were converted into .dta files using Extract_MSn (version 3.0) in Bioworks Cluster 3.2 (Thermo Scientific), and the SEQUEST algorithm (version 27, revision 12) was used to independently search all the MS/MS spectra against the mouse International Protein Index (IPI) database that had 51,489 total protein entries (version 3.35, released October 24, 2007). The false discovery rate (FDR) was estimated based on decoy-database searching methodology 19, 20. Search parameters and filtering criteria were applied to limit the FDR at the unique peptide level to <5%, as described previously18. Identified proteins were grouped to a non-redundant protein list using ProteinProphet™ software, after which one protein IPI number was randomly selected to represent each corresponding protein group that consisted of a number of database entries. Only those proteins or protein groups with two or more unique peptide identifications were considered as confident identifications.
Results
Reproducibility of the Tandem IgY7-SuperMix Separations
In this study, we evaluated the application of a mouse-specific IgY7 column and a SuperMix column as a tandem on-line immunoaffinity separation strategy. The mouse SuperMix column was produced using mouse plasma depleted of the seven most abundant proteins as an antigen mixture for generating IgY antibodies. Based on the theory that the abundance and immunogenicity of the proteins correspond to the antibody titers 21, the majority of proteins captured by the SuperMix column are assumed to be moderate abundance proteins, and most of the proteins passing through the SuperMix column would be of low abundance.
The design of the automated two-column separation system is presented in Figure 1(A–C). Protein patterns for all collected fractions were obtained using Coomassie Brilliant Blue G-250 staining on SDS-PAGE (Figure 1D). Most of the intensive protein bands in mouse plasma showed up in the IgY7 bound, IgY7 flow-through, or SuperMix bound, but not in the SuperMix flow-through. The appearance of some bands in the SuperMix flow-through fraction, but not in other fractions suggests that the detection of these relatively low abundance proteins was improved by the tandem separation.
Table 1 shows protein recoveries for the tandem separation process based on five independent experimental replicates. Starting with ~4.5 mg plasma proteins, 1155 ± 63 μg proteins in the IgY7 flow through fraction, 1039 ± 58 μg proteins in the SuperMix bound fraction and 45 ± 4 μg in the IgY-SuperMix flow through were recovered. These data illustrate the overall good reproducibility of the tandem separations and sample processing in terms of sample recovery. Specifically, the protein recovery rate (Table 1) for the SuperMix flow-through and SuperMix bound fractions were 23.3 ± 1.3% and 1.0 ± 0.1%, with a relative standard deviation (RSD) of 8.0% and 5.6%, respectively.
Table 1.
Protein recoveries for tandem IgY7 and SuperMix separations*
| Sample | μg protein | Recovery (%) |
|---|---|---|
| Mouse plasma | 4462 | |
| IgY7 flow-through | 1155 ±63 | 25.9 ± 1.4 |
| SuperMix bound | 1039 ± 58 | 23.3 ± 1.3 |
| SuperMix flow-through | 45 ± 4 | 1.0 ± 0.1 |
Based on five replicate experiments.
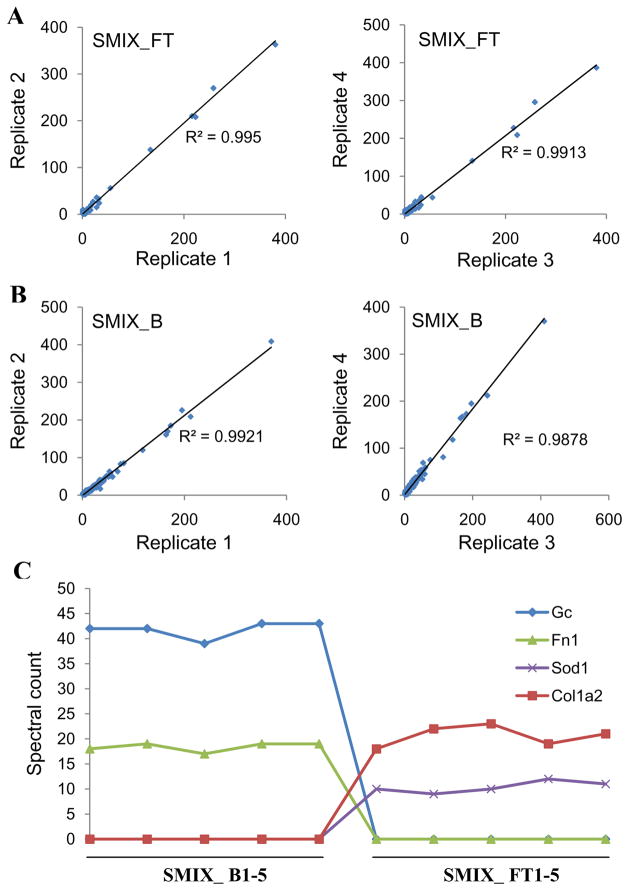
The reproducibility of tandem separations was also assessed on the basis of results obtained from 1D LC-MS/MS analyses. An average of 143 ± 3 proteins was confidently identified from the SuperMix bound fraction and 170 ± 13 proteins from the SuperMix flow-through samples. Comparison of spectral counts 8 for proteins among replicates (Figure 2A and 2B) illustrate the general reproducibility of the strategy; all R2 coefficients are >0.98. The reproducibility of four individual proteins, vitamin D-binding protein (Gc), fibronectin 1 (Fn1), superoxide dismutase (SOD1), and collagen alpha-2(I) chain (Col1a2), is also illustrated in Figure 2C. These proteins were either completely captured or not captured by the SuperMix column, and were identified with relatively reproducible spectral counts in all replicates with relative standard deviations less than 10%. This relatively good overall reproducibility is important for identifying differential protein abundances in biological and clinical applications.
