Abstract
To address the question of whether human T-cells are capable of recognizing novel isolates of influenza virus, in vitro responses to recombinant antigens and synthetic peptides derived from the sequences of H1, H3, and H5 were examined in a cohort of 64 individuals selected from a healthy blood donor population. Humans respond in vitro to H1 and H3 following exposure through natural infection and vaccination. Responses to H5 were well correlated with those to H1 or H3 and thus a significant repertoire of H5-responsive T-cells is present in many individuals; clear non-responders to H1, H3, and H5, however, do exist. Differences were observed in the cytokine responses to H1, H3, and H5; whereas both IL-2 and IFN-γ production characteristic of memory responses were observed for H1 and H3, H5-specific responses elicited primarily IL-2 and little or no IFN-γ consistent with a naïve T cell phenotype. Responses to all influenza HA were restricted by HLA-DR molecules. To address the structural basis for T-cell recognition of H1 and H5, overlapping synthetic peptides were used to identify epitopes and to determine whether recognition of H5 was limited to homologous sequences in H1, the most closely related HA phylogenetically. Although responses were generally correlated, no complete structural overlap was observed. These results suggest that helper T cell cross reactivity between different influenza strains may impart cross-protection to H5N1 strain of influenza.
Introduction
The potential risks surrounding a pandemic outbreak of influenza would apply to any new variant of the influenza virus, although, there is a significant clinical difference in the immunopathology of certain influenza strains when humans have been infected (1–5). In the case of H5N1, vaccine makers are scrambling to develop recombinant vaccine materials because H5N1 is lethal to eggs thus rendering conventional approaches inadequate (6–16); even ramping up production for a more conventional vaccine such as that for the new “swine flu” can be problematic. Little attention, however, has been given to the intrinsic ability of humans to mount an immune response against an emerging strain of influenza. Further, despite the availability of recombinant influenza viruses and corresponding sequence information, potential in vitro correlates of protective levels of responsiveness to influenza strains have not been established. We have examined in vitro T-cell responses to H5 antigen and compared them to those against H1 and H3 in order to ascertain whether normal healthy humans might be able to mount an immune response against newer isolate(s) of influenza in the absence of prior immunization.
Strain A/H5N1 was first identified in domestic chicken stocks in the Far East (5, 17–22). Only sporadic evidence of human infection was available early on but there has been a steady increase in the incidence and severity of small human outbreaks as well as an increasing possibility of a pandemic (5, 23). Although early vaccine trials are underway, whether or not current antigen constructs will prove to be protective under field conditions remains an open question. There has been additional speculation on the nature of the mutations that would be required for human to human spread (20, 24–34). While anywhere from a few to several different changes must occur in order for a human-adapted strain to emerge(30, 34, 35), reassortment in animal species and high viral mutability make such events likely. Assessment of responsiveness to naturally occurring variants or mutant viruses is important.
Significant overlap exists among sequences of H1, H3, and H5 influenza viral isolates (36–39). Whereas protective neutralizing antibodies are generally focused upon three major membrane distal epitopes of hemagglutinin (HA), there is evidence that antibodies can recognize conserved areas of HA as well as neuraminidase (40–44). The extent to which antibodies against conserved epitopes are able to confer protection against infection is unknown. Protection against infection, however, is not the only measure of an effective immune response against a pathogen; immune recognition of conserved epitopes by antibodies and, in particular, T-cells might be able to protect an individual from death due to infection.
Previous work has shown that both helper and cytotoxic T-cells are not only able to recognize conserved regions of the surface glycoproteins of influenza virus, but also internal, largely invariant viral proteins such as the matrix protein and ribonucleoprotein (42, 45–47). The extent to which cellular immune processes contribute to resistance to influenza viral infection has been examined extensively in mice, but their importance in human disease is less well understood compared to the significance of neutralizing antibodies. Using H5 as an archetypal example of an emerging pathogen, we reason that because antibody production and killer T-cell differentiation both depend upon strong T-cell helper responses, the demonstration of helper T-cell responses to H5 would form a solid basis for effective vaccination protocols. Activation of CD4 T cells would drive the appropriate production of antibodies by B cells and killing of virus infected cells by CTLs. In order to find those regions of the virus that T cells recognize we used recombinant HA antigens and synthetic peptides and show that responsiveness to H5 is well-correlated with a subject’s ability to respond to other influenza HA antigens such as H1 or H3. Human in vitro responses to H5 are qualitatively different, however, probably due to the lack of expansion of H5-specific CD4 memory T-cells that would occur following environmental or vaccine exposure. The fact that a viable H5-specific T-cell repertoire seems to exist at all in unexposed humans should provide some optimism for preventive vaccination strategies (48–50).
Materials and Methods
Methods
Subjects
A group of 64 healthy blood donors between the ages of 18 and 65 years provided repeat samples and follow-up histories. To mitigate immune response differences due to genetic heterogeneity, we arbitrarily used only patients positive for HLA-DR1 and/or DR4 in the study. Patients were excluded from the study if they had traveled out of the United States within one year of donation. Peripheral blood samples were obtained during routine donation and a leukocyte enriched fraction was provided for testing. Samples were diluted 1:4 in RPMI 1640, layered over Leukocyte Separation Medium, and centrifuged at 1200 g for 30 minutes to obtain peripheral blood mononuclear cells (PBMC). PBMC were suspended at 107/ml in RPMI 1640 containing 20% fetal calf serum (FCS) and 7.5% dimethylsulfoxide as a cryoprotectant and frozen in a control-rate liquid nitrogen freezer. All experiments were performed on thawed samples. This study was approved by the Blood Center of Wisconsin and University of Utah IRB committees.
Antigen preparations
Recombinant H1 (A/New Caledonia/1999), H3 (A/Phillipines/1992), and H5 (A/Vietnam/2004) were obtained from Protein Sciences Corporation (Meriden, CT). Antigens were dissolved in a drop (approximately 50 μl) of dimethyl sulfoxide (DMSO), then diluted to 1.0 mM in RPMI 1640 tissue culture medium. Synthetic peptides were obtained from Mimotopes Ltd (Melbourne, Australia) in sets consisting of H1 and H5 sequences of 18-mers overlapping by 11 amino acids covering the HA1 chain of both molecules. Where necessary two N-terminal lysines were added to promote solubility. In addition, 38 peptides corresponding to a limited set of known naturally occurring H5 variants from different isolates were synthesized; we have termed these “mutant” peptides throughout (Table 1 and Supplementary Table 1). Assuming a scale of synthesis of ~4 mg/peptide, each was solubilized in a drop of DMSO and brought up to 10 mM in phosphate buffered saline (PBS). For comparison, the amino acid sequences of H1, H3, and H5 are provided in Figure 1.
Table 1.
