Abstract
The CRTC1-MAML2 fusion oncogene underlies the etiology of mucoepidermoid salivary gland carcinoma (MEC) where it confers a favorable survival outcome as compared with fusion-negative MEC. While these analyses suggested that detection of CRTC1-MAML2 serves as a useful prognostic biomarker, we recently identified outlier cases of fusion-positive MEC associated with advanced-staged lethal disease. To identify additional genetic alterations that might cooperate with CRTC1-MAML2 to promote disease progression, we performed a pilot high-resolution oligonucleotide array CGH (aCGH) and PCR-based genotyping study on 23 MEC samples including14 fusion-positive samples for which we had clinical outcome information. Unbiased aCGH analysis identified inactivating deletions within CDKN2A as a candidate poor prognostic marker which was confirmed by PCR-based analysis (CDKN2A deletions in 5/5 unfavorable fusion-positive cases and 0/9 favorable fusion-positive cases). We did not detect either activating EGFR mutations, nor copy number gains at the EGFR or ERBB2 loci as poor prognostic features for fusion-positive MEC in any of the tumor specimens. Prospective studies with larger case series will be needed to confirm that combined CRTC1-MAML2 and CDKN2A genotyping will optimally stage this disease.
INTRODUCTION
MEC is the most common malignant salivary gland tumor characterized by variable histopathologic features and unpredictable clinical behavior. Although multiple phenotypic grading systems have been developed to better classify these tumors, (Clode et al., 1991; Hicks et al., 1995; Goode et al., 1998; Brandwein et al., 2001; Luna, 2006), their clinical utility has remained limited due to subjectivity and the biological heterogeneity within and between tumor grades.
In 2003, the CRTC1-MAML2 fusion oncogene was identified at the breakpoint of a recurrent translocation t(11;19) and was shown to be the pathogenic event that underlies the development of the majority of MEC cases (Tonon et al., 2003; Enlund et al., 2004). Evidence supporting CRTC1-MAML2 in the etiology of these tumors was i) the ability of CRTC1-MAML2 to transform rat RK3E cells in vitro and in vivo (Coxon et al., 2005), ii) the identification of the fusion transcript in MEC-like tumors arising from distinct tissues including major and minor salivary glands, bronchopulmonary tree, thyroid, breast, skin, and cervix (Behboudi et al., 2005; Kazakov et al., 2007; Tirado et al., 2007; Achcar et al., 2009; Lennerz et al., 2008), and iii) tumor growth suppression by RNAi targeted to the fusion transcript exclusively in tumor cells carrying the t(11;19) translocation (Komiya et al., 2006).
Over 150 cases of MEC have been tested for the CRTC1-MAML2 transcript by RT-PCR or fluorescent in situ hybridization (FISH) with a 55% detection rate overall (Martins et al., 2004; Behboudi et al., 2006; Okabe et al., 2006; Tirado et al., 2007; Fehr et al., 2008a,b). Undifferentiated MEC, however, rarely expressed CRTC1-MAML2 while over 80% of low or intermediate grade mucoepidermoid cases tested were fusion-positive. This suggested that CRTC1-MAML2 is a reliable diagnostic marker in differentiating subtypes of MEC and that some high-grade fusion-negative tumors may represent a misclassification of a non-MEC aggressive carcinoma not otherwise specified (NOS). Accordingly, retrospective survival analyses of different series of these tumors demonstrated that patients with fusion-negative tumors had a significantly worse survival as compared with fusion-positive cases (Behboudi et al., 2006; Okabe et al., 2006; Tirado et al., 2007), suggesting that CRTC1-MAML2 may serve a specific diagnostic and prognostic molecular marker for MEC.
While most fusion-positive tumors were cured following surgical resection, a few outlier cases initially presented with, or subsequently developed, lethal stage 4 disease (Tirado et al., 2007; Kazakov et al., 2009), suggesting that these unfavorable CRTC1-MAML2 positive tumors may have somatically acquired additional genetic alterations that conferred enhanced invasiveness or tumor survival properties. To address this important issue, we collected all available cases of fusion-positive primary MEC tumors and performed global, high-resolution aCGH to compare the copy number variation (CNV) genotype between samples collected from good and poor prognosis fusion-positive cases.
MATERIALS AND METHODS
Patient Samples
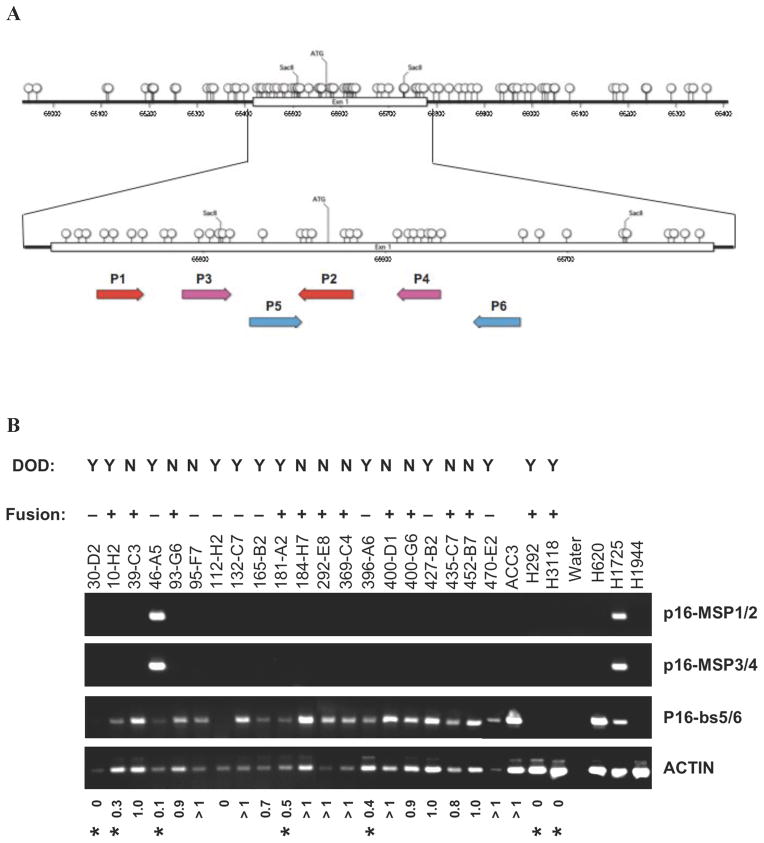
Primary MEC samples were obtained from the Head and Neck Section of the Department of Pathology, MD Anderson Cancer Center. All primary tumor cases were reviewed by two pathologists and samples were collected under approved Institutional review. In addition, we studied two MEC tumor cell lines (H292 and H3118) that were isolated from patients at the National Naval Medical Center who died of complications from metastatic stage 4 MEC (Tonon et al., 2003) as well as 4 non-MEC control tumor cell lines (ACC3, H620, H1725, H1944) (Otterson et al., 1995). CRTC1-MAML2 status was obtained by RT-PCR from microdissected sections as previously described (Tirado et al., 2007).