Figure 2.
Reproducibility of tandem IgY7-SuperMix separation evaluated by spectral count information from LC-MS/MS analyses. (A) SuperMix flow-through fraction (B) SuperMix bound fraction. (C) The spectral counts of selected proteins from five replicate analyses., SuperMix flow-through; SMIX_B, SuperMix bound; Gc, Vitamin D-binding protein; Fn1, fibronectin 1; SOD1, Superoxide dismutase; Col1a2, Collagen alpha-2(I) chain.
Quantification of Relative Protein Abundance Differences
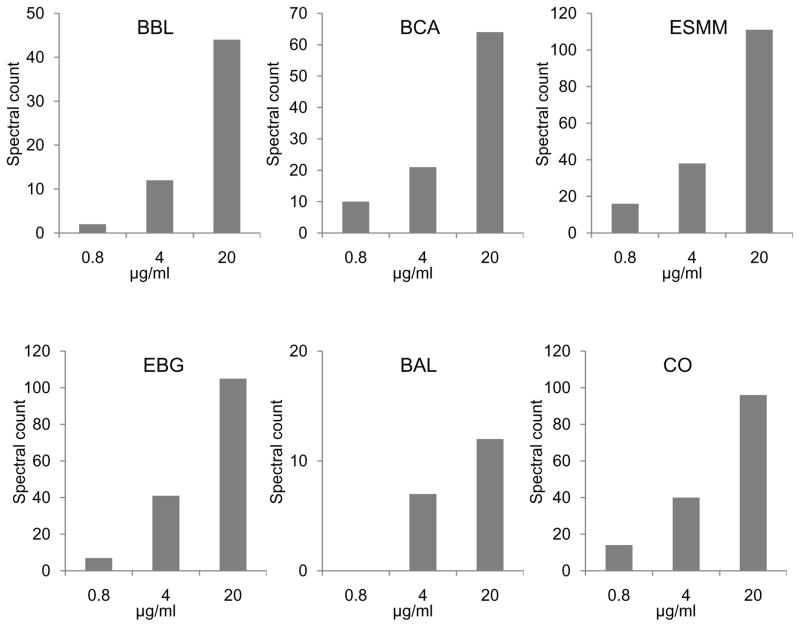
To assess the ability of this tandem immunoaffinity separation strategy for identifying relative protein abundance differences in different samples, six non-mouse standard proteins were spiked into the original mouse plasma at three different concentrations (i.e., 0.8, 4, 20 μg/ml). For each 1D LC-MS/MS analysis, the minimum amount of peptides injected for each standard protein was approximately 7 pg (or 0.2 femtomole) in the background of 0.5 μg total plasma digest. Figure 3 shows the spectral count data for each protein across the three spiked samples. Note that all six proteins were identified by LC-MS/MS in the SuperMix flow-through fraction only. The spectral counts for all proteins increased proportionally to the increase in protein concentrations, with the exception of bovine alpha-lactalbumin, which was not identified at the lowest spiking level. These data demonstrate that this tandem immunoaffinity separation system can reliably quantify abundance differences for relatively low abundance proteins.
Figure 3.
Spectral count quantification for six standard proteins spiked into mouse plasma at three different concentrations. All data were obtained using 1D LC-MS/MS. Bovine Beta-Lactoglobulin; BCA, bovine carbonic anhydrase 2; ESMM, Equine Skeletal Muscle Myoglobin; EBG, E.coli Beta-Galactosidase; BAL, Bovine Alpha-Lactalbumin; CO, chicken ovalbumin.
Capture Efficiency of Moderate Abundance Proteins by the SuperMix Column
SuperMix bound and flow-through samples were analyzed by 1D LC-MS/MS and the protein abundances were semi-quantitatively compared based on spectral count information to evaluate the binding or capturing efficiency of the moderate abundance proteins to the SuperMix column. A total of 161 proteins were identified by two or more unique peptides from the SuperMix bound fraction, 79 of which were not found in parallel flow-through samples. The capture efficiency of the SuperMix column was consistent among five replicates (Figure 2C). Table 2 lists 81 proteins with >99% capture efficiency and >5 spectral counts, including most of the top proteins identified in the IgY7 flow-through fraction. Note that some moderate abundance proteins were captured by the SuperMix column with an efficiency of <50%. For example, the capture efficiency of hemoglobin Y beta-like embryonic chain (Hbb-y), Hemoglobin subunit alpha (Hba-a), Hemoglobin subunit beta-1 (Hbb-b1), Isoform 1 of Gelsolin precursor (Gsn), and Apolipoprotein A-IV precursor (Apoa4) were 4.3%, 5.5%, 7.4%, 18.0%, and 18.3%, respectively. These results may be due to the relative low titration of corresponding antibodies generated by the chicken or to low reactivity between the antibody and antigen. Similar results were observed with the human SuperMix system 18.
Table 2.
List of moderate abundance proteins bound to the SuperMix column with relatively high capture efficiency. All proteins were identified with 2 or more unique peptides. Spectral counts were summed from 5 replicates of 1D LC-MS/MS analyses. SMIX_B, SuperMix bound fraction; SMIX_FT, SuperMix flow-through fraction.
| Spectral count |
Capture efficiency (%) | ||||
|---|---|---|---|---|---|
| IPI | Description | Gene | SMIX_B | SMIX_FT | |
| IPI00131830.1 | Serine protease inhibitor A3K precursor | Serpina3k | 1205 | 11 | >99 |
| IPI00624663.3 | Alpha-2-macroglobulin precursor | Pzp | 766 | 2 | >99 |
| IPI00323624.2 | Isoform Long of Complement C3 precursor | C3 | 620 | 3 | >99 |
| IPI00123223.2 | Murinoglobulin-1 precursor | Mug1 | 512 | 2 | >99 |
| IPI00869381.1 | apolipoprotein A-II | Apoa2 | 452 | 1 | >99 |
| IPI00129755.2 | Alpha-1-antitrypsin 1-2 precursor | Serpina1b | 276 | 1 | >99 |
| IPI00111315.1 | Apolipoprotein A-II precursor | Apoa2 | 268 | 2 | >99 |
| IPI00138342.3 | Liver carboxylesterase N precursor | Es1 | 257 | 1 | >99 |
| IPI00136642.1 | Antithrombin-III precursor | Serpinc1 | 235 | 1 | >99 |
| IPI00271262.6 | Murinoglobulin-2 precursor | Mug2 | 234 | 0 | >99 |
| IPI00126184.7 | Vitamin D-binding protein precursor | Gc | 209 | 0 | >99 |
| IPI00475157.1 | Serpina1b protein | Serpina1b | 201 | 0 | >99 |
| IPI00128484.1 | Hemopexin precursor | Hpx | 182 | 0 | >99 |
| IPI00124725.1 | Inter-alpha-trypsin inhibitor heavy chain H3 precursor | Itih3 | 163 | 0 | >99 |
| IPI00677290.3 | similar to Murinoglobulin 1 | EG640530 | 146 | 0 | >99 |
| IPI00227834.7 | Inter-alpha-trypsin inhibitor heavy chain H2 precursor | Itih2 | 146 | 0 | >99 |
| IPI00312711.2 | PK-120 | Itih4 | 135 | 0 | >99 |
| IPI00605187.4 | ceruloplasmin isoform a | Cp | 123 | 0 | >99 |
| IPI00113539.2 | Fibronectin precursor | Fn1 | 120 | 0 | >99 |
| IPI00129225.1 | Isoform LMW of Kininogen-1 precursor | Kng1 | 120 | 1 | >99 |
| IPI00322867.2 | Itih1 protein | Itih1 | 111 | 0 | >99 |
| IPI00128249.1 | Alpha-2-HS-glycoprotein precursor | Ahsg | 110 | 0 | >99 |
| IPI00469387.1 | GUGU alpha | Fetub | 106 | 0 | >99 |
| IPI00119818.1 | Adult male liver cDNA, RIKEN full-length enriched library | Itih4 | 98 | 0 | >99 |
| IPI00407657.2 | Complement component 8, alpha polypeptide | C8a | 94 | 0 | >99 |
| IPI00352163.3 | fibronectin 1 | Fn1 | 92 | 0 | >99 |
| IPI00322936.1 | Plasminogen precursor | Plg | 77 | 0 | >99 |
| IPI00230718.3 | complement component 9 | C9 | 75 | 0 | >99 |
| IPI00406302.2 | Alpha-1-antitrypsin 1-1 precursor | Serpina1a | 71 | 0 | >99 |
| IPI00121055.1 | Complement factor H- related protein | Cfh | 70 | 0 | >99 |
| IPI00857383.1 | Truncated ceruloplasmin | Cp | 64 | 0 | >99 |
| IPI00117831.1 | Ceruloplasmin precursor | Cp | 63 | 0 | >99 |
| IPI00139788.2 | Serotransferrin precursor | Trf | 60 | 0 | >99 |
| IPI00320675.1 | Complement factor I precursor | Cfi | 55 | 0 | >99 |
| IPI00123927.1 | Alpha-1-antitrypsin 1-5 precursor | Serpina1e | 53 | 0 | >99 |
| IPI00136266.1 | Apolipoprotein C-II precursor | Apoc2 | 53 | 0 | >99 |
| IPI00123924.1 | Alpha-1-antitrypsin 1-4 precursor | Serpina1d | 45 | 0 | >99 |
| IPI00123411.1 | Spi2 proteinase inhibitor (Fragment) | Serpina3g | 44 | 0 | >99 |
| IPI00831484.1 | liver carboxylesterase N-like | LOC382044 | 44 | 0 | >99 |
| IPI00135547.1 | Serum amyloid A-4 protein precursor | Saa4 | 43 | 0 | >99 |
| IPI00121274.2 | Isoform 1 of Complement component C8 beta chain precursor | C8b | 42 | 0 | >99 |
| IPI00121190.1 | Epidermal growth factor receptor precursor | Egfr | 40 | 0 | >99 |
| IPI00225715.7 | glycosylphosphatidylinositol specific phospholipase D1 | Gpld1 | 40 | 0 | >99 |
| IPI00137987.1 | Zinc-alpha-2- glycoprotein precursor | Azgp1 | 39 | 0 | >99 |
| IPI00130654.6 | Isoform 1 of Afamin precursor | Afm | 35 | 0 | >99 |
| IPI00322304.5 | Histidine-rich glycoprotein HRG | Hrg | 35 | 0 | >99 |
