Abstract
Sam68 has been implicated in a variety of important cellular processes such as RNA metabolism and intracellular signaling. We have recently shown that Sam68 cytoplasmic mutants induces stress granules (SG) and inhibit HIV-1 nef mRNA translation [1]. These findings prompted us to investigate the possibility and the underlying mechanisms of the wild-type counterpart Sam68 SG recruitment. Herein, we revealed that Sam68 was significantly recruited into cytoplasmic SG under oxidative stress. We then demonstrated that domain aa269–321 and KH domain were both essential for this recruitment. Nevertheless, Sam68 knockdown had no effects on SG assembly, indicating that Sam68 is not a constitutive component of the SG. Moreover, we showed that Sam68 cytoplasmic mutants-induced SG formation was independent of eIF2α phosphorylation. Lastly, we demonstrated that Sam68 was complexed with T-cell intracellular antigen-1 (TIA-1), a core SG component, and that the complex formation was correlated with Sam68 SG recruitment. Taken together, these results provide direct evidence for the first time that Sam68 is recruited into SG through complexing with TIA-1 in response to oxidative stress and suggest that cytoplasmic SG recruitment of Sam68 and ensuing changes in Sam68 physiological functions are part of the host response to external stressful conditions.
Keywords: stress granules, Sam68, oxidative stress, TIA-1 binding, stress response, relocalization
INTRODUCTION
Sam68 was initially identified to be associated with the p120-RasGTPase-activating protein (Ras-GAP) in cells transformed by tyrosine kinase oncogenes including v-Src and, and it was called p62 for a tyrosine-phosphorylated protein of 62 kDa [2, 3]. Human p62 was later purified by affinity chromatography, and the cDNA was subsequently cloned [4]. Since p62 was found to migrate at 68 kDa and was a Src substrate during mitosis, it was then renamed Src-associated protein in mitosis or Sam68 [5, 6]. Human Sam68 gene contains nine exons, its cDNA encodes an open reading frame of 443 amino acids. The primary structure of Sam68 has one heteronuclear ribonucleoprotein particle K homology (KH) domain, six proline-rich domains (P0–P5), one tyrosine-rich domain, one arginine-glycine (RG)-rich domain, and a nuclear localization signal (NLS), and some of them are overlapped [see reviews [7, 8]. KH domain is required for RNA binding and Sam68 self-association. Proline-rich domain consists of proline-rich amino acid sequences with a core consensus sequence PXXP, and it is the binding sites of Src homology 3 (SH3) proteins. Sam68 has 16 potential tyrosine residues, which can be phosphorylated by soluble tyrosine kinases, its phosphrylation leads to Sam68 association with SH2 proteins. Sam68 also contains RG-rich regions/RGG boxes, the potential sites for protein arginine methylation. The NLS is located in the last 24 C-terminal amino acids of Sam68.
Although much has been learned about the molecular biology of Sam68 over the last decade, Sam68 physiological functions are not well understood. As a prototype of the STAR (signal transduction and activators of RNA) protein family, Sam68 is involved in almost every aspect of RNA metabolism including transcription, splicing, polyadenylation, nuclear exportation, and translation [1, 9–13] as well intracellular signaling [see review [8]. In addition, Sam68 is also implicated in a number of other important cellular processes including cell cycle regulation, apoptosis, tumorigenesis, bone metabolism, and normal brain function [11, 14–16]. It is evident that some of these functions mandate Sam68 in the cytoplasm, despite the fact that Sam68 is predominantly localized in the nucleus.
Eukaryotic cells have evolved sophisticated response mechanisms to different external stresses through gene transcription and translation. One of the mechanisms is formation of cytoplasmic RNA-containing stress granules (SG). These granules are ribonucleoprotein macromolecular structures and contains 40S ribosomal subunits, translation initiation factors, poly(A)-binding protein (PABP), and multiple RNA-binding proteins (RBP), some of which are indispensable for SG assembly [17]. Initially thought to be a passive compartment for mRNA storage, SG are now believed to be a dynamic triage center that sort mRNA for storage, decay, or re-initiation during stressful conditions [18–20], allowing the cells to prioritize the translation of specific mRNA for proteins that are involved in stress responses, e.g. molecular chaperone [18–20]. The assembly of SG can be induced by eIF2α phosphorylation such as oxidative stress, osmotic stress, ER stress, and ischemia [18, 21, 22]; eIF2α phosphorylation-independent inhibition of ribosome recruitment such as pateamine A and poliovirus infection [23, 24]; or over-expression of certain RBP [20, 25, 26].
To date, little is known about the role of Sam68 in host response to external stress. Several lines of indirect evidence suggest that Sam68 could be recruited into SG in response to various stresses. Sam68 translocates and accumulates in cytoplasmic granules under stressful conditions such as poliovirus infection, ischemia, and puromycin treatment, which are also known to promote SG assembly [24, 27–30]. Treatment of urate crystals results in Sam68 cytoplasmic localization in neutrophils [31]. Sam68 localizes into dendrite granules of hippocampal and cortex neurons, where it associates with polysomes [32–34] and regulates the translation efficiency of eEFA1 mRNA [35]. Sam68 is detected with translation initiation factors in the cytoplasm [30, 35] and with polysomes in mouse spermatocytes [36]. GLD-1, a member of the extended STAR family, is targeted to cytoplasmic particles that resemble SG and processing body (PB) in C. elegans [37]. Several previously characterized SG components are complexed with Sam68 in a direct (e.g., hnRNP A1, FAST, PABP) or indirect (KSRP) manner [11, 38–40]. Meanwhile, we have recently found that Sam68 cytoplasmic mutants are capable of inducing SG formation and inhibiting HIV-1 Nef translation through sequestration of Nef mRNA in SG [1]. Moreover, independent studies have shown that Sam68 cytoplasmic mutant Δ410 lacking the NLS accumulates in RNA granules and selectively represses mRNA translation [35, 41]. Thus, in the current study, we aimed to investigate the possibility of Sam68 recruitment into SG in host response to external stress and the underlying mechanisms.
MATERIALS AND METHODS
Plasmids
HA- and GFP-tagged Sam68, Δ410, Δ321, Δ269, Δ269–321, and 269–321, as well as RFP.Sam68 and GFP-hdcp-1α were described elsewhere [1, 9, 42, 43]. HA- and GFP-tagged ΔKH and Δ410ΔKH were constructed in the context of respective parental plasmids HA/GFP.Sam68 or HA/GFP.Δ410 using a Stratagene QuickChange Site-Directed Mutagenesis kit (La Jolla, CA) with primers 5′-CCT GTC AAG CAG TAT CCC AAG CTT CGC AAA GGT GGA GAC CC-3′ and 5′-GGG TCT CCA CCT TTG CGA AGC TTG GGA TAC TGC TTG ACA GG-3′. GFP.TIA-1, GFP.hnRNP A1, GFP.PABP, and GFP.G3BP plasmids were kindly provided by Dr. Sonia Guil and Dr. Javier Caceres [44].
Cell culture and transfection
293T cells were purchased from the American Tissue Culture Collection (Vanassas, VA) and maintained in Dulbeco’s Modified Eagles Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C and 10% CO2. 293T cells were transfected by the standard calcium phosphate precipitation method, which usually gives rise to 80% or higher transfection efficiency. Annealed oligonucleotide duplex control siRNA and Sam68 siRNA were purchased from Dharmacon (Lafayette, CO). Sam68 siRNA targeting sequence was 5′-CGG AUA UGA UGG AUG AUA U-3′ [45]. siRNA was transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The siRNA transfection efficiency was estimated to be 95% or higher in 293T cells using a fluorescence-labeled control siRNA in the transfection.
Cell lysates and Western blot analysis
Cells were washed twice with ice cold phosphate-buffered saline (PBS) and then harvested. Cell pellets were suspended in 2 vol. of cell lysis buffer (30 mM Tris.HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 10 mM EDTA, 10 mM sodium pyrophosphate, 160 mM sodium fluoride, 1 mM sodium orthovanadate, and protease inhibitors) and incubated on ice for 30 min. Whole cell lysates were obtained by centrifugation and removal of the cell debris, and the protein concentration was determined using a BioRad DC protein assay kit (BioRad, Hercules, CA). Cell lysates of an equal amount of protein were separated by SDS-PAGE, blotted and probed using an ECL system (WVR International, West Chester, PA). The sources of the antibodies were: α-β-actin (1:3500), α-HA (1:1000), and α-phosphotyrosine (1:1000) antibodies from Santa Cruz Biotechnologies (Santa Cruz, CA), α-GFP antibody (1:250) from Clontech (Mountain View, CA), α-phospho-ERK1/2 (1:1000), α-ERK1 (1:1000), α-phospho-eIF2α (Ser51) (1:1000) and α-eIF2α antibodies (1:1000) from Cell Signaling technologies (Danvers, MA), α-phosphothreonine and α-phosphoserine were from Stressgen Bioreagents (Ann Arbor, MI), α-JR-CSF Nef antibody (1:750) from the NIH AIDS Research and Reference Reagent Program (donated by Dr. K. Krohn and Dr. V. Ovod).
Soluble and insoluble cell fractions
Soluble and insoluble cell fractions were prepared as previously described [46]. Briefly, cells (2 × 106) were washed in ice-cold PBS and suspended in 250 μl of ice-cold cell lysis buffer (20 mM Tris.HCl, pH 7.5, 5 mM EDTA, 1% Triton X-100, 150 mM NaCl and protease inhibitors) and incubated on ice for 30 min. The samples were then centrifuged at 20,000 g at 4oC for 20 min. The supernatant was removed and saved as the soluble fraction. The pellet was washed three times in cell lysis buffer, then suspended in 70 μl of 4X SDS-PAGE sample buffer (8% SDS, 0.4 M DTT, 0.25 M Tris.HCl, pH 6.8, 40% glycerol and 0.1% bromophenol blue) and incubated at 60oC for 30 min. An equal volume of soluble and insoluble fractions was boiled for 5 min, separated by SDS-PAGE, followed by Western blot analysis.
Immunoprecipitation assay
Cells were washed with ice-cold PBS, suspended in 2 vol. of a low stringency cell lysis buffer (50 mM Tris.HCl, pH 7.5, 120 mM NaCl, 0.25% NP40, 4 mM sodium fluoride, 1 mM sodium orthovanadate, 0.2 mM EDTA, 0.2 mM EGTA, 10% glycerol, and protease inhibitors), and incubated on ice for 30 min. Cell lysates were obtained by centrifugation and removal of the cell debris. Cell lysates of about 0.8–1.2mg total cellular protein were incubated with 1 μg normal IgG (Sigma, St, Louis, MO), polyclonal α-Sam68 (Santa Cruz), or goat α-TIA-1 (Santa Cruz) at 4oC for 16 hr with constant rocking. Then, 25 μl protein A agarose beads (Upstate Biotechnologies, Temecula, CA) were added to the lysates and continued to incubate for additional 2 hr. The agarose beads were then recovered and washed twice with the low stringency buffer and suspended in 60 μl of 4X SDS-PAGE sample buffer, followed by SDS-PAGE and Western blot analysis.
Immunofluorescence staining
Cells were fixed in 4% paraformaldehyde at room temperature (RT) for 25 min and permeabilized in 0.3% Triton X-100 for 5 min. The cells were then blocked in 5% FBS/1% BSA at RT for 30 min, followed by incubation with: rabbit polyclonal α-Sam68 (1:50, Santa Cruz), goat polyclonal α-TIA-1 (1:50, Santa Cruz), goat polyclonal α-eIF3η (1:50, Santa Cruz), or mouse monoclonal α-G3BP (1:75, BD transduction laboratories) at RT for 16 hr. Next, the cells were incubated with phycoerythrin (PE)-conjugated rabbit α-mouse IgG (1:175, Santa Cruz), FITC-conjugated goat α-mouse (1:50, Santa Cruz), fluorescein isothiocyanate (FITC)-conjugated donkey α-goat (1:50), rhodamine (Rh)-conjugated mouse α-rabbit (1:75, Santa Cruz), or Rh-conjugated goat α-rabbit (1:50, