Introductory paragraph
DNA-double strand break (DSB) repair involves complex interactions between chromatin and repair proteins, including the Tip60 tumor suppressor1. Tip60 is an acetyltransferase which acetylates both histones2-5 and the ATM kinase6, 7. Inactivation of Tip60 leads to defective DNA repair2-4 and increased cancer risk8-11. However, how DNA damage activates Tip60’s acetyltransferase activity is not known. Here, we show that direct interaction between Tip60’s chromodomain and histone H3 trimethylated on lysine 9 (H3K9me3) at DSBs activates Tip60’s acetyltransferase activity. Depletion of intracellular H3K9me3 blocks activation of Tip60’s acetyltransferase activity, resulting in defective ATM activation and widespread defects in DSB repair. In addition, the ability of Tip60 to access H3K9me3 is dependent on the DNA damage induced displacement of HP1β from H3K9me3. Finally, we demonstrate that the mre11-rad50-nbs1 complex targets Tip60 to H3K9me3, and is required to activate Tip60’s acetyltransferase activity. These results reveal a new function for H3K9me3 in co-ordinating activation of Tip60-dependent DNA repair pathways, and imply that aberrant patterns of histone methylation may contribute to cancer by altering the efficiency of DSB repair.
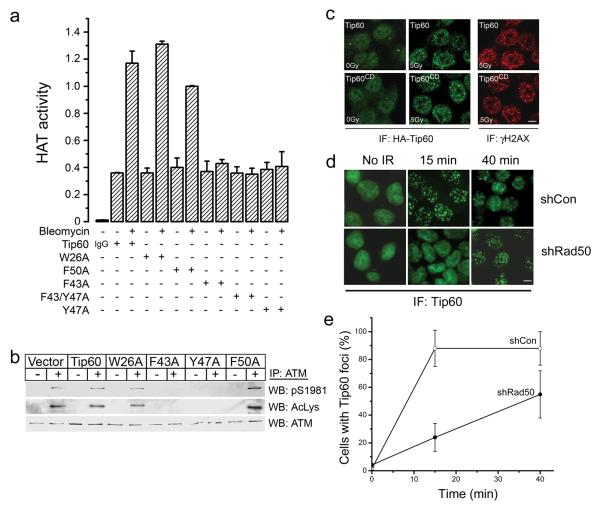
The Tip60 acetyltransferase is a tumor suppressor gene whose expression is altered in both breast and prostate tumors8-11. Tip60 acetylates key proteins involved in the DNA damage response2-7, 12 and plays a critical role in the detection and repair DNA double strand breaks (DSBs)2-5, 7. Currently, the mechanism by which Tip60’s acetyltransferase activity is increased by DNA damage is not known. Here, we examined if interaction between Tip60’s chromodomain13 and histones adjacent to DSBs regulate Tip60 acetyltransferase activity. Chromodomains are specialized binding modules containing conserved hydrophobic amino-acids which interact with methyl groups on methylated lysine residues13-15. For example, the chromodomain of HP1α interacts with histone H3 trimethylated on lysine 9 (H3K9me3)14,15. Sequence alignment of the chromodomains of Tip60 and HP1α demonstrates that Tip60 contains the conserved aromatic amino-acids required for HP1α binding to H3K9me3 (figure S1a), implying that Tip60’s chromodomain functions as a methyl-lysine binding domain. To examine this possibility, point mutations were introduced into Tip60’s chromodomain, and the ability of the DNA-damaging agent bleomycin to activate Tip60’s acetyltransferase activity examined. The acetyltransferase activity of immunopurified Tip60 was quantitated by measuring the ability of Tip60 to acetylate a peptide derived from histone H4 (figure S1b), one of Tip60’s in vivo substrates2. DNA damage increases the acetyltransferase activity of both endogenous and transfected Tip60 (figure S1c and S1d)6, 7, but does not alter the overall level of Tip60 protein (figure S2b)6, 7. The Tip60 proteins with mutations in the chromodomain were stably expressed in HeLaS3 cells (figure S2a), and their intrinsic acetyltransferase activity measured. Mutation of phenylalanine-43 (Tip60F43A) or tyrosine-47 (Tip60Y47A) in Tip60’s chromodomain abolished activation of Tip60’s acetyltransferase activity, whereas mutations in tryptophan-26 (Tip60W26A) or phenylalanine-50 (Tip60F50A) had little or no impact (figure 1a). Activation of Tip60 by DSBs leads to the Tip60-dependent acetylation and activation of the ATM kinase6, 7. Mutations in the chromodomain which abolished activation of Tip60’s acetyltransferase activity (Tip60F43A and Tip60Y47A) also blocked the Tip60-dependent acetylation and autophosphorylation of the ATM protein (figure 1b). A more detailed analysis of cells expressing the Tip60Y47A mutant (referred to as Tip60CD) revealed a profound defect in both ATM activation and ATM-dependent phosphorylation of chk2 (figure S2b), indicating that Tip60’s chromodomain is essential for activation of the ATM signaling pathway.
Figure 1. Tip60’s chromodomain is required for acetyltransferase activity.
(a) HeLaS3 cells expressing HA-Tip60 or HA-Tip60 with the indicated mutations were exposed to bleomycin (5μM for 40 minutes) as indicated. Tip60 was immunopurified with HA antibody or IgG, and the associated acetyltransferase activity measured as described in methods. Each data point represents the average of 3 independent immunoprecipitation assays, results ± SD. (b) HeLaS3 cells expressing vector, Tip60 or the indicated Tip60 chromodomain mutants were untreated (−) or irradiated (5Gy: +), and ATM immunoprecipitated. Western blot analysis to detect ATM, phospho-ATM (pS1981) or Tip60-dependent acetylation of ATM (AcLys) was then carried out. (c) HeLaS3 cells expressing either HA-Tip60 or HA-Tip60CD (Tip60Y47A) were untreated (0Gy) or irradiated (5Gy). Cells were then fixed and immunofluorescent (IF) staining with antibodies to detect γH2AX or HA-Tip60 carried out. Scale bar equals 10μM. (d) HCT116 cells expressing either a non-targeting shRNA (shCon) or shRNA targeting Rad50 (shRad50) were transfected with HA-Tip60. Cells were either unirradiated (No IR) or exposed to 0.5Gy. Cells were fixed 15 or 40 minutes post-irradiation and immunofluorescent staining with antibodies to detect HA-Tip60 carried out. Scale bar equals 10μM. (e) Quantification of the images in (e). The number of cells with Tip60 foci (defined as cells with > 5 foci) was measured visually. Results ± SD, based on analyzing at least 80 cells per slide.