Whole Genome Amplification
Ten nanograms of genomic DNA was amplified using the GenomePlex Whole Genome Amplification (WGA-2) kit (Sigma-Aldrich, St. Louis, MO) and purified using the QIAprep Spin Miniprep kit (Qiagen, Valencia, CA). For purification, five volumes of Buffer PB was added directly to the amplification products, applied to QIAprep Spin Miniprep Columns, and centrifuged at maximum speed for one minute. The spin column was then washed with 75 μl PE Buffer. An additional two-minute centrifugation step was performed to remove residual wash buffer. The purified DNA was eluted with 50 μl Buffer EB and quantified using the NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE).
Array CGH
For each aCGH hybridization, 2500 ng of amplified DNA from the reference control (female gDNA, Promega, UK) and MEC sample was directly labeled with Cy3-dUTP and Cy5-dUTP, respectively, using the BioPrime Array CGH Genomic Labeling Module (Invitrogen, Carlsbad, CA) and the manufacturer’s recommended protocol. Labeled products were purified two times with 450 μl TE and concentrated using a Vivaspin concentrator (Sartorius, Germany). Labeled reference and experimental samples were combined to a final volume of 79 μl and mixed with 25 μl of human COT1 DNA (Invitrogen, Carlsbad, CA), 26 μl Agilent 10X Blocking Agent, and 130 μl Agilent 2X Hybridization Buffer (Agilent Technologies, Santa Clara, CA). The hybridization mixture was denatured at 95°C for 5 minutes, incubated at 37°C for 30 minutes, and applied to commercially available Human Genome CGH Microarray Kit 105A arrays (Agilent Technologies, Santa Clara, CA), which contain 99,000+ coding and non-coding human sequences. Following 48-hour rotating incubation at 65°C in an Agilent microarray chamber, the arrays were washed in Oligo aCGH Wash Buffer 1 at room temperature for 5 minutes followed by a second wash for 1 minute in Oligo aCGH Wash Buffer 2 prewarmed to 37°C. The arrays were scanned at 5 μm resolution using an Agilent G2505C DNA microarray scanner and the data were normalized using Feature Extraction software (version 10.5.1, Agilent Technologies, Santa Clara, CA).
aCGH Data Analysis
The data were visualized and analyzed using CGH Analytics (version 6.0, Agilent Technologies, Santa Clara, CA) or Nexus 4 software (version 8.0, BioDiscovery Inc, El Segundo, CA). To optimize aberration calls and minimize background-related gains and losses using Nexus 4, the Rank Segmentation algorithm with a significance threshold of 1.0 × 10−10 was used. The settings for aberration calls for all but three of the samples were 0.8 for amplification, 0.4 for gain, −0.4 for loss, and −0.8 for homozygous deletion. For three samples (184H7, 396A6, and 400D1), the settings for aberration calls were 0.85 for amplification, 0.43 for gain, −0.43 for loss, and −0.85 for homozygous deletion.
CDKN2A Methylation Analysis
Bisulfite conversion of DNA was performed as described previously to create a template for methylation-specific PCR (MS-PCR) (Chen et al., 2005). Briefly, 200 ng genomic DNA from each sample was denatured by NaOH at a final concentration of 0.2 M in a volume of 50 μL at 37 °C for 15 minutes. 30 μL of 10 mM freshly prepared hydroquinone and 520 μL of freshly prepared 3.0 M NaHSO3, pH 5.0 (Sigma, St. Louis, MO) was added and the mixture was incubated at 56 °C for 16 hours. Bisulfite-modified DNA was purified using the Wizard DNA Cleanup kit (Promega, Madison, WI). The DNA was desulfonated by incubation with NaOH at a final concentration of 0.3 M at room temperature for 15 min and further neutralized by adding ammonium acetate, pH 7.0, to a final concentration of 3 M. The DNA was precipitated with ethanol and resuspended in distilled water to a final concentration of 2 ng/μL. Bisulfite-treated DNA was used as the template for methylation specific-PCR, as described previously (Chen, et al. 2005). Briefly, 5 μL of bisulfite-converted genomic DNA served as the PCR template in a 25 μL reaction containing 0.19 mM each dNTP, 1.5 mM MgCl2, 400 nM of forward and reverse primers, and 1.25 U of AmpliTaq Gold. Two different primer pairs to detect CDKN2A methylation were employed as follows: forward amplification primer1 (5′-TATTCGGTGCGTTGGGTAGCGTTTTC-3′) and reverse amplification primer2 (5′-CGACGAAAAACAACATAAAACCGACGACGA-3′), forward primer3 (5′-TTTTTTATTCGATTTCGGGTCGCGGTC-3′) and reverse primer4 (5′-AACCGCGTACGCTCGACGACTACG-3′), respectively. The PCR cycling parameters were as follows: hot start at 94 °C for 9 minutes to inactive the inhibitors of AmpliTaq Gold, followed by 45 cycles of 94°C (30 seconds), 66 °C (30 seconds), and 72 °C (45 seconds), then 72 °C for10 minutes, and 10 °C to cool. A separate PCR reaction with forward primer5 (5′-AATCAACCAAAAACTCCATACTACTCCCC-3′) and reverse primer6 (5′-AGGAAGAAAGAGGAGGGGTTGGTTGG-3′) was carried out to detect the presence of either methylated or unmethylated CDKN2A exon 1. The exon 6 region of β-actin (ACTB1) was amplified as genomic DNA control using forward primer (ACT2118(AS)bsF), 5′-TCCTAACCTCACTATCCACCTTCCAAC-3′ and reverse primer (ACT2297(AS)bsR), 5′-CAGTATGAGGTGTGTGTATTTGTTAGGGGT-3′. All the PCR products were separated by 3% agarose gel electrophoresis with 1X TAE, and the DNA intensities were analyzed by using InGenius LHR system (Cambridge, United Kingdom).