18 mer overlapping peptides spanning the influenza A hemagglutinin (HA) protein
| Peptides | H1 Sequence | H5 sequence | Peptides | Mutant sequence |
|---|---|---|---|---|
| A1 | MKAKLLVLLCTFTATYAD | KKMEKIVLLFAIVSLVKSDQ | H10 | KKMEKIVLLLAIVSLVKSDQ |
| B1 | LLCTFTATYADTICIGYH | KKFAIVSLVKSDQICIGYHA | ||
| C1 | TYADTICIGYHANNSTDT | KSDQICIGYHANNSTEQV | A7 | KSDQICIGYHANNWTEQV |
| D1 | IGYHANNSTDTVDTVLEK | GYHANNSTEQVDTIMEKN | B7 | GYHANNWTEQVDTIMEKN |
| E1 | STDTVDTVLEKNVTVTHS | TEQVDTIMEKNVTVTHAQ | ||
| F1 | VLEKNVTVTHSVNLLEDS | MEKNVTVTHAQDILEKKH | C7 | MEKNVTVTHAQDILEKTH |
| G1 | VTHSVNLLEDSHNGKLCL | THAQDILEKKHNGKLCDL | D7 | THAQDILEKTHNGKLCDL |
| H1 | LEDSHNGKLCLLKGIAPL | EKKHNGKLCDLDGVKPLI | E7 | EKTHNGKLCDLNGVKLIL |
| A2 | KLCLLKGIAPLQLGNCSV | KKLCDLDGVKPLILRDCSVA | G10 | KKLCDLNGVKPLILRDCSVA |
| B2 | IAPLQLGNCSVAGWILGN | KKKPLILRDCSVAGWLLGNP | ||
| C2 | NCSVAGWILGNPECELLI | KKCSVAGWLLGNPMCDEFIN | ||
| D2 | ILGNPECELLISKESWSY | KKLGNPMCDEFINVPEWSYI | ||
| E2 | ELLISKESWSYIVETPNP | EFINVPEWSYIVEKANPV | F7 | EFINVPEWSYIVEKANPA |
| F2 | SWSYIVETPNPENGTCYP | WSYIVEKANPVNDLCYPG | G7 | WSYIVEKANPANDLCYPG |
| G2 | TPNPENGTCYPGYFADYE | ANPVNDLCYPGDFNDYEE | H7 | ANPANDLCYPGDFNDYEE |
| H2 | TCYPGYFADYEELREQLS | CYPGDFNDYEELKHLLSR | ||
| A3 | ADYEELREQLSSVSSFER | DYEELKHLLSRINHFEKI | ||
| B3 | EQLSSVSSFERFEIFPKE | LLSRINHFEKIQIIPKSS | A8 | LLSRINHFEKIQIIPKNS |
| C3 | SFERFEIFPKESSWPNHT | FEKIQIIPKSSWSSHEAS | B8 | FEKIQIIPKNSWSSHEAS |
| D3 | FPKESSWPNHTVTGVSAS | PKSSWSSHEASLGVSSAC | C8 | PKNSWSSHEASLGVSSAC |
| D8 | PKSSWLSHEASLGVSSAC | |||
| B11 | PKSSWSSHEVSLGVSSAC | |||
| E3 | PNHTVTGVSASCSHNGKS | HEASLGVSSACPYQGKSS | E8 | HEASLGVSSVCPYQGKSS |
| F8 | HEASLGVSSACPYQRKSS | |||
| F3 | VSASCSHNGKSSFYRNLL | SSACPYQGKSSFFRNVVW | G8 | SSVCPYQGKSSFFRNVVW |
| H8 | SSACPYQRKSSFFRNVVW | |||
| G3 | NGKSSFYRNLLWLTGKNG | GKSSFFRNVVWLIKKNST | A9 | RKSSFFRNVVWLIKKNST |
| B9 | GKSSFFRNVVWLIKKNNA | |||
| H3 | RNLLWLTGKNGLYPNLSK | NVVWLIKKNSTYPTIKRS | C9 | NVVWLIKKNNAYPTIKRS |
| A4 | GKNGLYPNLSKSYVNNKE | KNSTYPTIKRSYNNTNQE | D9 | KNNAYPTIKRSYNNTNQE |
| B4 | NLSKSYVNNKEKEVLVLW | IKRSYNNTNQEDLLVLWG | ||
| C4 | NNKEKEVLVLWGVHHPPN | TNQEDLLVLWGIHHPNDA | ||
| D4 | LVLWGVHHPPNIGDQRAL | VLWGIHHPNDAAEQTKLY | E9 | VLWGIHHPNDAAEQTRLY |
| D11 | VLWGIQHPNDAAEQTKLY | |||
| E4 | HPPNIGDQRALYHTENAY | PNDAAEQTKLYQNPTTYI | F9 | PNDAAEQTKLYQNPTTYV |
| E11 | PNDAAEQTRLYQNPTTYI | |||
| F4 | QRALYHTENAYVSVVSSH | TKLYQNPTTYISVGTSTL | G9 | TKLYQNPTTYVSVGTSTL |
| G4 | ENAYVSVVSSHYSRRFTP | TTYISVGTSTLNQRLVPR | H9 | TTYVSVGTSTLNQRLVPR |
| H4 | VSSHYSRRFTPEIAKRPK | TSTLNQRLVPRIATRSKV | A10 | TSTLNQRLVPKIATRSKV |
| F11 | TSTLNQRSVPKIATRSKV | |||
| A5 | RFTPEIAKRPKVRDQEGR | LVPRIATRSKVNGQSGRM | B10 | LVPKIATRSKVNGQNGRM |
| B5 | KRPKVRDQEGRINYYWTL | RSKVNGQSGRMEFFWTIL | C10 | RSKVNGQNGRMEFFWTIL |
| C5 | QEGRINYYWTLLEPGDTI | SGRMEFFWTILKPNDAIN | D10 | NGRMEFFWTILKPNDAIN |
| G11 | SGRMEFFRTILKPNDAIN | |||
| D5 | YWTLLEPGDTIIFEANGN | WTILKPNDAINFESNGNF | ||
| E5 | GDTIIFEANGNLIAPWYA | DAINFESNGNFIAPEYAY | ||
| F5 | ANGNLIAPWYAFALSRGF | NGNFIAPEYAYKIVKKGD | ||
| G5 | PWYAFALSRGFGSGIITS | EYAYKIVKKGDSTIMKSE | E10 | EYAYKIVKKGDSAIMKSE |
| H5 | F10 | |||
| SRGFGSGIITSNAPMDEC | KKGDSTIMKSELEYGNCN | KKGDSAIMKSELEYGNCN | ||
| A6 | IITSNAPMDECDAKCQTP | MKSELEYGNCNTKCQTPM | ||
| B6 | MDECDAKCQTPQGAINSS | GNCNTKCQTPMGAINSSM | ||
| C6 | CQTPQGAINSSLPFQNVH | QTPMGAINSSMPFHNIHP | ||
| D6 | INSSLPFQNVHPVTIGEC | NSSMPFHNIHPLTIGECP | ||
| E6 | QNVHPVTIGECPKYVRSA | NIHPLTIGECPKYVKSNR | ||
| F6 | IGECPKYVRSAKLRMVTG | GECPKYVKSNRLVLATGL | ||
| G6 | VRSAKLRMVTGLRNIPSI | KSNRLVLATGLRNSPQRE | ||
| H6 | MVTGLRNIPSIQSRGLFG | LATGLRNSPQRERRRKKR |
H1: A/New Caledonia/20/99. (H1N1) H5: A/Viet Nam/1203/2004. (H5N1)
Forty-eight peptides (18 aa overlapping by 11 aa) were synthesized according to the HA1 segments of the A/New Caledonia H1 and the A/Vietnam H5 hemagglutinins. Red underlined amino acids represent changes in amino acids for the H5 mutant sequences relative to the H5 peptides from the A/Vietnam reference strain. See Supplementary Table 1 for NCBI accession numbers.
Figure 1.
Comparison of full-length HA (HA1 and HA2 chains) from isolates of H1, H3, and H5 strain A influenza viruses. Gray highlights represent sequences that stimulated T cell responses to both H1 and H5 peptides. Bold letters above the sequence represent the synthetic peptide label as indicated in Table 1.
Proliferation assays
To measure proliferative responses, PBMC from subjects were re-suspended at a concentration of 1×06/ml in RPMI 1640 tissue culture medium containing 25 mM HEPES, 2.0 mM L-glutamine, 1.0 mM Na-pyruvate, 10 U/ml heparin sodium, 100 U/ml penicillin, 100 mg/ml streptomycin, 5.0 mg/ml gentamycin and 10% pooled human serum. To 96-well round-bottom plates, 100 μl aliquots of PBMC were added to antigens of influenza virus at indicated concentrations in 100 μl 10% PHS tissue culture medium. Controls consisted of medium without antigens. Cultures were incubated at 37°C in 5% humidified CO2 for six days, pulsed overnight with 1.0 μCi/well tritiated thymidine (3H-TdR), and harvested onto glass fiber filters. Radioactive label incorporation was measured by gas scintillation spectroscopy. Results are represented as the mean ± SEM of at least triplicate cultures.