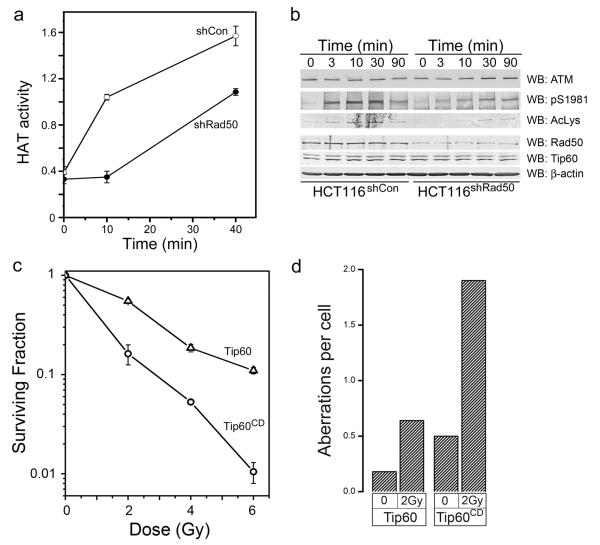
Chromodomains function as methyl-lysine binding proteins13-15, implying that Tip60’s chromodomain may be essential to recruit and retain Tip60 at DSB. However, the Tip60CD mutant was efficiently recruited to sites of DNA damage (figure 1c), indicating that the chromodomain is not required to target Tip60 to DSBs. This implies that an alternative mechanism must exist to recruit Tip60 to sites of DNA damage. Previously, we demonstrated that Tip60 and ATM exist as a unique complex in cells 7, 12, and that, in the absence of ATM, Tip60 recruitment to DSBs is severely compromised 7, 12. In addition to Tip60, the efficient recruitment of ATM to DSBs and activation of ATM’s kinase requires the mre11-rad50-nbs1 (MRN) complex 16-18. This suggests that Tip60 is recruited to MRN at DSBs as part of an ATM-Tip60 complex. To test this possibility, we first demonstrated that the interaction between ATM and Tip60 was not altered by mutations of Tip60’s chromodomain (figure S2a). Next, we examined the role of the MRN complex in Tip60 activation using cells in which the rad50 component of MRN was reduced by shRNA19 (figure 2b). Cells in which rad50 was depleted failed to recruit Tip60 to DSBs within 15 minutes of exposure to ionizing radiation (figure 1d and 1e), although there was a slow accumulation of Tip60 at DSBs at later times. Formation of γH2AX foci was unaffected by either rad50 depletion or mutational inactivation of Tip60’s chromodomain (figure S3a). However, activation of Tip60’s acetyltransferase activity (figure 2a) and the Tip60-dependent acetylation and activation of the ATM kinase (figure 2b) were reduced in cells lacking functional MRN complex. This is consistent with previous reports indicating that cells with compromised MRN function exhibit reduced or attenuated activation of the ATM kinase in response to DSBs 16-18. These results demonstrate that both Tip60 and the MRN complex make critical contributions to the activation of ATM at endogenous DSBs. Cells in which the chromodomain of Tip60 is mutated should therefore exhibit a defective DNA damage response. Consistent with this, cells expressing the Tip60CD mutant exhibited increased sensitivity to ionizing radiation (figure 2c), and contained significantly higher levels of chromosome aberrations after exposure to ionizing radiation (figure 2d and S3b). These results indicate a model in which the previously characterized ATM-Tip60 complex 7, 12 is recruited to the MRN complex as an inactive ATM-Tip60 complex. Activation of Tip60’s acetyltransferase (through a mechanism involving the chromodomain) then leads to acetylation of ATM and activation of ATM’s kinase.
Figure 2. MRN is essential for Tip60 activation.
(a) HCT116 cells expressing either a non-targeting shRNA (shCon) or shRNA targeting Rad50 (shRad50) were exposed to bleomycin (5μM) for the indicated times. Tip60 was then immunopurified, and the associated HAT activity measured. Each data point represents the average of 3 independent assays, results ± SD. (b) HCT116 cells expressing either a non-targeting shRNA (shCon) or shRNA targeting Rad50 (shRad50) were irradiated (2Gy) and cell extracts prepared. ATM was immunoprecipitated, and ATM levels, phospho-ATM (pS1981) and ATM acetylation (AcLys) measured by western blot analysis. Levels of Rad50, Tip60 and β-actin in the original lysate are shown. (c) HeLaS3 cells expressing either HA-Tip60 (Δ) or HA-Tip60CD (○) were irradiated at the indicated dose, and the number of surviving colonies measured 12 days later. Each data point represents the average of 3 independent assays, results ± SD. (d) Tip60 and Tip60HD cells were irradiated (2Gy) and allowed to recover for 14hr. Metaphase spreads were subsequently scored for chromosome aberrations. Results expressed as number of aberrations per cell (n = 50 cells).
