Abstract
The dleu2 tumor suppressor locus encodes two microRNAs, miR-15a and miR-16, which are thought to play an important role in B-cell neoplasms. However, relatively little is known about proteins that regulate or are regulated by this microRNA cluster. Here we demonstrate that the Pax5 oncoprotein downregulates the dleu2 gene and at the same time boosts expression of its own heterodimeric partner c-Myb. Interestingly, c-Myb upregulation occurs primarily at a post-transcriptional level, suggesting that it might be a target for microRNAs such as miR-15a/16. Indeed, miR-15a/16 have predicted binding sites in the c-Myb 3′-UTR and through them diminish protein output in luciferase sensor assays. Moreover, forced overexpression of miR-15a/16 reduces endogenous c-Myb levels and compromises Pax5 function. Conversely, restoration of c-Myb levels partly alleviates tumors suppressive effects of miR-15a/16, suggesting that c-Myb is a key downstream target of this microRNA cluster.
Keywords: Myb, Pax5, microRNAs, lymphoma, leukemia, tumor suppressors, oncogenes
Introduction
Pax5, a paired box transcription factor, is frequently overexpressed, translocated or hypermutated in human follicular (FL), diffuse large B cell (DLBCL), and some other non-Hodgkin lymphomas. Although the hallmark of B cell acute lymphocytic leukemia (B-ALL) is loss-of-function mutations in Pax5, there is considerable circumstantial evidence to suggest that Pax5 promotes B lymphomagenesis in more mature B-cells.1,2
To obtain direct evidence in support of this idea, we utilized our previously described cell line (Myc5)3 derived from a p53-null, Myc-overexpressing lymphoma.4 The original Myc5 tumor was Pax5-positive, but upon culturing in vitro Pax5 expression was spontaneously extinguished and B cell markers were lost. Instead, the cultured cells readily engulfed latex beads and provided T-cell help like bona fide macrophages.5 To analyze the immediate effects of Pax5 on B-lymphomagenesis, we generated Myc5 subclones expressing the hydroxytamoxifen (4OHT)-inducible variant of Pax5 (Pax5-ER). Upon re-injection into syngeneic recipients, these clones indeed formed much larger tumors in 4OHT-treated than in control mice.6
It is widely accepted that gene regulation by Pax5 underlies its ability to promote neoplastic growth. However, to bind to DNA with high affinity, Pax5 needs to interact with other transcription factors, such as c-Myb. c-Myb, along with Ets1, is encoded by the acutely transforming E26 avian retrovirus7 and regulates gene expression.8 Moreover, there is solid evidence that Pax5 activity is enhanced through interactions with Myb9,10 which further brings in LEF1.9 Interestingly, c-Myb was not expressed at detectable levels in Myc5 cells prior to transduction with the Pax5 retrovirus. Therefore, in order to establish and sustain its protein network, Pax5 must upregulate its expression. Herein we demonstrate that transduction with a Pax5 retrovirus leads to re-expression of c-Myb. This occurs primarily at a post-transcriptional level, suggesting that it might be a target for one of several microRNAs repressed by Pax5. Indeed, one of these microRNA clusters (miR-15a/16) is a direct regulator of c-Myb, which has important implications for B-cell neoplasms.