EGFR PCR and sequencing
Using 10 ng genomic DNA from each sample as template, the exon19 and exon21 regions of EGFR gene were amplified in a 50 μl volume with forward primer (EGFR-E19F), 5′-ACCATCTCACAATTGCCAGTTAACGTC-3′ and reverse primer (EGFR-E19R), 5′-ACATCGAGGATTTCCTTGTTGGCTTTC-3′, and forward primer (EGFR-E21F), 5′-GGCATGAACTACTTGGAGGACCGTC-3′ and reverse primer (EGFR-E21R), 5-CTGCATGGTATTCTTTCTCTTCCGCAC-3′, respectively. The PCR cycling parameters were as follows: hot start at 94 °C for 9 minutes to inactive the inhibitors of AmliTaq Gold, followed by 40 cycles of 94°C (30 seconds), 65 °C (30 seconds), and 72 °C (45 seconds), then 72 °C for10 minutes, and 10 °C to cool. The PCR products were analyzed by 2% agarose gel electrophoresis with 1X TAE and subjected to DNA sequencing.
RESULTS
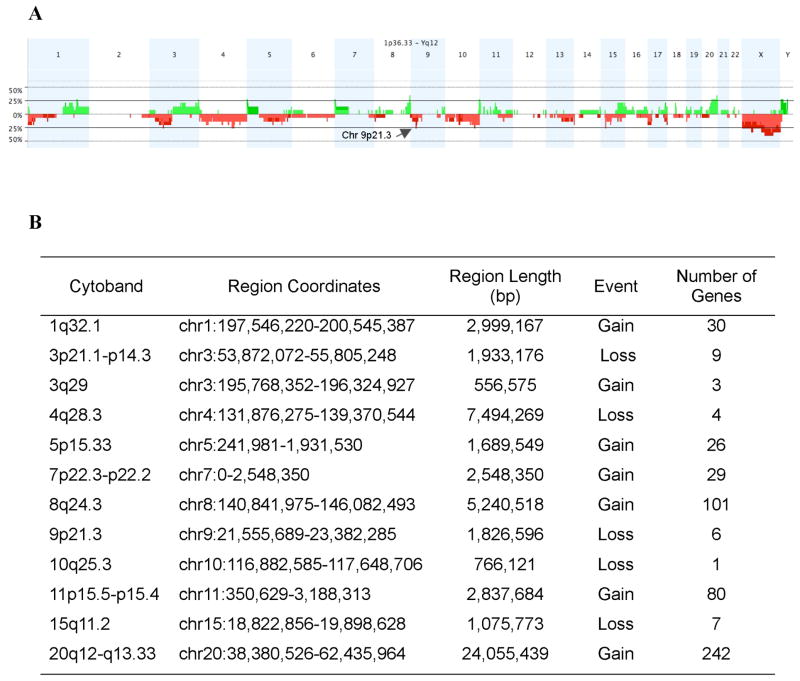
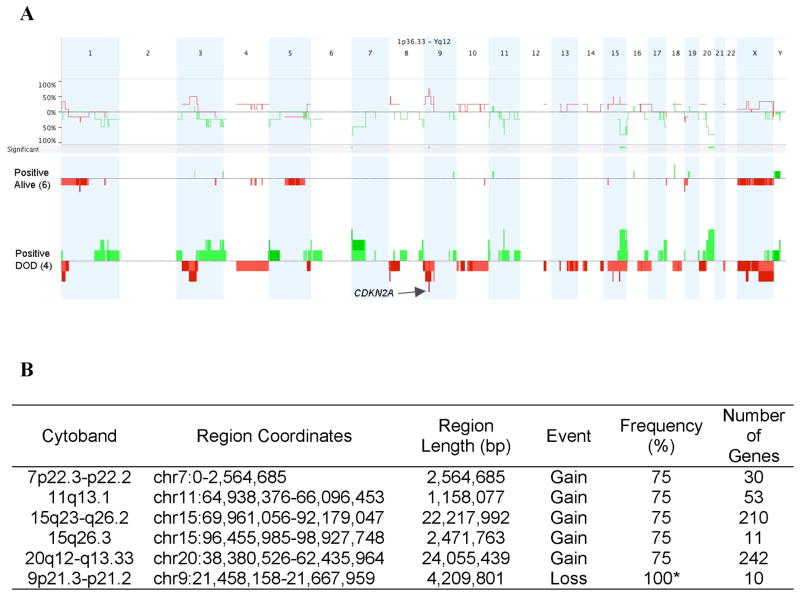
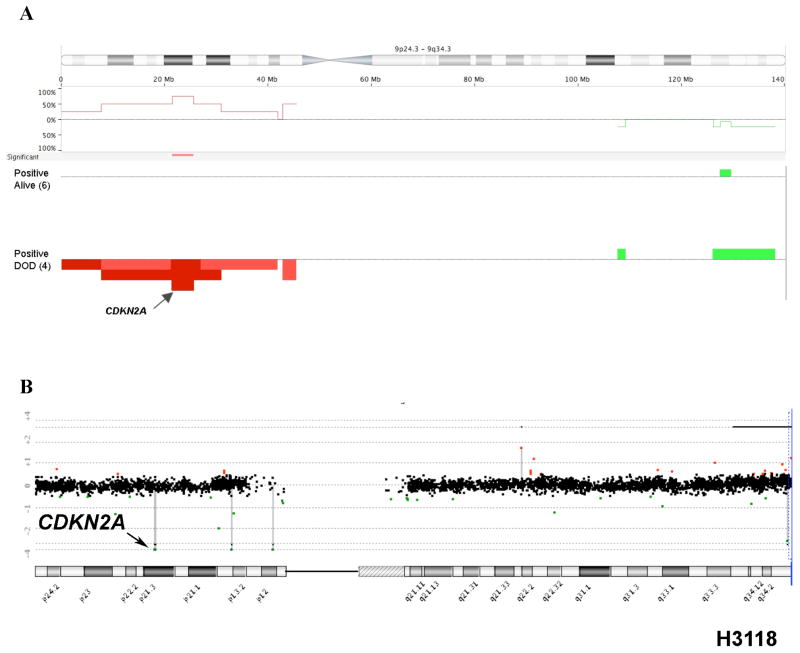
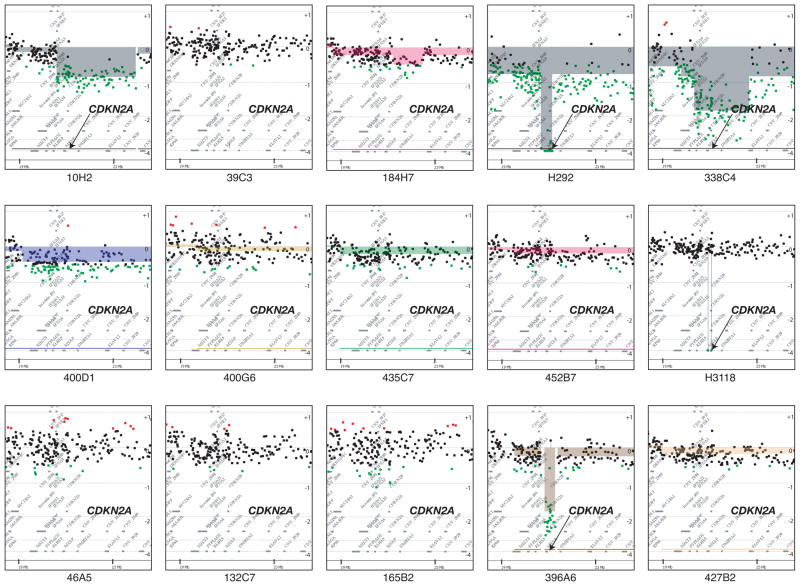
We performed a pilot study using high-resolution aCGH on tumors with known CRTC1-MAML2 status to conduct a global unbiased search for candidate gene loci exhibiting recurrent CNV gains or losses in MEC tumors. Of the 23 MEC samples in this dataset, 14 were CRTC1-MAML2 fusion-positive and 9 were fusion-negative (Table 1). As previously reported (Behboudi et al., 2006; Okabe et al., 2006; Tirado et al., 2007), patients with fusion-negative MEC samples showed an inferior prognosis with 8 of 9 subjects that succumbed to lethal disease and the sole fusion-negative survivor had a completely resected tumor localized to the thyroid gland (Table 1). In contrast, the majority of fusion-positive MEC patients did not die of their disease (9/14). Therefore, we have focused this pilot study on identifying candidate CNV loci within this more homogeneous subset and initially tested 15 genomic samples with the highest quality DNA (10 fusion-positive and 5 fusion-negative MEC samples) by aCGH to identify all chromosomal loci that showed a discrete segment with a common region showing gain or loss in at least 4 independent samples (Figure 1A). This analysis identified 12 loci on 11 different chromosomal arms with either gains or losses, including a region of DNA loss in chromosome band 9p21.3 that spanned the CDKN2A gene (Figure 1B). Using Nexus CGH software, we then searched for regions of significant copy number differences between favorable and unfavorable CRTC1-MAML2 fusion-positive cases (Figure 2). Of interest, fusion-positive favorable cases showed fewer CNV alterations by aCGH than unfavorable cases which is consistent with the hypothesis that aggressive tumors have acquired a degree of genomic instability with additional genetic alterations that characterizes advanced disease in other malignancies. This comparative analysis identified 6 chromosomal regions with significant differences (5 with gains and 1 with loss) between the two groups (Figure 2B). Detailed inspection of the 9p21.3 chromosomal region showed that it overlapped precisely