Abstract
The fungal neurotoxin α-cyclopiazonic acid (CPA), a nanomolar inhibitor of Ca2+-ATPase with a unique pentacyclic indole tetramic acid scaffold is assembled by a three enzyme pathway CpaS, CpaD and CpaO in Aspergillus sp. We recently characterized the first pathway-specific enzyme CpaS, a hybrid two module polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) that generates cyclo-acetoacetyl-L-tryptophan (cAATrp). Here we report the characterization of the second pathway-specific enzyme CpaD that regiospecifically dimethylallylates cAATrp to form β-cyclopiazonic acid. By exploring the tryptophan and tetramate moieties of cAATrp, we demonstrate that CpaD discriminates against free Trp but accepts tryptophan-containing thiohydantoins, diketopiperazines and linear peptides as substrates for C4-prenylation and also acts as regiospecific O-dimethylallyltransferase (DMAT) on a tyrosine-derived tetramic acid. Comparative evaluation of CpaDs from A. oryzae RIB40 and A. flavus NRRL3357 indicated the importance of the N-terminal region for its activity. Sequence alignment of CpaD with eleven homologous fungal Trp-DMATs revealed five regions of conservation suggesting the presense of critical motifs that could be diagonostic for discovering additional Trp-DMATs. Subsequent site-directed mutagenesis studies identified five polar/charged residues and five tyrosine residues within these motifs that are critical for CpaD activity. This motif characerization will enable a gene probe-based approach to discover additional biosynthetic Trp-DMATs.
INTRODUCTION
Prenylated indole alkaloids represent a large family of natural products that are produced by various terrestrial and marine organisms including plants, fungi, cyanobacteria and actinomyces (1). Fungi are the most prolific producers that biosynthesize an array of prenylated indole alkaloids with complex molecular architectures and profound biological activities, including postpartum haemorrhage prevention drugs ergometrine and ergotamine (2), antitumor agents tryprostatins (3) and stephacidins (4), antiparasitic agents paraherquamides (5), and mycotoxins aflatrem (6) and cyclopiazonic acid (7).
Except for indole-diterpene alkaloids that use geranylgeranyl pyrophosphate as prenyl donor (8), dimethylallyl pyrophosphate (DMAPP) is the predominant prenyl source for fungal prenyl indole alkaloids. Dimethylallylation critically functionalizes indole alkaloid backbones by introducing structural complexity and profoundly influencing biological activity. Regioselective dimethylallylation of didemethylasterriquinone D with bisindolylbenzoquinone scaffold leads to a variety of asterriquinones including terrequinone A (9), cochliodinol (10) and asterriquinone CT5 (11) with distinct biological activities. Biosynthetic dimethylallylation provides a functional alkene and a set of allylic hydrogens that constitute the reactive center for subsequent enzymatic oxidatations and cyclizations en route to the complex polycyclic frameworks observed in many prenylated indole alkaloids (12, 13).
Dimethylallylation of scaffolds in fungal prenylated indole alkaloids is catalyzed by tryptophan dimethylallyltransferases (Trp-DMATs). Initially isolated from Claviceps purpurea (14), the characterization and sequencing of a C-4 Trp-DMAT (DmaW) involved in ergot alkaloid biosynthesis set the foundation for discovery and study of functionally related Trp DMATs (15, 16). By a combination of recent advances in fungal genome sequencing with in silico bioinformatic and in vitro biochemical analyses six additional pathway-specific Trp-DMATs have been identified. These include FgaPT1 and FgaPT2 in fumigaclavine C biosynthesis (17), FtmPT1 and FtmPT2 in fumitremorgin B biosynthesis (18), TdiB in terriquinone A biosynthesis (19) and AnaPT in acetylaszonalenin biosynthesis (20). Analogous efforts have resulted in the identification of three homologs, 7-DMATS (21), CdpNPT (22) and MaPT (23), in currently undefined biosynthetic pathways. All characterized fungal Trp-DMATs lack the conserved (N/D)DxxD and N(Q/D)xxDxxxD sequences for pyrophosphate binding in trans-polyprenyl pyrophosphate synthases and membrane bound aromatic prenyl transferases respectively (24). The combined biochemical results indicate that Trp-DMAT catalyzed dimethylallyl transfer does not require Mg2+, a characteristic that distinguishes the Trp-DMAT family of prenyl transferases.
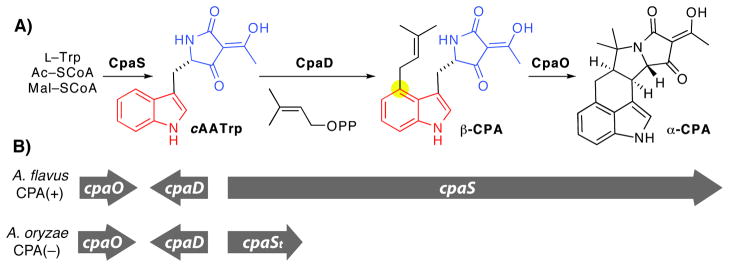
The latest addition to the family of fungal prenylated indole alkaloid gene clusters is for biosynthesis of cyclopiazonic acid (CPA or α-CPA), a potent neurotoxin with a rigid pentacyclic indole tetramate scaffold that acts as a nanomolar inhibitor of the sarcoplasmic Ca2+ ATPase (25). The CPA biosynthetic genes cpaS, cpaD and cpaO were recently identified in Aspergillus sp. by a combined bioinformatic and genetic approach (26, 27) and correlate well with previous predictions that the CPA scaffold is derived from two intermediates, cyclo-acetoacetyl L-tryptophan (cAATrp or L-cAATrp) and β-CPA, which are assembled from tryptophan, two molecules of acetate, and DMAPP (Figure 1) (28). We recently reported the detailed study of CpaS, a hybrid two module polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) that incorporates acetyl CoA, malonyl CoA, and tryptophan and utilizes a C-terminal redox-incompetent reductase domain to make and release the tryptophan tetramic acid, cAATrp, as the first intermediate in the pathway (29).
Figure 1.
Cyclopiazonic acid (CPA) biosynthesis in Aspergillus sp. (A) Functions of CPA biosynthetic genes CpaS, CpaD and CpaO (B) CPA gene cluster organization in CPA producing/CPA(+) strain A. flavus NRRL3357 and CPA nonproducing/CPA(−) strain A. oryzae RIB40.
CpaD is proposed to be a C-4 Trp-DMAT that tailors cAATrp with DMAPP to generate β-CPA in the second step of the pathway (Figure 1A). Since tetramic acid moieties in related natural products often serve as the pharmacophore for recognition by a range of biological targets (30), we speculated that this ring would be an important recognition element for CpaD as well. Here we validate the role of CpaD in the CPA biosynthetic pathway by showing that while it is essentially inactive with free tryptophan, it can act as a C-4 DMAT towards Trp-containing thiohydantoins, diketopiperazines and linear peptides, and as an O-DMAT on a tyrosine-derived tetramic acid. Comparison of CpaD with eleven other fungal Trp-DMATs led us to a detailed mutagenesis study in which we identified ten critical residues within five conserved motifs that are catalytically consequential to the activity of CpaD.
EXPERIMENTAL PROCEDURES
General methods and materials
Standard molecular biology procedures were performed as described (31). Oligonucleotide primers were synthesized by Integrated DNA Technologies (Coralville, IA). PCR was performed either using KAPA HiFi DNA polymerase (KAPA Biosystems) or Fusion high fidelity DNA polymerase (New England Biolabs) under suggested conditions on a BioRAD MyCycler™ thermocycler. For general cloning, E. coli NovaBlue(DE3) cells (Novagen) were used. DNA sequencing was performed at the Molecular Biology Core Facilities of the Dana Farber Cancer Institute (Boston, MA). The strain and genomic DNA from Aspergillus oryzae RIB40 (ATCC 42149) were obtained from American Type Culture Collection (ATCC). The strain and genomic DNA from Aspergillus flavus NRRL3357 were obtained from Dr. K. McCluskey (FGSC) and Dr. J. Yu (USGA). Penicillium cyclopium (i.e. Penicillium griseofulvum Dierckx) strain NRRL 3523 was obtained from Dr. S. Peterson (USGA). Dimethylallylpyrophosphate (DMAPP) and related isoprenoid pyrophosphates were obtained from Isoprenoids LC. Tryptophan-containing dipeptides and thiohydantoins were purchased from Research Organics and Chem-Impex, Inc. Tryptophan-containing oligopeptides were synthesized by Peptide 2.0 (purity > 98%). All other chemicals were obtained from Sigma-Aldrich. HPLC was performed on a Beckman System Gold (Beckman Coulter) instrument using Phenomenex Luna C18 columns (250 × 4.5 mm, for analytical purpose; 250 × 21.6 mm, for preparative purpose). The eluant was monitored for absorption at 280 nm for all LC analysis of tryptophan derivatives. A 3%–97% acetonitrile/water gradient over 30 min was used for analytical HPLC assay, unless otherwise noted. LC-MS analysis was carried out on a Shimadzu LCMS-QP8000a (low resolution) and an Agilent 6520 QTOF-LCMS (high resolution) machines. 1H NMR spectra were recorded on a Varian 600 MHz spectrometer. Chemical shifts are reported in ppm with the solvent resonance resulting from incomplete deuteration as the internal standard (CDCl3 δ7.26, CD3OD δ3.31). Data are reported as follows: chemical shift, multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, p = pentet, br = broad, m = multiplet), coupling constants (Hz), and integration.
Cloning, overproduction and purification of CpaD
The protein-coding exons of cpaD genes from A. oryzae RIB40 were amplified individually from genomic DNA using the oligonucleotide primers listed in Table S1 (Supporting information). The amplified exons were gel-purified, overlap-extended to yield the full-length cpaD gene and subsequently ligated into the BamHI and NotI sites of expression vector pQTev (Protein Structure Factory) using DNA ligation kit Ver 2.1 (Takara). This gave the expression vector pQTev::cpaD, which was subsequently sequence-verified and transferred into E. coli DE3 (Star) competent cells for protein expression. Cells were cultured in Luria Bertani medium containing 100 μg/mL of Ampicillin. A fresh overnight culture from a single colony was used to inoculate 2 × 1L medium. Cells were grown at 37 °C in a shaker at 200 rpm till mid-log phase (A600 ≈ 0.5). The cultures were then cooled to 16 °C and induced by isopropyl β-D-1-thiogalactopyranoside with a final concentration of 0.25 mM. The cultures were allowed to further grow at 16 °C for 12–16 h. The cells were harvested by centrifugation and cell pellets from the 2L culture were resuspended in a buffer (50 mL) containing 20 mM Tris-HCl pH 8, 500 mM NaCl, 20 mM imidazole, 1mM DTT, 0.5% (v/v) Tween-20, and homogenized. The homogenized cells were lysed by passing through a cell disruptor (Avestin EmulsiFlex-C5) three times at 5000–10,000 psi, and the lysate was clarified by ultracentrifugation (35,000 rpm × 35 min). The supernatant was incubated with 2 mL of Ni-NTA-agarose resin (Qiagen) at 4 °C for 1 h and loaded to a column. The resin was then washed with a buffer (20 mL) containing 20 mM Tris-HCl pH 8, 500 mM NaCl, 20 mM imidazole. His7-tagged CpaD was eluted with 20 mL of 200 mM imidazole in 20 mM Tris-HCl pH 8, 500 mM NaCl. The protein was further purified by gel filtration chromatography using a HiLoad 26/60 Superdex 200 column (GE Biosciences) with a buffer containing 20 mM Tris-HCl pH 7.5, 50 mM NaCl, 1mM DTT, 5% glycerol. Fractions containing CpaD were pooled and concentrated using Amicon Ultra centrifugal filter device (Millipore). The protein concentration was determined by Bradford assay and the protein’s absorbance at 280 nm with molar extinction coefficient 72310 M−1cm−1 (predicted using ProtParam http://ca.expasy.org/tools/protparam.html). Proteins were flash-frozen in aliquots in liquid nitrogen and stored at −80 °C for further usages. The overproduction yield was ca. 25 mg/L for CpaD from A. oryzae RIB40. The cloning of the original and revised cpaD genes from A. flavus NRRL3357 were carried out in a similar fashion with the oligonucleotides listed in Table S1 (supporting information). Subsequent protein overexpression gave purified CpaD in a yield 0.2 mg/L for originally assigned CpaD from A. flavus NRRL3357 and 20 mg/L for revised CpaD from A. flavus NRRL3357.