Proliferation was also assessed by staining cells with carboxyfluorescein diacetate succinimidyl ester (CFSE) and analyzing by flow cytometry. Cells were stimulated with antigens at indicated concentrations, stained with 0.5 μM CFSE as recommended by Quah, et al (50) and analyzed at 7 days. This approach enables simultaneous phenotyping of cells using fluorescently labeled monoclonal antibodies for CD4, CD8, and CD45RO as compared to medium only controls or irrelevant antigen (HCV-NS3). Analysis was done using FlowJo software (TreeStar Inc., Ashland, OR).
Elispot assays
PBMC were assayed for spot forming cells (SFC) producing IFNγ, IL-2, IL-4, and IL-5, IL-6, and IL-10 in the presence of H1, H3, H5, and media. Ninety-six-well plates with a polvinylidene difluoride filter base (Millipore, Bedford, MA) were coated with 5 μg/ml capture Ab IFNy, IL2, IL4, IL5, IL10 (BD Biosciences San Diego, CA) and 10 mu;g/ml of IL-6 (R&D Systems) in sterile coating buffer (1 ×Phosphate Buffered Saline [PBS]) overnight at 4°C. The plates were washed 3 times for 5 minutes each with 200 μl PBS/well and blocked with 200 μl/well of RPMI-1640, 10% Fetal Bovine serum(FBS), 25 mM HEPES, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM L-glutamine for 2 hour at room temperature. PBMC were plated at 1×105 cells/well with 1μg/ml of recombinant antigen, in a total volume of 200 μl/well for 44–48 hours at 37°C in 5% CO2. The plates were washed with 0.05% Tween-20/PBS. A solution of 100 μl of biotinylated anti-human detection antibody specific for IFNγ, IL-2, IL-4, IL-5, IL-6, or IL-10 was added at 2 μg/ml in PBS containing 10% FBS to each well for 2 hours at room temperature. Enzyme conjugate (100 μl/well streptavidin-HRP diluted 1:1000) was added to each well and incubated at RT for 1hr and washed as above. Color substrate (3-amino-9-ethyl-carbazole) was added at 100 μl/well for 10min to 1 hour at room temperature; for IL-6 the assay was developed with BCIP/NBT Chromagen (R&D systems) for 30 minutes in the dark. Color development was stopped with water and the membranes were dried overnight in the dark. SFC were counted on the Immunospot Analyzer (CTL Analyzers, LLC). Positive controls were specific for the individual cytokine tested and consisted of the following: For IFNγ, PBMC were cultured at 5×103/well, stimulated with 5ng/ml phorbol myristic acid (PMA, Sigma) and 500 ng/ml ionomycin (Io, Sigma) for 24 hours. For IL-2, PBMC were cultured at 2.5×103/well, stimulated with 5ng/ml PMA and 500 ng/ml Io for 24 hours. For IL-4 and IL-5, PBMC were cultured at 1×104/well after pre-stimulation with immobilized anti-human CD3 antibody (10μg/ml for plate coating), 2μg/ml soluble anti-human CD28, 10ng/ml recombinant human IL-2, and 50ng/ml recombinant human IL-4 for 48 hours. The cells were washed, then cultured in medium containing 10 ng/ml recombinant human IL-2 and 50 ng/ml recombinant human IL-4 for another 48 hours. Finally, the cells were harvested, washed, and re-stimulated with 5ng/ml PMA and 500ng/ml Io for 15 hours. For IL-10, PBMC were cultured at 1×105/well and stimulated with 1μg/ml LPS (Sigma) for 24 hours and enumerated as above.
Results
Initial screening with recombinant antigens
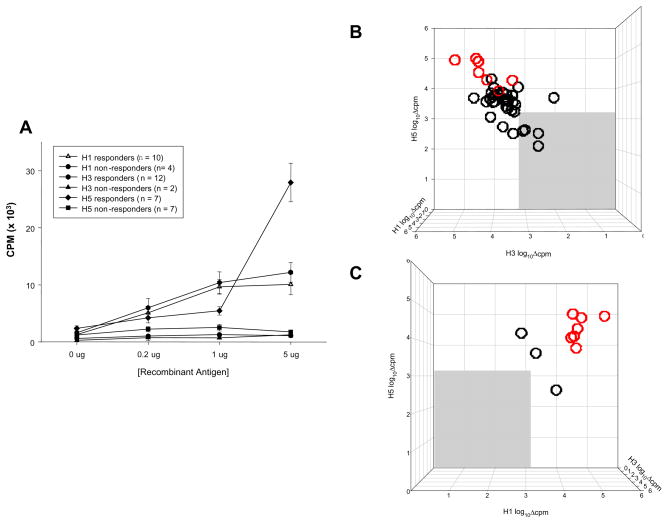
To compare human T-cell responses to recombinant HA isolates, recall responses were measured in vitro. PBMC from a subset (n=14) of healthy blood donors, all of whom shared either or both HLA-DR1 and DR4, were stimulated in proliferation assays with recombinant H1, H3, or H5 antigens at indicated doses (Figure 2A). Indicative of cross-reactivity in responding subjects, T cell proliferative responses to H1 and H3 generally could be measured at lower antigen concentrations than those to H5 (Figure 2A). Responses were quite variable and ranged from those not significantly above negative controls to more than 10-fold over background. Because of individual variation in assays with human PBMC, we transformed the data by designing an algorithm that could compensate for such wide ranging background proliferation while not distorting individual responsiveness. Typically, others have used the Stimulation Index (SI) to accomplish this: SI=[X̄exp−X̄bkg]/X̄bkg, where X̄ is the mean triplicate value of experimental (exp) or background (bkg) cultures. Such a transformation normalizes the magnitude of individual differences such that an X̄exp response of 1,000 cpm over an X̄bkg of 100 cpm yields an SI of 9, which is identical to another individual with an X̄exp response of 100,000 cpm over an X̄bkg of 10,000 cpm. Because the SI neglects differences in the magnitude of responses by different subjects, we used an alternative transformation to reduce the contribution of individual variability: log10Δcpm=log10[X̄exp−X̄bkg], not to be confused with the geometric mean. Using the above example, the log10Δcpm transformation yields values of 2.95 and 4.95, or 900 and 90,000 respectively, and preserves the linearity of the relationship between different subjects. This transformation is used for all pair-wise comparisons of responsiveness to H1, H3, and H5 recombinant antigens shown herein. Responders were considered those producing responses greater than 2 standard deviations above X̄bkg. Figures 2B and 2C show a pair-wise comparison of responses to H1, H3, and H5 where the medium background was subtracted from the maximum triplicate response (based on an antigen dose titration) to give a Δmax value plotted on a log10 scale. While a cohort of subjects failed to respond to H1, H3, or H5 antigens (Figure 2B, gray area), significant responses to the H5 antigen correlated with responses to either H1 or H3 recombinant antigens in a few high responders (Figure 2C), that is, if a subject responded to H5 they also responded to H1 or H3; seven subjects responded to all three (red circles). The results suggest significant cross-reactivity may exist in the repertoire of any given individual.
Figure 2.
Subjects responding to all three antigens are represented by red circles.
