Abstract
Reversible methylation of histone tails serve as either positive signals recognized by transcriptional assemblies or negative signals that result in repression 1–4. Invading viral pathogens that depend upon the host cell’s transcriptional apparatus are also subject to the regulatory impact of chromatin assembly and modifications5–8. Here we show that infection by the α-herpesviruses HSV and VZV results in the rapid accumulation of chromatin bearing repressive histone H3-lysine 9 methylation. To enable expression of viral immediate early (IE) genes, both viruses use the cellular transcriptional coactivator HCF-1 to recruit the demethylase LSD1 to the viral immediate early promoters. Depletion of LSD1 or inhibition of its activity with MAO inhibitors results in the accumulation of repressive chromatin and a block to viral gene expression. As HCF-1 is a component of the Set1 and MLL1 histone H3 lysine 4 methyl-transferase complexes 9,10, it thus coordinates modulation of repressive H3-lysine 9 methylation levels with addition of activating H3-lysine 4 trimethylation marks. Strikingly, MAO inhibitors also block the reactivation of HSV from latency in sensory neurons, indicating that the HCF-1 complex is a critical component of the reactivation mechanism. The results support pharmaceutical control of histone modifying enzymes as a strategy for controlling herpesvirus infections.
The cellular transcriptional coactivator, HCF-1, is essential for expression of the immediate early genes (IE) of the α-herpesviruses HSV and VZV11. Both viruses utilize virion-encapsidated activators to recruit HCF-1-Set/MLL1 histone methyl-transferase (HMT) complexes9,12 to the IE promoters, resulting in histone H3-lysine 4 (H3K4) trimethylation and initiation of gene transcription12,13. HCF-1 depletion results in increased levels of repressive histone H3-lysine 9 (H3K9) methylation, suggesting a central role for HCF-1 in modulating chromatin modifications that determine viral gene expression.
To investigate histone methylation in herpesvirus gene expression, we assessed the state of H3K4 and H3K9 methylation by chromatin immunoprecipitation (ChIP) assays using a model VZV IE promoter-reporter (Fig. 1a). In the absence of the VZV IE activator (IE62), repressive H3K9 methylation accumulated on the promoter while in its presence, H3K9 methylation was reduced and positive H3K4 trimethylation was enhanced. This indicated that, in addition to the Set1/MLL1 H3K4 methyl-transferase, an H3K9 demethylase would also be required to modulate the levels of repressive chromatin.
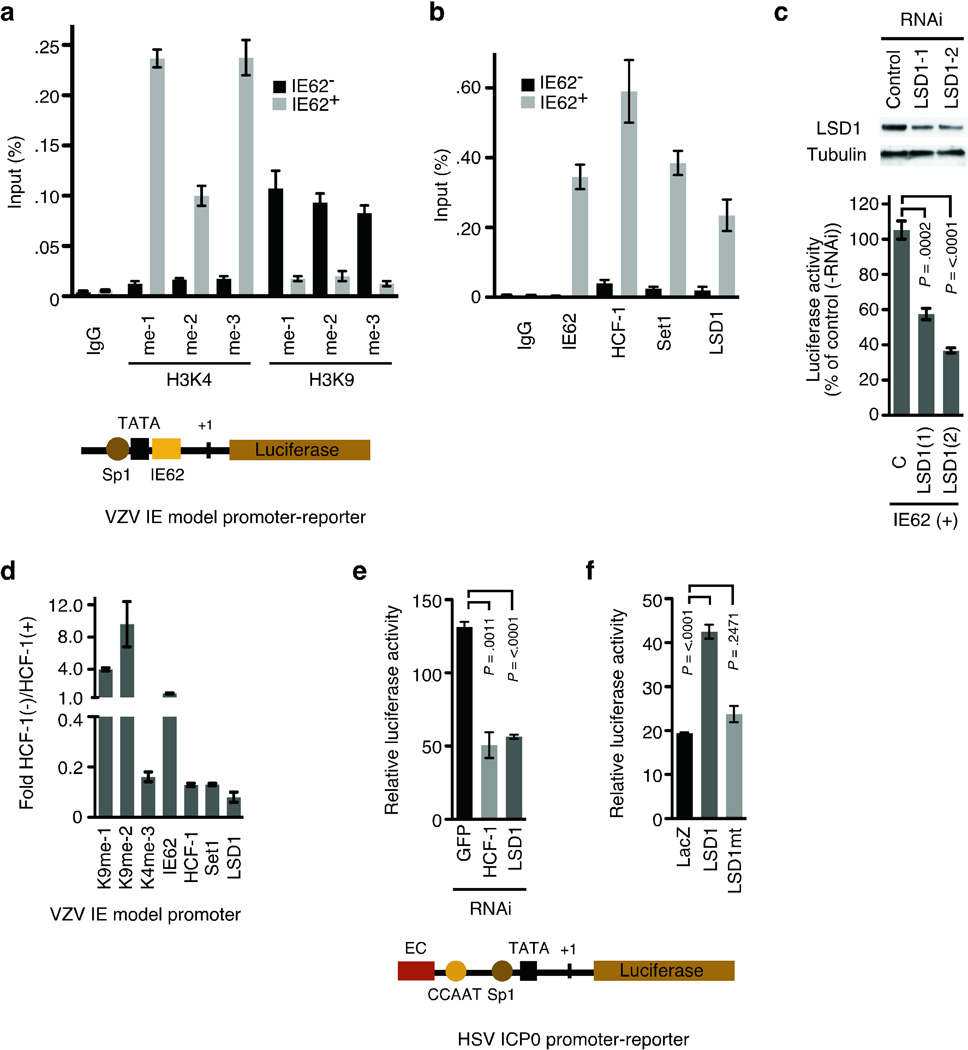
Figure 1. LSD1 is critical for viral activator mediated transcription of VZV IE and HSV IE model promoters.