Synthetic substrates preparation
cyclo-Acetoacetyl L-tryptophan (L-cAATrp) was prepared as previously described (29). The corresponding stereoisomer cyclo-acetoacetyl D-tryptophan (D-cAATrp) was synthesized in the same manner, using D-tryptophan methylester hydrochloride as the starting material instead of L-tryptophan methylester hydrochloride. All spectral data of D-cAATrp matched those of cyclo-L-cAATrp. [α]D24 = 118.2 (c = 0.36, MeOH). L-tyrosine derived tetramic acid 10 was also synthesized in an analogous fashion as illustrated in Scheme S1 (Supporting Information). Briefly, O-tert-butyl-L-tyrosine methyl ester hydrochloride was acetoacetylated using S-tert-butyl acetothioacetate and silver trifluoroacetate in THF. Treatment of the derived product with sodium methoxide afforded the tetramate moiety, and deprotection of the tert-butyl tyrosine hydroxyl was removed by exposure to pure trifluoroacetic acid. [α]D24 = −168 (c = 1.05, MeOH). 1H NMR (600 MHz, CD3OD) δ 7.16 (d, 2H, J = 7.8 Hz), 6.89 (d, 2H, J = 7.8 Hz), 4.81 (dd, 1H, J = 7.8, 5.4 Hz), 3.31 (dd, 1H, J = 14.0, 5.4 Hz), 3.08 (dd, 1H, J = 14.0, 7.8 Hz), 2.03 (s, 3H). HRMS (ESI): Calcd for C13H13N2O4 [M – H]−: 246.0766, Found: 246.0761.
Generation of cAATrp, β-CPA and α-CPA standards from P. cyclopium culture
The presence of cAATrp and β-CPA as the biosynthetic intermediates of α-CPA in the culture medium of P. cyclopium was previously observed (32). Since our primary focus was to obtain β-CPA as a reference material for the subsequent enzymatic assay, the described condition was optimized to maximize the production of β-CPA. Both cAATrp and α-CPA were isolated as minor products. Complete synthetic medium as described by McGrath et al. (32) was used for culturing P. cyclopium and prepared as follows: glucose (60 g), NaNO3 (4.5 g), K2HPO4 (1 g), MgSO4•7H2O (0.5 g), KCl (0.5 g), FeSO4•7H2O (10 mg), ZnSO4•7H2O (17.6 mg), Na2B4O7•10H2O (0.7 mg), (NH4)6Mo7O24•4H2O (0.5 mg), CaSO4•5H2O (0.3 mg), and MnSO4•H2O (0.11 mg) were dissolved in 1 L of water. The pH was adjusted to 5.5 and then autoclaved for 20 min. A 4-day-old potato dextrose agar plate of P. cyclopium was flooded with a sterile aqueous solution of 0.001% Triton-X. The spore suspension obtained in this way was used to inoculate 30 mL of the complete synthetic medium in a 250 mL Erlenmyer flask. The culture was maintained on a rotary shaker (170 rpm) at 25 °C for 3.5 days (longer than 4 days resulted in predominant production of α-CPA). The mycelia were removed by filtration, and the filtrate was acidified to pH ≈ 2 using concentrated HCl and extracted with chloroform. The combined organic layers were washed with water, dried over Na2SO4 and evaporated to dryness to give a mixture containing cAATrp, β-CPA and a-CPA as shown in Figure 2B (lane 1). The identities of each metabolite were further verified by LC-MS analysis: cAATrp, [M+H]+ cal for C15H14N2O3: 271.1083, obs: 271.1081; β-CPA, [M+H]+ cal for C20H22N2O3: 339.1709, obs: 339.1711; α-CPA, [M+H]+ cal for C20H20N2O3: 337.1552, obs: 337.1553.
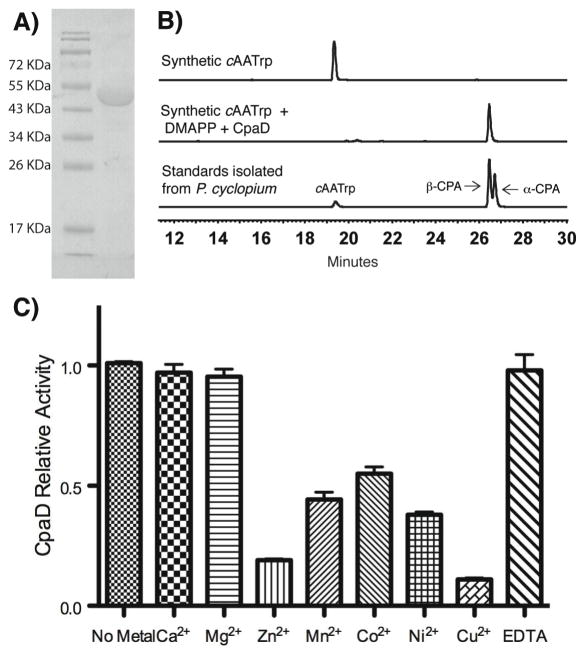
Figure 2.
In vitro validation of the function of CpaD. (A) Heterologously overexpressed and purified CpaD from A. oryzae RIB40. (B) HPLC traces that indicate CpaD is able to covert cAATrp to β-CPA in the presence of DMAPP. (C) Comparison of CpaD activity with or without addition of divalent metals and EDTA.
Assays for CpaD activity
The initial assay to verify the function of CpaD was carried out in 100 μL in 50 mM Tris•HCl (pH 7.5) buffer containing 0.25 mM cAATrp, 0.30 mM DMAPP and 0.1 μmol of purified CpaD. The reaction mixture was incubated at 30 °C for 1 h and stopped by the addition of MeOH (100 μL). The quenched mixture was mixed thoroughly and centrifuged at the maximum speed using a bench top centrifuge for 10 min at 4 °C. The supernatant (160 μL) was subjected to HPLC and LC-MS analysis. For evaluating the effect of divalent metal on the activity of CpaD, the assays were carried out in 50 μL in 50 mM Tris•HCl (pH 7.5) buffer containing 0.25 mM cAATrp, 0.25 mM DMAPP with selected divalent ion solution or EDTA (5 mM). The assay was initiated by adding 20 nmol of purified CpaD and quenched after 10 min with MeOH (50 μL). Assays were carried out in duplicate, and the conversion rate was calculated based by HPLC analysis and compared to the assay using CpaD enzyme without any exogenous divalent ion present.
Substrate specificity of CpaD
For evaluating the substrate specificity of CpaD toward isoprenoid pyrophosphates other than DMAPP, initial assay conditions were used as above but with replacement of DMAPP with IPP, GPP, FPP and GGPP individually. Assays were conducted for an extended period (up to 16 h) to ensure even low turnover would be detected. For study of CpaD activity on tryptophan-containing thiohydantoins, DKPs, dipeptides and tyrosine-derived tetramic acid, the assay was carried out in 50 μL in 50 mM Tris•HCl (pH 7.5) buffer containing 0.25 mM tryptophan derivative or tyrosine-derived tetramic acid, 0.30 mM DMAPP and 1 μmol of purified CpaD. The reaction mixture was incubated at 30 °C for 15–16 h and stopped by the addition of MeOH (50 μL). After centrifugation, the supernatant was subjected to HPLC and high resolution LC-MS analysis to verify the identities of the products.
Structural characterization of dimethylallylated trytophan containing thiohydantoins, DKPs, dipeptides and tyrosine tetramic acids
To verify the regioselectivity of CpaD-mediated dimethylallylation of tryptophan-containing thiohydantoins (1–3), DKPs (4–8), dipeptides (9–14) and tyrosine tetramic acid 15, assays using representative substrates 1, 5, 9, 15 were scaled up and the corresponding products 1a, 5a, 9a, 15a were isolated for 1H NMR analysis. Briefly, the assays were carried out in a 4 × 1mL scale in 50 mM Tris•HCl (pH 7.5) buffer containing 0.5 mM cAATrp, 0.6 mM DMAPP and 2 μmol of purified CpaD. The reaction mixture was incubated at 30 °C for 24 h and stopped by the addition of MeOH (4 × 1mL). The reaction mixture was subjected to preparative HPLC and peaks were collected for 1H NMR analysis. For 1a, 1H NMR (600 MHz, CDCl3) δ 7.95 (brs, 1H), 7.21 (d, 1H, J = 7.5 Hz), 7.18 (s, 1H), 7.13 (d, 1H, J = 7.5 Hz), 6.96 (app. t, 1H, J = 7.5 Hz), 5.96 (brs, 1H), 5.75 (app. t, 1H, J = 6.5 Hz), 4.01 (dd, 1H, J = 7.8, 5.4 Hz), 3.35 (s, 1H), 3.31 (d, 1H, J = 14.0, 5.4 Hz), 3.07 (d, 1H, J = 14.0, 5.4 Hz), 1.83 (s, 3H), 1.71 (s, 3H). For 5a, 1H NMR (600 MHz, CDCl3) δ 8.01 (brs, 1H), 7.41 (m, 2H), 7.30 (m, 2H), 7.28 (m, 1H), 7.21 (d, 1H, J = 7.5 Hz), 7.17 (s, 1H), 7.09 (d, 1H, J = 7.5 Hz), 6.92 (app. t, 1H, J = 7.5 Hz), 5.96 (brs, 1H), 5.75 (app. t, 1H, J = 6.5 Hz), 4.16 (dd,, J = 8.2, 5.6 Hz), 4.01 (dd, 1H, J = 7.8, 5.4 Hz), 3.42 (d, 1H, J = 14.2, 5.6 Hz), 3.33 (d, 1H, J = 14.0, 5.4 Hz), 3.42 (d, 1H, J = 14.2, 5.6 Hz), 3.05 (d, 1H, J = 14.0, 5.4 Hz), 1.83 (s, 3H), 1.71 (s, 3H). For 9a, 1H NMR (600 MHz, CD3OD) δ 7.22 (d, 1H, J = 7.5 Hz), 7.17 (s, 1H), 7.12 (d, 1H, J = 7.5 Hz), 6.99 (app. t, 1H, J = 7.5 Hz), 5.75 (app. t, 1H, J = 6.5 Hz), 4.39 (q, 1H, J = 6.8 Hz), 4.00 (dd, 1H, J = 7.8, 5.4 Hz), 3.31 (m, 1H), 3.02 (d, 1H, J = 14.0, 5.4 Hz), 1.83 (s, 3H), 1.71 (s, 3H), 1.42 (d, 3H, J = 6.8 Hz). For 15a, 1H NMR (600 MHz, CD3OD) δ 7.21 (d, 2H, J = 7.8 Hz), 6.99 (d, 2H, J = 7.8 Hz), 5.35 (t, 1H, J = 6.3 Hz), 4.82 (dd, 1H, J = 7.8, 5.4 Hz),4.69 (d, 2H, J = 6.6 Hz), 3.33-3.30 (m, 1H), 3.09 (dd, 1H, J = 14.0, 7.8 Hz), 2.03 (s, 3H), 1.84 (s, 3H), 1.70 (s, 3H).
