Summary
The mouse homeobox gene, Gbx2, is expressed in discreet domains in the neural tube and plays a key role in forebrain and hindbrain development. Previous studies have demonstrated that mutual inhibition between Gbx2 and Otx2, which are respectively expressed in the anterior and posterior parts of the neural plate, positions the prospective midbrain-hindbrain junction. We describe here a conditional Gbx2 gain-of-function transgenic mouse line, Gbx2-GOF, which expresses Gbx2 and red fluorescence protein, mCherry, upon Cre-mediated recombination. In the absence of Cre, β-galactosidase is broadly expressed in mouse embryos and adult brains carrying the transgene. By combining Gbx2-GOF and En1Cre knock-in allele, we activated expression of Gbx2 and mCherry throughout the mesencephalon (mes) and rhombomere 1 (r1). The ectopic expression of Gbx2 causes an anterior shift of the mes/r1 junction at embryonic day 10.5. Interestingly, we found that persistent expression of Gbx2 throughout the mes/r1 region largely abolishes expression of the isthmic organizer gene Fgf8, leading to deletion of the midbrain and cerebellum at later stages. Our data suggest that the juxtaposition of the expression domains of Gbx2 and Otx2 within the mes/r1 area is essential for the maintenance of Fgf8 expression. Furthermore, the Gbx2-GOF transgenic line is suitable for functional study of Gbx2 during development.
Keywords: Gbx2, Fgf8, transcription factor, Cre, conditional gain of function
The mouse homeobox gene, Gbx2 (gastrulation and brain homeobox gene) displays a complex and dynamic expression profile in the posterior mesoderm, limb buds and the neural tube during development (Bouillet et al., 1995; Bulfone et al., 1993; Chen et al., 2009). Gbx2 is initially expressed in the posterior part of the mouse embryo during gastrulation (Bouillet et al., 1995; Wassarman et al., 1997). Genetic studies have demonstrated that mutual inhibition between Gbx2 and Otx2, which is expressed in the anterior part of the embryo, positions a signaling center, known as the isthmic organizer, at the prospective mid-hindbrain junction (Joyner et al., 2000). Mutation of Gbx2 leads to a posterior expansion of the expression domain of Otx2 and deletion of the entire cerebellum (Li and Joyner, 2001; Martinez-Barbera et al., 2001; Millet et al., 1999; Wassarman et al., 1997). Conversely, ectopic expression of Gbx2 in the mesencephalon (mes) in chick embryos by electroporation, or in mouse embryos by a transgene driven by regulatory elements of Wnt1, which is expressed in the mes, results in an anterior shift of the isthmus (Katahira et al., 2000; Millet et al., 1999). At later stages, however, the formation of the midbrain and cerebellum are relatively normal in the above Gbx2 misexpression experiments (Katahira et al., 2000; Millet et al., 1999). By contrast, ectopic expression of Otx2 in r1 leads to complete transformation of r1 into a midbrain fate (Broccoli et al., 1999; Katahira et al., 2000). The ectopic expression of Gbx2 by electroporation or using a Wnt1-Gbx2 transgene is transient (Katahira et al., 2000; Millet et al., 1999), which precludes an assessment of whether Gbx2 is sufficient to respecify the mes. Therefore, it remains unclear whether persistent expression of Gbx2 is required to respecify the mes into a cerebellar fate.