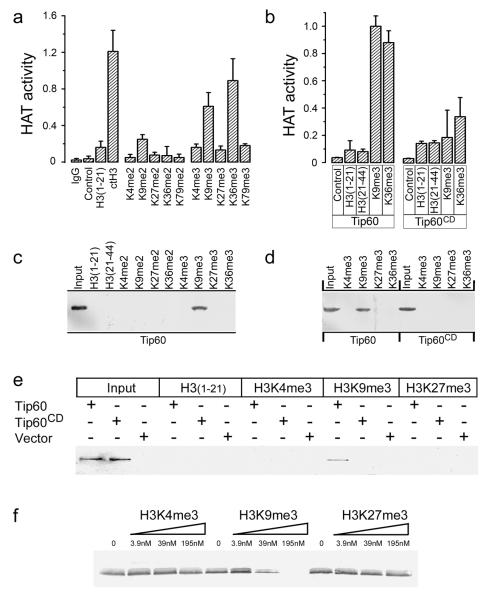
Next, we determined how Tip60’s chromodomain contributes to the regulation of Tip60’s acetyltransferase activity. Histone H3 is methylated in vivo on lysines 4, 9, 27, 36 and 79, making H3 a potential binding partner for Tip60’s chromodomain 20, 21. Calf thymus H3 (which is post-translationally modified) and recombinant H3 which was methylated in vitro activated the acetyltransferase activity of Tip60 (figure S4a and S4b), but not the Tip60CD mutant (figure S4c). This implies that methylated histone H3 activates Tip60’s acetyltransferase activity through interaction with Tip60’s chromodomain. To identify the methylation sites on H3, peptides corresponding to the major histone H3 methylation sites (supplementary table 1) were examined for their ability to stimulate Tip60’s acetyltransferase activity in vitro. H3 peptides di- or tri-methylated at lysines 4, 27 and 79 did not stimulate Tip60’s acetyltransferase activity (figure 3a), whereas H3K9me3 and H3K36me3 peptides activated Tip60’s acetyltransferase activity. H3K9me2 had a small stimulatory effect on Tip60 acetyltransferase activity. Tip60 showed a concentration dependent increase in acetyltransferase activity when incubated with increasing concentrations of H3K9me3 peptide, but not with H3K4me3 (figure S5a). Importantly, peptides containing either H3K9me3 or H3K36me3 did not significantly activate the acetyltransferase activity of the Tip60CD chromodomain mutant (figure 3b), implying specific interaction between Tip60’s chromodomain and H3K9me3 or H3K36me3. To further examine these interactions, nuclear extracts containing Tip60 were incubated with methylated H3 peptides immobilized on affinity resin. Tip60 was specifically bound to H3K9me3 (figure 3c and S5b), whereas the Tip60CD mutant failed to bind to H3K9me3 (figure 3d and S5c). However, no significant binding of Tip60 to H3K36me3 was seen under standard binding conditions (figure 3b), although weak binding of H3K36me3 to Tip60 was detected under low stringency binding conditions (figure S5d). This implies that the interaction between Tip60 and H3K36me3 is significantly weaker than the interaction with H3K9me3, implying that H3K9me3 is the primary binding site for Tip60’s chromodomain. To further confirm this conclusion, the interaction between purified in vitro translated (IVT) Tip60 protein and H3K9me3 was examined. IVT Tip60, but not IVT Tip60CD, bound specifically to H3K9me3, but not H3K4me3, H3K27me3 (figure 3e) or to H3K36me3 (figure S5e). Further, Tip60 was the only detectable polypeptide associated with the H3K9me3 affinity column, indicating that Tip60’s chromodomain interacts directly with H3K9me3 (figure S6a and S6b). When IVT Tip60 was prebound to immobilized H3K9me3 affinity columns, Tip60 was specifically eluted by H3K9me3 peptide, but not by either H3K4me3 or H3K27me3 (figure 3f). Finally, the acetyltransferase activity of purified IVT Tip60 (but not IVT Tip60CD) was specifically activated by the H3K9me3 peptide (figure S6c). Figure 3 therefore demonstrates that direct interaction between H3K9me3 and Tip60’s chromodomain specifically activates Tip60’s acetyltransferase activity, implying that the chromodomain of Tip60 functions as an allosteric regulator of Tip60’s acetyltransferase activity.
Figure 3. Tip60’s chromodomain interacts with H3K9me3 in vitro.
(a) Immunopurified HA-Tip60 was incubated with buffer (control), calf thymus H3 (ctH3), unmodified H3 peptide (amino-acids 1-21), or H3 peptide di- or trimethylated at the indicated lysine. Peptide sequences listed in supplementary table 1. Excess peptide was removed by washing, and associated Tip60 acetyltransferase activity measured. Mock immunopurification with IgG shown. Each data point represents the average of 3 independent assays, results ± SD. (b) Immunopurified HA-Tip60 or HA-Tip60CD were incubated with buffer (control), unmodified peptides H3(1-21) or H3(21-44), corresponding to amino-acids 1-21 or 21-44 of H3 respectively, or peptides trimethylated on lysine 9 (H3K9me3) or lysine 36 (H3K36me3). Each data point represents the average of 3 independent assays, results ± SD. (c) Biotinylated peptides were pre-bound to sepharose-avidin beads and incubated with nuclear extracts from cells expressing HA-Tip60. Bound Tip60 was eluted and detected by western blot using HA antibody. Unmodified peptides H3(1-21) and H3(21-44), di- or trimethylated H3K4, H3K9, H3K27, H3K36 and input Tip60 is shown. Polyacrylamide gels were stained with Coomasie Blue to confirm loading (figure S3b). (d) Biotinylated peptides were pre-bound to sepharose-avidin beads and incubated with nuclear extracts from cells expressing HA-Tip60 or HA-Tip60CD. Bound Tip60 was eluted and detected by western blot analysis using HA antibody. Polyacrylamide gels were stained with Coomasie Blue to confirm loading (figure S3c). (e) Biotinylated H3(1-21), H3K4me3, H3K9me3 or H3K27me3 peptides were pre-bound to sepharose-avidin beads and incubated with in vitro translated HA-Tip60, in vitro translated HA-Tip60CD or mock translation reactions (vector). Bound Tip60 was eluted and detected by western blot analysis using HA antibody. Input in vitro translates as indicated. (f) In vitro translated HA-Tip60 was prebound to biotinylated H3K9me3 peptide immobilized on sepharose-avidin beads. Following washing, increasing concentrations of competitor peptides H3K4me3, H3K9me3 or H3K27me3 were added and incubated for 30 minutes. The remaining HA-Tip60 was eluted from the column, separated by SDS-PAGE and detected by western blot analysis using HA antibody.