Results and Discussion
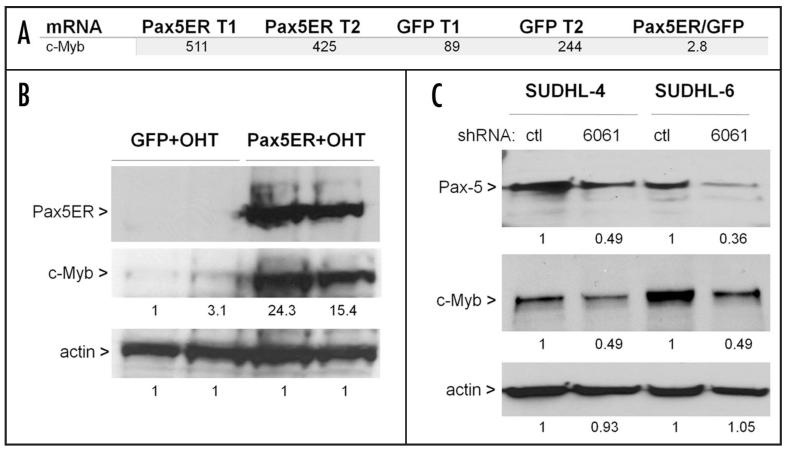
To determine expression pattern of c-Myb in Pax5-reconstituted Myc5 cells, we first re-analyzed the Pax5-ER microarray data.6 To rule out possible effects of the estrogen on gene expression, we chose to compare 4OHT-treated GFP- vs. Pax5ER tumors, not treated vs. untreated Pax5ER tumors. The mRNA encoding c-Myb was detectable in all samples and slightly upregulated in tumors with activated Pax5-ER fusion, less than 3-fold on average (Fig. 1A). This upregulation was later confirmed using qPCR (Figure 2b). Then the same tumor samples were analyzed using immunoblotting. We found that Pax5-ER tumors had much higher protein levels of c-Myb (20-fold on average) than more modest increases in mRNA levels might suggest (Fig. 1B). To determine whether Pax5 also controls c-Myb expression in human B-cell lymphomas, we measured their protein levels in the DLBCL lines SUDHL-4 and -6 with stable knockdown of Pax5.6 Despite relatively modest effects of lentivirus-encoded hairpins on Pax5 expression, a commensurate decrease in c-Myb protein levels was readily detectable (Fig. 1C).
Figure 1.
Pax5 increases c-Myb expression. (A) Microarray analysis of gene expression in Pax5-ER and control GFP-only tumors. Probesets corresponding to c-Myb are shown for 2 tumors (T1 and T2) of each type. Ratio in the last column reflects average expression levels. (B) Immunoblotting demonstrating protein expression levels in samples from A. Band intensities were quantitated using the ImageJ software. Numbers under lanes refer to expression levels relative to those observed in the GFP T1 tumor. (C) Pax5 and c-Myb protein levels in human diffuse large cell lymphoma lines SUDHL-4 and -6 following lentiviral transduction with a control hairpin and the hairpin targeting Pax5 expression (6061). Numbers under lanes refer to expression levels relative to those observed in control shRNA-expressing cells. In the last two panels, beta actin was used as a loading control.
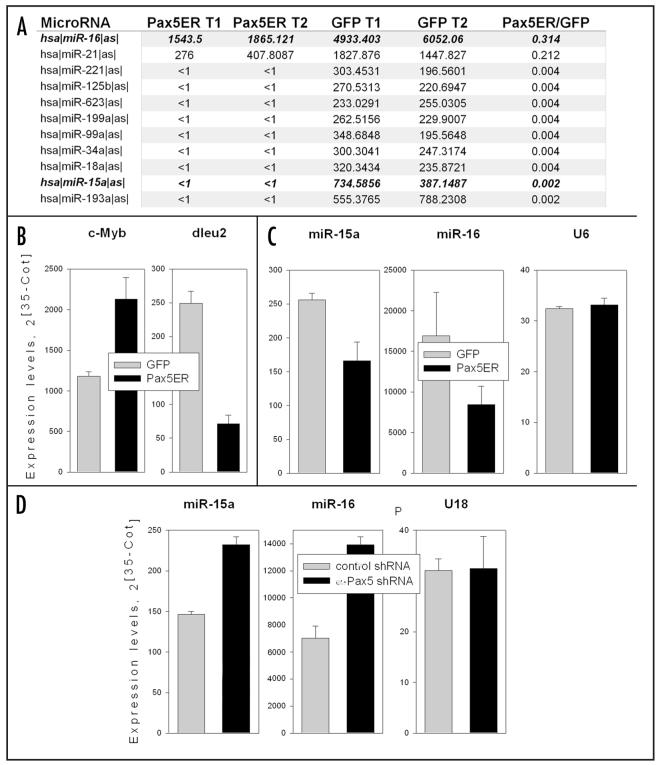
Figure 2.
Pax5 down-regulates the dleu2 microRNA cluster. (A) MicroRNA profiling of Pax5-ER and control GFP-only tumors. Probesets corresponding to all differentially expressed miRNAs are shown for 2 tumors (T1 and T2) of each type (same as in Figure 1). Ratios in the last column reflect average expression levels. Highlighted in bold are two members of the dleu2 cluster, miR-16 and miR-15a. (B) Quantitation of the c-Myb and dleu2 primary transcripts in tumors from A using qPCR. Expression levels were adjusted for beta-actin. (C) Quantitation of mature miR-15a and -16 in tumors from A using qPCR. U6 (or sno202 in case of miR-15a) RNAs were used as internal controls. (D) Quantitations of mature miR-15a and -16 in SUDHL-4 cells with Pax5 knock-down from panel 1C. U18 RNA was used as an internal control. In all panels error bars denote SD.
That Pax5 increases mRNA-, but primarily protein levels of its major heterodimeric partner could be suggestive of microRNA-mediated coupled post-transcriptional/translational regulation. To determine if this was indeed the case, we profiled microRNAs from 2 Pax5-ER and 2 control tumors using custom-made CombiMatrix arrays. As seen previously for c-Myc-overexpressing cells,11 the primary effect of Pax5 overexpression was systemic downregulation of miRNA levels. No upregulated miRNAs fulfilled the criteria for being included: 3-fold difference in average expression levels and flagged as “present” in both samples in at least one group. Individual miRNAs whose levels were decreased upon Pax5 re-expression are listed in Figure 2A. Of particular interest was downregulation of miR-16 and -15a, because they are encoded by a single genomic locus, dleu2, which is a tumor suppressor in chronic lymphocytic leukemia (CLL)12 and is also repressed by c-Myc.11
To confirm that miR-15a/16 are indeed repressed in Pax5-reconstituted neoplasms, we assessed the levels of its host transcript, dleu2, using qPCR. While c-Myb mRNA levels were elevated in the presence of active Pax5 (consistent with microarray data in Figure 1a), levels of dleu2 were decreased (Figure 2b). Moreover, Taqman-based qPCR assay demonstrated that levels of mature miR-15a and -16 are also consistently lower in Pax5-ER tumors (Figure 2c.) Conversely, miR-15a and -16 levels in SUDHL cells with Pax5 knock-down were consistently higher than in control cells (Figure 2d and similar data for SUDHL-6 not shown.) Collectively, these data suggest that Pax5, either directly or indirectly, down-regulates miR-15a/16 expression.
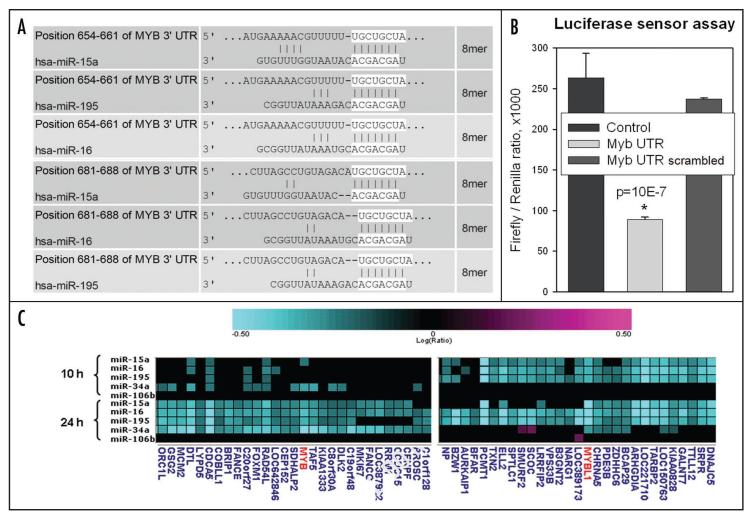
Interestingly, these two related microRNAs, as well as another family member miR-195, are predicted to target c-Myb mRNA directly, according to the MiRanda algorithm13 (Fig. 3A). To test whether this is indeed the case, we generated several pGL3-based firefly luciferase sensor vectors, wherein the luciferase coding sequence is followed by the two predicted miR-15a/16 binding sites (MiBS) from the c-Myb 3′-UTR. As controls, we used pGL3 variants with scrambled MiBS or an unrelated sequence. The resultant constructs were cotransfected into 293 cells along with miR-15a/16 and renilla luciferase expression vectors, the latter to adjust for transfection efficiency. Consistent with MiRanda predictions, inclusion of the two adjacent miR-15a/16 MiBS from Myb 3′UTR decreased luciferase output (Fig. 3B). This suggested that c-Myb is a direct target of miR-15a/16.
Figure 3.
c-Myb is a direct target for miR-15a/16. (A) Predicted miR-15a, -16 and-195 binding sites in the 3′-UTR of c-Myb. Predictions are based on the MiRanda algorithm. 8-nt long seed sequences are highlighted. (B) Luciferase sensor assay. Constructs tested were pGL3 derivatives containing MiBSs from 3′-UTR of c-Myb (intact or scrambled) as well as an unrelated sequence lacking homology to miR-15a/16 (control). All data were normalized for renilla luciferase activity. Asterisk denotes statistical significance. (C) Decreased levels of c-Myb and Myb-L1 mRNA in Dicer-deficient HCT116 cells transfected with miR-15a family mimics. mRNAs were harvested either 10 or 24 h post-transfection and hybridized to human Agilent arrays. miR-106b was used as an irrelevant miRNA control.
To corroborate this notion and to establish the time course of c-Myb downregulation by miR-15a/16, we utilized HCT116 cells with homozygous disruption of exon 5 encoding the helicase domain of Dicer, the key ribonuclease responsible for miRNA processing. Consequently, in Dicerex5 cells basal levels of many miRNA including miR-15a/16 are very low.14 We transfected HCT116 Dicerex5 cells with synthetic miRNAs corresponding to different members of the miR-15a/16 family, namely miR-15a, -16, -195 and -34a (known to target c-Myb) as described previously.15 Unrelated miR-106b was used as a negative control. Expression profiles were measured 10 and 24 h later, when secondary effects due to target protein depletion are minimal.16 Different members of the family, which share the seed sequence, gave nearly identical expression profiles, which included downregulation of both c-Myb and its close relative MybL1 (a.k.a. A-Myb) (Fig. 3C). Notably, at least miR-15a caused downregulation of these two transcripts as early as 10 h post transfection, consistent with the idea that they represent direct miR-15a/16 targets.
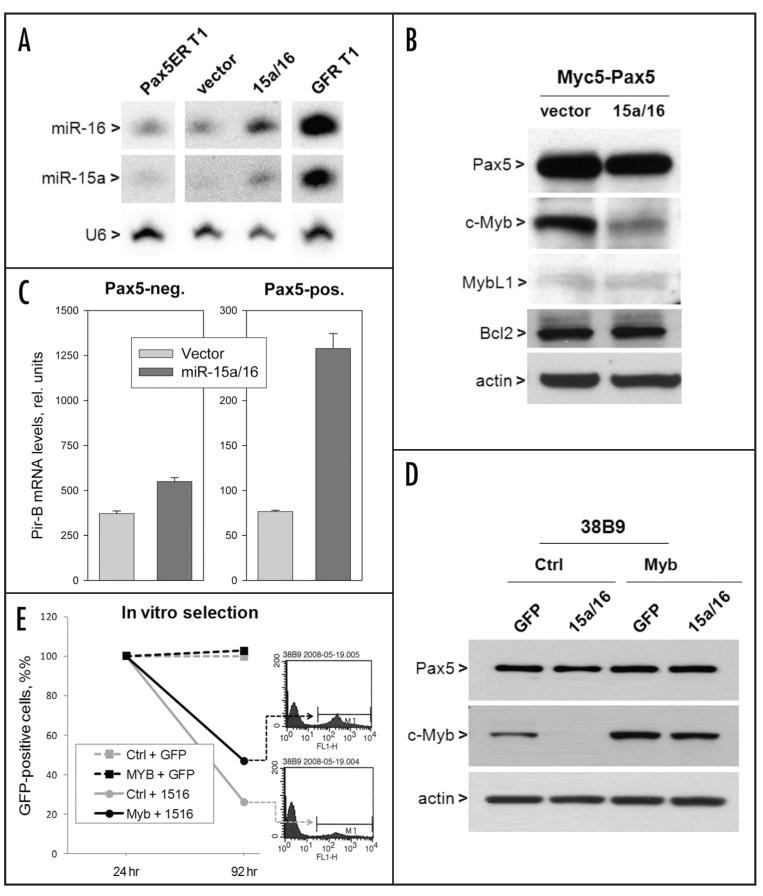
We were further interested in determining whether miR-15a/16-dependent decrease in c-Myb levels compromises Pax5 function. Towards this end, Pax5-expressing and parental Myc5 cells were infected with a retrovirus co-expressing miR-15a/16 and GFP.11 High transduction rates (60–80%) were observed immediately after infection for both miR-15a/16 and control retroviruses, allowing for direct analysis of transduced cultures. At that time point, expression of miR-15a and miR16 was clearly elevated. Importantly, the elevated levels were still physiological, i.e., did not exceed those observed in Pax5-negative tumors (Northern blotting data in Fig. 4A and qPCR data not shown). Yet this modest overexpression was sufficient to bring down c-Myb protein levels, while MybL1 was virtually undetectable in either culture (Fig. 4B). Interestingly, levels of Bcl-2 were unchanged, suggesting that in this setting any biological effects of miR-15a/16 would be independent of Bcl-2 downregulation observed in other cell types.17
Figure 4.
c-Myb levels correlate with Pax5 function and cell expansion. (A) miRNA levels in Pax5-sufficient cells after infection with a miR-15a/16 retrovirus. Northern blotting was used to determine steady-state levels of mature miRNAs. Pax5-positive and -negative tumors were used for comparison. U6 was used as a loading control. (B) c-Myb, MybL1 and Bcl2 levels in miR-15a/16-transduced cells. Western blotting was performed as in Figure 1B. (C) Quantitation of PirB mRNAs in vector- and miR-15-a/16-transduced Pax5-negative and -positive cells. (D) Immunoblotting of Pax5 and Myb in cultures with and without constitutive c-Myb expression following infection with a GFP-miR-15a/16 retrovirus. (E) Flow cytometric quantitation of GFP-positive cells in the same cultures. They were enumerated 24h and 96h post infection and normalized to the former.
To determine the effects of transient miR-15a/16 re-expression on Pax5 transcriptional activity, we chose to study Pax5-repressed genes, since in this case pre-existing molecules would not significantly affect mRNA steady-state levels, as measured by qRT-PCR. Specifically, we chose CD22 and PirB, as these two genes are rapidly down-regulated by Pax5 (reviewed in ref. 6 and data not shown). By an unknown mechanism, CD22 mRNA levels were increased following miR-15a/16 overexpression even in the absence of Pax5, and no further de-repression was observed in Pax5-reconstituted cells, making this system non-informative (data not shown) However, in the case of PirB, miR-15a/16 had very minor effect on its expression in Pax5-deficient cells, but sharply increased its steady state levels in Pax5-sufficient cells (Fig. 4C) This observation indicates that the ability of Pax5 to deregulate at least some of its target genes could be curtailed through reconstitution of miR-15a/16 levels.
Finally, we asked whether c-Myb is an essential downstream effector of miR-15a/16 and whether it alleviates tumor suppressive effects of this microRNA cluster. We employed the previously developed selection approach, wherein miR-15a/16-overexpressing cells are steadily eliminated from acutely infected cultures following passaging in vitro.11 38B9 pro-B-cells were transduced with a c-Myb-encoding retrovirus and used, along with control cells, in the selection assay. 24 hours after infection with the miR-15a/16 retrovirus, endogenous c-Myb was sharply downregulated in control cells, but c-Myb levels were maintained, despite miR-15a/16 production, when the corresponding cDNA (lacking the 3′-UTR) was expressed by a retrovirus (Fig. 4D). 96 h after infection, only 20% of GFP-positive cells remained in miR-15a/16-transduced cultures lacking c-Myb. In contrast, over 40% of GFP-positive cells remained in cultures with enforced c-Myb expression (grey and black plots and the corresponding flow cytometric profiles, respectively). We thus concluded that tumor suppressive effects of miR-15a/16 could be partly attributed to the downregulation of its bona fide target c-Myb.
The miR-15a/16 locus was first identified as a classical two-hit tumor suppressor gene in chronic lymphocytic leukemia over five years ago,12 firmly implicating microRNAs in the control of cell fate. In recent months, the key roles of microRNAs in controlling many cellular phenotypes, including those pertaining to hematopoietic lineages, were rigorously established by means of mouse genetics (reviewed in refs. 18-22). Yet definitive identification of their essential target genes has proven difficult. Interestingly, one notable exception is the finding that overexpression of miR-150, which also targets c-Myb, affects the balance between B1 and B2 types of B-cells in a manner similar to that observed in c-myb heterozygous mice where levels of c-Myb are reduced two-fold.23 This observation suggests that miR-150 might exert all of its B-cell specific effects through c-Myb but doesn’t rule out the possibility that c-Myb might be an essential target for other microRNAs.
Herein we demonstrate that c-Myb is a bona fide target of the miR-15a/16 locus and that enforcing its levels attenuates, albeit doesn’t abolish, the detrimental effect of these microRNAs on neoplastic B-cell expansion. Interestingly, these effects are apparent even in the absence of changes in the levels of Bcl2, which in other cell types might be a key downstream target of miR-15a/16, responsible for increased cell survival.17 On the other hand, Myb family members are strongly implicated in the control of both cell proliferation and apoptosis (reviewed in refs. 24 and 25), although the corresponding genes are seldom mutated in tumors. This makes them attractive targets for epigenetic regulation by microRNAs. Moreover, c-Myb is an essential heterodimeric partner for Pax5,9,10 further implicating it in both normal B-cell development and B-cell neoplasms. Insofar as Pax5 is overtly oncogenic in pre-B-cells, it might need to sustain c-Myb levels, and one means to this end appears to be downregulation of the dleu2 gene. To our knowledge, this is the first example whereby oncogenic transcription factors regulate and are reciprocally regulated by a microRNA locus with tumor suppressor properties.
Materials and Methods
MicroRNA profiling using Combimatrix arrays
Pax5-ER and control neoplasms6 were used as a source of total RNA. RNA labeling, hybridization and analysis were carried out as described earlier.11
Cell culture
Myc5 cells were maintained in RPMI 1640 (Gibco) supplemented with 10% fetal bovine serum (Sigma), 100 U/ml penicillin, 100 μg/ml streptomycin (Gibco), and lipopolysaccharide from Escherichia coli (10 μg/ml; Sigma-Aldrich). DLBCL cell line SUDHL-4,26 was maintained as above but without lipopolysaccharide. 38B9, a pro-B cell line transformed by v-Abl,27 was maintained in RPMI 1640 supplemented with 25 mM HEPES (Gibco), 0.1% 2-mercaptoethanol (Gibco), 10% fetal bovine serum (Sigma), 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco). HEK293T and GP293, human embryonic kidney cell lines, were grown in Dulbecco modified Eagle medium supplemented with 4.5 g/L D-glucose (Gibco), 10% fetal bovine serum (Hyclone) 100 U/ml penicillin, and 100 μg/ml streptomycin. All cells were incubated at 37°C in 5% CO2.
Retroviral infection
Full-length miR15a/16-1 cluster was cloned into a retroviral vector MSCV-PIG expressing GFP.11 Pax5 and c-Myb coding sequences were inserted into MIGR1 and MSCVpuro retroviruses, respectively. Retroviral constructs were transfected into GP293 packaging cells using Lipofectamine2000 (Invitrogen). Medium was changed 16 hr post transfection (Day 0). Virus-containing supernatant was harvested on Day 1 and 2. The supernatant was supplemented with polybrene (Sigma) and was used to infected target cells at a ratio of 0.5 × 106 target cells/3 ml virus. Flow cytometric analysis of infected cultures was performed as described in.28
Pax5 knockdown in DLBCL lines
Pax5 knockdown in DLBCL cell lines using lentivirus-encoded hairpins has been described previously.6
Western blotting
Total whole cell lysates (50 μl) with 0.5 × 106 cells were boiled for 10 min followed by 5 min incubation on ice and the samples were subject to electrophoresis and immunoblotting. The primary antibody against Pax5 was a kind gift from Dr. Michael L. Atchison (University of Pennsylvania). Other primary antibodies used include mouse monoclonal anti-Myb (05–175, Upstate), rabbit polyclonal anti-Bcl2 (Santa Cruz), and mouse monoclonal anti-β-actin antibody (A3854 Sigma). Appropriate secondary anti-bodies conjugated to horseradish peroxidase (Amersham Pharmacia Biotech, Piscataway, NJ) were used, followed by ECL Plus Western Blotting Detection System (Amersham).
RNA isolation, northern blotting, reverse transcription and qPCR
Total RNA was extracted using Tri® Reagent (Sigma) according to the manufacturer’s protocol. DNase (Roche) treatment was performed on RNA samples in order to remove genomic DNA carry-over. RNA blotting for miR15a and miR16-1 was performed as described previously.11 RT reaction was performed using SuperScript™ First-Strand Synthesis System (Invitrogen). Real-time PCR was performed using a 2 × SyBr Green PCR Master Mix (Applied Biosystem; Piscataway, NJ) containing 20 ng of cDNA mixed with pairs of primers (10 μM) specific for either murine CD22, Pirb or β-actin in a 20 μl volume. The primer sequences and PCR profiles are available upon request. All reactions were performed in triplicates.
Taqman RT-PCR for miR-15a/16-1 expression
For analysis of miR15a/16-1 expression, total RNA was extracted as described. 20 ng of total RNA was reverse transcribed using the Reverse Transcription System (ABI) according to the manufacturer’s instructions. Primer sequences were labeled with a 5′-reporter dye FAM (6-carboxy-fluorescein) and a 3′-quencher dye TAMRA (6-carboxy-N,N,N’,N’-tetramethyl-rhodamine). Primers directed against human U18 or murine U6 or sno202 were used as the endogenous controls (ABI). Detection of PCR products were performed on an ABI7000 prism detection system (ABI) as described in.29
Luciferase reporter constructs and assays
Oligonucleotides containing the predicted binding sites of miR-15a/16 in the 3′UTR of human c-myb gene (intact or mutated at three positions within the seed sequence) were synthesized with ends compatible with XbaI and ApaI. To facilitate cloning of the putative miR-15a/16 binding sites, a StyI fragment of the polylinker from pSL1180 was inserted into the pGL3 control vector (Promega) at the XbaI site 3′ of the luciferase gene. Annealed oligos above were ligated into the pGL3-1180PL construct digested with XbaI and ApaI. All constructs were confirmed by sequencing. Oligonucleotide sequences were as follows (seed sequences are underlined):
Myb-wt sense
CTA GAA TGA AAA ACG TTT TTT GCT GCT AT GGT CTT AGC CTG TAG ACA TGC TGC TAG TAT CGG GCC
Myb-wt a/sense
TTA CTT TTT GCA AAA AAC GAC GAT ACC AGA ATC GGA CAT CTG TAC GAC GAT CAT AGC
Myb-mut sense
CTA GAA TGA AAA ACG TTT TTC GCC GCC ATG GTC TTA GCC TGT AGA CAC GCC GCC AGT ATC GGG CC
Myb-mut a/sense
TTA CTT TTT GCA AAA AGC GGC GGT ACC AGA ATC GGA CAT CTG TGC GGC GGT CAT AGC
HEK293T cells were transfected with the luciferase reporter constructs described above (100 ng), pRL-CMV (5 ng, Promega) and either MSCV-PIG empty vector or MSCV-PIG-miR-15a/16 (800 ng) using Expressfect (Denville Scientific). After 48 h, cells were washed and lysed with passive lysis buffer (Promega) and f-luc and Renilla luciferase (r-luc) activities were determined using the dual-luciferase reporter assay system (Promega) and a luminometer. The relative reporter activity was obtained by normalization to the r-luc activity. Parallel dishes were analyzed by Taqman qPCR to confirm transient expression of miR-15/16 in transfected cells.
MicroRNA gain of function gene expression analyses
Synthetic RNA duplexes (25 nM), corresponding to mature miRNAs and designed as described previously,30 were transfected into HCT116 Dicerex5 cells.14 RNA was isolated 10 and 24 h after transfection for microarray analysis as described previously.16
Statistical analysis
All transfection studies were performed in duplicates. Taqman RT-PCR and quantitative RT-PCR were performed in triplicates. The results were expressed as mean S.D. from at least 3 independent experiments. All statistical analysis was performed with two-tailed Student’s t test. Data were considered statistically significant when p < 0.05.
Acknowledgements
We thank Dr. Alan M. Gewirtz (University of Pennsylvania) for providing us with the full-length human c-myb cDNA and Dr. Michael Atchison (University of Pennsylvania) for many helpful comments. This work was supported by grants from the National Institutes of Health (R01CA120185 to J.T.M., and R01CA122334 and R01CA102709 to A.T.-T.).
Footnotes
Notes While this manuscript was in production, down-regulation of c-Myb by miR-15a has been also observed in normal human hematopoietic cells where it is comprises part of an autoregulatory feedback loop postulated to be important in myelo- and erythropoiesis.31
References
- 1.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–70. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 2.Thomas-Tikhonenko A, Cozma D. PAX5 and B-cell neoplasms: transformation through presentation. Future Oncol. 2008;4:5–9. doi: 10.2217/14796694.4.1.5. [DOI] [PubMed] [Google Scholar]
- 3.Yu D, Allman D, Goldscmidt M, Atchison M, Monroe JG, Thomas-Tikhonenko A. Oscillation between B-lymphoid and myeloid lineages in Myc-induced hematopoietic tumors following spontaneous silencing/reactivation of the EBF/Pax5 pathway. Blood. 2003;101:1950–5. doi: 10.1182/blood-2002-06-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu D, Thomas-Tikhonenko A. A non-transgenic mouse model for B-cell lymphoma: in vivo infection of p53-null bone marrow progenitors by a Myc retrovirus is sufficient for tumorigenesis. Oncogene. 2002;21:1922–7. doi: 10.1038/sj.onc.1205244. [DOI] [PubMed] [Google Scholar]
- 5.Hodawadekar S, Yu D, Freedman B, Sunyer JO, Atchison M, Thomas-Tikhonenko A. B-lymphoma cells with epigenetic silencing of Pax5 trans-differentiate into macrophages, but not other hematopoietic lineages. Exp Cell Res. 2007;313:331–40. doi: 10.1016/j.yexcr.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cozma D, Yu D, Hodawadekar S, Azvolinsky A, Grande S, Tobias JW, Metzgar MH, Paterson J, Erikson J, Marafioti T, Monroe JG, Atchison ML. Thomas-Tikhonenko A. PAX5 promotes lymphomagenesis through the stimulation of B-cell receptor signaling. J Clin Inv. 2007;117:2602–10. doi: 10.1172/JCI30842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nunn MF, Seeburg PH, Moscovici C, Duesberg PH. Tripartite structure of the avian erythroblastosis virus E26 transforming gene. Nature. 1983;306:391–5. doi: 10.1038/306391a0. [DOI] [PubMed] [Google Scholar]
- 8.Dudek H, Tantravahi RV, Rao VN, Reddy ES, Reddy EP. Myb and Ets proteins cooperate in transcriptional activation of the mim-1 promoter. Proc Natl Acad Sci USA. 1992;89:1291–5. doi: 10.1073/pnas.89.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin ZX, Kishi H, Wei XC, Matsuda T, Saito S, Muraguchi A. Lymphoid enhancer-binding factor-1 binds and activates the recombination-activating gene-2 promoter together with c-Myb and Pax-5 in immature B cells. J Immunol. 2002;169:3783–92. doi: 10.4049/jimmunol.169.7.3783. [DOI] [PubMed] [Google Scholar]
- 10.Kishi H, Jin ZX, Wei XC, Nagata T, Matsuda T, Saito S, Muraguchi A. Cooperative binding of c-Myb and Pax-5 activates the RAG-2 promoter in immature B cells. Blood. 2002;99:576–83. doi: 10.1182/blood.v99.2.576. [DOI] [PubMed] [Google Scholar]
- 11.Chang TC, Yu D, Lee YS, Arking DE, West CM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by c-Myc promotes tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and downregulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, Raymond CK, Roberts BS, Juhl H, Kinzler KW, Vogelstein B, Velculescu VE. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687–92. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, Chau N, Cleary M, Jackson AL, Carleton M, Lim L. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–52. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–7. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 17.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–74. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 20.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–9. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 21.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 22.O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–94. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–59. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 24.Lipsick JS. synMuv verite—Myb comes into focus. Genes Dev. 2004;18:2837–44. doi: 10.1101/gad.1274804. [DOI] [PubMed] [Google Scholar]
- 25.Sala A. B-MYB, a transcription factor implicated in regulating cell cycle, apoptosis and cancer. Eur J Cancer. 2005;41:2479–84. doi: 10.1016/j.ejca.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 26.Gururajan M, Chui R, Karuppannan AK, Ke J, Jennings CD, Bondada S. c-Jun N-terminal kinase (JNK) is required for survival and proliferation of B-lymphoma cells. Blood. 2005;106:1382–91. doi: 10.1182/blood-2004-10-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alt F, Rosenberg N, Lewis S, Thomas E, Baltimore D. Organization and reorganization of immunoglobulin genes in A-MULV-transformed cells: rearrangement of heavy but not light chain genes. Cell. 1981;27:381–90. doi: 10.1016/0092-8674(81)90421-9. [DOI] [PubMed] [Google Scholar]
- 28.Yu D, Cozma D, Park A, Thomas-Tikhonenko A. Functional validation of genes implicated in lymphomagenesis: an in vivo selection assay using a Myc-induced B-cell tumor. Annals NY Acad Sci. 2005;1059:145–59. doi: 10.1196/annals.1339.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–5. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 31.Zhao H, Kalota A, Jin S, Gewirtz AM. The c-myb protooncogene and microRNA (miR)-15a comprise an active autoregulatory feedback loop in human hematopoietic cells. Blood. 2008 doi: 10.1182/blood-2008-01-136218.; in press.