with the CDKN2A locus in all unfavorable case (4/4) while none of the six favorable fusion-positive cases showed genomic CDKN2A loss (Figure 3A). One MEC tumor cell line (H3118) showed a discrete, single oligonucleotide probe homozygous deletion by aCGH that could not be detected with the Nexus software using the default settings for segmentation but could be detected with the CGH analytics software (Figure 3B), and homozygous deletion of CDKN2A was confirmed in this sample using semi-quantitative PCR and by absent protein expression in immunoblot analysis (data not shown). In addition, we observed that the genomic deletion in H3118 mapped exclusively to CDKN2A and flanking intronic sequence, leaving intact the adjacent CDKN2B/p15 and p14arf coding sequences which suggests that CDKN2A is selectively targeted in the 9p21.3 region for inactivation in these tumors.
Table 1.
Clinicopathologic characterization of MEC patients
| case | age | sex | site | grade | size (cm) | metastasis | DOD | Follow-up Period | Crtc1-Maml2 |
|---|---|---|---|---|---|---|---|---|---|
| 10H2 | 33 | F | Parotid | High | N/A | Lymph node | yes | 4 months | Positive |
| 39C3 | 41 | F | Parotid | Intermediate | 2 | No | no | 16 years | Positive |
| 93G6 | 72 | M | Parotid | High | N/A | No | no | 7 months | Positive |
| 181A2 | 67 | F | mandible | Intermediate | 2.8 | Lungs, Cervical, Bone | yes | 2 months | Positive |
| 184H7 | 61 | F | Lung | Intermediate | 5.3 | No | no | 11 years | Positive |
| 292E8 | 90 | F | Thyroid | Intermediate | 9 | No | no | 4 months | Positive |
| H292 | 32 | F | Lung | Intermediate | N/A | Diffuse metastases | yes | 3 months | Positive |
| 338C4 | 54 | M | Parotid | Intermediate | 4 | unknown | died unknown | 3 years | Positive |
| 369C4 | 51 | F | parotid | Intermediate | 3 | NED | no | 19 years | Positive |
| 400D1 | 73 | M | BOT | Intermediate | 2 | No | no | 6 years | Positive |
| 400G6 | 54 | F | BOT | Intermediate | 1.8 | No | no | 7 years | Positive |
| 435C7 | 29 | F | BOT | High | 1.5 | No | no | 5 years | Positive |
| 452B7 | 47 | F | BOT | Intermediate | 1.5 | No | no | 4 years | Positive |
| H3118 | 29 | F | Parotid | High | 5 | Diffuse metastases | yes | 2 years | Positive |
| 30D2 | 50 | F | thyroid, lobe | Inter | N/A | Cervical/Lungs | yes | 55 months | Negative |
| 46A5 | 68 | M | Maxillary Sinus | Intermediate | 4.2 | Lung and Brain | yes | 6 months | Negative |
| 95F7 | 54 | F | Parotid | Intermediate | N/A | No | no | 14 years | Negative |
| 112H2 | 84 | M | tongue, NOS | Intermediate | 4 | Cervical/Lungs | yes | 11 months | Negative |
| 132C7 | 49 | F | Oral Cavity | High | 3 | Lung | yes | 25 months | Negative |
| 165B2 | 51 | F | Parotid | High | 4.5 | Diffuse metastases | yes | 5 years | Negative |
| 396A6 | 56 | F | Maxilla | High | 5 | Lung and bone | yes | 18 months | Negative |
| 427B2 | 73 | F | Thyroid | Intermediate | 10 | No | yes | 24 months | Negative |
| 470E2 | 79 | M | parotid | High | 2 | Brain/Lungs | yes | 9 months | Negative |
Figure 1.