| IPI00116105.1 | Corticosteroid-binding globulin precursor | Serpina6 | 35 | 0 | >99 |
| IPI00666034.2 | Apolipoprotein B homolog | Apob | 35 | 0 | >99 |
| IPI00407541.2 | Isoform 2 of Leukemia inhibitory factor receptor precursor | Lifr | 33 | 0 | >99 |
| IPI00330833.3 | Complement C5 precursor | Hc | 31 | 0 | >99 |
| IPI00330843.2 | Factor XII | F12 | 29 | 0 | >99 |
| IPI00321666.1 | H-2 class I histocompatibility antigen, Q10 alpha chain precursor | H2-Q10 | 29 | 0 | >99 |
| IPI00314141.5 | Serine protease inhibitor A3N precursor | Serpina3n | 25 | 0 | >99 |
| IPI00314270.3 | Complement component 6 | C6 | 24 | 0 | >99 |
| IPI00223231.2 | Isoform 1 of Sulfhydryl oxidase 1 precursor | Qsox1 | 24 | 0 | >99 |
| IPI00400202.1 | kininogen 2 isoform 1 | Kng2 | 21 | 0 | >99 |
| IPI00322463.3 | Beta-2-glycoprotein 1 precursor | Apoh | 20 | 0 | >99 |
| IPI00309214.1 | Serum amyloid P-component precursor | Apcs | 19 | 0 | >99 |
| IPI00129240.1 | Vitronectin precursor | Vtn | 19 | 0 | >99 |
| IPI00331551.3 | Coagulation factor X, full insert sequence | F10 | 17 | 0 | >99 |
| IPI00127856.1 | Alpha-1-acid glycoprotein 2 precursor | Orm2 | 17 | 0 | >99 |
| IPI00113057.1 | Plasma kallikrein precursor | Klkb1;Cyp4v3 | 17 | 0 | >99 |
| IPI00122122.1 | Complement component C8 gamma chain precursor | C8g | 16 | 0 | >99 |
| IPI00317356.10 | Serum paraoxonase/arylesterase 1 | Pon1 | 15 | 0 | >99 |
| IPI00118457.1 | Serum amyloid A-2 protein precursor | Saa2 | 15 | 0 | >99 |
| IPI00122977.1 | Plasma protease C1 inhibitor precursor | Serping1 | 15 | 0 | >99 |
| IPI00134808.1 | C4b-binding protein precursor | C4bp | 14 | 0 | >99 |
| IPI00113227.1 | Heparin cofactor 2 precursor | Serpind1 | 13 | 0 | >99 |
| IPI00128260.2 | Carboxypeptidase N catalytic chain precursor | Cpn1 | 13 | 0 | >99 |
| IPI00153258.1 | Isoform 1 of Protein Z- dependent protease inhibitor precursor | Serpina10 | 11 | 0 | >99 |
| IPI00120192.1 | Isoform Long of Complement C2 precursor (Fragment) | C2 | 11 | 0 | >99 |
| IPI00133222.1 | Vitamin K-dependent protein Z precursor | Proz | 10 | 0 | >99 |
| IPI00118130.1 | Alpha-1-acid glycoprotein 1 precursor | Orm1 | 10 | 0 | >99 |
| IPI00127352.2 | AMBP protein precursor | Ambp | 8 | 0 | >99 |
| IPI00137599.1 | Selenoprotein P precursor | Sepp1 | 7 | 0 | >99 |
| IPI00118994.1 | Apolipoprotein C-IV precursor | Apoc4 | 7 | 0 | >99 |
| IPI00474340.3 | Complement C1s-B subcomponent precursor | EG317677 | 7 | 0 | >99 |
| IPI00133500.3 | Phosphatidylcholine- sterol acyltransferase precursor | Lcat | 6 | 0 | >99 |
| IPI00314309.3 | Apolipoprotein D precursor | Apod | 6 | 0 | >99 |
| IPI00108003.2 | Macrophage colony- stimulating factor 1 receptor precursor | Csf1r | 5 | 0 | >99 |
| IPI00230412.7 | Vascular non- inflammatory molecule 3 precursor | Vnn3 | 5 | 0 | >99 |
Note: The capture efficiency of each protein was evaluated by dividing the spectral count for each protein in the SuperMix bound fraction (SMIX_B) by the sum of spectral counts from the SuperMix bound fraction and flow-through fraction (SMIX_FT), i.e. SMIX_B/(SMIX_B+SMIX_FT). The estimated capture efficiency will be less accurate for proteins with only a few spectral counts.
Improved Proteome Coverage
1D and 2D LC-MS/MS analyses of IgY7 flow-through and SuperMix flow-through fractions and 1D LC-MS/MS analyses of SuperMix bound portion were performed to assess the improvement in proteome coverage by the SuperMix separations. For 2D LC-MS/MS analyses, protein digests were further separated into 25 fractions by off-line SCX. Results indicated that 808 proteins were identified in the SuperMix flow-through and bound fractions combined (523 were confidently identified with at least 2 unique peptides) compared to 391 proteins (263 with at least 2 unique peptides) identified in the IgY7 flow-through, a 2-fold improvement in proteome coverage after SuperMix separation. (All of the identified proteins and peptides are listed in supplemental Tables S1 and S2).