Kinetic parameters of CpaD toward L–cAATrp, D-cAATrp, L–Trp, D-Trp and other substrates (1–15)
For calculating the kinetic parameters of CpaD towards its native substrate, L-cAATrp, the assay was conducted in 50 μL in 50 mM Tris-HCl (pH 7.5) with 250 μM DMAPP and varied concentration of L-cAATrp (5–500 μM). The reaction was initiated at 30 °C by the addition of 10 nmol of purified CpaD (from A. oryzae). The reaction was quenched after 10 min with MeOH (100 μL). The quenched mixture was mixed thoroughly and centrifuged at the maximum speed using a bench top centrifuge for 10 min at 4 °C. The supernatant (80 μL) was subjected to HPLC analysis. Product quantification was based on integration of the 280 nm absorption peaks. Assays were run in duplicate and the data fit to the Michaelis-Menten equation using GraphPad Prism software. For D-cAATrp, the assay was carried out in the same manner except 100 nmol of purified CpaD were used to initiate the reaction. The relative turnover numbers of CpaD towards L–Trp and D-Trp were estimated based on the conversion rate using 250 μmol L–Trp or D-Trp with 12 μmol CpaD after a 16 h incubation. For calculating the kinetic parameters of CpaD towards substrates 1–15, the assays were carried out in a similar fashion as that for L-cAATrp, except 1 μmol CpaD was used to initiate the reaction and substrates (1–15) concentration were varied up to 1 mM. For 9–14, saturation kinetics could not be achieved even with 1 mM substrate concentration; therefore the initial apparent maximum velocities were obtained when using 0.25 mM substrate, 0.25 mM DMAPP, 1 μM CpaD at 30 °C.
Generation of CpaD mutant proteins and their enzymatic activity assays
The mutants of cpaD were generated according to the protocol described in Stratagene QuikChange® Multi Site-Directed Mutagenesis Kit, using plasmid pQTev::cpaD as the template and primers listed in Table S2 (supporting information). Primer oligonucleotides were in situ 5′-phosphorylated using T4 polynucleotide kinase (New England Biolab). Correct mutant plasmids were verified by sequencing. The mutant proteins were overproduced and purified in the same manner as that described for the wild type CpaD. The protein concentrations were measured using a Nanodrop device (Thermo Scientific) based on the absorbance at 280 nm with the predicted molar extinction coefficient. Except for mutants CpaD W33F, W33L, D119L, all other mutant proteins were overproduced as highly soluble as the wild type protein and isolated in ca. 15–30 mg/mL yields. Mutants W33F was isolated in ca 0.2 mg/mL, whereas W33L and D119L were not soluble. To compare the activities of mutant proteins with wild type, assays were carried out in 50 μL in 50 mM Tris-HCl (pH 7.5) with 250 μM L-cAATrp and 250 μM DMAPP at 30 °C. The reaction was initiated by the addition of 50 nmol of purified CpaD wild type or mutant protein. The reaction was quenched at an optimal time point in order to obtain the maximum initial velocity. All assays were run in duplicate.
RESULTS
Cloning and overexpression of CpaD
The gene that codes DMAT CpaD responsible for β-CPA biosynthesis was initially disclosed in A. oryzae A1560 strain (33). BLAST analysis revealed an identical gene in A. oryzae RIB40 strain and subsequently led to the discovery of truncated and full CPA gene clusters in A. oryzae RIB40 and A. flavus NRRL3357, respectively (26, 27). We first cloned the cpaD gene from A. oryzae RIB40 genomic DNA in an effort to correlate its function with CPA biosynthesis in vitro and the overproduced CpaD A. oryzae RIB40 protein is used throughout this study unless otherwise noted. The two protein coding exon fragments (1154 bp and 157 bp) were amplified individually and overlap-extended to generate the full length cpaD (1311 bp) followed by insertion into the pQTev vector to afford the N-terminal His7-tagged CpaD expression construct. Overexpression in E. coli gave highly soluble CpaD (see methods), which was subsequently purified to apparent homogeneity using successive nickel-affinity and gel filtration chromatographies (Figure 2A). CpaD appears homodimeric in solution based on its retention time on size exclusion chromatography.
Validation of the function of CpaD
To assess the function of CpaD, we chemically synthesized the putative substrate L-cAATrp and isolated an authentic sample of β-CPA mixed with minor amount of α-CPA and L-cAATrp from the native producer, Penicillium cyclopium NRRL 3523, as the reference material (see methods). Incubation of L-cAATrp and DMAPP with CpaD at 30 °C led to the rapid formation of a new peak by HPLC analysis that corresponds to β-CPA (Figure 2B). The identity of this new peak was assured to be β-CPA by co-elution with the isolated standard and high resolution MS analysis ([M+H]+ cal for C20H22N2O3: 339.1709, obs: 339.1713). We further examined if the activity of CpaD depends on the presence of divalent ion, a hallmark of cis- and trans- prenyltransferases (34–36). At 5 mM concentrations, we did not observe any statistically significant rate enhancement by inclusion of Ca2+ and Mg2+ (Figure 2C), as occasionally observed for other tryptophan DMATs (24). Other examined divalent ions, including Zn2+, Mn2+, Co2+, Ni2+ and Cu2+, had moderate to profound inhibitory effects on the activity of CpaD (Figure 2C). Furthermore, the activity of CpaD was not influenced by the addition of EDTA (5 mM), ruling out the possible presence of endogenous metal cofactor. Consequently, all the assays using CpaD involved in this study were carried out without addition of divalent ions.
Activity of CpaD towards D-cAATrp, L-/D-Trp and other isoprenoid pyrophosphates
With the basic function of CpaD verified, we compared the kinetic profiles of CpaD towards its native substrate, L-cAATrp, and its stereoisomer, D-cAATrp. The KM value for L-cAATrp was determined to be 109±16 μM with kcat 53±12 min−1 (Table 1). This corresponds to ca. 0.48 μM−1 min−1 in catalytic efficiency, a value comparable to most of the tryptophan DMATs characterized to date with their genuine substrates (24). In comparison, the catalytic efficiency of CpaD with D-cAATrp is about 24 fold lower than that with L-cAATrp, in accordance with other characterized Trp-DMATs that prefer L- over D-isomers (24). CpaD had extremely low activity towards the free amino acid L-Trp. Since we could not obtain saturation for L-Trp even at 1 mM concentration, the approximate turn over number was estimated to be 4.1 ×10−3 min−1, which implies the catalytic efficiency of CpaD for L-Trp is at least 105 fold less in comparison with L-cAATrp, and clearly not a physiologic substrate. The dramatic difference in catalytic efficiency of CpaD between L-cAATrp and L-Trp exceeds that observed for FtmPT1, the first committed Trp-DMAT involved in fumitremorgin B biosynthesis (18). FtmPT1 is a genuine C-2 Trp-DMAT for diketopiperzine cyclo-L-Trp-L-Pro (brevianamide F), but still can accept L-Trp with 675 fold decrease in catalytic efficiency (37). These combined observations indicate the tetramic acid portion of the L-cAATrp scaffold is an important recognition element for CpaD. Not surprisingly, the activity of CpaD towards D-Trp is even lower (Table 1). We also examined if CpaD utilizes other naturally occurring isoprenoid pyrophosphates including IPP, GPP, FPP and GGPP as prenyl donors. Even after an extended incubation time and increased enzyme concentration, no turnover was detected using these pyrophosphate substrates (data not shown), suggesting CpaD strictly utilizes DMAPP as the sole prenyl donor.
Table 1.
Kinetic parameters of CpaD with its native substrate L-cAATrp, the enantiomer D-cAATrp, L-Trp and D-Trp.
| Substrate | Product Identification | Kinetic profile | |||
|---|---|---|---|---|---|
| [M+H]+cal. | [M+H]+obs. | Km (μM) | kcat (min−1) | kcat/Km (min−1 μM−1) | |
| L-cAATrp | 339.1709 | 339.1713 | 109 ± 16 | 53 ± 12 | 0.486 |
| D-cAATrp | 339.1709 | 339.1712 | 382 ± 29 | 8.2 ± 1.9 | 0.021 |
| L-Trpa | 273.1603 | 273.1601 | >1000 | 4.1×10−3 | < 4.1 × 10−6 |
| D-Trpa | 273.1603 | 273.1605 | >1000 | 7.8×10−4 | < 7.8 × 10−7 |
Activity of CpaD towards tryptophan-containing thiohydantoins and diketopiperazines (DKPs)
Our initial experiments indicated that CpaD, unlike other characterized C-4 Trp-DMATs including FgaPT2 (38), DmaW (39) and MaPT (23), kinetically disfavors L-Trp and its simple derivatives such as L-abrine and methyl substituted L-Trp as substrates (data not shown). We envisioned CpaD might have a distinctive substrate preference that could be useful for chemoenzymatic preparations of more complex dimethylallylated tryptophan containing molecules and sought to explore it in a systematic manner.
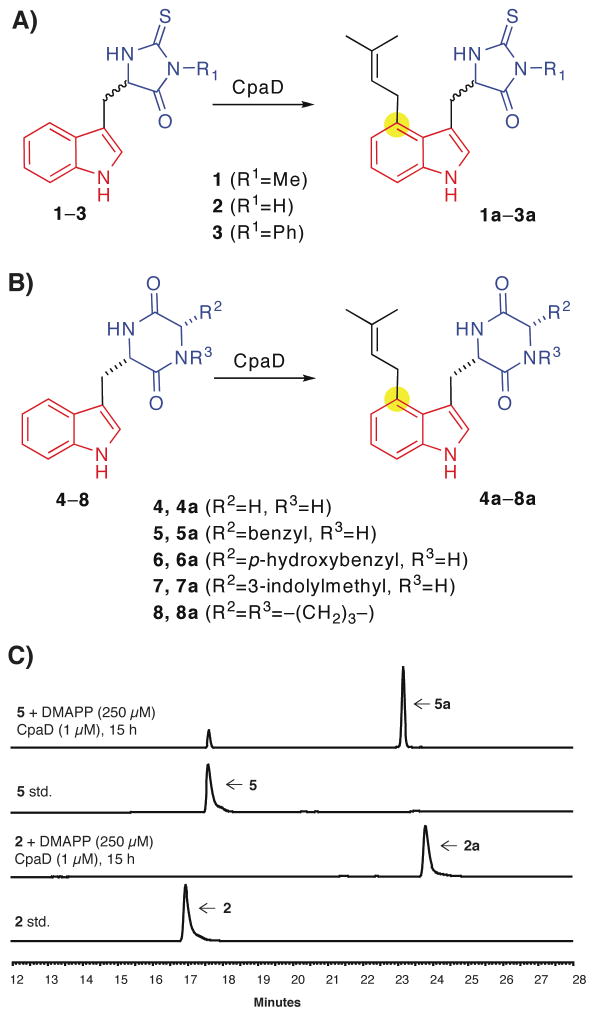
Bearing in mind that the tetramic acid portion of L-cAAtrp is a key motif recognized by CpaD, we turned our initial attention to the readily available tryptophan-containing thiohydantoin derivatives 1–3 (Figure 3A), a family of necroptosis inhibitors (40). We speculated the 5-membered heterocyclic thiohydantoins would serve as a tetramate mimic (Figure 3A). Incubation of 1 with equal molar amount of DMAPP and 0.4 mol% CpaD overnight led to the clean formation of a new peak that corresponds to the dimethylallylated product 1a (Figure 3C, lane 1–2). The dimethylallylation site was unequivocally assigned to be the C-4 position of indole ring by comparing the 1H NMR of 1 with 1a (see methods). Analogously, CpaD also accepts tryptophan thiohydantoins 2 and 3 as substrates and showed slight overall preference for 2 (Table 2, entries 1–3). Subsequent kinetic profiling indicated the KM values of CpaD for 1–3 increased only 3–5 fold in comparison with the native substrate L-cAATrp, but the corresponding kcat values drop 150–380 fold, resulting in a 500–2000 fold decrease in catalytic efficiency (Table 2, entries 1–3). While 1–3 are all racemic, based on our and others’ observations that Trp-DMATs strongly prefer the L-isomer, 1–3 derived from L-Trp are most likely the preferred substrates for CpaD.