Class II restricted, helper T-cell proliferation
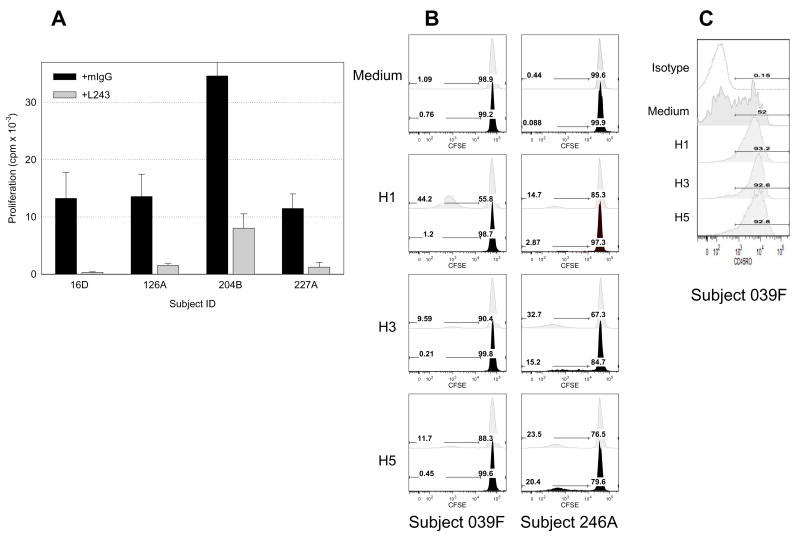
To determine whether such proliferative responses were due to class II restricted helper (CD4+) T-cell responses, four typical responders were selected for assays in which proliferation was blocked with the monoclonal antibody L243, specific for all human DR molecules (Fig. 3A). As a negative control, murine IgG was added in parallel cultures; each antibody was added at 2 μg/ml. In response to 2.5 μM recombinant H5 antigen, without exception, the presence of L243 reduced proliferation significantly. Flow cytometric analysis of proliferating T-cells stained with CFSE revealed that the vast majority of dividing cells are derived from the class II-restricted CD4+ population (Figure 3B, Table 2). The CD4+ population consisted mainly of memory T-cells (approximately 90% CD45RO+) as shown in one subject in Figure 3C and generalized for six subjects in Table 2). As with the results above, different subjects responded to H1, H3, or H5 to varying degrees; responses to an HCV recombinant NS3 control peptide were comparable to medium (data not shown).
Figure 3.
Responses to H5 are class II restricted. (A) Proliferative responses in all cases could be blocked by the L243 monoclonal antibody specific for the DRβ chain of all human class II MHC molecules in 4 subjects (gray) in comparison to IgG control (black). (B) Representative figures of CFSE staining and cytofluorometric phenotyping indicates the majority of proliferating cells were CD4-positive (light gray) more than 85% were CD45RO-positive as shown in (C). (B) Limited proliferation was observed in the CD8 population of T-cells for the majority of subjects tested (black).
Table 2.
CD4 or CD8 T cells proliferative response to recombinant HA antigens as shown as frequency of CFSElow and the % of cells that are CD45RO+
| Protein |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Medium |
H1 |
H3 |
H5 |
||||||
| Subject | CD4 | CD8 | CD4 | CD8 | CD4 | CD8 | CD4 | CD8 | |
| 003C | Freq. | 1.36 | 2.94 | 32.2 | 7.04 | 90.9 | 92.9 | 20.6 | 1.97 |
| %CD45RO | 46.6 | 26.8 | 92 | 66.9 | 90.5 | 70.8 | 90.5 | 46.9 | |
| 025C | Freq. | 3.14 | 19.2 | 86.7 | 0.85 | 53.4 | 2.89 | 26.7 | 20.8 |
| %CD45RO | 77.7 | 98.8 | 95.6 | 98.9 | 97.2 | 98.8 | 95.1 | 99.6 | |
| 039F | Freq. | 1.09 | 0.76 | 44.2 | 1.2 | 9.59 | 0.21 | 11.7 | 0.45 |
| %CD45RO | 52 | 38.9 | 93.2 | 77.1 | 91.6 | 63.6 | 92.8 | 76.2 | |
| 237A | Freq. | 0.88 | 0.28 | 2.25 | 0.54 | 7 | 0.74 | 31 | 10.6 |
| %CD45RO | 18.7 | 50 | 77.5 | 48.8 | 89.4 | 51.7 | 93.3 | 87.1 | |
| 246A | Freq. | 0.44 | 0.06 | 14.7 | 2.87 | 32.7 | 15.2 | 23.5 | 20.4 |
| %CD45RO | 52 | 100 | 93.2 | 68.2 | 92.6 | 78.4 | 92.8 | 90 | |
| 255A | Freq. | 1.34 | 0.37 | 7.7 | 0.43 | 13.4 | 0.65 | 25.6 | 2.04 |
| %CD45RO | 89.7 | 54.2 | 95.7 | 53.8 | 95.7 | 55.2 | 89.2 | 68.7 | |
Frequency of CD45RO+ CD4+ or CD8+ T cells in CFSElow lymphocytes population when stimulated with recombinant influenza antigens. PBMC were incubated with either H1, H3, or H5 at 1μg/ml for 7 days and analyzed by flow cytometry.
Cytokine production
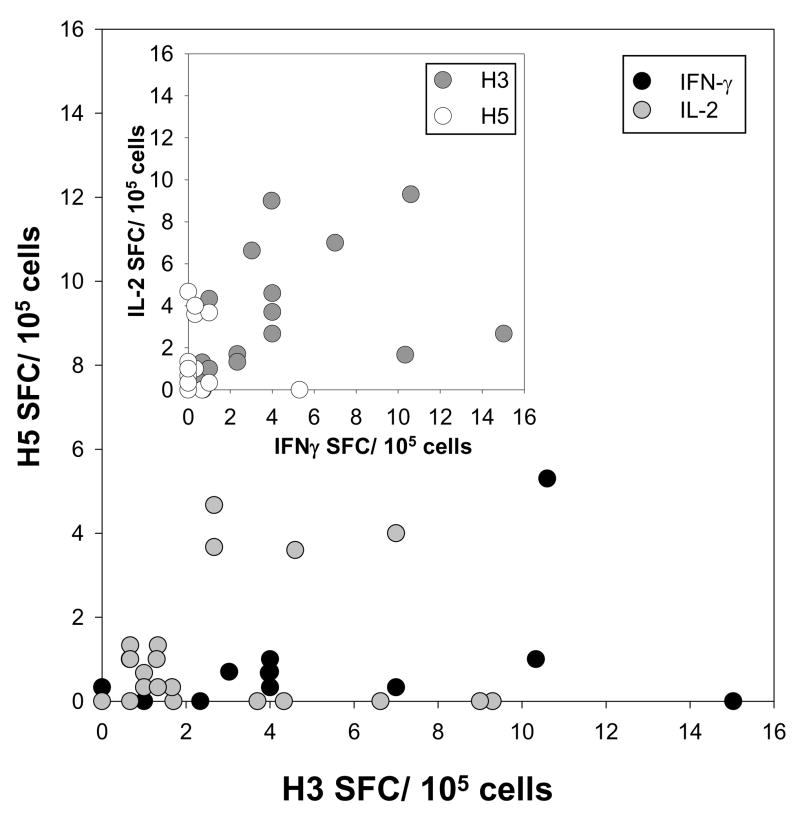
To address the qualitative nature of the helper T-cell response to HA antigens, ELISPOT analysis was performed looking at cytokine production in response to stimulation by H1, H3, or H5 recombinant antigens. In general, responders produced IL-2, IL-4, IL-5, and IFN-γ in response to H1 and H3 strains of HA (data not shown). In contrast, H5 elicited little or no IL-4 or IL-5, while producing limited quantities of IL-2 or IFN-γ (Figure 4). In this limited cohort (n=26), only one H5-responder produced significant levels of IFN-γ spot-forming cells (SFC) and 5 subjects produced moderate levels of IL-2 SFC, but essentially no IFN-γ.
Figure 4.
ELISPOT analysis of IL-2/Ifn-γ spot forming cells. Responses to H5 were characterized by modest levels of IL-2 and low levels of Ifn-γ producing T-cells consistent with an immature phenotype. Inset shows relatedness between IL-2 and Ifn-γ production in those subjects that responded well to H3, but not H5. Cells were stimulated with 1μg/ml of protein.