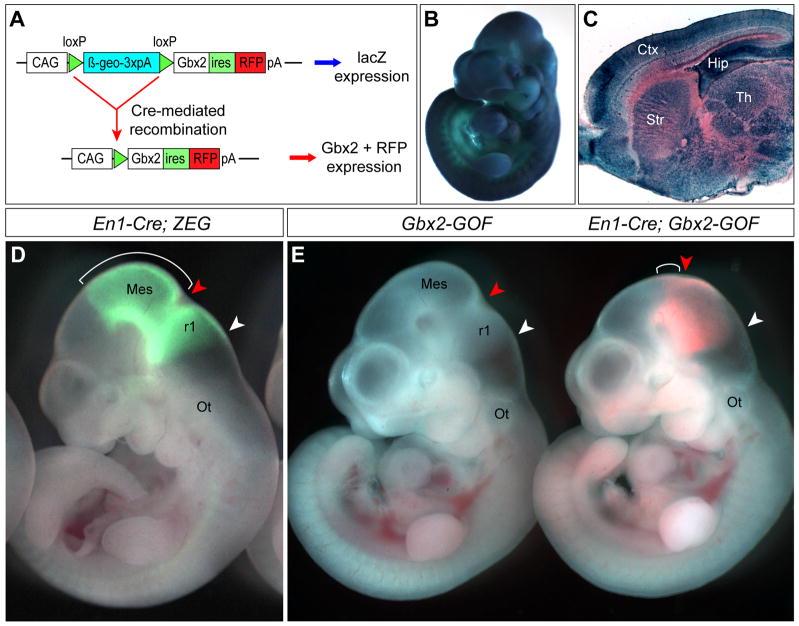
In order to further study Gbx2 function and develop a tool useful for controlled spatiotemporal expression of Gbx2, we generated a transgenic mouse line, called Gbx2-GOF. The transgene is driven by the CAGGS promoter, which contains a chicken β-actin promoter with upstream cytomegalovirus enhancer (Niwa et al., 1991). In transgenic mice, transgene-expressing cells produce β-geo, a fusion protein with neomycin-resistance and β-galactosidase (β-gal) activity (Friedrich and Soriano, 1991). Cre-mediated recombination will remove the β-geo and render cells able to express a bicistronic mRNA producing both Gbx2 and red fluorescent protein (RFP), mCherry (Shaner et al., 2004) (Fig. 1A). In mice carrying the Gbx2-GOF transgene, β-gal activity was initially detected at embryonic day 9.5 (E9.5) (data not shown). Broad and robust expression of β-gal was seen throughout the embryo at E10.5 and in the central nervous system (CNS) at E18.5 and adult stages (Fig. 1B, C and 4D). Therefore, the Gbx2-GOF transgene is widely expressed in the CNS of embryos and adult mice. The transgenic mice are viable and display no detectable abnormality.
Figure 1. Generation and characterization of a conditional Gbx2 gain-of-function transgenic mouse line.
(A) Schematic representation of the Gbx2-GOF transgene. The CAGG promoter drives expression of the β-geo gene, which is followed by a triple polyadenylation sequence (3xpA). Cre-mediated recombination deletes β-geo, allowing expression of both Gbx2 and red fluorescent protein, mCherry (RFP) from a bicistronic mRNA containing an internal ribosome entry site (ires). (B–C) Histochemical analysis of β-gal activity by X-gal in E10.5 embryos (B) or sagittal sections of E18.5 brains (C) hemizygous for Gbx2-GOF. (D) Analysis of Cre activity of En1Cre by green fluorescent protein (GFP) using a Cre-reporter line, ZEG. Bracket marks the mesencephalon. (E) Analysis of RFP fluorescence following Cre-mediated recombination in En1Cre/+; Gbx2-GOF embryos at E10.5 (right). No fluorescence is present in embryos carrying only the Gbx2-GOF transgene (left). The isthmus and the caudal limit of r1 are marked by red and white arrowheads, respectively. Note that the isthmic constriction is shifted rostrally in En1Cre/+; Gbx2-GOF embryos, and the mesencephalon (bracket) is significantly reduced. The isthmic constriction is more evident in controls. Abbreviations: Ctx, cortex; Hip, hippocampus; Mes, mesencephalon; Ot, otic vesicle; r1, rhombomere 1; Str, striatum; Th, Thalamus.