(a) The VZV IE model promoter-reporter is illustrated with Sp1, TATA, and IE62 binding sites. ChIP assay showing H3K4 and H3K9 methylation and (b) activator/coactivator occupancy at the VZV IE model promoter in the presence and absence of the VZV IE62 activator. IgG, control immunoglobulin, me-1, monomethyl; me-2, di-methyl; me-3, tri-methyl. (c) Western blot of LSD1 and control (tubulin) showing depletion of LSD1 (LSD-1, LSD-2) relative to cells transfected with control scrambled RNAi (C). VZV IE promoter-luciferase reporter activity in cells transfected with IE62 and LSD1 or control RNAi(s) relative to cells transfected with no RNAi. LSD1 depletions ranged from 42–57%. (d) ChIP assay showing H3K4 and H3K9 methylation and activator/coactivator occupancy on the model VZV IE promoter in cells transfected with control shRNA (HCF-1+) or HCF-1 shRNA (HCF-1−). Occupancy is expressed as the ratio of that in HCF-1-depleted cells to that in control HCF-1+ cells. (e) The HSV-1 ICP0 promoter-reporter is schematically illustrated with the enhancer core (EC) element that nucleates the assembly of the HCF-1 protein enhancer complex, CCAAT, TATA, and Sp1 binding sites. ICP0 promoter-luciferase reporter activity in cells transfected with HCF-1, or LSD1 RNAi relative to control (GFP) RNAi. (f) Activity of ICP0 promoter-luciferase reporter in cells expressing control β-galactosidase (LacZ), wild-type LSD1, or an LSD1 catalytic mutant (LSD1 K661A).
Recently it has been demonstrated that the H3K9 demethylase activity of Lysine Specific Demethylase1 (LSD1) is important for nuclear hormone receptor-dependent transcription14–16 and cell fate determination17. Therefore, we investigated the role of this enzyme in viral IE transcription. As shown in Fig. 1b, LSD1 occupied the VZV IE promoter-reporter with HCF-1 and Set1, in the presence but not absence of the viral activator. Furthermore, depletion of LSD1 resulted in reduced induction of the reporter, demonstrating that LSD1 was important for IE62-mediated activation (Fig. 1c).
We next asked if LSD1 recruitment was dependent upon the coactivator HCF-1. In cells depleted of HCF-1, Set1 and LSD1 occupancy was reduced and correlated with a reduction in H3K4 trimethylation and an enhancement of H3K9 methylation (8–9 fold, Fig. 1d). In contrast, occupancy of the activator IE62 was not affected.
The α-herpesviruses VZV and HSV-1 share similar regulatory mechanisms, including the recruitment of HCF-1 by the respective viral IE activators11. As shown in Fig. 1e, LSD1 depletion also reduced the viral-induced expression of an HSV IE reporter gene. Additionally, exogenous expression of wild-type LSD1 stimulated the reporter expression while a catalytic mutant had no significant impact (Fig. 1f).
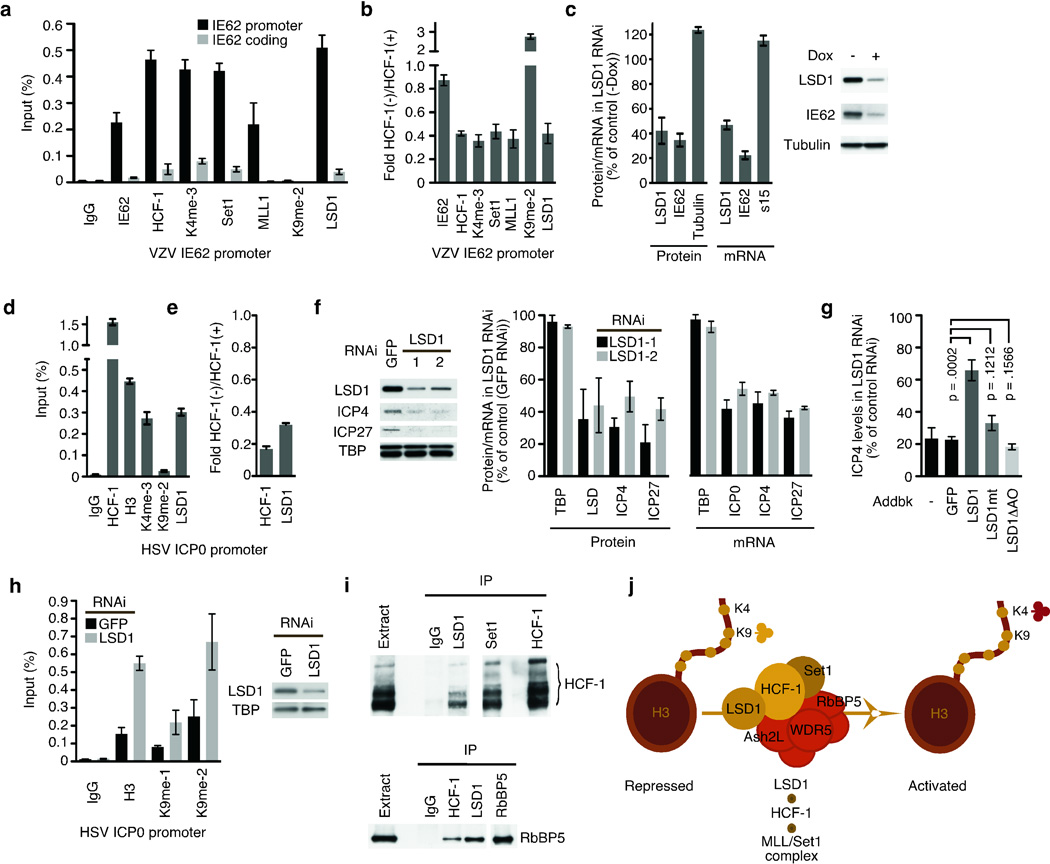
To investigate the role of these factors during viral infection, HCF-1 depleted cells were infected with VZV. In non-depleted cells (Fig. 2a), promoter occupancy by Set1, MLL1, and LSD1 were substantial with a high level of H3K4 trimethylation and near background level of H3K9 methylation. In contrast, promoter occupancy by Set1, MLL, and LSD1 were decreased in HCF-1 depleted cells with a correlating decrease in H3K4 trimethylation and increase in repressive H3K9 methylation (Fig. 2b). The requirement for LSD1 was addressed by infection of an inducible LSD1-RNAi cell line (Fig. 2c). Depletion of 60% of the cellular LSD1 reduced the levels of the IE protein by 66% and mRNA by 78%; indicating that LSD1 was critical for IE gene expression during viral infection.
Figure 2. An HCF-1/LSD1 complex is essential for α-herpesvirus IE gene transcription.
(a) ChIP assay showing H3K4 and H3K9 methylation and activator/coactivator occupancy on the VZV genomic IE62 promoter and coding sequences at 4 hrs post infection. (b) ChIP assay showing H3K4 and H3K9 methylation and activator/coactivator occupancy on the VZV IE promoter in cells transfected with HCF-1 shRNA (HCF-1−) relative to that in cells transfected with control shRNA (HCF-1+). (c) Western blot of LSD1, IE62, and control (tubulin) in VZV infected MCF7 cells inducibly expressing LSD1 shRNA in the presence and absence of doxycycline induction. S15, ribosomal subunit mRNA. (d) ChIP assay showing H3K4 and H3K9 methylation and coactivator occupancy on the genomic HSV ICP0 promoter at 4 hours post HSV-1 infection. H3, histone H3. (e) ChIP assay showing HCF-1 and LSD1 occupancy on an HSV IE promoter in cells transfected with control or HCF-1 siRNA (HCF-1−). (f) Western blot of HSV IE proteins (ICP4, ICP27), LSD1, and control TATA-binding protein (TBP) in control cells (GFP RNAi) and cells depleted for LSD1 (LSD1-1, LSD1-2). (g) The levels of the HSV-1 IE protein ICP4 in cells depleted for LSD1 and transfected with plasmids expressing no protein (−), control GFP, wild-type LSD1, an LSD1 catalytic mutant (K661A), or an LSD1 mutant lacking the amine oxidase domain (LSD1ΔAO). The results of 2-tailed t tests are shown representing 4 independent experiments. (h) ChIP assay showing histone H3 and H3K9 methylation on the HSV ICP0 promoter in cells depleted of LSD1 (LSD1 RNAi) and control cells (GFP RNAi). The data is normalized to the levels of H3 at the cellular GAPDH promoter in the appropriate GFP RNAi cells or LSD1 RNAi cells. Western blot showing depletion of LSD. (i) HCF-1 western blot of HCF-1, LSD1, Set1, and control IgG immunoprecipitates (top panel). Western blot of the Set1/MLL1 HMT core subunit RbBP5 from LSD1, HCF-1, and control IgG immunoprecipitates (bottom panel). (j) Model of the HCF-1-Set1-LSD1 complex representing HCF-1 coupled demethylase (LSD1) and methyltransferase (Set1) activities. RbBP5, Ash2L, and WDR5 are core subunits of the Set1 HMT complex.
In an analogous manner, HCF-1 and LSD1 occupied the HSV-1 IE promoter during infection with the correlating high level of H3K4 and low level of H3K9 methylation (Fig. 2d). Furthermore, HCF-1 depletion resulted in a concomitant decrease in the recruitment of LSD1 (Fig. 2e). Importantly, depletion of LSD1 resulted in reduced viral IE proteins and mRNAs (Fig. 2f) and the levels of IE gene expression were recovered by exogenous expression of wild-type LSD1 but not an LSD1 catalytic mutant or a mutant lacking the amine oxidase domain (Fig. 2g). Strikingly, depletion of LSD1- resulted in accumulation of nucleosomes bearing repressive H3K9 methylation on the viral IE promoter (Fig. 2h). Together, the results demonstrate that the HCF-1-dependent recruitment of LSD1 plays an important role in the initiation of both VZV and HSV-1 infection, likely via modulation of the levels of repressive H3K9 chromatin marks at the IE promoters.
Recruitment of LSD1 is dependent on HCF-1 (Fig. 1d, Fig. 2b, and 2e). Therefore, we determined if this reflected an interaction by coimmunoprecipitation assays. Both Set1- and LSD1-specific antibodies efficiently coimmunoprecipitated HCF-1 (Fig. 2i, top). In addition, immunoprecipitation of either HCF-1 or LSD1 resulted in coimmunoprecipitation of RbBP5 (Fig. 2i, bottom), a common core subunit of the Set/MLL HMTs18; suggesting that LSD1 was associated with the HCF-1/HMT complex.
LSD1 has both repressive (H3K4 demethylation) and activating (H3K9 demethylation) activities. Demethylation of H3K4 is mediated by the CoREST/LSD1 complex and targeting or specificity is determined by the associated components19,20. As shown in Supplementary Fig. 1, CoREST factors were present in the LSD1 immunoprecipitate but absent from the HCF-1/LSD1 complex. Based on these data, we propose that HCF-1 couples the demethylase LSD1 with the Set1/MLL1 HMT in a novel complex, providing both specificities to promote IE gene transcription (Fig. 2j).
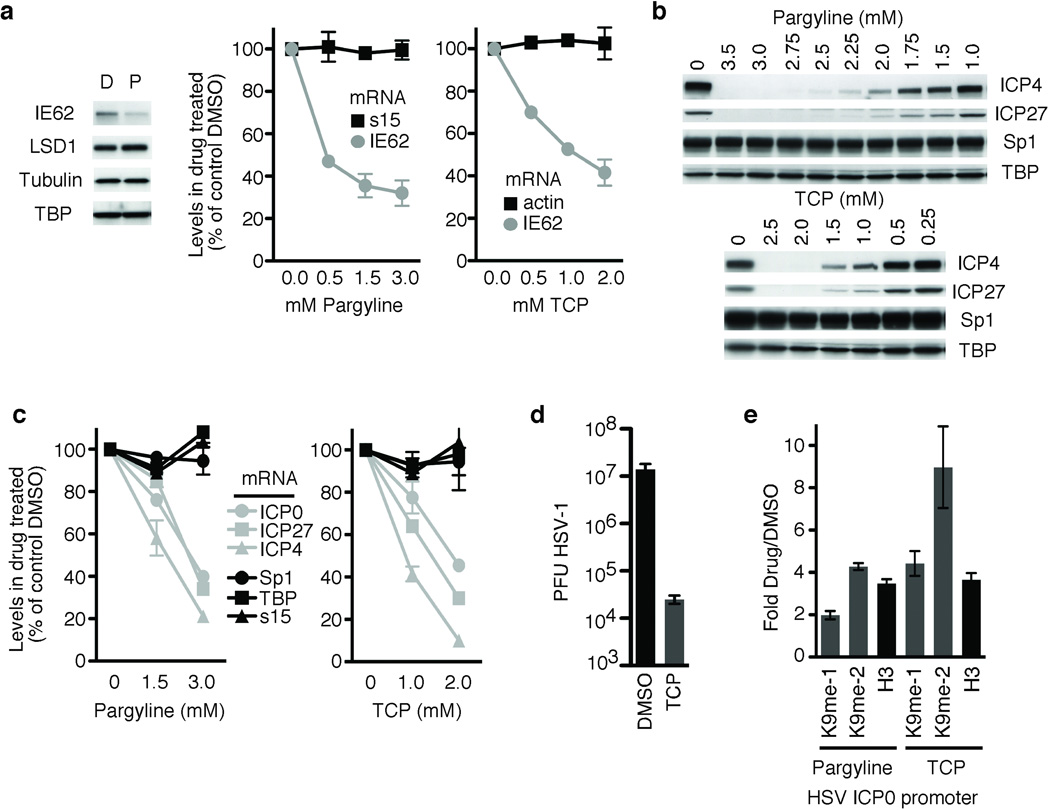
Mechanistically, LSD is unique relative to other identified demethylases, including the members of the Jmjd family, as it demethylates lysine residues via a flavin-adenine-dinucleotide-dependent reaction21,22. This reaction is inhibited by monoamine oxidase inhibitors (MAOIs)15,23–25; pharmaceuticals used in the treatment of psychiatric disorders (clinical depression, anxiety), Parkinson’s, and migraines. Furthermore, select MAOIs inhibit LSD1 at levels comparable to inhibition of the clinical mitochondrial MAO targets23,24. Therefore, we investigated the impact of the MAOIs Pargyline and Tranylcypromine (TCP) on the initiation of VZV and HSV infection. For both viruses, treatment with either MAOI resulted in a dose dependent decrease in viral IE mRNA and proteins (Fig. 3a–c). The levels of cellular protein controls were unaffected and no significant cellular toxicity was seen (Supplementary Fig. 2). TCP also reduced viral yields from HSV-infected cells nearly 1000-fold (Fig. 3d). Strikingly, analogous to LSD1 RNAi-mediated depletions (Fig. 2h), nucleosomes bearing repressive marks accumulated on the HSV-1 IE promoter in the presence of either MAOI (Fig. 3e). The results support the model whereby LSD1 prevents accumulation of H3K9 methylation, thereby allowing productive infection by both α-herpesviruses.
Figure 3. Inhibition of LSD1 with MAOIs blocks α-herpesviral lytic gene expression.
(a) Western blot showing Pargyline (P) mediated inhibition of VZV IE gene expression (IE62) in cells infected with VZV for 4 hrs relative to control DMSO (D). LSD1, Tubulin, and TBP control proteins are shown. qRT-PCR of IE62 and control (s15, actin) mRNA levels in cells infected with VZV for 4 hrs in the presence of increasing amounts of Pargyline or Tranylcypromine (TCP). The results are graphed as the percent of levels in control treated cells. (b) Western blot showing inhibition of HSV IE protein expression (ICP4, ICP27) at 4 hours post HSV-1 infection in cells treated with increasing concentrations of either Pargyline or TCP. Control proteins (Sp1, TBP) are shown. (c) qRT-PCR analyses of mRNA levels of HSV IE genes and controls (Sp1, TBP, s15) in cells treated with selected concentrations of Pargyline or TCP. (d) Viral yields from cells infected with 0.1 plaque forming units (PFU) HSV-1 per cell in the presence of 2 mM TCP or control DMSO for 24 hrs. (e) ChIP assay showing histone H3 and H3K9 methylation on the HSV-1 IE0 promoter in the presence of 3 mM Pargyline or 2 mM TCP at 4 hrs post HSV-1 infection. The results are shown as ratios of occupancy in drug treated cells to those in control DMSO treated cells. The data is normalized to the ratio of total H3 in drug treated/DMSO treated cells at the cellular actin promoter. me-1, mono-methyl; me-2, di-methyl; H3, total histone H3.
In addition to mono- and di-methyl H3K9, inhibition of LSD1 resulted in increased H3K9-trimethylation and occupancy by the heterochromatin protein 1 (Supplementary Fig. 3a). As LSD1 only removes mono- and di-methyl modifications, the increased H3K9 trimethylation in the absence of LSD1 may reflect (i) increased levels of dimethyl substrates that accumulate during chromatin assembly or (ii) the requirement for an additional H3K9 demethylase(s) of the Jmjd family, in conjunction with LSD1, to provide the specificity required to remove tri-methylation26,27. The latter is supported by the observation that even in the presence of LSD1, H3K9 trimethylation was detected on the IE promoters during initial stages of HSV-1 infection (Supplementary Fig. 3b). Irrespective, the requirement for LSD1 to promote viral IE expression identifies it as an essential control component and a target for inhibition of α-herpesvirus infection.
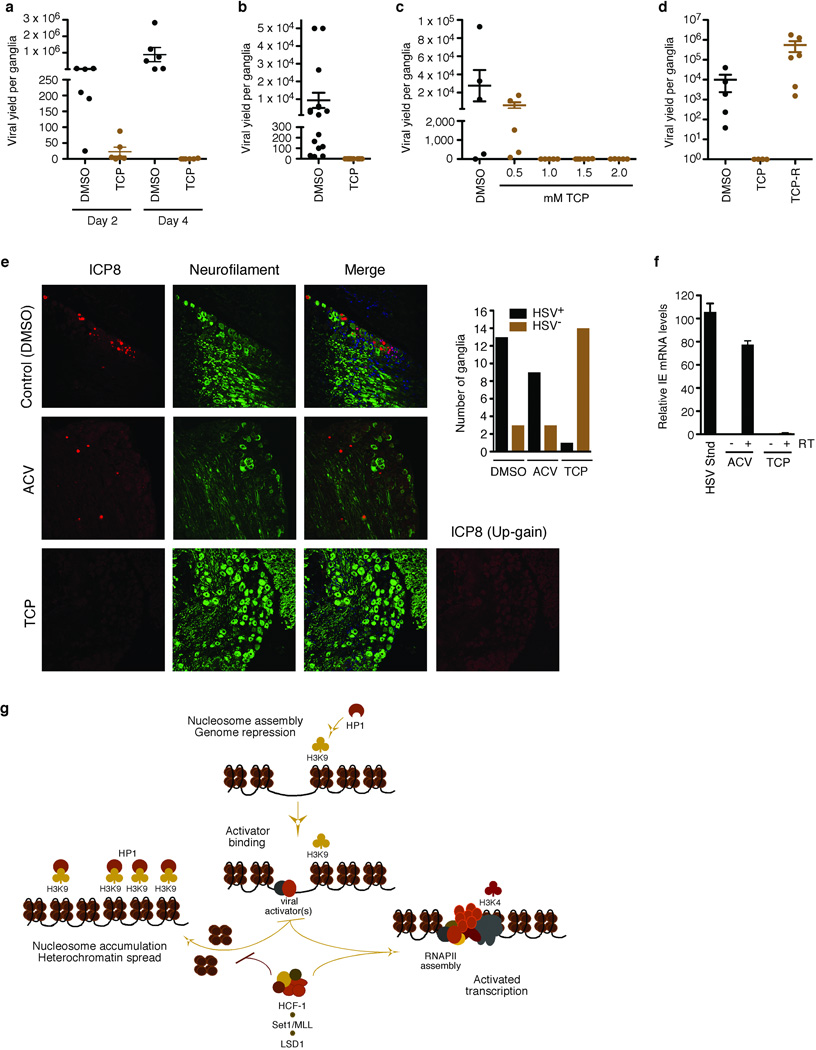
In addition to lytic replication, α-Herpesviruses establish latent infections and cycles of reactivation in sensory neurons. In HSV-1, latency and reactivation correlate with alterations in chromatin modifications on the viral IE promoters5,28–31. Significantly, HCF-1 is (i) sequestered in the cytoplasm of unstimulated sensory neurons; (ii) rapidly transported to the nucleus upon reactivation stimuli32,33; and (iii) recruited to the viral IE promoters at the outset of reactivation34. Therefore, given the impact of LSD1 on viral IE gene expression and its association with HCF-1, we investigated the potential role of LSD1 in viral reactivation by tissue explant of HSV latently infected trigeminal ganglia (TGs) in the presence and absence of TCP (Fig. 4a). Strikingly, TCP significantly reduced the reactivation of HSV-1 (P = 0.0043 and 0.0011 for days 2 and 4, respectively). Due to variance in viral loads of individual animals, these results were confirmed by studies in which each half of a TG was explanted in the presence and absence of TCP (P = 0.0002; Fig. 4b). Moreover, reduced levels of the inhibitor also effectively blocked viral reactivation (Fig. 4c). Finally, as shown in Fig. 4d, potential TCP toxicity was not responsible for the suppression of viral reactivation as high viral titers were recovered following drug reversal.
Figure 4. Inhibition of LSD1 with MAOIs blocks HSV-1 reactivation from latency.
(a) Viral yield from ganglia explanted in the absence (DMSO) or presence of 2 mM TCP for 2 days or 4 days. Day 2 P = 0.0043; day 4 P = 0.0011; n=6 for each sample set. (b) Viral yield from paired explanted ganglia in the absence (DMSO) or presence of 2 mM TCP for 2 days. P = 0.0002; n=16 for each sample set. (c) Viral yield from explanted ganglia in the presence or absence (DMSO) of various concentrations of TCP. P < 0.05 for 1.0, 1.5, and 2.0 mM TCP while P > 0.05 for 0.5 mM TCP; n=5 for control, 0.5, 1.0, and 2.0 mM; n=6 for 1.0 mM. (d) Viral yield from explanted ganglia in the absence (DMSO) or presence (TCP) of 2 mM TCP for 2 days followed by incubation in the absence for 3 days (TCP-R). P < 0.025 for TCP vs TCP-R; n=4 for TCP; n=6 for TCP-R. Details of all statistical analyses are in Supplementary Methods. (e) Immunofluorescent staining of HSV-1 latently infected ganglia explanted for 48 hrs in the presence of control (DMSO), 100 uM ACV, or 2 mM TCP. ICP8, HSV single stranded DNA binding protein. For each condition, the number of HSV-1+ and HSV-1− ganglia is shown (P =.00002). (f) Nested RT-PCR analyses of HSV ICP27 IE mRNA from ganglia explanted in the presence of 2 mM TCP or control ACV for 12 hrs. cDNA samples were normalized to the levels of the cellular Sp1 mRNA as determined by qPCR. HSV Stnd represents is the signal from an equivalent amount of control cDNA produced from HSV infected 3T3 cells at 6.4×10−5 pfu/cell. Quantitation was as described in Supplementary Methods. −RT, +RT denote the absence or presence of the reverse transcriptase in the cDNA synthesis reaction. (g) An HCF-1 complex couples LSD1 with Set1/MLL1 to promote α-herpesvirus IE gene transcription. Upon infection, the genomes of infecting α-herpesviruses are subject to the accumulation of repressive chromatin (H3K9 methylation and HP1 occupancy). For productive infection, α-herpesviruses recruit an HCF-1-dependent modification complex containing LSD1 and the H3K4 HMTs Set1/MLL1 to promote the installation of positive transcriptional marks. Failure to recruit this complex results in continued accumulation of nucleosomes bearing repressive H3K9 methylation that silences the viral genome. RNAPII, RNA polymerase II; HP1, heterochromatin protein 1.
These studies suggested that MAOI inhibition of LSD1 prevented viral reactivation. However, it remained possible that TCP inhibited lytic spread of the virus but not the initiating reactivation events. Therefore, ganglia were explanted in the presence of DMSO (control), Acyclovir (to prevent viral DNA replication/spread35,36), or TCP. Sections were probed with antibodies to an HSV lytic antigen (ICP8) (Fig. 4e and Supplementary Fig. 4a). In control treated ganglia, clusters of ICP8+ neurons were detected in multiple sections of 13 of 16 ganglia, representing initiating neurons as well as infected neurons and support cells from lytic spread. In the presence of Acyclovir (ACV), distinct ICP8+ neurons were detected in sections of 9 of 12 ganglia, representing primary neurons undergoing viral reactivation. Strikingly, in the presence of TCP, only a single ICP8+ neuron was detected in 1 of 15 ganglia, clearly demonstrating that TCP inhibited the initiation of reactivation rather than inhibiting lytic spread (P = .00002). As additional evidence that TCP inhibits IE gene expression and consequently, reactivation of HSV from latency, viral IE mRNAs could be readily detected by nested RT-PCR from ganglia explanted in the presence of DMSO or ACV but were not detected in the presence of TCP (Fig. 4f and Supplementary Fig. 4c).
Together the data support the model (Fig. 4g) whereby the genomes of infecting α-herpesviruses are subject to cell-directed accumulation of repressive chromatin. For productive infection, α-herpesviruses recruit HCF-1-dependent modification complexes containing LSD1 and the H3K4 HMTs Set1/MLL1 to prevent the accumulation of repressive chromatin marks and install positive marks. It should be noted that the encapsidated HSV-1 genome is devoid of nucleosomes which are deposited during the initial stage of infection37,38. As positive chromatin marks are installed and viral gene transcription is activated, the levels of associated nucleosomes decrease; likely due to targeted chromatin remodeling. In contrast, inhibition of the HCF-1 complex components results in accumulation of nucleosomes bearing H3K9 methylation and repression of viral gene expression.
As LSD1 can demethylate both histone H3K4 and H3K9, the coupling of this protein in the HCF-1 Set/MLL methyltransferase complex may enhance H3K9 demethylation or preferentially target it to this substrate; although additional histone modifications and modification activities may also contribute to the H3K4 or H3K9 recognition and specificity. Moreover, the presence of the Set1/MLL1 H3K4 methyltransferase components in the complex could function to maintain the levels of H3K4 methylation, even in the presence of LSD1 H3K4 demethylase activity.
The recruitment of HCF-1 complex(es) during the initiation of infection emphasizes the mechanism by which these viruses escape the host cell-directed assembly of repressive chromatin. Interestingly, LSD1 has also been recently shown to demethylate non-histone proteins39,40, raising the possibility that components involved in the viral IE gene transcription machinery may also be modulated by LSD1-dependent demethylation.
With respect to the cycle of latency and reactivation established by these viruses, signals that lead to viral reactivation result in rapid nuclear transport32,33 and occupancy of viral IE promoters by HCF-134. Coupled with the data presented here that inhibition of the HCF-1 associated demethylase LSD1 blocks viral reactivation, the observations strongly support the model that HCF-1 modification complexes play a critical role in determining the latency-reactivation state of these viruses.
The dependence of viral pathogens on host cell chromatin machinery highlights a potential for therapeutic intervention. As LSD1 is a well defined target of MAOIs and these pharmaceuticals are widely used therapeutically, these observations identify a novel therapeutic target for herpesvirus infections and enhances the importance of ongoing efforts to develop additional LSD1 inhibitors.
METHODS SUMMARY
Cell culture and virus
HeLa, BS-C-1, HEK293, Vero, MeWo and VZV (Ellen) stocks were obtained from American Type Culture Collection. Viral infections with HSV-1 and cell-associated VZV were done according to standard protocols.
Latently infected mice and trigeminal ganglia
Balb/c mice were infected with 5 × 105 PFU HSV-1 (strain 17) per eye after corneal scarification. Latently infected mice were sacrificed 30 days post clearance of the primary infection and trigeminal ganglia were rapidly explanted into culture in the presence or absence of TCP or control (DMSO or ACV). Post explant incubation as indicated, the ganglia were homogenized and briefly sonicated. The reactivated viral yield of each ganglia was determined by titering the clarified supernatant on Vero cells. For analyses of viral reactivation by immunofluorescence, ganglia were explanted in the presence of control vehicle (DMSO), ACV (100 uM) or TCP (2 mM) for 48 hrs, fixed in 4% paraformaldehyde, and embedded in paraffin. Ganglia sections were subjected to citric acid treatment for antigen retrieval, stained with the indicated antibodies (Supplemental Materials), and visualized using a Leica TCA SP5 confocal microscope. RT-PCR detection of viral IE and cellular control mRNAs in ganglia explanted in the presence of ACV or TCP was done as described in Supplementary Methods. All animal care and handling was done in accordance with the US National Institutes of Health Animal Care and Use Guidelines and as approved by the NIAID Animal Care and Use Committee.
Statistical analyses
Statistical comparisons were made using two-tailed t test (reporter assays) with a statistical significance of <0.05; Wilcoxon signed rank test (paired ganglia) with a statistical significance of <0.05; Kruskal-Wallis test with post hoc Dunn’s multiple comparison test (drug titration and reversal) with a statistical significance of <0.025; Mann-Whitney U test with Dunn’s post hoc adjustment (non-paired ganglia timecourse) with a statistical significance of <0.025; or Fischer’s Exact Test with a statistical significance of <.05. Analyses were made using Prism (V5.0a) and are expressed as the mean +/− s.e.m. Details of statistical analyses are given in Supplemental Methods.
Chromatin Immunoprecipitations and qPCR
Chromatin immunoprecipitations from control, HCF-1 depleted, and LSD1 depleted cell extracts were done as described12. Recovered DNAs were analyzed, in triplicate, by qPCR using ABI Sybr Green PCR Master Mix. In each case, the ChIP data are the means +/− s.e.m. from at least two independent experiments. The sequence of primer sets, qPCR conditions, and the antibodies used are in Supplementary Methods.
qRT-PCR
Oligo dT primed cDNA was produced from total RNA using RNAqueous-4PCR and RETROscript (Ambion) according to the manufacturer’s recommendations. cDNAs were quantitated by qPCR. Data are the means +/− s.e.m. from at least two independent experiments. Primer sets are listed in Supplementary Methods.
Reporter assays
The VZV model IE promoter-reporter (pIE62P-61), IE62 expression plasmid (pCMV-IE62), and luciferase reporter assays have been described12. The HSV IE promoter-luciferase reporter contained the promoter sequences required for the HSV IE activator (VP16) mediated induction (−171 to +57 relative to the ICP0 transcription initiation site) in pGL4.18 (pICP0-171). Transfections and RNAi mediated depletions are described in Supplementary Methods. Luciferase reporter activity was measured and analyzed 24 hrs later as described12. The data are the means +/− s.e.m. from at least three independent experiments.
Coimmunoprecipitations
Nuclear extracts of HEK293 cells transfected with epitope tagged LSD and HCF expression plasmids were incubated with FLAG-M2 agarose beads in NP40 buffer. Endogenous coimmunoprecipitations were done from nuclear extracts of HeLa cells using the indicated antibodies. Western Blots of resolved extracts and immunoprecipitates were developed using Pierce SuperSignal West Dura. Details are provided in the Supplementary Methods.
Supplementary Material
Acknowledgements
We thank J. Skinner (Bioinformatics and Computational Biosciences Branch, National Institute of Allergy and Infectious Diseases) for expert statistical analysis of the experimental data; B. Moss, J. Yewdell, T. Pierson, Z. Whitlow, and A. McBride for critical discussions and comments on this manuscript; members of the Molecular Genetics Section of the Laboratory of Viral Diseases for discussion, advice, and technical assistance; the NIAID Bld33 Animal Care Facility staff; N. Fraser (University of Pennsylvania School of Medicine) for HSV-1 strain 17, W. Ruyechan (University at Buffalo, SUNY) for anti-ICP8 sera, and X. Chen (University of California at Davis) for MCF7 inducible LSD shRNA cells. These studies were supported by the Laboratory of Viral Diseases, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, US National Institutes of Health.