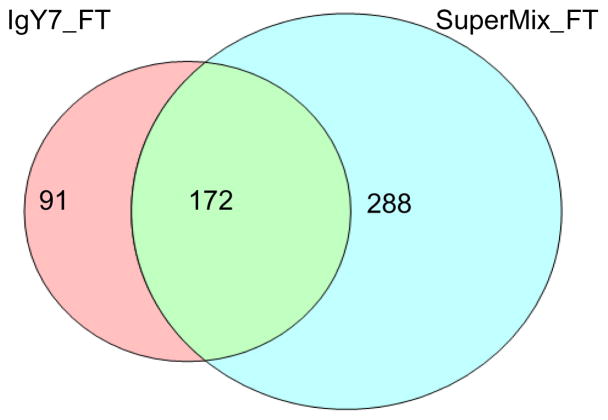
Figure 4 shows a side-by-side comparison of proteome coverage obtained for IgY7 and SuperMix flow-through samples. Following 2D LC-MS/MS analysis, 263 proteins were confidently identified (at least 2 unique peptides) in the IgY7 flow-through and 460 proteins in the SuperMix flow-through. 288 proteins were identified in the SuperMix flow-through, but not in the IgY flow-through, and 91 proteins were identified in the IgY7 flow-through only. 76 out of the 91 proteins not identified in the SuperMix flow-through were identified in 1D LC-MS/MS analyses of the SuperMix bound fraction. The rest of 15 proteins absent in the SuperMix flow-through fraction could also be captured in the SuperMix bound fraction, but were below the detection limit of 1D LC-MS/MS analysis due to their relatively low abundances. This assumption is further supported by the results that all 15 proteins were also not observed by 1D LC-MS/MS analyses of the IgY7 flow-through (Supplemental table 1).
Figure 4.
Comparison of proteome coverage of IgY7 flow-through and SuperMix flow-through samples, as analyzed by 2D LC-MS/MS.
Table 3 further illustrates the improvement in the detection of low abundance proteins following tandem IgY7-SuperMix separation. All of the selected proteins were identified in SuperMix flow-through by 2D LC-MS/MS with at least two unique peptides. Several proteins in the list were previously reported as potential disease biomarkers, including osteopontin (Spp1) for breast cancer 22, mimecan (Ogn) for pituitary cancer 23, alpha-synuclein (Snca) and DJ-1 (Park7) for Parkinson’s disease 24.
Table 3.
A selected list of known cytokines, growth factors, and other low abundance proteins identified from 2D LC-MS/MS analyses. Only proteins identified with at least 2 unique peptides are listed.
| Descript | Gene | IPA category | Peptide number | spectral count |
|
|---|---|---|---|---|---|
| IgY7_FT | SMIX_FT | ||||
| Osteopontin precursor | Spp1 | cytokine | 8 | 2 | 10 |
| Uteroglobin precursor | Scgb1a1 | cytokine | 2 | 5 | |
| Guanine deaminase | Gda | enzyme | 19 | 52 | |
| Transitional endoplasmic reticulum ATPase | Vcp | enzyme | 32 | 56 | |
| Isoform IGF-IA of Insulin-like growth factor I precursor | Igf1 | growth factor | 2 | 10 | 16 |
| Mimecan precursor | Ogn | growth factor | 3 | 3 | |
| Regenerating islet-derived protein 3 beta precursor | Pap | growth factor | 4 | 16 | |
| Regenerating islet-derived protein 3 gamma precursor | Reg3g | growth factor | 4 | 7 | |
| Phosphoglycerate kinase 1 | Pgk1 | kinase | 16 | 1 | 59 |
| Programmed cell death protein 5 | Pdcd5 | NA | 2 | 2 | |
| Pancreatic alpha-amylase precursor | OTTMU SG0000 0022462 | NA | 12 | 27 | |
| C-reactive protein precursor | Crp | other | 18 | 6 | 160 |
| Epithelial cadherin precursor | Cdh1 | other | 4 | 1 | 9 |
| Insulin-like growth factor-binding protein 5 precursor | Igfbp5 | other | 4 | 2 | 3 |
| Isoform 1 of Alpha-synuclein | Snca | other | 5 | 1 | 8 |
| Macrophage-capping protein | Capg | other | 3 | 3 | |
| Neuropeptide Y precursor | Npy | other | 3 | 3 | |
| Protein DJ-1 | Park7 | other | 9 | 16 | |
| Secretogranin-1 precursor | Chgb | other | 4 | 5 | |
| Stress-induced-phosphoprotein 1 | Stip1 | other | 2 | 2 | |
| Transforming growth factor-beta- induced protein ig-h3 precursor | Tgfbi | other | 24 | 6 | 60 |
| Cathepsin L precursor | Ctsl | peptidase | 3 | 3 | |
| Insulin-degrading enzyme | Ide | peptidase | 7 | 8 | |
| Proteasome subunit beta type-1 precursor | Psmb1 | peptidase | 17 | 1 | 39 |
| Serine/threonine-protein phosphatase 2A regulatory subunit B′ | Ppp2r4 | phosphatase | 6 | 10 | |
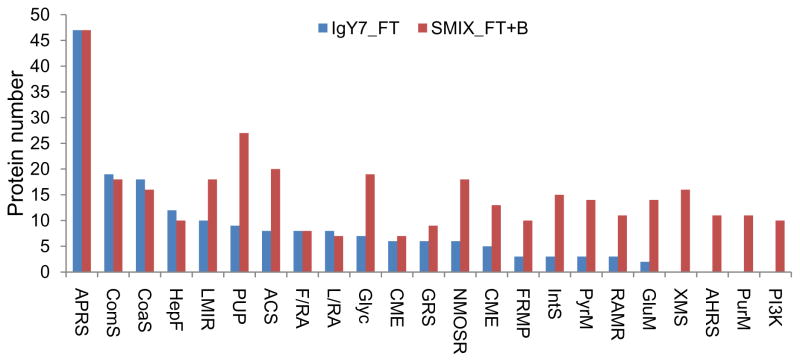
The enhanced detection of low abundance proteins also led to significantly improved coverage of a number of canonical pathways mapped by our data based on Ingenuity Pathways Analysis Tool 25. In Figure 5, the protein numbers in each pathway are compared between IgY7 flow-through and the combined results from SuperMix flow-through and bound fractions. Note that proteins were identified by at least 2 unique peptides. No obvious differences were observed for acute phase response signaling, complement system, coagulation system and hepatic fibrosis/hepatic stellate cell activation pathways involving relatively abundant proteins. However, the coverage of pathways such as clathrin-mediated endocytosis, Fcγ receptor-mediated phagocytosis in macrophages and monocytes, integrin signaling, pyruvate metabolism, regulation of actin-based motility by Rho and glutathione metabolism improved by >2-fold. This improvement suggests that many of the low abundance protein components within these pathways become detectable following the SuperMix separation. Some pathways were only identified in SuperMix flow-through but not in IgY7 flow-through; for example, PI3K/AKT signaling, which is involved in many human diseases such as diabetes, cancer and neurodegenerative diseases 26. A list of mapped canonical pathways is provided in Supplemental Table S3.
Figure 5.
Improved coverage of canonical pathways by tandem IgY7-SuperMix separation. Blue bars, IgY7 flow-through; Red bars, SuperMix flow-through and SuperMix bound combined. APRS, Acute Phase Response Signaling; ComS, Complement System; CoaS, Coagulation System; HepF, Hepatic Fibrosis / Hepatic Stellate Cell Activation; LMIRF, LPS/IL-1 Mediated Inhibition of RXR Function; PUP, Protein Ubiquitination Pathway; ACS, Actin Cytoskeleton Signaling; F/RA, FXR/RXR Activation; L/RA, LXR/RXR Activation; Glyc, Glycolysis/Gluconeogenesis; CME, Caveolar-mediated Endocytosis; GRS, Glucocorticoid Receptor Signaling; NMOSR, NRF2-mediated Oxidative Stress Response; CME, Clathrin-mediated Endocytosis; FRMP, Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes; IntS, Integrin Signaling; PyrM, Pyruvate Metabolism; RAMR, Regulation of Actin-based Motility by Rho; GluM, Glutathione Metabolism; XMS, Xenobiotic Metabolism Signaling; AHRS, Aryl Hydrocarbon Receptor Signaling; PurM, Purine Metabolism; PI3K, PI3K/AKT Signaling.
Discussion
Although blood plasma represents the most promising specimen for biomarker discovery, the vast heterogeneity across human populations presents a major challenge for confidently identifying candidate biomarkers for diseases. Mouse models are commonly employed to minimize the effect of heterogeneity, offering high quality matched samples when genetic background, breeding conditions, and sampling standards are controlled. In recent years, mice have played a critical role in biomarker discovery and drug tests of human diseases, i.e., the majority of full-scale tests on humans are preceded by tests on mice. Because many human diseases can only be diagnosed at a late stage when it is often too late for effective treatment 10, 27, the relatively shorter time course of disease development in mice and availability of mouse models at defined disease stages makes discovery of candidate biomarkers for early diagnosis possible. Thus, proteomic biomarker discovery using mouse plasma is increasingly attractive.
In this work, we demonstrated an automated tandem IgY7-SuperMix separation strategy for effectively enriching low abundance proteins from mouse plasma, thus enhancing LC-MS(/MS)-based proteomic detection of low abundance proteins in mouse plasma or serum. Our results illustrate nearly a two-fold improvement in proteome coverage, i.e., 523 proteins were confidently identified after SuperMix fractionation compared to only 263 proteins identified in the IgY7 flow-through. The enhanced detection of low-abundance proteins is further illustrated by the improved coverage of proteins in many mapped canonical signaling pathways. Automation of the tandem two column separations resulted in good reproducibility, decreased costs in labor and materials, and simplified experimental procedures, all of which may be critical for large-scale experiments.
There are also some limitations of this current strategy that should be noted. First, some moderate abundance proteins were partially captured by the SuperMix column in both mouse and human plasma, such as Apoa4 (18.3%) 18, which is likely due to the low titration of a corresponding antibody and the low binding affinity between the antibody and antigen. However, because of the reproducibility of the system, these proteins are still quantifiable. Although we optimized the amount of sample to load onto the columns and the capture efficiency of some proteins improved when a smaller amount of sample was loaded, the reduced amount in recovered proteins may present a sensitivity challenge for down-stream sample processing and LC-MS analysis. Second, some proteins of potential interest in the plasma may be bound to the higher abundance proteins 28, 29 and hard to be separated from those high abundance proteins. Analyzing both the SuperMix bound and flow-through fractions may address such issues.
Combining the current strategy with other protein separation methods, such as SDS-PAGE, SCX, and/or reverse-phase fractionation may result in even greater depth of coverage for the mouse plasma proteome. Moreover, combining this strategy with techniques to enrich modified proteins/peptides may provide insight into plasma protein modifications, such as phosphorylation, acetylation, ubiquitination, sumolation, oxidation, and glycosylation that are involved in many biological processes and diseases. The incorporation of data from plasma with the repository of data for mouse tissues 30 strengthens future biomarker discovery efforts.
Conclusion
A tandem IgY7-SuperMix immunoaffinity separation strategy designed to specifically enrich low abundance proteins from mouse plasma improves the proteome coverage and enhances detection of low abundance proteins by LC-MS/MS. Results also demonstrate reproducibility for effectively determining differential abundances. Considering the overall reproducibility and sensitivity of this automated tandem separation system, we anticipate broad applications of this strategy for biomarker discovery using mouse models.
Supplementary Material
Acknowledgments
Portions of this research were supported by National Institutes of Health grants R01 DK074795 and RR018522. Experimental work was performed in the Environmental Molecular Sciences Laboratory, a U. S. Department of Energy (DOE) Office of Biological and Environmental Research national scientific user facility on the Pacific Northwest National Laboratory (PNNL) campus. PNNL is multi-program national laboratory operated by Battelle for the DOE under Contract No. DE-AC05-76RLO 1830.
Abbreviations
- SCX
strong cation exchange
- 1D
one-dimensional
- 2D
two-dimensional
- IgY
immunoglobulin yolk
- SPE
solid-phase extraction
- LTQ
linear ion trap quadrupole
- IPI
International Protein Index
- Xcorr
cross-correlation score
- ΔCn
delta correlation
- FDR
false discovery rate
- SMIX_B
SuperMix bound fraction
- SMIX_FT
SuperMix flow-through fraction
Footnotes
SUPPORTING INFORMATION AVAILABLE
A complete list of identified proteins and peptides and mapped canonical pathways along with corresponding spectral count information is available in a Microsoft Excel worksheet. All files are downloadable free of charge at http://pubs.acs.org.
References
- 1.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452 (7187):571–9. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 2.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1 (11):845–67. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 3.Qian WJ, Jacobs JM, Liu T, Camp DG, 2nd, Smith RD. Advances and challenges in liquid chromatography-mass spectrometry-based proteomics profiling for clinical applications. Mol Cell Proteomics. 2006;5 (10):1727–44. doi: 10.1074/mcp.M600162-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Issaq HJ, Xiao Z, Veenstra TD. Serum and plasma proteomics. Chem Rev. 2007;107 (8):3601–20. doi: 10.1021/cr068287r. [DOI] [PubMed] [Google Scholar]
- 5.Pieper R, Su Q, Gatlin CL, Huang ST, Anderson NL, Steiner S. Multi-component immunoaffinity subtraction chromatography: an innovative step towards a comprehensive survey of the human plasma proteome. Proteomics. 2003;3 (4):422–32. doi: 10.1002/pmic.200390057. [DOI] [PubMed] [Google Scholar]
- 6.Zolotarjova N, Martosella J, Nicol G, Bailey J, Boyes BE, Barrett WC. Differences among techniques for high-abundant protein depletion. Proteomics. 2005;5 (13):3304–13. doi: 10.1002/pmic.200402021. [DOI] [PubMed] [Google Scholar]
- 7.Huang L, Harvie G, Feitelson JS, Gramatikoff K, Herold DA, Allen DL, Amunngama R, Hagler RA, Pisano MR, Zhang WW, Fang X. Immunoaffinity separation of plasma proteins by IgY microbeads: meeting the needs of proteomic sample preparation and analysis. Proteomics. 2005;5 (13):3314–28. doi: 10.1002/pmic.200401277. [DOI] [PubMed] [Google Scholar]
- 8.Liu T, Qian WJ, Mottaz HM, Gritsenko MA, Norbeck AD, Moore RJ, Purvine SO, Camp DG, 2nd, Smith RD. Evaluation of multiprotein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol Cell Proteomics. 2006;5 (11):2167–74. doi: 10.1074/mcp.T600039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters LL, Robledo RF, Bult CJ, Churchill GA, Paigen BJ, Svenson KL. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat Rev Genet. 2007;8 (1):58–69. doi: 10.1038/nrg2025. [DOI] [PubMed] [Google Scholar]
- 10.Whiteaker JR, Zhang H, Zhao L, Wang P, Kelly-Spratt KS, Ivey RG, Piening BD, Feng LC, Kasarda E, Gurley KE, Eng JK, Chodosh LA, Kemp CJ, McIntosh MW, Paulovich AG. Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J Proteome Res. 2007;6 (10):3962–75. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