Structural basis for H1 and H5 responsiveness
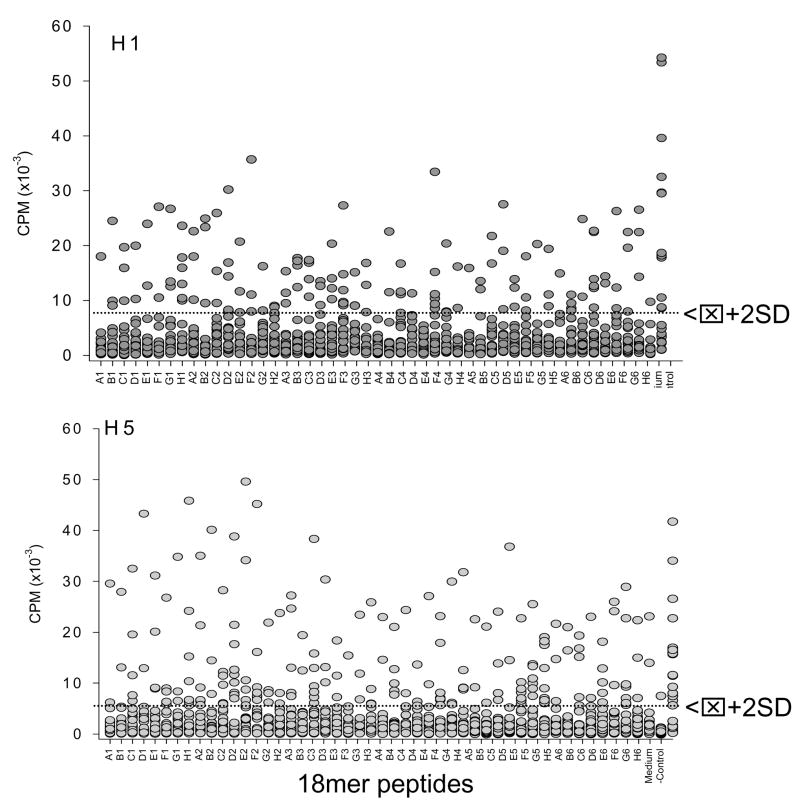
Because H5 is phylogenetically most closely related to H1, two sets of synthetic peptides were constructed (18-mers overlapping by 11 aa) based on the A/New Caledonia/1999 and A/Vietnam/2004 sequences along with recent mutants consisting of H5 (Figure 1 and Table 1). Responses were evaluated in 24 subjects to address the possibility of a structural basis for H1/H5 cross-reactivity (Figure 5). Every peptide was recognized by T-cells from at least one subject; conversely, no peptide was recognized universally. Thus, responders and non-responders were observed for each peptide, but the same subjects were not always responders or non-responders to H1 and H5 peptides.
Figure 5.
Proliferative responses by 24 subjects to 48 peptides each from H1 and H5. Negative controls consisted of medium alone and positive controls consisted of the respective recombinant protein antigen, either H1 or H5. Dotted line indicates positive threshold (mean plus 2 standard deviations of negative controls).
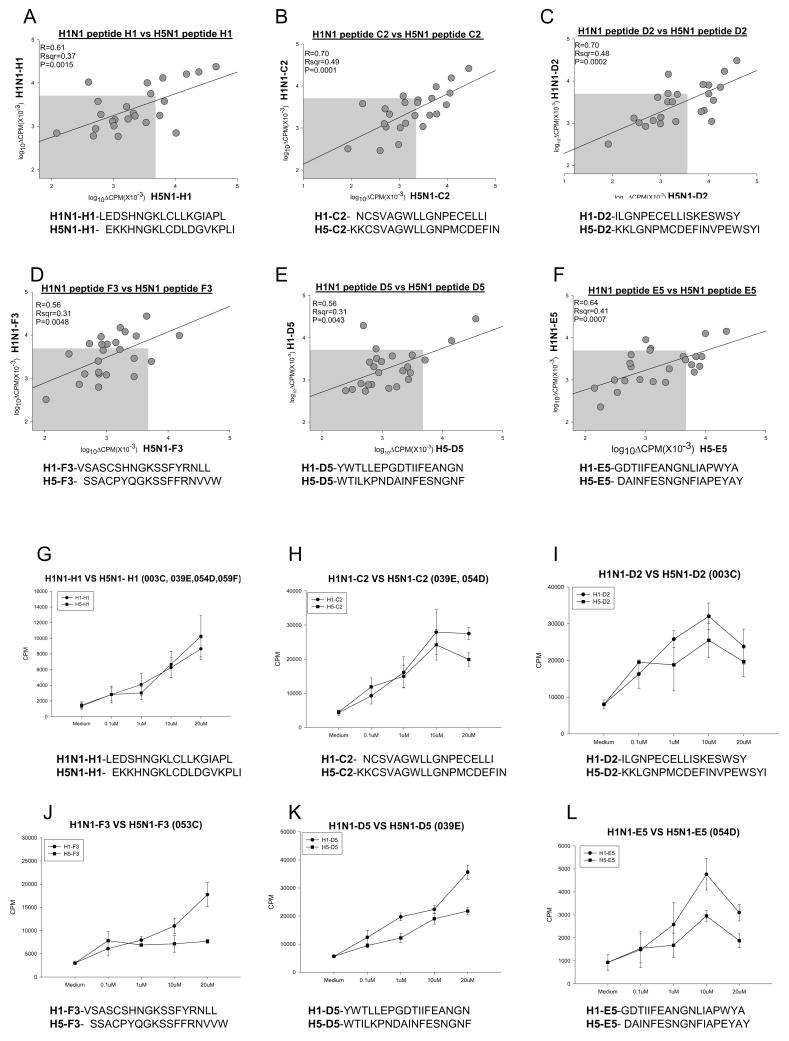
Responses to 48 homologous pairs of 18-mer peptides from the H1 and H5 sequences were compared (Figure 6A–F and supplementary 1). Linear regression (SigmaPlot 2000) was used to determine relatedness of each pair. Correlation coefficients were quite variable from a low of r=0.31 (p=0.134) for the A2 peptides to a high of r=0.83 (p<0.0001) for A6. Peptide A1 stimulated extremely strong responses in only one individual (Supplementary Figure 1). Peptide D2 was recognized by T-cells from 11 subjects with a slight bias towards H5 responsiveness (Table 1 and Supplementary Figure 1). This was in contrast to the responses observed with peptide F3 in which 11 subjects responded, but with an H1 bias (Fig. 6D). Dose dependent cross reactivity was observed with four subjects in response to the H1 peptide of both H1 and H5 hemagglutinins (Figure 6G). Such was not always true, however, as titrated responses to the H1-F3 peptide were observed, but not to H5-F3 (Figure 6J). In general, responses seemed biased towards the H1 peptides (Figures 6A–L), consistent with environmental and vaccine exposure to H1 strains in the population. One of the most closely related H1 and H5 sequences, peptide E5, produced relatively moderate responses with cells from 7 of 24 subjects and strong responses to either peptide from two (Fig. 6F, L). The peptides with a high degree of relatedness are similar to previous results (39, 51).
Figure 6.
T cell proliferative response to H1 and H5 18mer peptides are biased towards H1. Pair-wise comparison of peptide specific responses by 24 subjects; peptide sequences are indicated in the graph and in Table 1(A–F). Titration curves of H1 peptide response in comparison to corresponding H5 peptide (G–L). Subject identifiers are indicated in the heading of each graph. In panels G and H, responses by indicated subjects are presented as the mean ± SE of the pooled individual responses. Gray box indicates statistical cutoff for non-responders. Statistical values were calculated using SigmaPlot 2000.
In comparing T-cells from 24 subjects stimulated with peptides derived from various H5 mutants (Table 1, vs Fig. 7A–I), structural relatedness could not explain all responses. These were sometimes strikingly different (Fig. 7A–C), ranging from widely dispersed responses (E3 F8 in Fig. 7A) to those that were markedly linear (A5 vs B10 in Fig. 7B). The former peptides (E3 and F8) differ by only a single residue (R15G), whereas peptides A5 and B10 differ by two residues K4R and N15S, respectively (Table 1). Even a very conservative single amino acid substitution in the core of the peptide (K11R) produce significantly different results with cells from subjects 053C and 255A (H4 vs A10; Figure 7F,7I). Thus, single amino acid changes in H5 can convert a non-responder to a responder and visa versa. This has implications for pandemic spread of particular mutant strains.
Figure 7.
Comparison of the effects of mutations in H5 (underlined) and the index (A/Vietnam) sequence of H5; single amino acid mutations results in dramatic changes in T-cell proliferation (A–C). Individual T cell proliferative responses to each synthetic peptide (gray) are shown for 24 subjects compared to medium controls (black) (D–I). Responses to A/Vietnam index peptides are shown in panels D–F; responses to mutant peptides are shown in panels G–I. Note differences in responsiveness by subjects 053C and 255A to index peptide H4 (panel F) and mutant peptide A10 (panel I).
Discussion
In a group of healthy blood donors, we have shown that the H5 hemagglutinin of avian influenza virus is recognized in vitro and responses seem to correlate with responses to either the H1 or H3 strain antigens. The basis for the latter responses is thought to be vaccination and environmental exposure over recent decades; responses to H5, in contrast, are puzzling given that little epidemiologic evidence of exposure to H5 avian virus exists in the US population. The nature of H5-responsiveness is of relevance not only to vaccination efforts (Are they likely to be effective?), but also to concerns regarding the likelihood of a major avian influenza pandemic. Our focus has been on CD4, helper T-cells because of their central role in promoting both B-cell (neutralizing antibody) and CD8 T-cell (elimination of infection) responses. Responses to H5 and the most closely phylogenetically related H1, are distinguished solely by a somewhat lower frequency of IFN-γ producing cells, but IL-2 levels produced in response to H5 recombinant antigen appear comparable, indicative of at least the potential for an effective immune response. Using synthetic peptide from H1 and H5, we show that T-cells from most individuals within our study cohort were able to respond to at least one epitope in proliferative assays and responsiveness to any given H5 epitope seemed to be mostly correlated with the related H1 peptide, suggesting the possibility of cross recognition of phylogenetically conserved epitope sequences. Structural relatedness, on further analysis however, could not explain the qualitative and quantitative differences we observed on an epitope by epitope basis.
In studying human immune responses in vitro, no evidence has been uncovered that human T-cells can respond without previous exposure, either through vaccination or environmental contact. Thus, all data regarding human T-cell responses in vitro represent secondary immune responsiveness, which is consistent with secondary challenge experiments in animals. Therefore, whereas responsiveness to H5 recombinant antigen was not anticipated, the fact that it seemed correlated with responses in most cases to previously encountered H1 and H3 was reasonable. Higher level evaluation showed that the response to recombinant H5 was mediated in vitro by helper (CD4) T-cells as shown by CFSE staining and flow cytometric phenotyping; further, responding CD4 T-cells were of the memory subset (CD45RO+) and by ELISPOT could secrete reasonable levels of IL-2 and IFN-γ, even though, as noted above, the precursor frequency of the latter was lower relative to those found with T-cells stimulated with either H1 or H3 recombinant antigens. The response was class II (HLA-DR) restricted as shown by the fact that proliferation could be blocked by isotype-specific anti-HLA-DR antibody, L243, as expected. Despite the fact that all donors studied shared the HLA-DR1 and/or DR4 genes, no relationship could be observed between response patterns and HLA phenotype. Taken together, all of these data suggest that the response of human helper T-cells to H5, as measured in vitro, does not differ significantly from responses observed with other stimulatory antigens from influenza virus or other protein antigens such as tetanus toxoid. This was unexpected because the US population has not been environmentally challenged with H5 avian influenza virus convincingly.
Given the correlated responses to H1 and H5 antigens, we expected a structural basis for such comparability and sought to examine the epitope structures localized within each protein as a potential explanation. Based on the NCBI alignment of the HA1 segments of the A/New Caledonia H1 and the A/Vietnam H5, we synthesized 48 peptides, 18 amino acids in length and overlapping by 11 residues each. Peptide homology was not exact but was sufficient to provide the potential for highly similar epitopes to be found within the H5 and H1 counterparts. Significantly, there exists only a 56% homology within the HA1 region with the longest run of identity between H5 and H1 comprised of 10 amino acids. Incidentally, peptides B1, C1, and D1 comprised part or all of this conserved region and responses to these were undifferentiated from epitopes localized elsewhere. Although extensive homology is found within the HA2 segment reflecting greater conservation (81%), preliminary experiments with H1 and for which overlapping peptides were generated for the entire length of the molecule (both HA1 and HA2 segments) revealed few epitopes that stimulated strong proliferative responses. That is, most epitopes seemed to be localized to the distal HA1 domain wherein, perhaps coincidentally, are also located epitope sites for neutralizing antibodies. It is remarkable that the HA2 region is not more immunogenic in H5 as well as H1 or H3, especially given the degree of conservation, which implies lower levels of immune selection pressure on HA2-derived epitopes, most if not all of which should be available through understood mechanisms of antigen presentation. Even in HLA-DR1 transgenic mice challenged intranasally with A/New Caledonia (52), twice as many epitopes were found to be immunogenic within the HA1 sequence (30 per 342 aa) as compared to the HA2 segment (10 per 221 aa). Further, these DR1 mice could be pre-primed with an intranasal inoculation of A/New Caledonia/1999 and impart cross-reactivity to a 10μM concentration of H5N1 synthetic peptides in an in vitro ELISPOT assay counting IL-2 spot forming cells in this same region (51). In humans, Roti et al. (39) detected H5N1 HA-specific MHC class II tetramer positive CD4 T cells when stimulated with H5 peptides for 14 days with the addition of IL-2 at day 7 and then when needed. Although both studies, in combination, found similar CD4 T cell epitopes in accordance with our data in the HA region of both H1N1 and H5N1, both studies pre-primed or “enriched” the CD4 T cell population in order to evaluate the CD4 T cell responses to a naive influenza strain, in this case H5N1. The approach that we sought was to evaluate the CD4 T cell responses in a cohort of people without shifting the CD4 T cell repertoire towards either H1N1 or H5N1, in order to get an intrinsic portrayal of how cross-reactive CD4 T cells might be against a new strain of influenza. We speculate that lowered immune selection pressure, perhaps reflected in the lower phylogenetic variation in HA2, as well as fewer numbers of empirically determined T-cell epitopes in HA2 compromise what would otherwise provide an ideal focus for synthetic vaccine development (due to extensive conservation within HA2 among viral isolates), perhaps due to sequence overlap with mouse and human proteins that have engendered tolerant T-cells. Caveats to such approaches to ‘epitope counting’ include epitope representation within the peptide query set (ours includes every possible 11-mer and would miss longer sequences) and the representation of responding T-cells circulating in the peripheral blood.
We are left with a conundrum as to how H5 responsiveness has arisen. One possibility is that environmental priming has occurred contrary to the seroepidemiology. Perhaps more plausible is that exposure to other influenza viruses, other pathogens, or environmental antigens and H5 may share epitope structures and thus stimulate cross-reactive T-cell responsiveness. NCBI homology searches with H5 epitopes turned up avian and other influenza sequences, as might be expected, but also sequences associated primarily with gut flora. Thus, we postulate that the presence of anti-H5 T-cells may derive from environmental cross-priming, and the human in vitro T-cell response to H5 may be more robust than previously expected. Regardless of their origins, such results indicate that H5-responsive precursor cells pre-exist in the human population and as such, may prove conducive to vaccination efforts.
Supplementary Material
Acknowledgments
The authors thank Drs Matthew Williams, Jack Gorski, Bonnie Dittel, and Andrea Sant for helpful discussions along with Kristel Raelson, Christine Zabawa, and Rob Arao for technical assistance.