Figure 3.
Tryptophan thiohydantoin derivatives (1–3) and tryptophan containing diketopeperazines (DKPs) (4–8) as substrates for CpaD. Schemes illustrating the regioselective prenylation of tryptophan thiohydantoins 1–3 (A) and tryptophan DKPs 4–8 by CpaD (B). HPLC assay demonstrating the conversion of tryptophan thiohydantoin 2 and Trp-Phe DKP 5 to the corresponding prenylated products by CpaD (C).
Table 2.
Kinetic parameters of CpaD with tryptophan thiohydantoins 1–3 and tryptophan containing DKPs 4–8.
| Substrate | Product 1a–8a | Kinetic profile | |||
|---|---|---|---|---|---|
| [M+H]+cal. | [M+H]+obs. | Km (μM) | kcat (min−1) | kcat/Km (min−1 μM−1) | |
| 1 | 328.1484 | 328.1480 | 383 ± 32 | 0.27 ± 0.06 | 7.05 × 10−4 |
| 2 | 314.1327 | 314.1325 | 315 ± 28 | 0.34 ± 0.05 | 10.8 × 10−4 |
| 3 | 390.1640 | 390.1644 | 502 ± 36 | 0.14 ± 0.03 | 2.79 × 10−4 |
| 4 | 312.1712 | 312.1717 | 712 ± 38 | 0.18 ± 0.04 | 2.54 × 10−4 |
| 5 | 402.2182 | 402.2185 | 546 ± 42 | 0.29 ± 0.06 | 5.31 × 10−4 |
| 6 | 418.2131 | 418.2132 | 526 ± 34 | 0.32 ± 0.08 | 6.08 × 10−4 |
| 7 | 441.2291 | 441.2296 | 638 ± 46 | 0.21 ± 0.05 | 3.29 × 10−4 |
| 8 | 352.2025 | 352.2022 | 756 ± 52 | 0.12 ± 0.05 | 1.59 × 10−4 |
Given the above results using hydantoin substrates, the question arose of how the addition of one methylene unit to the 5 membered hydantoin scaffold, resulting in the 6-membered heterocycle diketopiperazine (DKP), would affect recognition by CpaD. The tryptophan containing DKP is one of the most prevalent scaffold elements in fungal prenylated indole alkaloids, including tryptostatin (3), echinullin (41) and fumitremorgin (42). However no natural product with C–4 prenylated Trp–containing DKP moieties have been discovered to date. The five Trp-derived DPKs 4–8 (Figure 3B) were analyzed for activity with CpaD (Table 2, entries 4–8). An HPLC assay using the Trp-Phe DKP 5 and 0.4 mol% CpaD (Figure 3C, lane 3–4) indicated ready conversion to the corresponding dimethylallylated product 5a; the identity of the product and the C-4 regioselectivity of the prenylation site was unambiguously determined by high resolution mass spectrometry (Table 2) and NMR spectroscopy (see methods). However, kinetic analysis indicates that CpaD disfavors DKP substrates over hydantonins as the catalytic efficiency towards enantiopure L-Trp-Gly DKP 4 is only quarter of that to racemic thiohydantoin 2 (Table 2). Introduction of aromatic residues to the inserted methylene unit, corresponding to the L-Trp-L-Phe 5, L-Trp-L-Tyr 6 and L-Trp-L-Trp 7 DKPs, increased the overall efficiency slightly to 110%, 140% and 30%, respectively, that of the L-Trp-Gly DKP 4 (Table 2, 5–7). The constrained DKP L-Trp-L-Pro 8 was the least favorable substrate for CpaD among the five DKPs tested. Although the overall catalytic efficiencies of CpaD toward Trp-derived DKPs are not high, they are slightly superior to FgaPT2, a genuine C-4 Trp-DMAT that was recently shown to accept DKPs as substrates. Another C-4 Trp-DMAT, MaPT, was also shown to accept DKP 8 as a substrate (23); however, we could not carry the comparison further as MaPT has not been kinetically characterized for 8.
Table 5.
Relative activity profiles of aromatic CpaD mutants targeted by examination of conserved motifs in the Trp-DMAT family.a
| Entry | CpaD protein | virel |
|---|---|---|
| 1 | Y193F | (5.3 ± 0.6) × 10−2 |
| 2 | Y262F | (3.6 ± 0.8) × 10−4 |
| 3 | Y346F | (4.9 ± 0.3) × 10−4 |
| 4 | Y410F | 0.12 ± 0.02 |
| 5 | Y414F | (2.8 ± 0.4) × 10−4 |
| 6 | Y193L | (4.3 ± 0.3) × 10−4 |
| 7 | Y410L | (7.2 ± 0.6) × 10−4 |
| 8 | W33F | 0.42 ± 0.06 |
| 9 | W33L | –– |
| 10 | W299F | 0.92 ± 0.07 |
| 11 | W299L | 0.38 ± 0.05 |
virel is the relative observed maximum initial velocity of each mutant CpaD to that of wild type, which is defined as 1. W33L could not be overproduced in soluble form for characterization.
Activity of CpaD towards tryptophan-containing peptides
In contrast to the prevalence of prenylated cyclic tryptophan containing peptides, mostly as DKPs, the occurrence of prenylated tryptophan-containing linear peptides are rather rare in nature (43). 1 Several characterized Trp-DMATs have been shown to have broad substrate specificities to tailor L-Trp and L-Trp-containing DKP scaffolds (44); however, and rather surprisingly, they all have considerable tryptophan aminopeptidase activity that render them unsuitable for chemoenzymatic preparation of L-Trp-containing linear peptides (45).
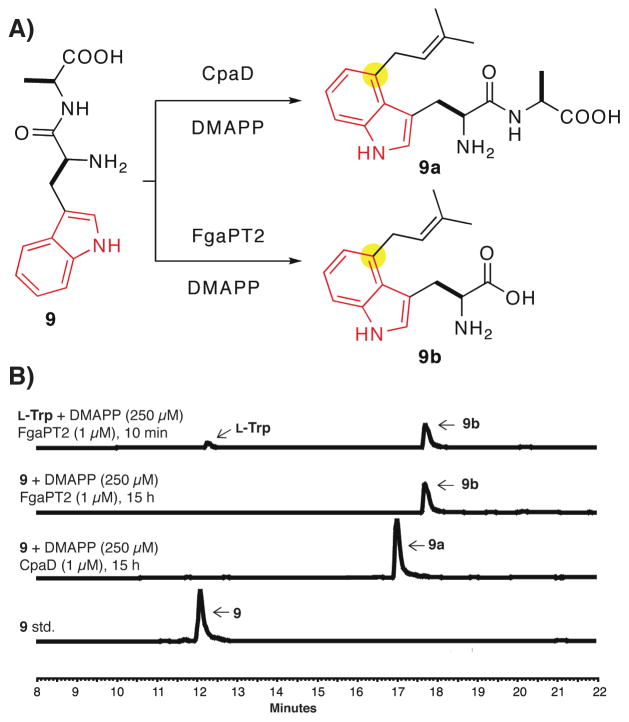
As an initial test, the dimethylallylation activity of CpaD was evaluated towards the dipeptide H-Trp-Ala-OH 9, a superior aminopepidase substrate for several Trp-DMATs characterized previously. With assay conditions analogous to those for Trp-containing thiohydantoins and DKPs, CpaD effected the quantitative conversion of H-Trp-Ala-OH 9 to the corresponding C-4 Trp dimethylallylated dipeptide 9b (Figure 4), which was verified by high resolution MS (Table 3) and 1H NMR (see methods). It is noteworthy that CpaD did not show measurable peptidase activity towards 9 as had been observed with other Trp-DMATs including 7-DMATS, FgaPT1, CdpNPT, and FtmPT1. To further examine if this is unique to CpaD as a C-4 Trp-DMAT, we cloned and overproduced FgaPT2, a known C-4 Trp-DMAT that accepts L-Trp as genuine substrate with negligible aminopeptidase activity as originally reported by Li and coworkers (45). Incubation of FgaPT2 with H-Trp-Ala-OH 9 under the identical conditions for CpaD resulted in a new peak by HPLC analysis that corresponds to C-4 dimethylallylated L-Trp 9b (Figure 4) as verified by MS analysis and HPLC co-elution with authentic 9b generated from L-Trp and DMAPP with FgaPT2. This result suggests FgaPT2 may also harbor aminopeptidase activity that degrades 9 to L-Trp, which is preferentially prenylated by FgaPT2.
Figure 4.
N-terminal tryptophan containing dipeptides 9–14 as substrates for CpaD. (A) Schemes illustrating the different reaction outcomes catalyzed by CpaD and FgaPT2 when using H-Trp-Ala-OH 9 as substrate. (B) HPLC traces showing the effective conversion of the dipeptide H-Trp-Ala-OH 9 to the corresponding prenylated product 9a by CpaD compared to FgaPT2 that gave 9b.
Table 3.
Activity profiles of CpaD with N-terminal Trp-containing dipeptides.
| Substrate | Product Identification | observed vi (min−1)a | |
|---|---|---|---|
| [M+H]+cal. | [M+H]+obs. | ||
| 9 (H-Trp-Ala-OH) | 344.1974 | 344.1977 | 0.34 ± 0.06 |
| 10 (H-Trp-Gly-OH) | 330.1818 | 330.1812 | 0.16 ± 0.05 |
| 11 (H-Trp-Val-OH) | 304.1661 | 304.1665 | 0.31 ± 0.07 |
| 12 (H-Trp-Leu-OH) | 386.2444 | 386.2449 | 0.18 ± 0.04 |
| 13 (H-Trp-Phe-OH) | 352.1661 | 352.1666 | 0.11 ± 0.03 |
| 14 (H-Trp-Tyr-OH) | 368.1610 | 368.1613 | 0.22 ± 0.05 |
Saturation kinetics could not be achieved even with 1 mM of 9–14 as substrates, therefore observed maximum initial velocity for these substrates is shown.