Footnotes
Supplementary Information including methods is available on the Nature Medicine website.
Author Contributions Y.L. performed ChIP, qPCR, IF, and animal reactivation studies; A.N. and H.P. performed ChIP and qPCR analyses; J.L.V. performed reporter and co-immunoprecipitation assays; T.M.K. designed the study, performed animal reactivation studies, and wrote the paper. J.V. and A.N. contributed equally to this study. All authors discussed the results and commented on the manuscript.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 2.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 3.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 4.Kouzarides T, Berger SL. Chromatin Modifications and Their Mechanism of Action. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2007. pp. 191–209. [Google Scholar]
- 5.Knipe DM, Cliffe A. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol. 2008;6:211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 6.Lieberman PM. Chromatin organization and virus gene expression. J Cell Physiol. 2008;216:295–302. doi: 10.1002/jcp.21421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mok HP, Lever AM. Chromatin, gene silencing and HIV latency. Genome Biol. 2007;8:228. doi: 10.1186/gb-2007-8-11-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinclair J. Chromatin structure regulates human cytomegalovirus gene expression during latency, reactivation and lytic infection. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbagrm.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokoyama A, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kristie TM. Early events pre-initiation of alphaherpesvirus viral gene expression. In: Arvin A, et al., editors. Human Herpesviruses Biology, Therapy, and Immunoprophylaxis. New York: Cambridge University Press; 2007. pp. 112–127. [Google Scholar]
- 12.Narayanan A, Ruyechan WT, Kristie TM. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc Natl Acad Sci U S A. 2007;104:10835–10840. doi: 10.1073/pnas.0704351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, et al. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J Virol. 2006;80:5740–5746. doi: 10.1128/JVI.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Bassets I, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metzger E, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 16.Perillo B, et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- 18.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 19.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 20.Shi YJ, et al. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 21.Forneris F, Binda C, Vanoni MA, Mattevi A, Battaglioli E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS letters. 2005;579:2203–2207. doi: 10.1016/j.febslet.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 22.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 23.Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem Biol. 2006;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt DM, McCafferty DG. trans-2-Phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry. 2007;46:4408–4416. doi: 10.1021/bi0618621. [DOI] [PubMed] [Google Scholar]
- 25.Yang M, et al. Structural basis for the inhibition of the LSD1 histone demethylase by the antidepressant trans-2-phenylcyclopropylamine. Biochemistry. 2007;46:8058–8065. doi: 10.1021/bi700664y. [DOI] [PubMed] [Google Scholar]
- 26.Shin S, Janknecht R. Activation of androgen receptor by histone demethylases JMJD2A and JMJD2D. Biochemical and biophysical research communications. 2007;359:742–746. doi: 10.1016/j.bbrc.2007.05.179. [DOI] [PubMed] [Google Scholar]
- 27.Wissmann M, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- 28.Amelio AL, Giordani NV, Kubat NJ, O'Neil JE, Bloom DC. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J Virol. 2006;80:2063–2068. doi: 10.1128/JVI.80.4.2063-2068.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubat NJ, Tran RK, McAnany P, Bloom DC. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J Virol. 2004;78:1139–1149. doi: 10.1128/JVI.78.3.1139-1149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neumann DM, Bhattacharjee PS, Giordani NV, Bloom DC, Hill JM. In vivo changes in the patterns of chromatin structure associated with the latent herpes simplex virus type 1 genome in mouse trigeminal ganglia can be detected at early times after butyrate treatment. J Virol. 2007;81:13248–13253. doi: 10.1128/JVI.01569-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang QY, et al. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci U S A. 2005;102:16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolb G, Kristie TM. Association of the cellular coactivator HCF-1 with the Golgi apparatus in sensory neurons. J Virol. 2008 doi: 10.1128/JVI.01174-08. doi:10.1128/JVI.01174-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kristie TM, Vogel JL, Sears AE. Nuclear localization of the C1 factor (host cell factor) in sensory neurons correlates with reactivation of herpes simplex virus from latency. Proc Natl Acad Sci U S A. 1999;96:1229–1233. doi: 10.1073/pnas.96.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitlow Z, Kristie TM. Recruitment of the transcriptional coactivator HCF-1 to viral immediate-early promoters during initiation of reactivation from latency of herpes simplex virus type 1. J Virol. 2009;83:9891–9595. doi: 10.1128/JVI.01115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pesola JM, Zhu J, Knipe DM, Coen DM. Herpes simplex virus 1 immediate-early and early gene expression during reactivation from latency under conditions that prevent infectious virus production. J Virol. 2005;79:14516–14525. doi: 10.1128/JVI.79.23.14516-14525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawtell NM, Thompson RL, Haas RL. Herpes simplex virus DNA synthesis is not a decisive regulatory event in the initiation of lytic viral protein expression in neurons in vivo during primary infection or reactivation from latency. J Virol. 2006;80:38–50. doi: 10.1128/JVI.80.1.38-50.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh J, Fraser NW. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J Virol. 2008;82:3530–3537. doi: 10.1128/JVI.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Placek BJ, et al. The histone variant H3.3 regulates gene expression during lytic infection with herpes simplex virus type 1. J Virol. 2009;83:1416–1421. doi: 10.1128/JVI.01276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang J, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nature genetics. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.