A) Genome-wide frequency plot of gains (green) and losses (red) in MEC samples as a percentage of the total group (n=15). Arrow depicts the 9p21 locus spanning the CDKN2A gene. B) Summary of discrete chromosomal segment gains or losses observed in ≥ 4 independent MEC samples.
Figure 2.
A) Genome-wide comparison between CRTC1-MAML2 fusion-positive cases that died of disease (DOD) versus fusion-positive cases that did not DOD. Regions of p<0.05 are marked by horizontal bars of gains (green) and losses (red) on the significance track (and tabulated in the lower panel). The frequency plot is shown for each subgroup as vertical bars and the arrow depicts the CDKN2A locus in chromosome band 9p21.3. B) Regions of significant difference (p<0.05) between fusion-positive died of disease (DOD) versus fusion-positive alive samples. Dataset included six alive samples and four DOD samples. Frequency indicates percentage in positive, DOD samples. * The frequency of loss at 9p21 is 100% because one sample (H3118) had a discrete oligo deletion which was not detected using the default settings for segmentation, but was demonstrated by inspection of the CGH analytics pattern (see text and Fig 3).
Figure 3.
A) Difference in frequency of chromosome 9 copy number gains or losses in fusion-positive DOD versus fusion-positive did not DOD. Area of p<0.05 is marked by a red (loss) horizontal bar on the significance track and gains and losses are shown in vertical green and red bars, respectively. For each group, values above the 0% baseline indicate a higher frequency in the fusion-positive, DOD group whereas values below the 0% baseline represent a higher frequency in the fusion-positive, alive MEC group. The location of CDKN2A is indicated (arrow). B) Identification of a discrete CDKN2A deletion in fusion-positive MEC sample H3118.
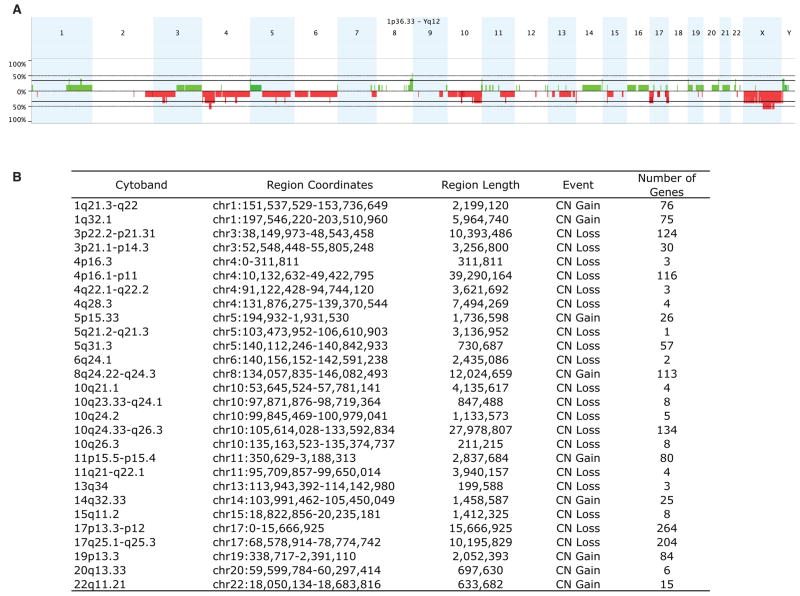
While it was not a focus of this study, we also analyzed separately the aCGH patterns for the CRTC1-MAML2 fusion-negative MEC samples (Figure 4A). We generated a frequency plot table for discrete chromosomal regions with CNVs that were detected in at least 2 MEC samples (Figure 4B) and, as expected, we observed a high degree of genomic variability in each of these poor prognosis cases (Figure 4). However, although we detected evidence for a CDKN2a deletion in 1/5 primary tumor samples, the number of cases tested were too small for any further analysis.
Figure 4.
A) Genome-wide frequency plot of gains (green) and losses (red) in the group of fusion-negative MEC samples as a percentage of the total group (n=5). B) Summary of discrete chromosomal segment gains or losses observed in ≥ 2 independent MEC samples.
Although inactivation of CDKN2A in salivary gland tumors has been inconsistently reported (Cerilli et al., 1999; Guo et al., 2007), deletion or hypermethylation of CDKN2A has been previously identified as an early event, as well as a poor prognostic marker, for several other tumor types, including lung cancer and oral squamous tumors (Rocco and Sidransky 2001; Brock et al., 2008; Sailasree et al., 2008). Therefore, since both somatic deletion and/or hypermethylation are common events to inactivate CDKN2A (Herman et al., 1995; Otterson et al., 1995), we subjected genomic DNA from the MEC samples to metabisulfite treatment followed by methylation-specific PCR using two different sets of methylation specific CDKN2A primers (Figure 5A). We also included three lung cancer cell lines with defined CDKN2A status as controls: H620 (unmethylated CDKN2A); H1755 (hypermethylated CDKN2A); and H1944 (deleted CDKN2A) as previously reported (Otterson et al., 1995) and the ACC3 non-MEC cell line. We observed a methylation signal in the H1725 control lung cancer samples, but detected evidence for CDKN2A methylation in only 1/9 fusion-negative and in none of the fusion-positive MEC samples (Figure 5B), suggesting that hypermethylation is an uncommon event in MEC tumors.
Figure 5.
A) Cartoon depicting the CpG island flanking regulatory CDKN2A exon 1 sequences (Otterson, et al. 1995) and the approximate locations for methylation-specific primer pairs MSP1/2 and MSP3/4 and non-methylation sensitive primer pairs bs 5/6. B) Semi-quantitative PCR reactions were performed at the same time using linear amplification. Control non-MEC samples for unmethylated (ACC3 and H620), methylated (H1725), and deleted (H1944) CDKN2A.