Footnotes
This work was supported by grant U19 AI062627 from the National Institute for Allergy and Immunology Diseases, National Institutes of Health.
References
- 1.The year of bird flu. Nature immunology. 2006;7:115. doi: 10.1038/ni0206-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Influenza. Bird flu moves west, spreading alarm. Science (New York, NY) 2006;311:1084. doi: 10.1126/science.311.5764.1084b. [DOI] [PubMed] [Google Scholar]
- 3.Too little, too late? Nature medicine. 2007;13:225. doi: 10.1038/nm0307-225. [DOI] [PubMed] [Google Scholar]
- 4.Abbott A, Pearson H. Fear of human pandemic grows as bird flu sweeps through Asia. Nature. 2004;427:472–473. doi: 10.1038/427472a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abdel-Ghafar AN, Chotpitayasunondh T, Gao Z, Hayden FG, Nguyen DH, de Jong MD, Naghdaliyev A, Peiris JS, Shindo N, Soeroso S, Uyeki TM. Update on avian influenza A (H5N1) virus infection in humans. The New England journal of medicine. 2008;358:261–273. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- 6.Katz JM, Plowden J, Renshaw-Hoelscher M, Lu X, Tumpey TM, Sambhara S. Immunity to influenza: the challenges of protecting an aging population. Immunologic research. 2004;29:113–124. doi: 10.1385/IR:29:1-3:113. [DOI] [PubMed] [Google Scholar]
- 7.Fauci AS. Seasonal and pandemic influenza preparedness: science and countermeasures. The Journal of infectious diseases. 2006;194(Suppl 2):S73–76. doi: 10.1086/507550. [DOI] [PubMed] [Google Scholar]
- 8.Hoelscher MA, Singh N, Garg S, Jayashankar L, Veguilla V, Pandey A, Matsuoka Y, Katz JM, Donis R, Mittal SK, Sambhara S. A broadly protective vaccine against globally dispersed clade 1 and clade 2 H5N1 influenza viruses. The Journal of infectious diseases. 2008;197:1185–1188. doi: 10.1086/529522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hampton T. Drug, vaccine research target avian flu. Jama. 2007;297:1179–1180. doi: 10.1001/jama.297.11.1179. [DOI] [PubMed] [Google Scholar]
- 10.Bresson JL, Perronne C, Launay O, Gerdil C, Saville M, Wood J, Hoschler K, Zambon MC. Safety and immunogenicity of an inactivated split-virion influenza A/Vietnam/1194/2004 (H5N1) vaccine: phase I randomised trial. Lancet. 2006;367:1657–1664. doi: 10.1016/S0140-6736(06)68656-X. [DOI] [PubMed] [Google Scholar]
- 11.Ehrlich HJ, Muller M, Oh HM, Tambyah PA, Joukhadar C, Montomoli E, Fisher D, Berezuk G, Fritsch S, Low-Baselli A, Vartian N, Bobrovsky R, Pavlova BG, Pollabauer EM, Kistner O, Barrett PN. A clinical trial of a whole-virus H5N1 vaccine derived from cell culture. The New England journal of medicine. 2008;358:2573–2584. doi: 10.1056/NEJMoa073121. [DOI] [PubMed] [Google Scholar]
- 12.Poland GA. Vaccines against avian influenza--a race against time. The New England journal of medicine. 2006;354:1411–1413. doi: 10.1056/NEJMe068047. [DOI] [PubMed] [Google Scholar]
- 13.Horimoto T, Takada A, Fujii K, Goto H, Hatta M, Watanabe S, Iwatsuki-Horimoto K, Ito M, Tagawa-Sakai Y, Yamada S, Ito H, Ito T, Imai M, Itamura S, Odagiri T, Tashiro M, Lim W, Guan Y, Peiris M, Kawaoka Y. The development and characterization of H5 influenza virus vaccines derived from a 2003 human isolate. Vaccine. 2006;24:3669–3676. doi: 10.1016/j.vaccine.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Itoh Y, Ozaki H, Tsuchiya H, Okamoto K, Torii R, Sakoda Y, Kawaoka Y, Ogasawara K, Kida H. A vaccine prepared from a non-pathogenic H5N1 avian influenza virus strain confers protective immunity against highly pathogenic avian influenza virus infection in cynomolgus macaques. Vaccine. 2008;26:562–572. doi: 10.1016/j.vaccine.2007.11.031. [DOI] [PubMed] [Google Scholar]
- 15.Smith KA, Colvin CJ, Weber PS, Spatz SJ, Coussens PM. High titer growth of human and avian influenza viruses in an immortalized chick embryo cell line without the need for exogenous proteases. Vaccine. 2008;26:3778–3782. doi: 10.1016/j.vaccine.2008.04.048. [DOI] [PubMed] [Google Scholar]
- 16.Wack A, Baudner BC, Hilbert AK, Manini I, Nuti S, Tavarini S, Scheffczik H, Ugozzoli M, Singh M, Kazzaz J, Montomoli E, Del Giudice G, Rappuoli R, O’Hagan DT. Combination adjuvants for the induction of potent, long-lasting antibody and T-cell responses to influenza vaccine in mice. Vaccine. 2008;26:552–561. doi: 10.1016/j.vaccine.2007.11.054. [DOI] [PubMed] [Google Scholar]
- 17.Dudley JP. Human H5N1 influenza. The New England journal of medicine. 2007;356:1375–1376. author reply 1376–1377. [PubMed] [Google Scholar]
- 18.Fergus R, Fry M, Karesh WB, Marra PP, Newman S, Paul E. Migratory birds and avian flu. Science (New York, NY) 2006;312:845–846. doi: 10.1126/science.312.5775.845c. [DOI] [PubMed] [Google Scholar]
- 19.Gambotto A, Barratt-Boyes SM, de Jong MD, Neumann G, Kawaoka Y. Human infection with highly pathogenic H5N1 influenza virus. Lancet. 2008;371:1464–1475. doi: 10.1016/S0140-6736(08)60627-3. [DOI] [PubMed] [Google Scholar]
- 20.Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science (New York, NY) 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 21.Maines TR, Lu XH, Erb SM, Edwards L, Guarner J, Greer PW, Nguyen DC, Szretter KJ, Chen LM, Thawatsupha P, Chittaganpitch M, Waicharoen S, Nguyen DT, Nguyen T, Nguyen HH, Kim JH, Hoang LT, Kang C, Phuong LS, Lim W, Zaki S, Donis RO, Cox NJ, Katz JM, Tumpey TM. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. Journal of virology. 2005;79:11788–11800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shute N. Of birds and men. A deadly virus is brewing in Asia. Could this be the next killer pandemic? US. 2005;138:40–48. [PubMed] [Google Scholar]
- 23.Taubenberger JK, Morens DM, Fauci AS. The next influenza pandemic: can it be predicted? Jama. 2007;297:2025–2027. doi: 10.1001/jama.297.18.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glaser L, Stevens J, Zamarin D, Wilson IA, Garcia-Sastre A, Tumpey TM, Basler CF, Taubenberger JK, Palese P. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. Journal of virology. 2005;79:11533–11536. doi: 10.1128/JVI.79.17.11533-11536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Butler D. Alarms ring over bird flu mutations. Nature. 2006;439:248–249. doi: 10.1038/439248a. [DOI] [PubMed] [Google Scholar]
- 26.Cheung CL, Rayner JM, Smith GJ, Wang P, Naipospos TS, Zhang J, Yuen KY, Webster RG, Peiris JS, Guan Y, Chen H. Distribution of amantadine-resistant H5N1 avian influenza variants in Asia. The Journal of infectious diseases. 2006;193:1626–1629. doi: 10.1086/504723. [DOI] [PubMed] [Google Scholar]
- 27.Webster RG, Govorkova EA. H5N1 influenza--continuing evolution and spread. The New England journal of medicine. 2006;355:2174–2177. doi: 10.1056/NEJMp068205. [DOI] [PubMed] [Google Scholar]