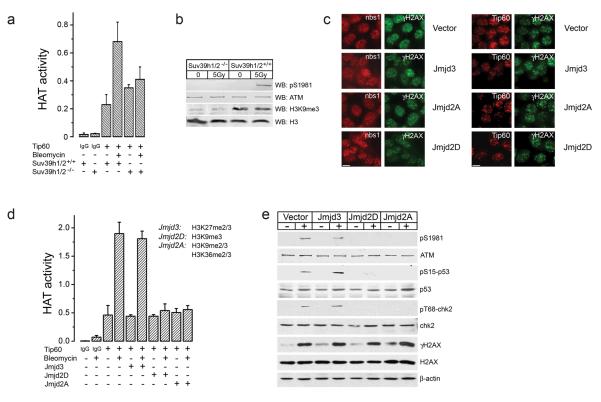
Next, we determined if the levels of H3K9me3 were important for Tip60 function in vivo. MEFS derived from mice in which the 2 main H3K9 methyltransferases, Suv39h1 and Suv39h2, are deleted were examined 22. Suv39h1/2−/− mice have reduced levels of H3K9me3 (figure 4b) and display increased genomic instability 22. Activation of Tip60’s acetyltransferase activity (figure 4a), and the Tip60-dependent activation of ATM (figure 4b) was significantly impaired in Suv39h1/2−/− MEFs immediately following exposure to ionizing radiation. However, weak ATM activation was detected at later time points (figure S7) in Suv39h1/2−/− MEFs. Reduced methylation of H3K9me3 therefore correlates with defective activation of Tip60 and ATM in Suv39h1/2−/− MEFs. To further manipulate H3K9 methylation in vivo, histone demethylases which specifically demethylate H3K9me3 (Jmjd2D/KDM4D; 23) or both H3K9me3 and H3K36me3 (Jmjd2A/KDM4A; 23, 24) were expressed in cells in order to reduce the levels of H3K9me3. Demethylation of H3K27me3 (Jmjd3/KDM6B; 25) was used as a control. The histone demethylases were efficiently expressed in cells (figure S8a), and specifically demethylated H3K9me3 and H3K27me3 (figure S8b and S8c). Demethylation of H3K9, H3K27 or H3K36 did not affect the formation of γH2AX foci, or the recruitment of the MRN complex to sites of DNA damage (figure 4c). Demethylation of histones does not, therefore, impose structural or other changes in the chromatin which inhibit DNA damage signaling events. In addition, Tip60 was recruited to DSBs in cells in which H3K9me3 levels were reduced (figure 4c), providing further evidence that interactions between methylated histones and Tip60’s chromodomain are not required to recruit Tip60 to DSBs. Demethylation of either H3K9me3 (by Jmjd2D/KDM4D) or both H3K9me3 and H3K36me3 (by Jmjd2A/KDM4A) inhibited the activation of Tip60’s acetyltransferase activity by bleomycin (figure 4d). Demethylation of H3K27me3 (by Jmjd3/KDM6B) had no effect. Demethylation of H3K9me3 (by either Jmjd2D/KDM4D or Jmjd2A/KDM4A) also blocked the Tip60-dependent activation of the ATM DNA damage response pathway, blocking ATM autophosphorylation, and preventing the subsequent phosphorylation of p53 and chk2 by ATM (figure 4e). However, phosphorylation of H2AX, which can be mediated by DNA-PKcs in the absence of ATM 26, was unaffected by demethylation of histone H3. Figure 4 therefore demonstrates that activation of Tip60’s acetyltransferase activity by DNA damage is inhibited when the levels of H3K9me3 in the cell are reduced. This is consistent with the in vitro binding data in figure 3, and indicates that interaction between Tip60’s chromodomain and H3K9me3 at DSBs mediates activation of Tip60’s acetyltransferase activity.
Figure 4. Demethylation of H3K9 blocks activation of Tip60 acetyltransferase activity.
(a) Wild type Suv39h1/2+/+ or Suv39h1/2−/− double knockout MEFs were untreated (−) or exposed to bleomycin (5μM for 30 minutes). Tip60 was immunopurified with Tip60 antibody, and associated acetyltransferase activity measured. Mock purification with IgG is shown for comparison. Each data point represents the average of 3 independent assays, results ± SD. (b). Wild type Suv39h1/2+/+ or Suv39h1/2−/− double knockout MEFs were untreated (−) or irradiated (5Gy). Levels of ATM, phospho-ATM (pS1981), histone H3 or H3 trimethylated on lysine 9 (H3K9me3) were monitored by western blot analysis. (c) 293T cells expressing HA-Tip60 were transiently transfected with vector, or expression vectors for the histone demethylases Jmjd3/KDM6B, Jmjd2A/KDM4A or Jmjd2D/KDM4D. Cells were irradiated (5Gy) and immunofluorescent staining used to detect either nbs1 and γH2AX (left hand panel) or FLAG-Tip60 and γH2AX (right hand panel). Scale bar equals 10μM. (d) 293T cells were transiently transfected with vector, Jmjd3/KDM6B, Jmjd2A/KDM4A or Jmjd2D/KDM4D. Exactly 30h post-transfection, cells were exposed to bleomycin (5μM for 30 minutes) as indicated. Tip60 was immunopurified, and the associated acetyltransferase activity measured. Each data point represents the average of 3 independent assays, results ± SD. (e) 293T cells were transiently transfected with vector, Jmjd3/KDM6B, Jmjd2A/KDM4A or Jmjd2D/KDM4D demethylases. Exactly 30h post-transfection, cells were untreated (−) or irradiated (5Gy) and analyzed by western blot analysis for ATM, phospho-ATM (pS1981), chk2 and phospho-chk2 (pT68-chk2), p53 and phospho-p53 (pS15-p53), and H2AX and phospho-H2AX (γH2AX).