Five additional N-terminal Trp-containing dipeptides 10–14 were evaluated and confirmed to be CpaD substrates by combined LC-MS analysis of the prenylated dipeptide products (Table 3), whereas FgaPT2 affords the amino acid monomer 9b as the dominant product in all cases. Further kinetic characterization showed that disruption of the cyclic DKP scaffold to the linear dipeptide clearly decreased the binding affinity to CpaD. Saturation kinetics could not be obtained even at 1 mM of the dipeptide substrates. The apparent turnover numbers were therefore estimated for comparing the relative catalytic efficiency of CpaD toward individual dipeptides (Table 3). Among the six substrates 9–14 tested, H-Trp-Ala-OH 9 was most efficiently converted to the corresponding product by CpaD with an observed kcat of 0.34 min−1, whereas H-Trp-Phe-OH 13 was least efficient with an observed kcat of 0.11 min−1. It is not clear from this set of experiments what is an optimized dipeptide sequence for CpaD and will be a subject for future investigation.
It appears that CpaD strictly requires the tryptophan residue in the N-terminus of the dipeptide for prenylation since H-Ala-Trp-OH, H-Gly-Trp-OH, H-Val-Trp-OH (Figure S1, supporting information), gave only trace amounts of dimethyallylated H-Ala-Trp-OH and H-Gly-Trp-OH by LC-MS analysis under the same assay conditions used for H-Trp-Ala-OH 9. There was no detectable conversion for H-Val-Trp-OH. Surprisingly, the dominant product observed for H-Ala-Trp-OH and H-Gly-Trp-OH assay with CpaD was the released monomeric L-Trp. This result is in contrast to the previous observation that dipeptides with a C-terminal tryptophan are not preferred substrates for the aminopeptidase activity of other Trp-DMAT (45). However, we cannot conclude that this observed activity is inherent in CpaD without further detailed investigation.
CpaD was also able to dimethylallylate N-terminal tryptophan-containing oligopeptides up to the pentapeptide H-Trp-(Ala)4-OH (data not shown). However, the catalytic efficiency is much lower than that for the dipeptide H-Trp-(Ala)-OH, and the reaction was accompanied by competitive degradation of the oligopeptides. Future investigation will be directed toward the detailed examination of the possible peptidase activity versus prenylation activity of CpaD, which may provide the opportunity to use CpaD as a promiscuous catalyst for labeling of peptide substrates for biomedical applications.
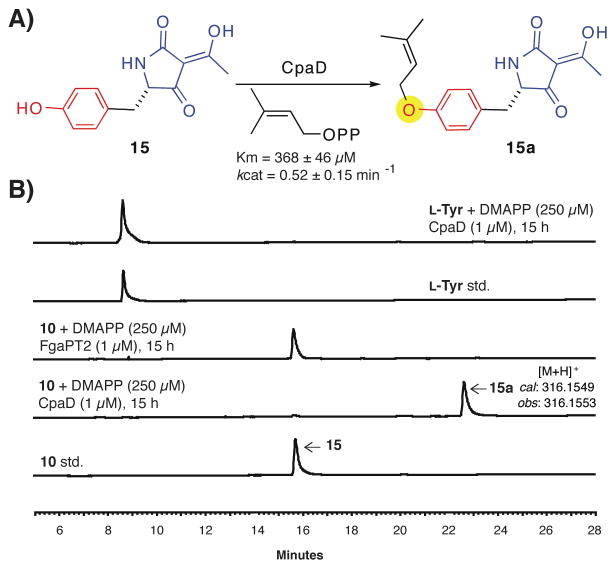
Activity of CpaD towards aromatic acid amino acid-derived tetramic acids
Since it is evident that the tetramic acid core in cAATrp is an important determinant for CpaD recognition, we turned to replacement of the tryptophan moiety in cAATrp with other aromatic scaffolds derived from natural amino acids. As the putative tyrosine O-DMAT gene sidD from sirodesmin PL biosynthesis shares good sequence homology (ca. 34%) with the Trp-DMAT 7-DMATS (21), we synthesized the tyrosine-derived tetramic acid 15 (see methods) and tested it as a substrate for CpaD (Figure 5). Incubation of 15 with an equal molar amount of DMAPP and 0.4 mol% CpaD overnight led to clean generation of a new peak by HPLC analysis (Figure 5B, lane 1–2), which was verified to be O-dimethylallylated 15a by high resolution MS and 1H NMR (see methods). Subsequent kinetic analysis indicates CpaD possesses reasonable kinetic parameters towards tyrosine tetramic acid 15 with KM ≈ 368 μM and kcat ≈ 0.52 min−1 (Figure 5A). The resulting catalytic efficiency is ca. 1.41 min−1 μM−1 and slightly superior to the racemic tryptophan thiohydantoin 2, which is the second best Trp-containing substrate for CpaD after cAATrp. The observed tyrosine O-dimethylallylation activity is unique to CpaD as FgaPT2 did not show any activity toward 15 under the identical assay condition (Figure 5B, lane 3–5). CpaD strictly requires the presence of the tetramic acid modification, as L-tyrosine alone is not a substrate for CpaD. In addition, we prepared the phenylalanine tetramic acid analogue, however it was not accepted as a substrate for C-prenylation by CpaD (data not shown).
Figure 5.
Tyrosine derived tetramic acid is a substrate for CpaD. (A) Scheme illustrating the CpaD–catalyzed O–prenylation of the tyrosine derived tetramic acid 15 to give 15a. (B) HPLC traces showing the effective conversion of 15 to 15a catalyzed by CpaD, but not FgaPT2. Unmodified L-tryrosine is not a substrate for CpaD.
The N-terminal domain of CpaD is crucial for dimerization and activity
The truncated and complete CPA biosynthetic genes cluster in A. oryzae RIB40 and A. flavus NRRL3357, respectively, were originally identified by a genome mining approach using a putative CpaD sequence from A. oryzae A1560 strain (26, 27). Our initial efforts to verify the function of CpaD by heterologous expression and in vitro reconstitution utilized the CpaD from A. oryzae RIB40 that is identical in amino acid sequence to that from A. oryzae A1560. However, examination of the CpaD sequence from A. flavus NRRL3357, (referred to here as CpaD_Af) as annotated in the Aspergillus Comparative Database at Broad Institute (Figure S2, supporting information), revealed that it shares 92% amino acid sequence identity to CpaD in A. oryzae RIB40 but is truncated by 43 amino acid residues from the N-terminus. This observation prompted us to question the consequence of the missing N-terminal sequence and if the originally assigned CpaD from A. flavus NRRL3357 is functional.
The overproduction of CpaD_Af in E. coli was lackluster (see methods). The dominant fraction of the overproduced CpaD_Af protein is insoluble and the yield of soluble protein is ca. 0.2 mg/L (Figure S3, supporting information), corresponding to less than 1% of that observed for the CpaD from A. oryzae RIB40 (CpaD_Ao). Moreover, the FPLC-purified CapD_Af appears monomeric in solution, in contrast to the dimeric CpaD_Ao, and did not shown any activity when incubated with synthetic cAATrp and DMAPP (Figure S3-B, supporting information). This outcome led us to speculate that the missing N-terminal sequence was required for activity and dimerization of CpaD. After careful examination of the 5′ region of cpaD in the A. flavus NRRL3357 genome, we found an adjacent nucleotide sequence that would encode for the missing 43 amino acid residues. Addition of this sequence to the original CpaD_Af resulted in overexpression and purification of a full length and functional CpaD that is dimeric in solution (Figure S3, supporting information).
The observation that deletion of the N-terminal 43 amino acids of CpaD disrupts proper folding/oligomerization in solution and results in inactive CpaD is noteworthy, as no report exists to date to correlate the oligomeric structures of fungal Trp-DMATs with their catalytic activities. To further correlate this observation to the entire family of characterized fungal Trp-DMATs, we initiated an effort to identify conserved motifs that are consequential to Trp-DMAT activity.
Identification of a conserved set of residues across the Trp-DMAT family that are consequential to CpaD activity
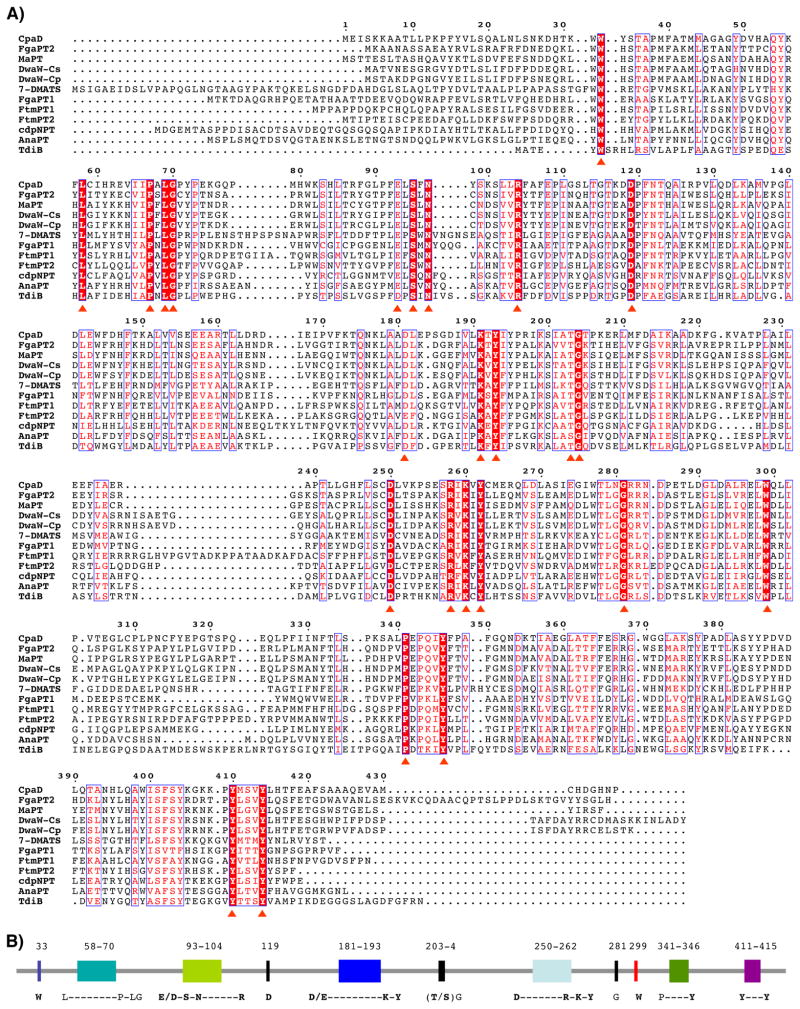
In order to identify possible conserved motifs within the fungal Trp-DMAT family, we carried out the sequence alignment of all in vitro characterized Trp-DMATs. Including CpaD, nine other Trp-DMATs have been characterized, which cover enzymatic dimethylallylation of indole backbones in L-Trp, L-Trp-derived DKP (24), -benzodiazepinone (20), -tetramic acid (29), and -benzoquinone (19, 46) at N-1, C-2, C-3, C-4, and C-7 positions. Although no C-5 and C-6 Trp-DMAT has been identified to date, the characterized Trp-DMATs represent sufficient diversity to allow identification of conserved motifs. The aromatic prenyltransferses Orf2 (47) and CloQ (48) were not included in this alignment effort, as their sequence homologies with Trp-DMATs are very low (sequence identity <15%). Previous mutagenesis studies in the context Trp-DMAT MaPT and FgaPT2 based on their homologies with Orf2 only generated limited results that two lysine residues are catalytically relevant to their activities (23, 49). Multiple alignments were carried out using ClustalW (http://www.ebi.ac.uk/clustalw/) and subsequently visualized with ESPript (http://espript.ibcp.fr/) (Figure 6A). The alignment results showed 24 strictly conserved amino acid residues that cluster into six recognizable regions of the Trp-DMAT sequences (Figure 6B).