- 28.Yamada S, Suzuki Y, Suzuki T, Le MQ, Nidom CA, Sakai-Tagawa Y, Muramoto Y, Ito M, Kiso M, Horimoto T, Shinya K, Sawada T, Kiso M, Usui T, Murata T, Lin Y, Hay A, Haire LF, Stevens DJ, Russell RJ, Gamblin SJ, Skehel JJ, Kawaoka Y. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature. 2006;444:378–382. doi: 10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- 29.Auewarakul P, Suptawiwat O, Kongchanagul A, Sangma C, Suzuki Y, Ungchusak K, Louisirirotchanakul S, Lerdsamran H, Pooruk P, Thitithanyanont A, Pittayawonganon C, Guo CT, Hiramatsu H, Jampangern W, Chunsutthiwat S, Puthavathana P. An avian influenza H5N1 virus that binds to a human-type receptor. Journal of virology. 2007;81:9950–9955. doi: 10.1128/JVI.00468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solorzano A, Pappas C, Cox NJ, Swayne DE, Palese P, Katz JM, Garcia-Sastre A. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science (New York, NY) 2007;315:655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- 31.Yang ZY, Wei CJ, Kong WP, Wu L, Xu L, Smith DF, Nabel GJ. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science (New York, NY) 2007;317:825–828. doi: 10.1126/science.1135165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bewley CA. Illuminating the switch in influenza viruses. Nature biotechnology. 2008;26:60–62. doi: 10.1038/nbt0108-60. [DOI] [PubMed] [Google Scholar]
- 33.Wu WL, Chen Y, Wang P, Song W, Lau SY, Rayner JM, Smith GJ, Webster RG, Peiris JS, Lin T, Xia N, Guan Y, Chen H. Antigenic profile of avian H5N1 viruses in Asia from 2002 to 2007. Journal of virology. 2008;82:1798–1807. doi: 10.1128/JVI.02256-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuiken T, Holmes EC, McCauley J, Rimmelzwaan GF, Williams CS, Grenfell BT. Host species barriers to influenza virus infections. Science (New York, NY) 2006;312:394–397. doi: 10.1126/science.1122818. [DOI] [PubMed] [Google Scholar]
- 35.Gamblin SJ, Haire LF, Russell RJ, Stevens DJ, Xiao B, Ha Y, Vasisht N, Steinhauer DA, Daniels RS, Elliot A, Wiley DC, Skehel JJ. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science (New York, NY) 2004;303:1838–1842. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 36.Ghedin E, Sengamalay NA, Shumway M, Zaborsky J, Feldblyum T, Subbu V, Spiro DJ, Sitz J, Koo H, Bolotov P, Dernovoy D, Tatusova T, Bao Y, St George K, Taylor J, Lipman DJ, Fraser CM, Taubenberger JK, Salzberg SL. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature. 2005;437:1162–1166. doi: 10.1038/nature04239. [DOI] [PubMed] [Google Scholar]
- 37.Chang S, Zhang J, Liao X, Zhu X, Wang D, Zhu J, Feng T, Zhu B, Gao GF, Wang J, Yang H, Yu J, Wang J. Influenza Virus Database (IVDB): an integrated information resource and analysis platform for influenza virus research. Nucleic acids research. 2007;35:D376–380. doi: 10.1093/nar/gkl779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Xu X, Wang J, Ma J, Fan Y, Rakestraw KM, Webster RG, Hoffmann E, Krauss S, Zheng J, Zhang Z, Naeve CW. Large-scale sequence analysis of avian influenza isolates. Science (New York, NY) 2006;311:1576–1580. doi: 10.1126/science.1121586. [DOI] [PubMed] [Google Scholar]
- 39.Roti M, Yang J, Berger D, Huston L, James EA, Kwok WW. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. J Immunol. 2008;180:1758–1768. doi: 10.4049/jimmunol.180.3.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerhard W, Braciale TJ, Klinman NR. The analysis of the monoclonal immune response to influenza virus. I. Production of monoclonal anti-viral antibodies in vitro. European journal of immunology. 1975;5:720–725. doi: 10.1002/eji.1830051013. [DOI] [PubMed] [Google Scholar]
- 41.Jou WM, Verhoeyen M, Devos R, Saman E, Fang R, Huylebroeck D, Fiers W, Threlfall G, Barber C, Carey N, Emtage S. Complete structure of the hemagglutinin gene from the human influenza A/Victoria/3/75 (H3N2) strain as determined from cloned DNA. Cell. 1980;19:683–696. doi: 10.1016/s0092-8674(80)80045-6. [DOI] [PubMed] [Google Scholar]
- 42.Anders EM, Katz JM, Jackson DC, White DO. In vitro antibody response to influenza virus. II. Specificity of helper T cell recognizing hemagglutinin. J Immunol. 1981;127:669–672. [PubMed] [Google Scholar]
- 43.Yamada A, Brown LE, Webster RG. Characterization of H2 influenza virus hemagglutinin with monoclonal antibodies: influence of receptor specificity. Virology. 1984;138:276–286. doi: 10.1016/0042-6822(84)90351-9. [DOI] [PubMed] [Google Scholar]
- 44.Bui HH, Peters B, Assarsson E, Mbawuike I, Sette A. Ab and T cell epitopes of influenza A virus, knowledge and opportunities. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:246–251. doi: 10.1073/pnas.0609330104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jameson J, Cruz J, Terajima M, Ennis FA. Human CD8+ and CD4+ T lymphocyte memory to influenza A viruses of swine and avian species. J Immunol. 1999;162:7578–7583. [PubMed] [Google Scholar]
- 46.Kreijtz JH, de Mutsert G, van Baalen CA, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. Cross-recognition of avian H5N1 influenza virus by human cytotoxic T-lymphocyte populations directed to human influenza A virus. Journal of virology. 2008;82:5161–5166. doi: 10.1128/JVI.02694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang M, Lamberth K, Harndahl M, Roder G, Stryhn A, Larsen MV, Nielsen M, Lundegaard C, Tang ST, Dziegiel MH, Rosenkvist J, Pedersen AE, Buus S, Claesson MH, Lund O. CTL epitopes for influenza A including the H5N1 bird flu; genome-, pathogen-, and HLA-wide screening. Vaccine. 2007;25:2823–2831. doi: 10.1016/j.vaccine.2006.12.038. [DOI] [PubMed] [Google Scholar]
- 48.Webby RJ, Webster RG. Are we ready for pandemic influenza? Science (New York, NY) 2003;302:1519–1522. doi: 10.1126/science.1090350. [DOI] [PubMed] [Google Scholar]
- 49.Check E. Bird flu not set for pandemic, says US team. Nature. 2006;442:490–491. doi: 10.1038/422490b. [DOI] [PubMed] [Google Scholar]
- 50.Quah BJ, Warren HS, Parish CR. Monitoring lymphocyte proliferation in vitro and in vivo with the intracellular fluorescent dye carboxyfluorescein diacetate succinimidyl ester. Nature protocols. 2007;2:2049–2056. doi: 10.1038/nprot.2007.296. [DOI] [PubMed] [Google Scholar]
- 51.Richards KA, Chaves FA, Sant AJ. Infection of HLA-DR1 transgenic mice with a human isolate of influenza A (H1N1) primes a diverse CD4 T cell repertoire that includes CD4 T cells with heterosubtypic cross-reactivity to avian (H5N1) influenza. J Virol. 2009 doi: 10.1128/JVI.00302-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richards KA, Chaves FA, Krafcik FR, Topham DJ, Lazarski CA, Sant AJ. Direct ex vivo analyses of HLA-DR1 transgenic mice reveal an exceptionally broad pattern of immunodominance in the primary HLA-DR1-restricted CD4 T-cell response to influenza virus hemagglutinin. Journal of virology. 2007;81:7608–7619. doi: 10.1128/JVI.02834-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.