To confirm that Tip60’s chromodomain and H3K9me3 interact in vivo, Tip60 immunoprecipitates were examined for the presence of H3. Figure 5a demonstrates that bleomycin induced a DNA-damage dependent interaction between Tip60 and histone H3 (figure 5a) and that this co-precipitating histone H3 was trimethylated on lysine 9. This interaction between Tip60 and H3 was significantly reduced in cells expressing the Tip60 chromodomain mutant (figure 5a). When H3K9me3 methylation was reduced by expression of the histone demethylases Jmjd2D/KDM4D or Jmjd2A/KDM4A, the DNA damage dependent interaction between Tip60’s chromodomain and histone H3 was significantly reduced (figure 5b). Further, isolation of intact Tip60-H3K9me3 complexes could be achieved without disrupting the chromatin (figure S8e), suggesting that the Tip60-H3 complexes are either mobile within the nucleoplasm, or are only weakly associated with the chromatin after DNA damage. The results are consistent with a model in which the recruitment of Tip60 to DSBs leads to direct interaction between Tip60’s chromodomain and H3K9me3, increasing Tip60’s acetyltransferase activity. Cells lacking H3K9me3 should therefore be sensitive to agents which generate DSBs. Indeed, demethylation of H3K9 (by Jmjd2D/KDM4D) but not H3K27 (by Jmjd3/KDM6B) sensitizes cells to ionizing radiation (figure 5c). H3K9me3 is therefore essential for activation of Tip60’s acetyltransferase activity and for cells to survive ionizing radiation exposure.
Figure 5. DNA damage regulates interaction between Tip60 and H3K9me3.
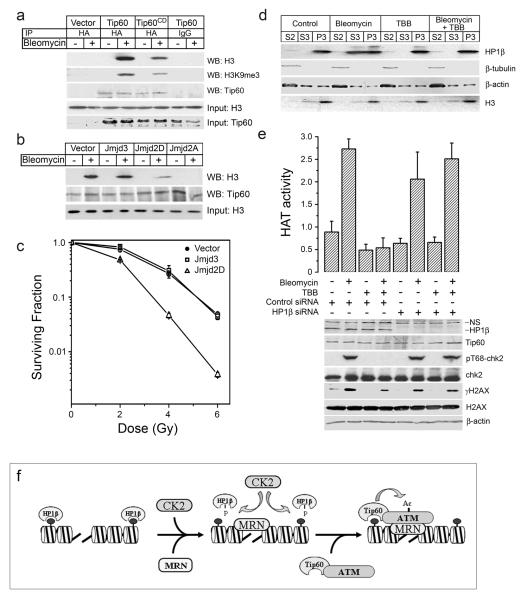
(a) HeLaS3 cells expressing vector, HA-Tip60 or HA-Tip60CD were untreated (−) or exposed to bleomycin (5μM for 40mins), and cells lyzed in the presence of benzonase to release chromatin associated proteins. Cell extracts were immunoprecipitated with IgG or HA antibody and associated histone H3, H3K9me3 and HA-Tip60 detected by western blot. Levels of input Tip60 and H3 shown. (b) HeLaS3 cells were transiently transfected with vector, Jmjd3/KDM6B, Jmjd2A/KDM4A or Jmjd2D/KDM4D. Exactly 30h post-transfection, cells were exposed to bleomycin (5μM for 40min) and lyzed in the presence of benzonase. FLAG-Tip60 was immunoprecipitated and Tip60 and histone H3 detected by western blot analysis. Levels of input H3 are shown. (c) 293T cells transiently expressing vector, Jmjd3/KDM6B or Jmjd2D/KDM4D were irradiated, and cell survival assays carried out. Each data point represents the average of 3 independent assays, results ± SD. (d) Cells were preincubated in TBB (75μM for 2hr), followed by bleomycin as indicated. Cytoplasmic (S2), nucleoplasmic (S3) and chromatin (P3) fractions were analyzed for HP1β expression. Levels of β-tubulin (cytoplasmic marker), β-actin and H3 shown. (e) 293T cells transfected with control or HP1β-specific siRNA specific were incubated with TBB (75μM for 2hr) followed by bleomycin (5μM for 30min). Top panel: Tip60 was immunopurified with HA antibody and the associated acetyltransferase activity measured. Each data point represents the average of 3 independent assays, results ± SD. Bottom panel: Levels of HP1β, Tip60, phospho-chk2 (pT68-chk2), chk2, phospho-H2AX (γH2AX), H2AX and β-actin measured by western blot analysis. (f) Model for ATM activation. Generation of DSBs leads to recruitment of MRN and CK2 to the break. CK2-dependent phosphorylation of HP1β releases HP1β from the chromatin. Subsequent recruitment of the ATM-Tip60 to MRN facilitates interaction between Tip60’s chromodomain and free H3K9me3. Tip60’s acetyltransferase activity is upregulated, leading to acetylation and activation of ATM’s kinase activity. The fully active ATM kinase then phosphorylates key target proteins and regulates ATM-dependent signaling events. Recruitment of CK2 and MRN to the DSB are independent events.
Interaction between Tip60 and H3K9me3 may occur either through DNA damage induced H3K9 methylation at DSBs, or through utilization of pre-existing H3K9 methylation sites by Tip60 at DSBs. The overall levels of H3K9me3, H3K27me3 and H3K36me3 are unaltered following DNA damage (figure S8d), indicating that interactions with pre-existing H3 methylation sites are involved. The majority of the cellular H3K9me3 is bound by the HP1 family of proteins 14, 15, potentially making it unavailable for interaction with Tip60. However, following DNA damage, the chromodomain of HP1β is phosphorylated by casein kinase 2, and this phosphorylation leads to release of HP1β from H3K9me3 27. This DNA damage induced release of HP1β from the chromatin may generate domains of H3K9me3 which can interact with Tip60’s chromodomain. Therefore, we examined if release of HP1β from the chromatin was required for activation of Tip60’s acetyltransferase activity. Chromatin fractionation studies confirmed that HP1β is located on the chromatin prior to DNA damage (figure 5d), and that a significant fraction of this HP1β is released to the nucleoplasm following exposure to bleomycin. Prior treatment of cells with 4,5,6,7-tetrabromobenzotriazole (TBB), a highly specific inhibitor of casein kinase 2 28, blocked release of HP1β from the chromatin (figure 5d), as previously shown 27. Inhibition of casein kinase II with TBB also blocked activation of Tip60’s acetyltransferase by bleomycin and prevented the ATM-dependent phosphorylation of chk2 (figure 5e). To determine if this effect was specific for HP1β, HP1β levels were depleted with siRNA, generating domains of H3K9me3 devoid of HP1β In the absence of HP1β the ability of TBB to block activation of Tip60’s acetyltransferase activity and prevent phosphorylation of chk2 was lost (figure 5e).
These results indicate that access of Tip60’s chromodomain to H3K9me3 is controlled through 2 key steps (figure 5f). First, the DNA-damage dependent ejection of HP1β from the chromatin is required to produce domains of H3K9me3 adjacent to the DSB. Second, association of the MRN complex with DSBs is required to efficiently recruit the inactive ATM-Tip60 complex to the DNA damage site. The localization of the ATM-Tip60 complex to the DSB facilitates productive interactions between Tip60’s chromodomain and adjacent chromatin domains containing H3K9me3. This interaction increases Tip60’s acetyltransferase activity, leading to the acetylation and activation of the ATM kinase, as well as the potential for Tip60 to directly acetylate other histones on the chromatin. The binding of H3K9me3 to Tip60’s chromodomain therefore functions as an allosteric regulator of Tip60’s catalytic activity, rather than as a binding module for recruiting Tip60 to DSB. In addition to Tip60, interactions between the MRN complex and ATM may also provide critical inputs for generating and maintaining ATM in its active form 18. These results therefore provide a mechanistic framework which links together previous observations on the relative contributions of the MRN complex 17, ATM autophosphorylation 29 and Tip60-dependent acetylation of ATM 7 to the activation of the ATM kinase. Further, H3K9me3 is predominantly located in the compacted, heterochromatic regions of the chromatin30, 31, where it is associated with HP1. This suggests that Tip60 may be preferentially activated by DSBs generated within heterochromatin, which would be consistent with reports that the repair of DSBs in heterochromatin involves ATM 32. However, because H3K9me3 can be found in non-heterochromatin regions 33, Tip60 function is unlikely to be entirely restricted to heterochromatic regions during DNA damage responses. These results also have important consequences for human disease. Histone methylation patterns differ between cell lineages and are frequently altered in human tumors20, 34. These altered histone methylation signatures may play a key role in the etiology of cancer by regulating the efficiency and extent of DNA repair at specific sites in the chromatin. The tumor suppressor functions of Tip60 are therefore intimately linked to the levels of H3K9me3, and to the methyltransferases and demethylases which regulate this histone modification.
Methods section
Cells
HeLaS3, 293T, HCT116 cells and Mouse Embryonic Fibroblasts were cultured as previously described7, 22. Cells were irradiated using a Cs137 irradiator and clonogenic cell survival monitored as in 7. For cell survival assays, averages were calculated from at least 3 independent plates, and expressed plus or minus the standard deviation (SD).
Immunoprecipitation and western blot analysis
Cell lysis buffers, immunoprecipitation and western blot analysis are as described by us 7, 12. Cells (1×107) were lyzed in ATM lysis buffer (20mM Hepes pH7.4; 150mM NaCl; 0.2% Tween 20; 1.5mM MgCl2; 1mM EGTA; 2mM DTT; 50mM NaF; 500μM NaVO4; 1mM PMSF; 1μg/ml aprotinin; 1μg/ml leupeptin) and cleared by centrifugation. Antibodies to ATM (PC116; EMD biosciences, CA) or Tip60 (HA or Tip60; Abcam, MA and Upstate Biotechnology, NY) were used for immunoprecipitation, and immune complexes collected on protein-A agarose beads. Immunoprecipitates were washed 3 times in ATM lysis buffer, and once each in High Salt Buffer (100mM Tris, pH7.4; 600mM NaCl; 1mM DTT; 1mM PMSF), and base buffer (10mM Hepes, pH7.4; 10mM MgCl2; 50mM NaCl; 1mM DTT; 1mM PMSF). For co-precipitation of Tip60 and histone H3, cells were lyzed in ATM lysis buffer supplemented with benzonase (Novagen, CA) to digest DNA, and then centrifuged (35Kg for 20min) to remove chromatin fragments and cell debris. Immunoprecipitation was carried out as described above. For histone extraction, cells were harvested, washed in phosphate buffered saline (PBS), resuspended in Buffer C (20mM Hepes pH7.9; 0.1% Triton X-100; 1.5mM MgCl2; 1mM PMSF; 1mM DTT) and incubated for 10 minutes. Lysates were centrifuged (1500g for 10 minutes), washed once in buffer C and the resulting pellet resuspended in buffer D (10mM Hepes pH7.9; 10mM KCl; 1.5mM MgCl2; 1mM DTT; 1mM PMSF) to a final volume of 600μl. Sulfuric acid (2.0N) was then added to a final concentration of 0.4N. Samples were incubated on ice for 30 minutes, and cleared by centrifugation (15Kg for 15 minutes). 1/5 volume of ice cold TCA (100%) was added to the supernatant and then incubated for 30 minutes on ice. Precipitates were collected by centrifugation (15kg for 15 minutes), and washed with ice-cold 0.1% HCl/ acetone followed by ice-cold acetone alone. Pellets were air dried and resuspended in 10mM Hepes pH 7.9. Antibodies used were: ATM 2C1 (Genetex, TX); phospho-Ser 1981 ATM (Rockland Biochemicals, PA); anti-acetyl-lysine, Tip60, H2AX, H3K9me3 (Upstate Biotechnology, NY); Tip60 (N17; Santa Cruz, CA); Flag M-2 (Sigma, MO); p53, phospho-Ser 15 p53, ATM antibody PC116 (EMD Biosciences, CA); β-actin, Chk2, phospho-Thr 68 chk2, γH2AX (Cell Signaling, MA); Nbs1 (Novus biologicals, CO); Histone H3, HA, H3K27me3, H3K36me3, CBX1 (HP1β) (Abcam, MA); Rad50 (Genetex, TX).
Transfection protocols
Mutagenesis was carried out using the QuikChange® II XL Site-Directed Mutagenesis Kits (Stratagene, CA). For stable cell lines, HeLaS3 cells were transfected with vector or Tip60 expression vectors using FUGENE-6, and clonal cell lines established by selection in neomycin (400μg/ml). For transient expression of histone demethylases in 293T cells, FUGENE-6 and plasmid DNA were mixed at a 3:1 ratio, and added directly to tissue culture media. Exactly 30hr post-transfection, cells were immediately used for experimental protocols. For siRNA experiments, cells were transfected using Lipofectamine 2000 (Invitrogen, CA). Control and HP1β (CBX1) specific siRNA were purchased from Dharmacon, IL (catalog numbers D-001810-10 and L-009716-00 respectively).
HAT assays
For HAT assays, Tip60 immunoprecipitates (prepared as described above) were washed twice in HAT assay buffer (50mM Tris pH 8.0; 10% Glycerol; 0.1mM EDTA; 1mM DTT), and incubated in HAT assay buffer (60μl) supplemented with acetyl-CoA (100μM), and biotinylated Histone H4 peptide (0.5μg) for 30min at 30°C. An aliquot of the reaction was immobilized onto 96-well streptavidin plates, and acetylation of the H4 peptide detected using a HAT ELISA according to the manufacturer’s instructions (Upstate Biotechnology, NY). HAT activity is expressed as the change in absorbance relative to the reference wavelength (450nm-540nm). Validation of the acetyltransferase assay, and details of procedures, are described in the supplementary figures and in6, 7, 12. For in vitro reactions to determine the ability of methylated histone H3 peptides to activate Tip60, Tip60 immunoprecipitates were incubated with peptide (500ng) or H3 (500ng) for 15 minutes, followed by 3 washes in HAT assay buffer to remove unbound peptide. HAT activity was measured as described above. Methylation of recombinant histone H3 was carried out as follows35: recombinant H3 (10μg) was incubated with freshly prepared NaBH4 (40 mg/ml) and formalin (5μl) on ice and aliquots removed every 10 minutes for western blot analysis using a pan methyl-lysine antibody. Histone H3 peptide sequences are listed in supplementary table 1. Statistical analysis of the HAT assay: For each data point, 3 independent immunopurification reactions were carried out, followed by assay of the HAT activity associated with each immunopurification. The resulting triplicate data points were averaged, and expressed plus or minus the standard deviation (SD). Individual experiments were repeated at least twice.
Peptide binding studies
Biotinylated peptides (5μg; supplementary table 1) were prebound to sepharose-avidin beads for 3 hr at 4°C, and washed extensively in binding buffer (20mM Hepes, pH7.5; 150mM NaCl; 2.5% glycerol; 0.05% Tween 20; 2mM DTT; 0.1μg/ml aprotinin; 0.1μg/ml leupeptin; 1mM PMSF). Nuclear extracts 36 were prepared from cells expressing either vector, HA-Tip60 or HA-Tip60CD. Extracts were precleared with sepharose-avidin beads, and incubated with peptide bound to sepharose-avidin beads for 2hr in binding buffer. Beads were washed with wash buffer (20mM Hepes pH7.9; 300mM KCl; 0.2% Tween 20; 1.5mM MgCl2; 2mM DTT; 50mM NaF; 500μM Na3VO4; 0.1μg/ml aprotinin; 0.1μg/ml leupeptin; 1mM PMSF), followed by base buffer (4mM Hepes pH7.9; 10mM NaCl; 1mM PMSF; 2mM DTT). Bound proteins were eluted from the resin with SDS sample buffer and analyzed by SDS PAGE.
In vitro translation
Tip60 or Tip60CD were in vitro translated using the TNT® T7 Coupled Reticulocyte Lysate System (Promega, WI) in a final volume of 500μl. For binding of in vitro translated Tip60 to H3K9me3, biotinylated peptides were prebound to sepharose-avidin beads as described in the previous section. In vitro translate (100μl) was diluted in binding buffer (20mM Hepes pH7.9; 150mM NaCl; 0.05% Tween 20; 2.5% Glycerol; 1.5mM MgCl2; 2mM DTT; 0.1μg/ml aprotinin; 0.1μg/ml leupeptin; 1mM PMSF) to a final volume of 500μl. In vitro translates were incubated with sepharose-avidin columns (4hr at 4°C), washed 5 times in binding buffer, once in base buffer (10mM Hepes, pH7.4; 10mM MgCl2; 50mM NaCl; 1mM DTT; 1mM PMSF) and proteins eluted in SDS-PAGE sample buffer.
Chromatin fractionation experiments
Chromatin fractionation was carried out as described in 37. 1×108 293T cells were resuspended in 200μl buffer A (10mM Hepes, pH7.9; 10mM KCl; 1.5mM MgCl2; 0.34M sucrose; 10% glycerol; 1mM DTT; 0.1μg/ml aprotinin; 0.1μg/ml leupeptin; 1mM PMSF). Triton X-100 was added to a final concentration of 0.05% followed by incubation on ice for 5 minutes. Nuclei were collected by centrifugation (15Kg for 5 minutes). The supernatant was cleared by centrifugation (20Kg for 20min) and designated S1. Nuclei were washed in buffer A, then lyzed in buffer B (3mM EDTA; 0.2mM EGTA; 1mM DTT) for 10 minutes on ice. The insoluble chromatin fraction was collected by centrifugation (2Kg for 4 mins). The supernatant was cleared by centrifugation (20Kg for 20min) and designated S2. Finally, the chromatin fraction was washed in buffer B, treated with benzonase (5μl at 1:10 dilution; Novagen, CA) to digest DNA, and the chromatin associated proteins solubilized in SDS-sample buffer.
Chromosome aberrations
HeLaS3 cells were irradiated (2Gy) and allowed to recover for 14hr. Colcemid was added, and the metaphase spreads prepared as described in 38. All spreads were counted blind.
Immunofluorescence
Cells (cultured on cover slides) were fixed in phosphate buffered saline (PBS) containing paraformaldehyde (2%), blocked with serum (2%) and permeabilized in Bovine Serum Albumen (0.2%) containing saponin (0.2%) for 10min. Cells were incubated with primary antibody for 1h, washed, followed by secondary antibody for 1h. Slides were mounted with Fluoromount-G (Southern Biotech, AL) and visualized with a Nikon Eclipse TE 2000 microscope.
Supplementary Material
Acknowledgements
We thank T. Jenuwein for providing Suv39h1/2 cells, D. Ferguson for providing rad50 deficient HCT116 cells, and H. Chan, J. Cote, D. Chowdhury and A. D’Andrea for critical discussions and reading of the manuscript. This work was supported by grants from the NCI (CA64585 and CA93602) and the DOD Breast Cancer Program to BDP, and by NCI training grants to YS and MKA (T32 CA09078). YX is supported by a U19 Center grant from NIAID (U19AI067751).
Footnotes
Author information: The authors declare that they have no competing financial interests.
References
- 1.Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006 doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Bird AW, et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–5. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 3.Downs JA, et al. Binding of Chromatin-Modifying Activities to Phosphorylated Histone H2A at DNA Damage Sites. Mol Cell. 2004;16:979–90. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Murr R, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–9. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 5.Kusch T, et al. Acetylation by Tip60 Is Required for Selective Histone Variant Exchange at DNA Lesions. Science. 2004 doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 6.Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol Cell Biol. 2007;27:8502–9. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102:13182–7. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorrini C, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–7. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- 9.ME LL, et al. New p53 related genes in human tumors: significant downregulation in colon and lung carcinomas. Oncol Rep. 2006;16:603–8. doi: 10.3892/or.16.3.603. [DOI] [PubMed] [Google Scholar]
- 10.Kim JH, et al. Transcriptional regulation of a metastasis suppressor gene by Tip60 and beta-catenin complexes. Nature. 2005;434:921–6. doi: 10.1038/nature03452. [DOI] [PubMed] [Google Scholar]
- 11.Halkidou K, et al. Expression of Tip60, an androgen receptor coactivator, and its role in prostate cancer development. Oncogene. 2003;22:2466–77. doi: 10.1038/sj.onc.1206342. [DOI] [PubMed] [Google Scholar]
- 12.Jiang X, Sun Y, Chen S, Roy K, Price BD. The FATC domains of PIKK proteins are functionally equivalent and participate in the Tip60-dependent activation of DNA-PKcs and ATM. J Biol Chem. 2006;281:15741–6. doi: 10.1074/jbc.M513172200. [DOI] [PubMed] [Google Scholar]
- 13.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–40. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–3. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 15.Nielsen PR, et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–7. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 16.Uziel T, et al. Requirement of the MRN complex for ATM activation by DNA damage. Embo J. 2003;22:5612–21. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–8. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 18.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–4. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 19.Zhong H, Bryson A, Eckersdorff M, Ferguson DO. Rad50 depletion impacts upon ATR-dependent DNA damage responses. Hum Mol Genet. 2005;14:2685–93. doi: 10.1093/hmg/ddi302. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 21.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Peters AH, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–37. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 23.Whetstine JR, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–81. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 24.Klose RJ, et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–6. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 25.Hong S, et al. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A. 2007;104:18439–44. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stiff T, et al. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–6. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 27.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–6. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 28.Ruzzene M, Penzo D, Pinna LA. Protein kinase CK2 inhibitor 4,5,6,7-tetrabromobenzotriazole (TBB) induces apoptosis and caspase-dependent degradation of haematopoietic lineage cell-specific protein 1 (HS1) in Jurkat cells. Biochem J. 2002;364:41–7. doi: 10.1042/bj3640041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 30.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 31.Regha K, et al. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol Cell. 2007;27:353–66. doi: 10.1016/j.molcel.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodarzi AA, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–77. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 33.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26:9185–95. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seligson DB, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–6. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 35.Pethe K, et al. Mycobacterial heparin-binding hemagglutinin and laminin-binding protein share antigenic methyllysines that confer resistance to proteolysis. Proc Natl Acad Sci U S A. 2002;99:10759–64. doi: 10.1073/pnas.162246899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu X, Stern DF. NFBD1/KIAA0170 is a chromatin-associated protein involved in DNA damage signaling pathways. J Biol Chem. 2003;278:8795–803. doi: 10.1074/jbc.M211392200. [DOI] [PubMed] [Google Scholar]
- 38.Yang Y, et al. Targeted disruption of the murine Fanconi anemia gene, Fancg/Xrcc9. Blood. 2001;98:3435–40. doi: 10.1182/blood.v98.12.3435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.