- 11.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O’Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420 (6915):520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 12.Faca VM, Song KS, Wang H, Zhang Q, Krasnoselsky AL, Newcomb LF, Plentz RR, Gurumurthy S, Redston MS, Pitteri SJ, Pereira-Faca SR, Ireton RC, Katayama H, Glukhova V, Phanstiel D, Brenner DE, Anderson MA, Misek D, Scholler N, Urban ND, Barnett MJ, Edelstein C, Goodman GE, Thornquist MD, McIntosh MW, DePinho RA, Bardeesy N, Hanash SM. A mouse to human search for plasma proteome changes associated with pancreatic tumor development. PLoS Med. 2008;5 (6):e123. doi: 10.1371/journal.pmed.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menon R, Zhang Q, Zhang Y, Fermin D, Bardeesy N, DePinho RA, Lu C, Hanash SM, Omenn GS, States DJ. Identification of novel alternative splice isoforms of circulating proteins in a mouse model of human pancreatic cancer. Cancer Res. 2009;69 (1):300–9. doi: 10.1158/0008-5472.CAN-08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pitteri SJ, Faca VM, Kelly-Spratt KS, Kasarda AE, Wang H, Zhang Q, Newcomb L, Krasnoselsky A, Paczesny S, Choi G, Fitzgibbon M, McIntosh MW, Kemp CJ, Hanash SM. Plasma proteome profiling of a mouse model of breast cancer identifies a set of up-regulated proteins in common with human breast cancer cells. J Proteome Res. 2008;7 (4):1481–9. doi: 10.1021/pr7007994. [DOI] [PubMed] [Google Scholar]
- 15.Hung KE, Kho AT, Sarracino D, Richard LG, Krastins B, Forrester S, Haab BB, Kohane IS, Kucherlapati R. Mass spectrometry-based study of the plasma proteome in a mouse intestinal tumor model. J Proteome Res. 2006;5 (8):1866–78. doi: 10.1021/pr060120r. [DOI] [PubMed] [Google Scholar]
- 16.Kumar SG, Rahman MA, Lee SH, Hwang HS, Kim HA, Yun JW. Plasma proteome analysis for anti-obesity and anti-diabetic potentials of chitosan oligosaccharides in ob/ob mice. Proteomics. 2009;9 (8):2149–62. doi: 10.1002/pmic.200800571. [DOI] [PubMed] [Google Scholar]
- 17.Ren Y, Wang J, Xia J, Jiang C, Zhao K, Li R, Xu N, Xu Y, Liu S. The alterations of mouse plasma proteins during septic development. J Proteome Res. 2007;6 (7):2812–21. doi: 10.1021/pr070047k. [DOI] [PubMed] [Google Scholar]
- 18.Qian WJ, Kaleta DT, Petritis BO, Jiang H, Liu T, Zhang X, Mottaz HM, Varnum SM, Camp DG, 2nd, Huang L, Fang X, Zhang WW, Smith RD. Enhanced detection of low abundance human plasma proteins using a tandem IgY12-SuperMix immunoaffinity separation strategy. Mol Cell Proteomics. 2008;7 (10):1963–73. doi: 10.1074/mcp.M800008-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4 (3):207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 20.Qian WJ, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Camp DG, Smith RD. Probability-Based Evaluation of Peptide and Protein Identifications from Tandem Mass Spectrometry and SEQUEST Analysis: The Human Proteome. J Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 21.Anderson NG, Willis DD, Holladay DW, Caton JE, Holleman JW, Eveleigh JW, Attrill JE, Ball FL, Anderson NL. Analytical techniques for cell fractions. XX. Cyclic affinity chromatography: principles and applications. Anal Biochem. 1975;68 (2):371–93. doi: 10.1016/0003-2697(75)90634-x. [DOI] [PubMed] [Google Scholar]
- 22.Rodrigues LR, Teixeira JA, Schmitt FL, Paulsson M, Lindmark-Mansson H. The role of osteopontin in tumor progression and metastasis in breast cancer. Cancer Epidemiol Biomarkers Prev. 2007;16 (6):1087–97. doi: 10.1158/1055-9965.EPI-06-1008. [DOI] [PubMed] [Google Scholar]
- 23.Hu SM, Li F, Yu HM, Li RY, Ma QY, Ye TJ, Lu ZY, Chen JL, Song HD. The mimecan gene expressed in human pituitary and regulated by pituitary transcription factor-1 as a marker for diagnosing pituitary tumors. J Clin Endocrinol Metab. 2005;90 (12):6657–64. doi: 10.1210/jc.2005-0322. [DOI] [PubMed] [Google Scholar]
- 24.Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson’s disease. Science. 2003;302 (5646):819–22. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 25.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF. A network-based analysis of systemic inflammation in humans. Nature. 2005;437 (7061):1032–7. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- 26.Franke TF. PI3K/Akt: getting it right matters. Oncogene. 2008;27 (50):6473–88. doi: 10.1038/onc.2008.313. [DOI] [PubMed] [Google Scholar]
- 27.Kelly-Spratt KS, Kasarda AE, Igra M, Kemp CJ. A mouse model repository for cancer biomarker discovery. J Proteome Res. 2008;7 (8):3613–8. doi: 10.1021/pr800210b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granger J, Siddiqui J, Copeland S, Remick D. Albumin depletion of human plasma also removes low abundance proteins including the cytokines. Proteomics. 2005;5 (18):4713–8. doi: 10.1002/pmic.200401331. [DOI] [PubMed] [Google Scholar]
- 29.Gong Y, Li X, Yang B, Ying W, Li D, Zhang Y, Dai S, Cai Y, Wang J, He F, Qian X. Different immunoaffinity fractionation strategies to characterize the human plasma proteome. J Proteome Res. 2006;5 (6):1379–87. doi: 10.1021/pr0600024. [DOI] [PubMed] [Google Scholar]
- 30.Karen S, Kelly-Spratt AEK, Igra Mark, Kemp Christopher J. A Mouse Model Repository for Cancer Biomarker Discovery. J Proteome Res. 2008;7 (8):3613–3618. doi: 10.1021/pr800210b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.