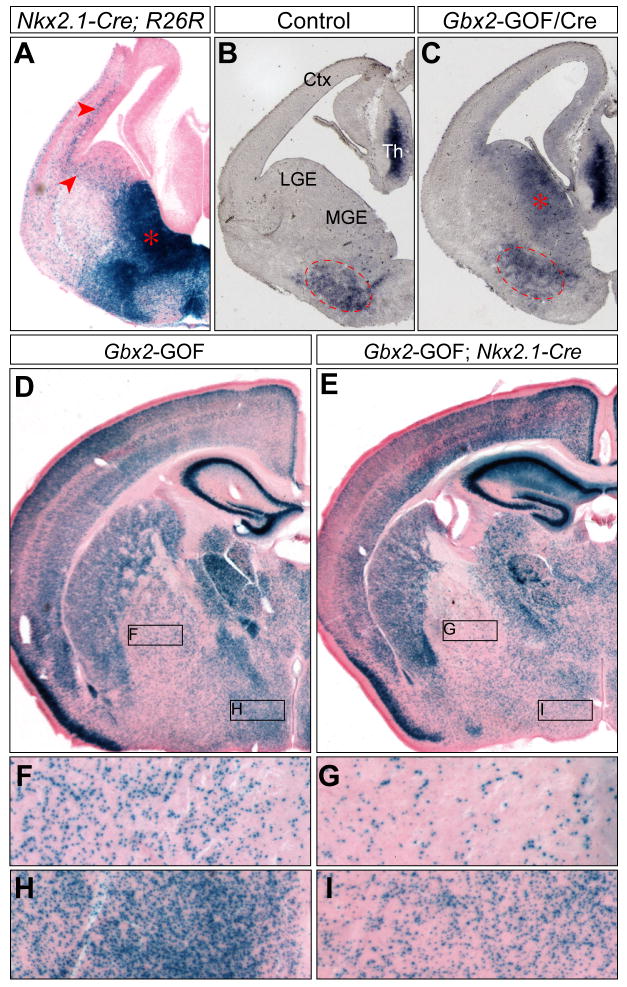
Figure 4. Misexpression of Gbx2 in the ventral telencephalon does not result in gross defects in neurons derived from the Nkx2.1 lineage.
(A) X-gal histochemistry of coronal sections of Nkx2.1-Cre; R26R embryos at E12.5. Note that Nkx2.1-expressing cells and their descendants contribute widely to the ventral telencephalon and the cortex (arrowheads). (B–C) In situ hybridization of Gbx2 on coronal sections of control (B) and Nkx2.1-Cre; Gbx2-GOF double transgenic (C) embryos at E12.5. Dashed circle marks the endogenous Gbx2 expression in MGE mantle zone; asterisk indicates ectopic expression of Gbx2 in MGE ventricular zone. (D–I) X-gal histochemical analysis on coronal sections of adult Gbx2-GOF hemizygous mice with and without the Nkx2.1-Cre transgene as indicated. Boxed areas in D and E corresponding to the globus pallidus and hypothalamus are magnified in F–I.
To assess the function of the recombined Gbx2-GOF transgene, we crossed mice carrying Gbx2-GOF and En1Cre, an En1 knock-in (KI) allele containing Cre (Kimmel et al., 2000). Using an EGFP reporter line, ZEG (Novak et al., 2000), to monitor the Cre activity from the En1Cre, we showed that the entire mes/r1 region is marked by EGFP (Fig. 1D). In En1Cre/+; Gbx2-GOF embryos at E10.5, RFP was detected in the mes/r1 area (Fig. 1E), consistent with previous reports (Li et al., 2002; Zervas et al., 2004). However, the RFP+ domain in En1Cre/+; Gbx2-GOF embryos was significantly smaller than the GFP+ domain in En1Cre/+; ZEG embryos, suggesting a reduction in the size of the mes/r1 region due to Gbx2 misexpression (Fig. 1D–E). Compared with that in control embryos, the isthmic constriction, which demarcates the prospective midbrain-hindbrain junction, was less prominent and apparently shifted rostrally in En1Cre/+; Gbx2-GOF embryos (Fig. 1D and E). These results demonstrate that Cre-mediated recombination leads to specific expression of Gbx2 and RFP in the mes/r1 region in En1Cre/+; Gbx2-GOF embryos. The anterior displacement of the isthmus observed in En1Cre/+; Gbx2-GOF embryos is in agreement with previous findings that ectopic expression of Gbx2 in the mes leads to an anterior shift of the isthmus (Katahira et al., 2000; Millet et al., 1999).
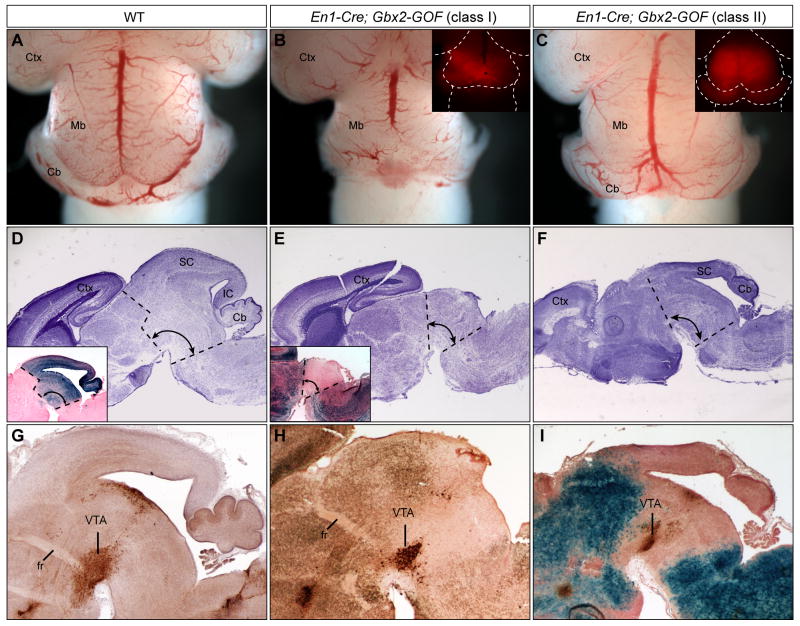
Given the apparent translocation of the isthmus in En1Cre/+; Gbx2-GOF embryos at E10.5, we expected that the misexpression of Gbx2 would enlarge the cerebellum at the expense of the midbrain. Surprisingly, both the midbrain and cerebellum were found truncated in En1Cre/+; Gbx2-GOF embryos at E18.5, although the extent of deletion varies among embryos (Fig. 2B and C). Based on the severity of the phenotype, the mutant embryos could be classified into two groups: group I (n=4/6) displayed almost complete deletion of the dorsal midbrain and the cerebellum (Fig. 2A–B and 2D–E); group II (2/6) exhibited deletion of the inferior colliculus, isthmus and the vermis of the cerebellum, while the superior colliculus and cerebellar hemispheres were largely unaffected (Fig. 2F and I). The variable severity of tissue loss may be caused by variations in the onset or levels of the expression of the Gbx2-GOF transgene on different genetic backgrounds. To examine the development of the ventral midbrain, we performed immunohistochemical analysis of tyrosine hydroxylase (TH), which labels dopaminergic neurons in the substantia nigra and ventral tegmental area (Fig. 2G). TH-positive neuron clusters were detected in the ventral midbrain in both classes of mutants, suggesting that the ventral midbrain is less affected compared with the dorsal structures following misexpression of Gbx2 (Fig. 2H–I).
Figure 2. Misexpression of Gbx2 in the mes/r1 region results in deletion of the midbrain and cerebellum.
(A–C) Dorsal views of E18.5 brains of the genotypes indicated. Insets in B and C show mCherry fluorescence in residual midbrain-hindbrain tissue. (D–F) Nissl staining of sagittal sections of E18.5 brains of the genotypes denoted. Inset in D shows X-gal labeling of a sagittal section of the brain of an E18.5 En1Cre/+; R26R+/− embryo. Note that in the En1Cre/+; R26R+/− line, the midbrain and the anterior hindbrain, including the cerebellum and pons, are marked by X-gal. Inset in E shows X-gal analysis of a section adjacent to E. Note that in the En1Cre/+; Gbx2-GOF embryos, X-gal staining is ubiquitous except in the region of Cre activity. Thus, the mid-hindbrain, which is delineated with dashed lines, is negative for X-gal stain (blue) and is significantly reduced in size in En1Cre/+; Gbx2-GOF embryos. (G–I) Tyrosine hydroxylase (TH) immunohistochemistry to mark dopaminergic neurons in the ventral tegmental area (VTA) on adjacent sections of D, E, and F, respectively. Sections in H and I are also stained with X-gal. Abbreviations: Cb, cerebellum; Ctx, cortex; fr, fornix; IC, inferior colliculus; SC, superior colliculus.
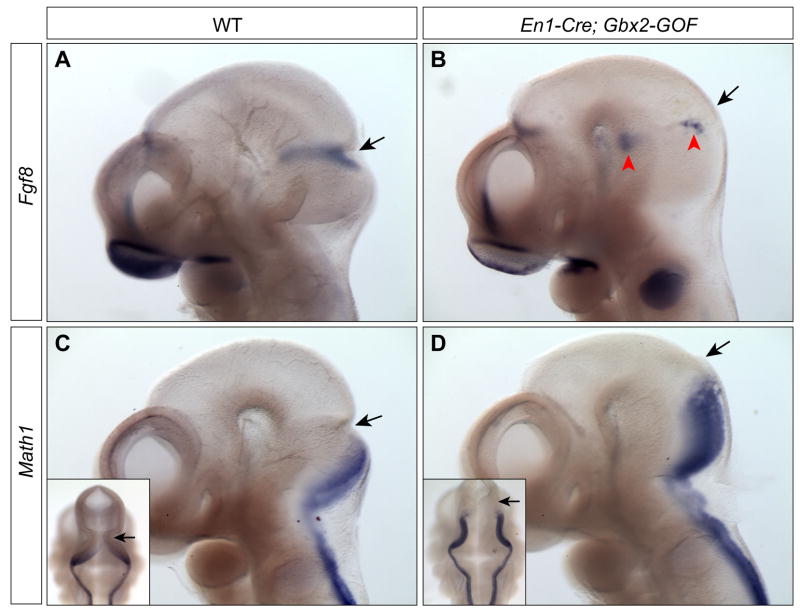
We next investigated the molecular basis of the loss of the midbrain and cerebellum in En1Cre/+; Gbx2-GOF embryos. As the phenotype of En1Cre/+; Gbx2-GOF embryos resembles that found in embryos with loss or reduction of Fgf8 signaling (Basson et al., 2008; Chi et al., 2003; Trokovic et al., 2003), we examined if the misexpression of Gbx2 disrupts Fgf8 expression. At E10.5, Fgf8 expression is localized in a transverse band corresponding to the isthmus (Fig. 3A). By contrast, in En1Cre/+; Gbx2-GOF embryos at E10.5, Fgf8 transcripts were only detected in a few clusters of cells in the mes/r1 area (Fig. 3B). Therefore, anterior expansion of Gbx2 expression leads to a failure to maintain Fgf8 expression in the isthmus. The disruption of Fgf8 expression may contribute to the deletion of the midbrain and cerebellum in En1Cre/+; Gbx2-GOF embryos.
Figure 3. The expression of Fgf8 in the isthmus is lost, and the roof plate of r1 is expanded due to misexpression of Gbx2 throughout the mes/r1 region.
(A–D) RNA in situ hybridization of Fgf8 (A–B) and Math1 (C–D) in controls and En1Cre/+; Gbx2-GOF embryos at E10.5. The genotypes are as indicated at the top of the panels. Insets in C and D are dorsal views of the embryos in C and D, respectively. The isthmic constriction (arrow) is shifted anteriorly and the roof plate of r1 is abnormally enlarged in En1Cre/+; Gbx2-GOF embryos. Red arrowhead indicates clusters of Fgf8-expressing cells.
Math1 encodes a basic helix-loop-helix transcription factor and plays an essential role in the development of the external granular layer of the cerebellum (Wang et al., 2005). At E10.5, Math1 is expressed in the rhombic lip of r1 and the posterior hindbrain, whereas Gbx2 is expressed in the ventricular zone of r1 outside of the Math1-positive domain (Fig. 3C and data not shown), suggesting that Gbx2 may normally restrict the expression of Math1 to the rhombic lip. We thus examined if ectopic expression of Gbx2 in the entire r1 abolishes Math1 expression and disrupt formation of the cerebellum in En1Cre/+; Gbx2-GOF embryos. Interestingly, we found that the expression of Math1 is maintained in the rhombic lip of r1 in En1Cre/+; Gbx2-GOF embryos at E10.5 (Fig. 3D). Furthermore, Math1 expression is significantly expanded anteriorly, indicating an anterior shift of the mes/r1 border in En1Cre/+; Gbx2-GOF embryos at E10.5 (Fig. 3D). Together, our results demonstrate that misexpression of Gbx2 throughout the mes/r1 leads to anterior shift of the isthmus, and Gbx2 does not repress Math1 expression.
To rule out the possibility that midbrain and cerebellum loss in En1Cre/+; Gbx2-GOF mice results from nonspecific cell death due to permanent expression of Gbx2, we mated Gbx2-GOF transgenic mice with Nkx2.1-Cre BAC transgenic mice, in which Cre is broadly expressed in the ventral telencephalon (Xu et al., 2008). As described previously (Xu et al., 2008), Cre activity was detected in the medial ganglionic eminence (MGE), anterior endopeduncular/preoptic area and cells originating from these structures in Nkx2.1-Cre embryos at E13.5 (Fig. 4A). In Nkx2.1-Cre; Gbx2-GOF embryos at E13.5, ectopic expression of Gbx2 was clearly seen in the ventricular zone of the MGE, in addition to the endogenous expression of Gbx2 in the thalamus and MGE mantle zone (Fig. 4B and C). In Nkx2.1-Cre; Gbx2-GOF mice on postnatal day 25, there was a significant reduction in the percentage of X-gal positive cells in the globus pallidus and hypothalamus, to which descendants of the Nkx2.1 lineage heavily contribute (Xu et al., 2008), thus confirming Cre-mediated recombination in these structures (Fig. 4D and E). Importantly, no tissue loss was detected in the forebrain of Nkx2.1-Cre; Gbx2-GOF mice, and these double transgenic mice were viable with no detectable gross defects. These data suggest that Cre-mediated activation of the Gbx2-GOF transgene does not lead to nonspecific cell death.
The phenotype of En1Cre/+; Gbx2-GOF embryos is clearly different from the results of transient misexpression of Gbx2 in the mes in chick or mouse embryos as described previously (Katahira et al., 2000; Millet et al., 1999). In those studies, particularly the one by Millet et al., transient misexpression of Gbx2 resulted in aberrant early patterning of the mes/r1 region but no obvious morphological changes at E12.5 and in the adult. Downregulation of the Wnt1-Gbx2 transgene likely led to a recovery of normal Gbx2/Otx2 limits with concomitant recovery of the normal MHB. In contrast to the persistent expression of Fgf8 in the translocated isthmus in embryos with transient Gbx2 misexpression, sustained ectopic expression of Gbx2 in En1Cre/+; Gbx2-GOF embryos largely abolishes Fgf8 expression in the isthmus, which likely contribute to deletion of the midbrain and cerebellum. Our results suggest that the juxtaposition of the expression domains of Gbx2 and Otx2 within the mes/r1 area is essential for the maintenance of Fgf8 expression and subsequent patterning and growth of the midbrain and cerebellum.
In summary, we here report generation of a novel conditional Gbx2 gain-of-function transgenic mouse line. We demonstrate that the Gbx2-GOF transgene is transcribed broadly from E10.5 onwards, making it particularly suitable for studies of CNS development. The transgene expresses Gbx2 in the CNS upon Cre-mediated recombination, and we show that Cre-induced expression of Gbx2 in the entire mes/r1 region results in loss of the midbrain and cerebellum. Therefore, our genetic study clearly demonstrates that the Gbx2-GOF transgene is functional. Additionally, our study provides insight into the maintenance of the mid-hindbrain border by Gbx2 and Otx2 interaction. The Gbx2-GOF mouse line will be an invaluable tool for further analysis of Gbx2 function in mouse development.
Materials and Methods
Mouse breeding and genotyping
Animals were housed in a facility with a 12-hour light/dark cycle (7am to 7pm) and allowed access to food and water ad libitum. All mutant mouse strains were maintained on a mixed genetic background. Noon of the day on which the vaginal plug was detected was designated as E0.5 in staging of embryos. Genotypes were determined by PCR analysis of embryonic yolk sac or tail DNA.
Generation of Gbx2-mCherry transgenic mice
To generate the Gbx2-GOF transgene, an ires-mCherry cassette was inserted into the XhoI/NotI sites of pCALL2 plasmids (a gift from Drs. Corrine Lobe and Caiyin Guo), and a Gbx2 full-length cDNA was inserted into the BglII/XhoI sites upstream of the ires-mCherry. Transgenic mice carrying the Gbx2-GOF were generated using standard protocol (Nagy, 2003). Four transgenic lines were analyzed for lacZ expression before Cre-mediated recombination, and one transgenic line that displayed broad expression of lacZ in the neural tube during embryogenesis was used in this study.
Histochemical and immunohistochemical analyses
Embryos were recovered in PBS and fixed immediately in 4% paraformaldehyde (PFA) at 4°C. Postnatal mice were deeply anesthetized and transcardially perfused with 4% PFA. Brains were postfixed in the same fixative overnight at 4°C. For frozen tissue sections, brains were cryoprotected in 30% sucrose in PBS before rapid freezing in OCT compound (TissueTek) and cryosectioned. Detailed protocols for RNA in situ hybridization in wholemount embryos, immunohistochemical assays, and β-gal activity analysis are described on the Li Lab website (http://www.genetics.uchc.edu/lilab/Pages/Protocols.html). The primary antibody was rabbit anti-TH (Pel-Freez Biologicals), and bound primary antibody was detected by Vectastain ABC kit (Vector Laboratory).
Acknowledgments
We are grateful to Drs. Corrinne Lobe and Caiying Guo for kindly providing the pCALL2 vector and Drs. Alexandra Joyner and Stewart Anderson for providing the En1Cre knock-in and Nkx2.1-Cre transgenic mice, respectively. We thank the gene targeting and transgenic facility at UConn Health Center for the generation of transgenic mice. J. Li is supported by grants from the NIH and March of Dimes foundation.
References
- Basson AE, Ntsala M, Martinson N, Tlale E, Corrigan GE, Shao X, Gray G, McIntyre J, Puren A, Morris L. Development of phenotypic HIV-1 drug resistance after exposure to single-dose nevirapine. J Acquir Immune Defic Syndr. 2008;49:538–543. doi: 10.1097/QAI.0b013e31818d5dcf. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Chazaud C, Oulad-Abdelghani M, Dolle P, Chambon P. Sequence and expression pattern of the Stra7 (Gbx-2) homeobox- containing gene induced by retinoic acid in P19 embryonal carcinoma cells. Dev Dyn. 1995;204:372–382. doi: 10.1002/aja.1002040404. [DOI] [PubMed] [Google Scholar]
- Broccoli V, Boncinelli E, Wurst W. The caudal limit of Otx2 expression positions the isthmic organizer. Nature. 1999;401:164–168. doi: 10.1038/43670. [DOI] [PubMed] [Google Scholar]
- Bulfone A, Puelles L, Porteus MH, Frohman MA, Martin GR, Rubenstein JL. Spatially restricted expression of Dlx-1, Dlx-2 (Tes-1), Gbx-2, and Wnt- 3 in the embryonic day 12.5 mouse forebrain defines potential transverse and longitudinal segmental boundaries. J Neurosci. 1993;13:3155–3172. doi: 10.1523/JNEUROSCI.13-07-03155.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Guo Q, Li JY. Transcription factor Gbx2 acts cell-nonautonomously to regulate the formation of lineage-restriction boundaries of the thalamus. Development. 2009;136:1317–1326. doi: 10.1242/dev.030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi CL, Martinez S, Wurst W, Martin GR. The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development. 2003;130:2633–2644. doi: 10.1242/dev.00487. [DOI] [PubMed] [Google Scholar]
- Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- Joyner AL, Liu A, Millet S. Otx2, Gbx2 and Fgf8 interact to position and maintain a mid-hindbrain organizer. Curr Opin Cell Biol. 2000;12:736–741. doi: 10.1016/s0955-0674(00)00161-7. [DOI] [PubMed] [Google Scholar]
- Katahira T, Sato T, Sugiyama S, Okafuji T, Araki I, Funahashi J, Nakamura H. Interaction between otx2 and gbx2 defines the organizing center for the optic tectum [In Process Citation] Mech Dev. 2000;91:43–52. doi: 10.1016/s0925-4773(99)00262-2. [DOI] [PubMed] [Google Scholar]
- Kimmel RA, Turnbull DH, Blanquet V, Wurst W, Loomis CA, Joyner AL. Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes Dev. 2000;14:1377–1389. [PMC free article] [PubMed] [Google Scholar]
- Li JY, Joyner AL. Otx2 and Gbx2 are required for refinement and not induction of mid-hindbrain gene expression. Development. 2001;128:4979–4991. doi: 10.1242/dev.128.24.4979. [DOI] [PubMed] [Google Scholar]
- Li JY, Lao Z, Joyner AL. Changing requirements for Gbx2 in development of the cerebellum and maintenance of the mid/hindbrain organizer. Neuron. 2002;36:31–43. doi: 10.1016/s0896-6273(02)00935-2. [DOI] [PubMed] [Google Scholar]
- Martinez-Barbera JP, Signore M, Boyl PP, Puelles E, Acampora D, Gogoi R, Schubert F, Lumsden A, Simeone A. Regionalisation of anterior neuroectoderm and its competence in responding to forebrain and midbrain inducing activities depend on mutual antagonism between OTX2 and GBX2. Development. 2001;128:4789–4800. doi: 10.1242/dev.128.23.4789. [DOI] [PubMed] [Google Scholar]
- Millet S, Campbell K, Epstein DJ, Losos K, Harris E, Joyner AL. A role for Gbx2 in repression of Otx2 and positioning the mid/hindbrain organizer. Nature. 1999;401:161–164. doi: 10.1038/43664. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo. 3. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Trokovic R, Trokovic N, Hernesniemi S, Pirvola U, Vogt Weisenhorn DM, Rossant J, McMahon AP, Wurst W, Partanen J. FGFR1 is independently required in both developing mid- and hindbrain for sustained response to isthmic signals. Embo J. 2003;22:1811–1823. doi: 10.1093/emboj/cdg169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang VY, Rose MF, Zoghbi HY. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron. 2005;48:31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Lewandoski M, Campbell K, Joyner AL, Rubenstein JL, Martinez S, Martin GR. Specification of the anterior hindbrain and establishment of a normal mid/hindbrain organizer is dependent on Gbx2 gene function. Development. 1997;124:2923–2934. doi: 10.1242/dev.124.15.2923. [DOI] [PubMed] [Google Scholar]
- Xu Q, Tam M, Anderson SA. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J Comp Neurol. 2008;506:16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- Zervas M, Millet S, Ahn S, Joyner AL. Cell behaviors and genetic lineages of the mesencephalon and rhombomere 1. Neuron. 2004;43:345–357. doi: 10.1016/j.neuron.2004.07.010. [DOI] [PubMed] [Google Scholar]