Figure 6.
Identification of conserved motifs in the Trp-DMAT family. (A) ClustalW alignment of all functionally validated Trp-DMATs. Red stars indicate exclusively conserved amino acid residues. (B) Representation of conserved motifs within the Trp-DMAT family. Numbers correspond to the amino acid residues in CpaD. The bold amino acid residues correspond to those subjected to mutagenesis.
In studies related to trans-/cis- prenyltransferases and terpene cyclases, the critical roles of conserved polar residues (Asp, Glu, Arg, Lys) in substrate pyrophosphate binding and aromatic residues (Trp, Phe, Tyr) in the stabilization and control of cyclization as well as determination of the prenyl product chain length, have been well documented and supported by crystallography and comprehensive mutagenesis (34–36, 50). Inspection of the Trp-DMAT family alignment revealed 11 conserved charged/polar residues and 7 conserved aromatic residues (Trp, Tyr). These residues thus became primary targets for subsequent mutagenesis studies in CpaD. We measured the initial maximum velocity of each mutant in comparison to wild type CpaD to determine if the mutation of the selected residue is consequential.
The eleven conserved polar/charged residues were first examined by mutating them to structurally neutral (D/E/N→L, S/T→A/V) or reverse charged (R/K→D) residues (as they may be involved in substrate binding of cAATrp and DMAPP). The targeted CpaD polar/charged residue mutants (Table 4) were overproduced as highly soluble in E. coli as CpaD wild type and dimeric in solution as estimated by size exclusion chromatography, with the exception of CpaD(D119L) which was totally insoluble. Among the ten isolated polar/charged CpaD mutants (Table 4), the activities of E93L, R104E, K191E, D250L and K260E dropped notably in a range of 20–3000 fold in comparison to wild type. These results imply that these residues are relevant to the activity of CpaD. The remaining five mutants, S95A, N97L, D181L, T203V, and R258E, did not show significant activity changes as compared to wild type.
Table 4.
Relative activity profiles of polar/charged CpaD mutants targeted by examination of conserved motifs in the Trp-DMAT family.a
| Entry | CpaD protein | virel |
|---|---|---|
| 1 | E93L | (6.8 ± 0.7) × 10−4 |
| 2 | S95A | 0.92 ± 0.11 |
| 3 | N97L | 0.63 ± 0.15 |
| 4 | R104E | (2.1 ± 0.3) × 10−2 |
| 5 | D119L | –– |
| 6 | D181L | 0.47 ± 0.03 |
| 7 | K191E | (2.7 ± 0.5) × 10−4 |
| 8 | T203V | 0.96 ± 0.08 |
| 9 | D250L | (5.1 ± 0.6) × 10−2 |
| 10 | R258E | 0.92 ± 0.12 |
| 11 | K260E | (3.2 ± 0.4) × 10−4 |
virel is the relative observed maximum initial velocity of each mutant CpaD to that of wild type, which is defined as 1. Mutants in bold are those with a notable decrease in relative activity. D119L could not be overproduced in soluble form for characterization.
In two separate studies by the groups of Sherman and Li, the MaPT D181, K191, K260 mutants (23), and the FgaPT2, FtmPT1 and 7-DMATS K191, D250, R258, and K260 mutants (49) were studied.2 Our observations on the critical roles of the two conserved lysine residues K191 and K260 and the irrelevant role of aspartate D181 in CpaD are in accordance with the observations of both Sherman and Li. However, the roles of D250 and R258 appeared somewhat different between CpaD, FgaPT2, FtmPT1 and 7-DMATS. While the CpaD mutant D250L showed ca. 5% activity of wild type, the corresponding D→H mutants in FtmPT1 and 7-DMATS showed ca. 30% activities of wild type, suggesting polarity at this position is important. In another case, the CpaD mutant R258E showed nearly identical activity to wild type, while the corresponding R→G mutants in FgaPT2, FtmPT1 and 7-DMATS showed ca. 5%, 48% and 37% activities compared to wild type, respectively. These discrepancies likely arise from the differences of the corresponding mutated residues limiting our ability to do a strict comparison. However, these discrepancies also highlight the limitations of using relative rate to describe the catalytic relevance of certain residues to enzyme activity. A more comprehensive approach with measurement of KM and kcat for all the substrates involved would likely be more informative albeit beyond the scope of this study.
Nevertheless, our observation that the E93L and R104E mutations in CpaD led to 1500 and 50 fold decreases in activity, repsectively, is noteworthy as these residues had not been identified before by sequence comparison with aromatic prenyltransferases Orf2 and CloQ. (23, 49) The magnitude of the decrease in activity of the E93L mutant is on par with those observed for the K191L and K260L mutants, suggesting E93 is of similar importance. Whether or not these residues are also relevant to other types of Trp-DMATs will need to be justified by further mutagenesis studies and/or crystal structure determination.
We next turned our attention to the seven conserved aromatic residues including two tryptophans and five tyrosines. The importance of aromatic residues in stabilization and control of the carbocation intermediates in enzyme-catalyzed transformations is well documented (51), in particular in the context of trans- and cis- prenyltransferases and related terpene cyclases (34–36, 50, 52–55). Enzymatic dimethylallylation of tryptophan has been characterized as an electrophilic aromatic substitution event mediated by Trp-DMAT (56), which may well require the presence of aromatic amino acid residues in the active site. Careful inspection of the crystal structure of Orf2 (47), a related aromatic prenyltransferase, suggests the presence of tyrosine residues in the pyrophosphate-binding pocket. With this hypothesis in mind, a two-step mutagenesis approach was designed. First, the conserved Tyr and Trp were mutated to Phe. Mutation of tyrosine residues that are directly involved in pyrophosphate binding should lead to a dramatic activity decrease, analogous to what was observed with the two conserved Lys residues. On the other hand, if the primary role of the conserved Trp and Tyr residues is to stabilize the cationic prenyl intermediate, we expect the Trp/Tyr→Phe will cause only moderate activity loss due to the comparable aromatic nature of Phe. As shown in Table 5, 2000–3000 fold activity loss was observed in comparison with wild type for CpaD mutants Y262F, Y346F, Y414F (entries 1–3). However, only 10–20 fold losses were observed in CpaD mutants Y193F and Y410F (entries 4–5), suggesting that Y193 and Y410 are involved in stabilizing the transient carbocation intermediate. As a second step, mutation of Y193 and Y410 to Leu (Table 5, entries 6–7) resulted in an additional >100 fold decrease in activity relative to wild type, implicating that the aromaticity of Y193 and Y410 is more critical to the activity of CpaD. While no significant changes occurred when mutating W33 and W299 to Phe and Leu (Table 5, entries 8–11), it is important to point out that the W33F mutant could not be overproduced with solubility equal to the other mutants (see methods). The W33L mutant was completely insoluble in E. coli and could not be isolated for characterization. (Table 5, entries 8–9).
DISCUSSION
Filamentous fungi are prolific natural product producers that generate an astonishing array of bioactive substances, ranging from therapeutically important penicillin antibiotics, cholesterol-lowering drug statins to notorious mycotoxins aflatoxin and cyclopiazonic acid (57). The prenylated indole alkaloid framework represents a distinctive branch of fungal natural products, which contain unique and complex molecular architectures with rich bioactive profiles. Regioselective dimethylallylation of indole scaffolds by Trp-DMATs is a key step in such fungal alkaloid biosynthesis (12). This modification can occur very early in the biosynthetic pathway; for example, C-4 dimethylallyation of free L-Trp is the first committed step in ergot alkaloid biosynthesis. More frequently, prenylation occurs during the later stages of biosynthesis after assembly of tryptophan-containing cyclic peptides or peptide-polyketide hybrids by NRPS and/or PKS, exemplified in the biosyntheses of fumitremorgin B (58), terriquione A (19) (46) and cyclopiazonic acid as described in this study. These resulting dimethylallyl decorations can further undergo enzyme-mediated oxidations and/or cyclizations to generate the condensed polycyclic scaffolds observed in many fungal prenylated natural products, such as the conversion of tricyclic β-CPA to pentacyclic α-CPA (Figure 1) (12).
Recent advances in fungal genome sequencing, combined with the knowledge gained during in vitro study of DmaW (15, 16), a C-4 Trp-DMAT involved in ergot alkaloid biosynthesis, has resulted in the subsequent identification and characterization of a number of fungal Trp-DMATs with substrate diversities ranging from free L-Trp to L-Trp-derived cyclic scaffolds arising from the NRPS assembly lines (24). While accumulating biochemical data on Trp-DMATs indicate their activities are independent of divalent ion cofactors that distinguish them as a distinct subset of prenyltransferases, fundamental questions remain on how they select Trp-containing substrates and perform regioselective dimethylallylations. Prior efforts using characterized Trp-DMATs as promiscuous biocatalysts have shown some promise for chemoenzymatic preparations of regiospecifically prenylated L-Trp and L-Trp- containing DKP derivatives (44); however, their apparently inherent aminopeptidase activity has limited their utility in dimethylallylated linear L-Trp peptides (45).
Cyclopiazonic acid (CPA) is a mycotoxin that selectively blocks the sarcoplasmic reticulum Ca2+ ATPase with nanomolar potency (25). Isolated from various Aspergillus and Penicillium species, the α-CPA end product has a rigid pentacyclic indole tetramic acid scaffold. α-CPA arises from a short three-enzyme pathway, making it unique both structurally and biosynthetically among the fungal prenylated alkaloids (26, 27). The first CPA pathway-specific enzyme is the hybrid PKS-NRPS CpaS, where the C- terminal redox-incompetent reductase serves as a Dieckmann cyclization catalyst to release L-cAATrp (29). Regioselective dimethylallylation of L-cAATrp at C4 of the indole ring by the action of the DMAT CpaD constitutes the second pathway-specific step, and the resulting β-CPA is further oxidatively tailored by the flavoenzyme CpaO to form two rings in one step.
In this study, we first cloned CpaD from A. oryzae RIB40, heterelogously overexpressed it in E. coli, and obtained soluble homodimeric enzyme in good yield. Synthetic L-cAATrp was validated as a substrate for CpaD with a KM of 109±16 μM and a kcat of 53±12 min−1. While CpaD showed clear kinetic preference for L-cAATrp over D-cAATrp in a 24 fold difference in catalytic efficiency, it virtually does not accept L-Trp as substrate with an estimated kcat/KM < 4 ×10−6 μM−1min−1, more than a 105 fold decrease from L-cAATrp. This observation distinguishes CpaD from the Trp-DMATs FtmPT1 and CdpNPT that utilize Trp-containing DKPs as their native substrate (18, 22) yet readily modify free L-Trp (37). This highlights the critical importance of the tetramic acid ring as a recognition element in cAATrp for CpaD catalysis.
Subsequent exploration of substrate promiscuity for CpaD started with Trp-containing thiohydantoins as mimics of the five-membered heterocyclic tetramic acid, then the Trp-containing DKPs with a six-membered cyclic peptide scaffold, and ultimately to linear Trp-containing dipeptides. Kinetic studies showed a trend of decreased catalytic efficiency on these substrate analogues for CpaD compared to the tetramic acid moiety in L-cAATrp. Despite the diminished efficiency of CpaD toward Trp-containg thiohydantoins, DKPs, and linear dipeptides, synthetically useful conversion rates (60–100%) were obtained when using 0.4 mol% CpaD over a 24-hour incubation period, making CpaD a candidate biocatalyst for chemoenzymatic preparations of C-4 dimethylallyated Trp-containing molecules with similar scaffolds. Moreover, CpaD showed negligible aminopeptidase activity toward N-terminal Trp-containing dipeptides in the absence of divalent cation, in contrast to other fungal Trp-DMATs, and thus allows access to several C-4 dimethylallyled Trp-containing dipeptides. It is also noteworthy that CpaD is able to dimethylallylate the pentapeptide H-Tpr-(Ala)4-OH albeit with competitive degradation of the starting peptide. Minimization of Trp-DMAT aminopeptidase activity will likely expand the utility of this family of enzymes. Indication of the tetramic acid as a substrate recognition element was demonstrated by the ability of CpaD to O-prenylate rather than C-prenylate during generation of the O-dimethylallyl tyrosine-derived tetramic acid.
We have shown that truncation of the N-terminal CpaD sequence is detrimental to activity in an effort to verify the function of the erroneously predicted sequence of CpaD from A. flavus NRRL3357. This subsequently led us to compare all characterized Trp-DMATs in search of conserved sequence motifs. A sequence alignment using ClustalW revealed 24 strictly conserved amino acid residues in six recognizable motifs across the Trp-DMAT family. Based on prior studies on the related trans-, cis-prenyltransferases and terpene cyclases, we chose eleven conserved polar/charged residues and seven conserved aromatic residues to mutagenize and assay for CpaD activity. CpaD mutants generated from five polar/charged residues (E93L, R104E, K191E, D250L and K260E) and five tyrosine residues (Y193L, Y262F, Y346F, Y410L, Y414F) led to 20–3000 fold reductions in activity compared to wild type, implicating these residues in binding with either substrate and/or stabilization of the carbocation intermediate. These ten residues are evenly distributed in five of the six conserved motifs and will serve as signatures for gene probes to identify new fungal Trp-DMATs that are involved in as of yet uncharacterized prenylated indole alkaloid biosynthesis.
During the preparation of this manuscript, the crystal structure of the C-4 Trp-DMAT FgaPT2 was reported (59). Our mutagenesis efforts leading to the identification of the above ten residues in CpaD largely correlate with predictions enabled by the FgaPT2 structure. E93 in CpaD is likely involved in the binding of the L-cAATrp Trp-N-1 atom analogous to the E89 residue observed for FgaPT2, whereas the remaining polar/charged residues R104, K191, D250, K260 and tyrosine residues Y193, Y262, Y346, Y410, Y414 are likely located in the binding pocket of DMAPP. The FgaPT2 structure and the comprehensive mutagenesis efforts described here provide insight into a common binding mode for the DMAPP prenyl donor. However, it is still unresolved how Trp-DMAT family members differentiate between tryptophan substrates, direct regioselectivity and selectively utilize the C5 prenyl donor, emphasizing the value of additional structural and biochemical characterization. In the related families of trans-, cis-prenyltransferases and terpene cyclases, such combined efforts have led to a deeper understanding of specificity and mechanism (34–36, 50, 60–63). Given the architectural complexity of the prenylated indole natural product scaffolds and their diverse biologic activities, a comparable set of insights into this subclass of prenyl transferases would prove useful during efforts to reprogram these biosynthetic pathways.
The intriguing substrate promiscuity of CpaD warrants a more detailed understanding of how CpaD functions at the molecular level, especially in comparison to other Trp-DMATs, which will enable future engineering efforts to improve its inherent utility as a biocatalyst. Additionally, further understanding of characteristics critical to Trp-DMATs as a family, such as the characterization of conserved sequence motifs described here, will facilitate the discovery of related prenylation enzymes and new natural products.
Supplementary Material
Acknowledgments
We thank Dr. Kevin McCluskey (FGSC) and Dr. Jiujiang Yu (USGA) for providing the NRRL3357 strain and its genomic DNA, Prof. Robert Williams (Colorado State University) for providing brevianamide F and Dr. Heidi Imker for proofreading.
Abbreviations
- CPA
cyclopiazonic acid
- cAATrp
cyclo-acetoacetyl-L-tryptophan
- PKS
polyketide synthase
- NRPS
nonribosomal peptide synthetase
- DMAPP
dimethylallyl pyrophosphate, IPP, isopentenyl pyrophosphate
- GPP
geranyl pyrophosphate
- FPP
farnesyl pyrophosphate
- GGPP
geranylgeranyl pyrophosphate
- DKP
diketopiperazine
- DMAT
dimethylallyltransferase
Footnotes
The only known example is the Bacillus subtilis quorum-sensing peptide pheromone ComX, where the C-3 position of Trp is farnesylated or geranylated (44).
The amino acid numbers quoted here are in reference to the positions in CpaD as shown in the alignment in Figure 6A.
This work was supported in part by National Institute of Health Grant GM20011 (to C.T.W.) and Swiss National Science Foundation and Ernst Schering Foundation (postdoctoral fellowships to X.L.).
Supporting Information Available: Tables S1–S3, Scheme S1 and Figures S1–S3. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Aniszewski T. Alkaloids - Secrets of Life: Alkaloid Chemistry, Biological Significance, Applications and Ecological Role. Elsevier Science; Amsterdam: 2007. [Google Scholar]
- 2.Haarmann T, Rolke Y, Giesbert S, Tudzynski P. Ergot: from witchcraft to biotechnology. Mol Plant Pathol. 2009;10:563–577. doi: 10.1111/j.1364-3703.2009.00548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui CB, Kakeya H, Okada G, Onose R, Osada H. Novel mammalian cell cycle inhibitors, tryprostatins A, B and other diketopiperazines produced by Aspergillus fumigatus. I. Taxonomy, fermentation, isolation and biological properties. J Antibiot. 1996;49:527–533. doi: 10.7164/antibiotics.49.527. [DOI] [PubMed] [Google Scholar]
- 4.Qian-Cutrone J, Huang S, Shu Y-Z, Vyas D, Fairchild C, Menendez A, Krampitz K, Dalterio R, Klohr SE, Gao Q. Stephacidin A and B: two structurally novel, selective inhibitors of the testosterone-dependent prostate LNCaP cells. J Am Chem Soc. 2002;124:14556–14557. doi: 10.1021/ja028538n. [DOI] [PubMed] [Google Scholar]
- 5.Shoop WL, Haines HW, Eary CH, Michael BF. Acute toxicity of paraherquamide and its potential as an anthelmintic. Am J Vet Res. 1992;53:2032–2034. [PubMed] [Google Scholar]
- 6.Gallagher RT, Clardy J, Wilson BJ. Aflatrem, a tremorgenic toxin from Aspergillus flavus. Tetrahedron Lett. 1980;21:239–242. [Google Scholar]
- 7.Holzapfel CW. Isolation and Structure of Cyclopiazonic Acid a Toxic Metabolite of Penicillium Cyclopium Westling. Tetrahedron. 1968;24:2101–2119. doi: 10.1016/0040-4020(68)88113-x. [DOI] [PubMed] [Google Scholar]
- 8.Saikia S, Nicholson MJ, Young C, Parker EJ, Scott B. The genetic basis for indole-diterpene chemical diversity in filamentous fungi. Mycol Res. 2008;112:184–199. doi: 10.1016/j.mycres.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 9.He J, Wijeratne EMK, Bashyal BP, Zhan J, Seliga CJ, Liu MX, Pierson EE, Pierson LS, Van Etten HD, Gunatilaka AAL. Cytotoxic and other metabolites of Aspergillus inhabiting the rhizosphere of Sonoran desert plants. J Nat Prod. 2004;67:1985–1991. doi: 10.1021/np040139d. [DOI] [PubMed] [Google Scholar]
- 10.Brewer D, Jerram WA, Meiler D, Taylor A. The toxicity of cochliodinol, an antibiotic metabolite of Chaetomium spp. Can J Microbiol. 1970;16:433–440. doi: 10.1139/m70-074. [DOI] [PubMed] [Google Scholar]
- 11.Mocek U, Schultz L, Buchan T, Baek C, Fretto L, Nzerem J, Sehl L, Sinha U. Isolation and structure elucidation of five new asterriquinones from Aspergillus, Humicola and Botryotrichum species. J Antibiot. 1996;49:854–859. doi: 10.7164/antibiotics.49.854. [DOI] [PubMed] [Google Scholar]
- 12.Williams R, Stocking E, Sanz-Cervera J. Biosynthesis of prenylated alkaloids derived from tryptophan. Top Curr Chem. 2000;209:97–173. [Google Scholar]
- 13.Williams RM, Cox RJ. Paraherquamides, brevianamides, and asperparalines: laboratory synthesis and biosynthesis. An interim report. Acc Chem Res. 2003;36:127–139. doi: 10.1021/ar020229e. [DOI] [PubMed] [Google Scholar]
- 14.Lee S-L, Floss HG, Heinstein P. Purification and properties of dimethylallylpyrophosphate:tryptopharm dimethylallyl transferase, the first enzyme of ergot alkaloid biosynthesis in Claviceps. sp. SD 58. Arch Biochem Biophys. 1976;177:84–94. doi: 10.1016/0003-9861(76)90418-5. [DOI] [PubMed] [Google Scholar]
- 15.Tsai HF, Wang H, Gebler JC, Poulter CD, Schardl CL. The Claviceps purpurea gene encoding dimethylallyltryptophan synthase, the committed step for ergot alkaloid biosynthesis. Biochem Biophys Res Commun. 1995;216:119–125. doi: 10.1006/bbrc.1995.2599. [DOI] [PubMed] [Google Scholar]
- 16.Gebler JC, Poulter CD. Purification and characterization of dimethylallyl tryptophan synthase from Claviceps purpurea. Arch Biochem Biophys. 1992;296:308–313. doi: 10.1016/0003-9861(92)90577-j. [DOI] [PubMed] [Google Scholar]
- 17.Unsold IA, Li SM. Reverse prenyltransferase in the biosynthesis of fumigaclavine C in Aspergillus fumigatus: Gene expression, purification, and characterization of fumigaclavine C synthase FGAPT1. ChemBioChem. 2006;7:158–164. doi: 10.1002/cbic.200500318. [DOI] [PubMed] [Google Scholar]
- 18.Grundmann A, Li SM. Overproduction, purification and characterization of FtmPT1, a brevianamide F prenyltransferase from Aspergillus fumigatus. Microbiology. 2005;151:2199–2207. doi: 10.1099/mic.0.27962-0. [DOI] [PubMed] [Google Scholar]
- 19.Schneider P, Weber M, Hoffmeister D. The Aspergillus nidulans enzyme TdiB catalyzes prenyltransfer to the precursor of bioactive asterriquinones. Fungal Genet Biol. 2008;45:302–309. doi: 10.1016/j.fgb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Yin WB, Grundmann A, Cheng J, Li SM. Acetylaszonalenin biosynthesis in Neosartorya fischeri. Identification of the biosynthetic gene cluster by genomic mining and functional proof of the genes by biochemical investigation. J Biol Chem. 2009;284:100–109. doi: 10.1074/jbc.M807606200. [DOI] [PubMed] [Google Scholar]
- 21.Kremer A, Westrich L, Li SM. A 7-dimethylallyltryptophan synthase from Aspergillus fumigatus: overproduction, purification and biochemical characterization. Microbiology. 2007;153:3409–3416. doi: 10.1099/mic.0.2007/009019-0. [DOI] [PubMed] [Google Scholar]
- 22.Ruan HL, Yin WB, Wu JZ, Li SM. Reinvestigation of a cyclic dipepticle N-prenyltransferase reveals rearrangement of N-1 prenylated indole derivatives. ChemBioChem. 2008;9:1044–1047. doi: 10.1002/cbic.200700723. [DOI] [PubMed] [Google Scholar]
- 23.Ding YS, Williams RM, Sherman DH. Molecular analysis of a 4-dimethylallyltryptophan synthase from Malbranchea aurantiaca. J Biol Chem. 2008;283:16068–16076. doi: 10.1074/jbc.M801991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steffan N, Grundmann A, Yin W-B, Kremer A, Li S-M. Indole prenyltransferases from fungi: a new enzyme group with high potential for the production of prenylated indole derivatives. Curr Med Chem. 2009;16:218–231. doi: 10.2174/092986709787002772. [DOI] [PubMed] [Google Scholar]
- 25.Seidler NW, Jona I, Vegh M, Martonosi A. Cyclopiazonic acid is a specific inhibitor of the Ca2+-ATPase of sarcoplasmic reticulum. J Biol Chem. 1989;264:17816–17823. [PubMed] [Google Scholar]
- 26.Chang PK, Horn BW, Dorner JW. Clustered genes involved in cyclopiazonic acid production are next to the aflatoxin biosynthesis gene cluster in Aspergillus flavus. Fungal Genet Biol. 2009;46:176–182. doi: 10.1016/j.fgb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Tokuoka M, Seshime Y, Fujii I, Kitamoto K, Takahashi T, Koyama Y. Identification of a novel polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) gene required for the biosynthesis of cyclopiazonic acid in Aspergillus oryzae. Fungal Genet Biol. 2008;45:1608–1615. doi: 10.1016/j.fgb.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Holzapfel CW, Wilkins DC. On the Biosynthesis of Cyclopiazonic Acid. Phytochemistry. 1971;10:351–358. [Google Scholar]
- 29.Liu X, Walsh C. Cyclopiazonic Acid Biosynthesis in Aspergillus sp.: Characterization of a Reductase-like R* Domain in Cyclopiazonate Synthetase That Forms and Releases cyclo-Acetoacetyl-l-tryptophan. Biochemistry. 2009;48:8746–8757. doi: 10.1021/bi901123r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schobert R, Schlenk A. Tetramic and tetronic acids: An update on new derivatives and biological aspects. Bioorg Med Chem. 2008;16:4203–4221. doi: 10.1016/j.bmc.2008.02.069. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. 3. Cold Spring Harbor Laboratory Press; Painview, NY: 2001. [Google Scholar]
- 32.Neethling DC, McGrath RM. Metabolic Development and Mitochondrial Changes during Cyclopiazonic Acid Production in Penicillium-Cyclopium. Can J Microb. 1977;23:856–872. doi: 10.1139/m77-128. [DOI] [PubMed] [Google Scholar]
- 33.Christensen BE, Mollgaard H, Kaasgaard S, Lehmbeck J. Methods for producing polypeptides in Aspergillus mutant cells. 7,241,614 US Patent. 2007
- 34.Takahashi S, Koyama T. Structure and function of cis-prenyl chain elongating enzymes. Chem Rec. 2006;6:194–205. doi: 10.1002/tcr.20083. [DOI] [PubMed] [Google Scholar]
- 35.Poulter CD. Farnesyl Diphosphate Synthase. A Paradigm for Understanding Structure and Function Relationships in E-polyprenyl Diphosphate Synthases. Phytochem Rev. 2006;5:17–26. [Google Scholar]
- 36.Liang P-H, Ko T-P, Wang AH-J. Structure, mechanism and function of prenyltransferases. Eur J Biochem. 2002;269:3339–3354. doi: 10.1046/j.1432-1033.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 37.Zou H, Zheng X, Li S-M. Substrate promiscuity of the cyclic dipeptide prenyltransferases from Aspergillus fumigatus. J Nat Prod. 2009;72:44–52. doi: 10.1021/np800501m. [DOI] [PubMed] [Google Scholar]
- 38.Steffan N, Unsold IA, Li SM. Chemoenzymatic synthesis of prenylated indole derivatives by using a 4-dimethylallyltryptophan synthase from Aspergillus fumigatus. ChemBioChem. 2007;8:1298–1307. doi: 10.1002/cbic.200700107. [DOI] [PubMed] [Google Scholar]
- 39.Markert A, Steffan N, Ploss K, Hellwig S, Steiner U, Drewke C, Li S-M, Boland W, Leistner E. Biosynthesis and accumulation of ergoline alkaloids in a mutualistic association between Ipomoea asarifolia (Convolvulaceae) and a clavicipitalean fungus. PLANT PHYSIOLOGY. 2008;147:296–305. doi: 10.1104/pp.108.116699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teng X, Degterev A, Jagtap P, Xing X, Choi S, Denu R, Yuan J, Cuny GD. Structure-activity relationship study of novel necroptosis inhibitors. Bioorg Med Chem Lett. 2005;15:5039–5044. doi: 10.1016/j.bmcl.2005.07.077. [DOI] [PubMed] [Google Scholar]
- 41.Inoue S, Murata J, Takamatsu N, Nagano H, Kishi Y. Synthetic studies on echinulin and related natural products. V. Isolation, structure and synthesis of echinulin-neoechinulin type alkaloids isolated from Aspergillus amstelodami. Yakugaku Zasshi. 1977;97:576–581. doi: 10.1248/yakushi1947.97.5_576. [DOI] [PubMed] [Google Scholar]
- 42.Yamazaki M, Fujimoto H, Kawasaki T. Chemistry of tremorogenic metabolites. I. Fumitremorgin A from Aspergillus fumigatus. Chem Pharm Bull. 1980;28:245–254. doi: 10.1248/cpb.28.245. [DOI] [PubMed] [Google Scholar]
- 43.Okada M, Yamaguchi H, Sato I, Tsuji F, Dubnau D, Sakagami Y. Chemical structure of posttranslational modification with a farnesyl group on tryptophan. Biosci Biotechnol Biochem. 2008;72:914–918. doi: 10.1271/bbb.80006. [DOI] [PubMed] [Google Scholar]
- 44.Li S. Applications of dimethylallyltryptophan synthases and other indole prenyltransferases for structural modification of natural products. Appl Microbiol Biotechnol. 2009;84:631–639. doi: 10.1007/s00253-009-2128-z. [DOI] [PubMed] [Google Scholar]
- 45.Kremer A, Li SM. Tryptophan aminopeptidase activity of several indole prenyltransferases from Aspergillus fumigatus. Chem Biol. 2008;15:729–738. doi: 10.1016/j.chembiol.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 46.Balibar CJ, Howard-Jones AR, Walsh CT. Terrequinone A biosynthesis through L-tryptophan oxidation, dimerization and bisprenylation. Nat Chem Biol. 2007;3:584–592. doi: 10.1038/nchembio.2007.20. [DOI] [PubMed] [Google Scholar]
- 47.Kuzuyama T, Noel JP, Richard SB. Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature. 2005;435:983–987. doi: 10.1038/nature03668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pojer F, Wemakor E, Kammerer B, Chen H, Walsh CT, Li S-M, Heide L. CloQ, a prenyltransferase involved in clorobiocin biosynthesis. Proc Natl Acad Sci USA. 2003;100:2316–2321. doi: 10.1073/pnas.0337708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stec E, Steffan N, Kremer A, Zou HX, Zheng XD, Li SM. Two lysine residues are responsible for the enzymatic activities of indole prenyltransferases from fungi. ChemBioChem. 2008;9:2055–2058. doi: 10.1002/cbic.200800237. [DOI] [PubMed] [Google Scholar]
- 50.Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chem Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 51.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Forcat S, Allemann RK. Dual role for phenylalanine 178 during catalysis by aristolochene synthase. Chem Commun. 2004:2094–2095. doi: 10.1039/b407932a. [DOI] [PubMed] [Google Scholar]
- 53.Deligeorgopoulou A, Taylor SE, Forcat S, Allemann RK. Stabilisation of eudesmane cation by tryptophan 334 during aristolochene synthase catalysis. Chem Commun. 2003:2162–2163. doi: 10.1039/b306867f. [DOI] [PubMed] [Google Scholar]
- 54.Calvert MJ, Ashton PR, Allemann RK. Germacrene A is a product of the aristolochene synthase-mediated conversion of farnesylpyrophosphate to aristolochene. J Am Chem Soc. 2002;124:11636–11641. doi: 10.1021/ja020762p. [DOI] [PubMed] [Google Scholar]
- 55.Calvert MJ, Taylor SE, Allemann RK. Tyrosine 92 of aristolochene synthase directs cyclisation of farnesyl pyrophosphate. Chem Commun. 2002:2384–2385. doi: 10.1039/b206517g. [DOI] [PubMed] [Google Scholar]
- 56.Gebler JC, Woodside A, Poulter CD. Dimethylallyltryptophan Synthase. An Enzyme-Catalyzed Electrophilic Aromatic Substitution. J Am Chem Soc. 1992;114:7354–7360. [Google Scholar]
- 57.Cole R, Schweikert M, Jarvis B. Handbook of Secondary Fungal Metabolites. Elsevier; San Diego: 2003. [Google Scholar]
- 58.Grundmann A, Kuznetsova T, Afiyatullov SS, Li SM. FtmPT2, an N-prenyltransferase from Aspergillus fumigatus, catalyses the last step in the biosynthesis of fumitremorgin B. ChemBioChem. 2008;9:2059–2063. doi: 10.1002/cbic.200800240. [DOI] [PubMed] [Google Scholar]
- 59.Metzger U, Schall C, Zocher G, Unsöld I, Stec E, Li S-M, Heide L, Stehle T. The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria. Proc Natl Acad Sci USA. 2009;106:14309–14314. doi: 10.1073/pnas.0904897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thulasiram HV, Erickson HK, Poulter CD. A common mechanism for branching, cyclopropanation, and cyclobutanation reactions in the isoprenoid biosynthetic pathway. J Am Chem Soc. 2008;130:1966–1971. doi: 10.1021/ja0771282. [DOI] [PubMed] [Google Scholar]
- 61.O’Maille PE, Malone A, Dellas N, Andes Hess B, Smentek L, Sheehan I, Greenhagen BT, Chappell J, Manning G, Noel JP. Quantitative exploration of the catalytic landscape separating divergent plant sesquiterpene synthases. Nat Chem Biol. 2008;4:617–623. doi: 10.1038/nchembio.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Christianson DW. Unearthing the roots of the terpenome. Curr Opin Chem Biol. 2008;12:141–150. doi: 10.1016/j.cbpa.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thulasiram HV, Erickson HK, Poulter CD. Chimeras of two isoprenoid synthases catalyze all four coupling reactions in isoprenoid biosynthesis. Science. 2007;316:73–76. doi: 10.1126/science.1137786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.