As expected, we detected a homozygous deletion of CDKN2A in the H1944 lung cancer control sample as well as in both MEC tumor cell lines (H292 and H3118) which also correlated with absent CDKN2A/p16 protein expression by immunoblot analysis in both MEC tumors (data not shown). In contrast to human tumor cell lines, however, primary MEC samples represent admixtures of cell types with variable degrees of contaminating normal tissue that makes the quantitative assessment of homozygous DNA deletions more difficult. To determine the presence of CDKN2A deletion, we performed semi-quantitative PCR on the data set under the same limited cycling conditions with linear amplification using primers for both CDKN2A and beta-actin. We observed a reduced CDKN2A/actin ratio for each patient sample where we had previously identified discrete CDKN2A deletions by aCGH. Conversely, we did not detect a reduced CDKN2A/actin ratio in samples without evidence for deletion by aCGH confirming these results and aCGH analyses corresponding to the chromosomal 9p21 region spanning CDKN2A are shown for all 15 MEC tested (Figure 6).
Figure 6.
Depiction of aCGH data at chromosome band 9p21.3 band in all 15 MEC tumors subjected to aCGH. Data were generated from CGH Analytics software using the ADM-2 algorithm at a threshold of 6. The normalized log2 ratios of each MEC to reference oligo array feature is represented as a single dot and plotted along the chromosome position on the vertical axis. Copy number gains (red) or losses (green) were defined as normalized log2 ratio > 0.5 or < −0.5, respectively. The position of CDKN2A is indicated by an arrow.
Finally, since activating mutations of EGFR (Dahse and Kosmehl 2008; Han et al., 2008), as well as amplification of ERBB1/EGFR and ERBB2, have been reported in subsets of MEC tumors, we subjected genomic DNA harvested from the 23 MEC cases to PCR amplification of EGFR exons 19–21 followed by resequencing and did not detect evidence for activating somatic mutations in any of the samples (data not shown). These, and other recent data (Macarenco et al., 2008; Rossi et al., 2009), suggest that activating EGFR kinase mutations are an exceedingly rare event in mucoepidermoid cancer. In addition, while we detected evidence for a high copy number ERBB2 amplicon in 1/15 samples, this patient presented with a favorable prognosis CRTC1-MAML2 fusion-positive tumor and was alive free of disease following surgery (case 93G6). These data, therefore, do not suggest that mutation or amplification of EGFR or ERBB2 is a poor prognostic marker for CRTC1-MAML2 positive MEC.
DISCUSSION
While MEC is the most common malignant salivary gland tumor, it has also been reported in a wide range of non-conventional salivary gland sites. The detection of the identical CRTC1-MAML2 fusion transcript in tumors from disparate organ sites including the oral cavity, lung, thyroid, breast, cervix, and skin, indicate this alteration represents an important unifying event in the tumorigenesis of MEC and suggests that the incidence of CRTC1-MAML2 related malignancies may be underestimated in current clinical practice (Behboudi et al., 2005; Kazakov et al., 2007; Tirado et al., 2007; Lennerz et al., 2008; Achcar et al., 2009). Traditionally, several grading systems based on light microscopic features have been used to guide the surgical and clinical management of these tumors (Scianna and Petruzzelli, 2007). However, recent retrospective studies have suggested that testing for the presence of the CRTC1-MAML2 rearrangement may complement histologic scoring to classify good prognosis patients to help avoid the late complications of combined modality treatment in these fusion-positive cases (Behboudi et al., 2006; Okabe et al., 2006; Tirado et al., 2007). Two obstacles for this approach, however, is the lack of prospective clinical studies demonstrating the usefulness of CRTC1-MAML2 testing, and the recent observation that a subset of patients with CRTC1-MAML2 positive MEC rapidly develop advanced stage lethal disease (Tirado et al., 2007; Kazakov et al., 2009).
To improve the ability to genotype MEC patients into more a homogeneous prognostic classification, we have undertaken a pilot study to perform a global, non-biased search for additional oncogenic targets that may cooperate with CRTC1-MAML2 expression to confer the unfavorable outlier phenotype. Strikingly, we identified deletions within the CDKN2A/p16 gene in all 5 poor prognosis CRTC1-MAML2-positive cases available to us which included 4 patients with a death due to documented metastatic MEC (10H2, 181A2, H292, H3118) and 1 case of a 54 yo male patient with a 4 cm neck tumor that died at home without available medical records for review (338C4). In contrast, no evidence for CDKN2A deletion or hypermethylation was detected in 9 fusion-positive MEC tumors that were alive free of disease or that were documented to have died of other causes without a MEC recurrence. Although not a focus of this study, we also noted that CRTC1-MAML2 fusion-negative tumors had a markedly inferior prognosis (8/9 died of disease) as previously reported (Behboudi et al., 2006; Okabe et al., 2006; Tirado et al., 2007) and several of these samples also showed evidence for either CDKN2A methylation or deletion. Similarly, deletion and/or hypermethylation of CDKN2A has been demonstrated to be an early tumorigenic event in many different types of carcinomas including squamous cell tumors of the head and neck and lung cancer (Rocco and Sidransky, 2001; Baylin and Ohm, 2006; Brock et al., 2008). However, in contrast to lung cancer where CDKN2A hypermethylation is a well described poor prognostic marker for early-stage disease (Brock et al., 2008) and where inactivation of the RB/CDKN2A pathway is detected in essentially all advanced stage cases (Kaye, 2002), there is less known about the role of CDKN2A in MEC or other subtypes of malignant salivary gland cancers (Cerilli et al., 1999; Li et al., 2005; Guo et al., 2007). For example, one study with patients from the United States detected loss of heterozygosity at polymorphic markers flanking the CDKN2A locus in 7/9 salivary duct carcinomas but rarely in MEC (Cerilli et al., 1999), while another study, with patients of Asian descent, observed evidence for CDKN2A deletions and methylation in 24% and 34% of cases, respectively (n=38 cases), using PCR analysis of archived genomic DNA (Guo et al., 2007). In this latter report, a higher rate of CDKN2A deletions was detected in high grade as compared to low or intermediate grade MEC, however none of these studies included an analysis for the CRTC1-MAML2 fusion transcript to allow for the assessment of the impact of CDKN2A inactivation on otherwise good prognosis fusion-positive cases. Further evidence suggesting a role for the sequential accumulation of CDKN2A mutations in the progression of invasive salivary gland cancer was the observation of a CDKN2A deletion in a case of carcinoma ex pleomorphic adenoma that was not present in the initial matched benign pleomorphic adenoma sample (Suzuki and Fujioka, 1998), as well as the finding of a higher rate of CDKN2A hypermethylation in a series of carcinoma ex pleomorphic adenoma as compared to pleomorphic adenoma (Augello et al., 2006).
In summary, progress in the classification and management of malignant salivary gland cancers has been hampered by the inclusion of a heterogeneous collection of distinct tumor subtypes within small clinical trials and case reports. The observation that both CRTC1-MAML2 and CDKN2A status may optimally define a more homogenous prognostic category for patients with MEC tumors may help in the design of future prospective studies to develop clinical guidelines and to search for new therapeutic agents.
References
- Achcar RD, Nikiforova MN, Dacic S, Nicholson AG, Yousem SA. Mammalian mastermind like 2 11q21 gene rearrangement in bronchopulmonary mucoepidermoid carcinoma. Hum Pathol. 2009;40:854–860. doi: 10.1016/j.humpath.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Augello C, Gregorio V, Bazan V, Cammareri P, Agnese V, Cascio S, Corsale S, Calo V, Gullo A, Passantino R, Gargano G, Bruno L, Rinaldi G, Morello V, Gerbino A, Tomasino RM, Macaluso M, Surmacz E, Russo A. TP53 and p16INK4A, but not H-KI-Ras, are involved in tumorigenesis and progression of pleomorphic adenomas. J Cell Physiol. 2006;207:654–659. doi: 10.1002/jcp.20601. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- Behboudi A, Enlund F, Winnes M, Andren Y, Nordkvist A, Leivo I, Flaberg E, Szekely L, Makitie A, Grenman R, Mark J, Stenman G. Molecular classification of mucoepidermoid carcinomas-Prognostic significance of the MECT1-MAML2 fusion oncogene. Genes Chromosomes Cancer. 2006;45:470–481. doi: 10.1002/gcc.20306. [DOI] [PubMed] [Google Scholar]
- Behboudi A, Winnes M, Gorunova L, van den Oord JJ, Mertens F, Enlund F, Stenman G. Clear cell hidradenoma of the skin-a third tumor type with a t(11;19)--associated TORC1-MAML2 gene fusion. Genes Chromosomes Cancer. 2005;43:202–205. doi: 10.1002/gcc.20168. [DOI] [PubMed] [Google Scholar]
- Brandwein MS, Ivanov K, Wallace DI, Hille JJ, Wang B, Fahmy A, Bodian C, Urken ML, Gnepp DR, Huvos A, Lumerman H, Mills SE. Mucoepidermoid carcinoma: a clinicopathologic study of 80 patients with special reference to histological grading. Am J Surg Pathol. 2001;25:835–845. doi: 10.1097/00000478-200107000-00001. [DOI] [PubMed] [Google Scholar]
- Brock MV, Hooker CM, Ota-Machida E, Han Y, Guo M, Ames S, Glockner S, Piantadosi S, Gabrielson E, Pridham G, Pelosky K, Belinsky SA, Yang SC, Baylin SB, Herman JG. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358:1118–1128. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- Cerilli LA, Swartzbaugh JR, Saadut R, Marshall CE, Rumpel CA, Moskaluk CA, Frierson HF., Jr Analysis of chromosome 9p21 deletion and p16 gene mutation in salivary gland carcinomas. Hum Pathol. 1999;30:1242–1246. doi: 10.1016/s0046-8177(99)90044-8. [DOI] [PubMed] [Google Scholar]
- Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L, Platzer P, Lu S, Dawson D, Willis J, Pretlow TP, Lutterbaugh J, Kasturi L, Willson JK, Rao JS, Shuber A, Markowitz SD. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124–1132. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- Clode AL, Fonseca I, Santos JR, Soares J. Mucoepidermoid carcinoma of the salivary glands: a reappraisal of the influence of tumor differentiation on prognosis. J Surg Oncol. 1991;46:100–106. doi: 10.1002/jso.2930460207. [DOI] [PubMed] [Google Scholar]
- Coxon A, Rozenblum E, Park YS, Joshi N, Tsurutani J, Dennis PA, Kirsch IR, Kaye FJ. Mect1-Maml2 fusion oncogene linked to the aberrant activation of cyclic AMP/CREB regulated genes. Cancer Res. 2005;65:7137–7144. doi: 10.1158/0008-5472.CAN-05-1125. [DOI] [PubMed] [Google Scholar]
- Dahse R, Kosmehl H. Detection of drug-sensitizing EGFR exon 19 deletion mutations in salivary gland carcinoma. Br J Cancer. 2008;99:90–92. doi: 10.1038/sj.bjc.6604430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enlund F, Behboudi A, Andren Y, Oberg C, Lendahl U, Mark J, Stenman G. Altered Notch signaling resulting from expression of a WAMTP1-MAML2 gene fusion in mucoepidermoid carcinomas and benign Warthin’s tumors. Exp Cell Res. 2004;292:21–28. doi: 10.1016/j.yexcr.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Fehr A, Roser K, Belge G, Loning T, Bullerdiek J. A closer look at Warthin tumors and the t(11;19) Cancer Genet Cytogenet. 2008a;180:135–139. doi: 10.1016/j.cancergencyto.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Fehr A, Roser K, Heidorn K, Hallas C, Loning T, Bullerdiek J. A new type of MAML2 fusion in mucoepidermoid carcinoma. Genes Chromosomes Cancer. 2008b;47:203–206. doi: 10.1002/gcc.20522. [DOI] [PubMed] [Google Scholar]
- Goode RK, Auclair PL, Ellis GL. Mucoepidermoid carcinoma of the major salivary glands: clinical and histopathologic analysis of 234 cases with evaluation of grading criteria. Cancer. 1998;82:1217–1224. doi: 10.1002/(sici)1097-0142(19980401)82:7<1217::aid-cncr2>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Guo XL, Sun SZ, Wang WX, Wei FC, Yu HB, Ma BL. Alterations of p16INK4a tumour suppressor gene in mucoepidermoid carcinoma of the salivary glands. Int J Oral Maxillofac Surg. 2007;36:350–353. doi: 10.1016/j.ijom.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Han SW, Kim HP, Jeon YK, Oh DY, Lee SH, Kim DW, Im SA, Chung DH, Heo DS, Bang YJ, Kim TY. Mucoepidermoid carcinoma of lung: potential target of EGFR-directed treatment. Lung Cancer. 2008;61:30–34. doi: 10.1016/j.lungcan.2007.11.014. [DOI] [PubMed] [Google Scholar]
- Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- Hicks MJ, el-Naggar AK, Flaitz CM, Luna MA, Batsakis JG. Histocytologic grading of mucoepidermoid carcinoma of major salivary glands in prognosis and survival: a clinicopathologic and flow cytometric investigation. Head Neck. 1995;17:89–95. doi: 10.1002/hed.2880170203. [DOI] [PubMed] [Google Scholar]
- Kaye FJ. RB and cyclin dependent kinase pathways: defining a distinction between RB and p16 loss in lung cancer. Oncogene. 2002;21:6908–6914. doi: 10.1038/sj.onc.1205834. [DOI] [PubMed] [Google Scholar]
- Kazakov DV, Ivan D, Kutzner H, Spagnolo DV, Grossmann P, Vanecek T, Sima R, Kacerovska D, Shelekhova KV, Denisjuk N, Hillen U, Kuroda N, Mukensnabl P, Danis D, Michal M. Cutaneous hidradenocarcinoma: a clinicopathological, immunohistochemical, and molecular biologic study of 14 cases, including Her2/neu gene expression/amplification, TP53 gene mutation analysis, and t(11;19) translocation. Am J Dermatopathol. 2009;31:236–247. doi: 10.1097/DAD.0b013e3181984f10. [DOI] [PubMed] [Google Scholar]
- Kazakov DV, Vanecek T, Belousova IE, Mukensnabl P, Kollertova D, Michal M. Skin-type hidradenoma of the breast parenchyma with t(11;19) translocation: hidradenoma of the breast. Am J Dermatopathol. 2007;29:457–461. doi: 10.1097/DAD.0b013e318156d76f. [DOI] [PubMed] [Google Scholar]
- Komiya T, Park Y, Modi S, Coxon A, Oh H, Kaye FJ. Sustained expression of Mect1-Maml2 is essential for tumor cell growth in salivary gland cancers carrying the t(11;19) translocation. Oncogene. 2006;25:6128–6132. doi: 10.1038/sj.onc.1209627. [DOI] [PubMed] [Google Scholar]
- Lennerz JK, Perry A, Mills JC, Huettner PC, Pfeifer JD. Mucoepidermoid Carcinoma of the Cervix: Another Tumor With the t(11;19)-associated CRTC1-MAML2 Gene Fusion. Am J Surg Pathol. 2009;33:835–843. doi: 10.1097/PAS.0b013e318190cf5b. [DOI] [PubMed] [Google Scholar]
- Li J, El-Naggar A, Mao L. Promoter methylation of p16INK4a, RASSF1A, and DAPK is frequent in salivary adenoid cystic carcinoma. Cancer. 2005;104:771–776. doi: 10.1002/cncr.21215. [DOI] [PubMed] [Google Scholar]
- Luna MA. Salivary mucoepidermoid carcinoma: revisited. Adv Anat Pathol. 2006;13:293–307. doi: 10.1097/01.pap.0000213058.74509.d3. [DOI] [PubMed] [Google Scholar]
- Macarenco RS, Uphoff TS, Gilmer HF, Jenkins RB, Thibodeau SN, Lewis JE, Molina JR, Yang P, Aubry MC. Salivary gland-type lung carcinomas: an EGFR immunohistochemical, molecular genetic, and mutational analysis study. Mod Pathol. 2008;21:1168–1175. doi: 10.1038/modpathol.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins C, Cavaco B, Tonon G, Kaye FJ, Soares J, Fonseca I. A study of MECT1-MAML2 in mucoepidermoid carcinoma and Warthin’s tumor of salivary glands. J Mol Diagn. 2004;6:205–210. doi: 10.1016/S1525-1578(10)60511-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe M, Miyabe S, Nagatsuka H, Terada A, Hanai N, Yokoi M, Shimozato K, Eimoto T, Nakamura S, Nagai N, Hasegawa Y, Inagaki H. The Mect1-Maml2 fusion transcript defines a favorable subset of mucoepidermoid carcinoma: a molecular and clinicopathological study of 71 cases. Clin Cancer Res. 2006;12:3902–3907. doi: 10.1158/1078-0432.CCR-05-2376. [DOI] [PubMed] [Google Scholar]
- Otterson GA, Khleif SN, Chen W, Coxon AB, Kaye FJ. CDKN2 gene silencing in lung cancer by DNA hypermethylation and kinetics of p16INK4 protein induction by 5-aza 2′deoxycytidine. Oncogene. 1995;11:1211–1216. [PubMed] [Google Scholar]
- Rocco JW, Sidransky D. p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res. 2001;264:42–55. doi: 10.1006/excr.2000.5149. [DOI] [PubMed] [Google Scholar]
- Rossi G, Sartori G, Cavazza A, Tamberi S. Mucoepidermoid carcinoma of the lung, response to EGFR inhibitors, EGFR and K-RAS mutations, and differential diagnosis. Lung Cancer. 2009;63:159–160. doi: 10.1016/j.lungcan.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Sailasree R, Abhilash A, Sathyan KM, Nalinakumari KR, Thomas S, Kannan S. Differential roles of p16INK4A and p14ARF genes in prognosis of oral carcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17:414–420. doi: 10.1158/1055-9965.EPI-07-0284. [DOI] [PubMed] [Google Scholar]
- Scianna JM, Petruzzelli GJ. Contemporary management of tumors of the salivary glands. Curr Oncol Rep. 2007;9:134–138. doi: 10.1007/s11912-007-0011-6. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Fujioka Y. Deletion of the p16 gene and microsatellite instability in carcinoma arising in pleomorphic adenoma of the parotid gland. Diagn Mol Pathol. 1998;7:224–231. doi: 10.1097/00019606-199808000-00007. [DOI] [PubMed] [Google Scholar]
- Tirado Y, Williams MD, Hanna EY, Kaye FJ, Batsakis JG, El-Naggar AK. CRTC1/MAML2 fusion transcript in high grade mucoepidermoid carcinomas of salivary and thyroid glands and Warthin’s tumors: implications for histogenesis and biologic behavior. Genes Chromosomes Cancer. 2007;46:708–715. doi: 10.1002/gcc.20458. [DOI] [PubMed] [Google Scholar]
- Tonon G, Modi S, Wu L, Kubo A, Coxon AB, Komiya T, O’Neil K, Stover K, El-Naggar A, Griffin JD, Kirsch IR, Kaye FJ. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet. 2003;33:208–213. doi: 10.1038/ng1083. [DOI] [PubMed] [Google Scholar]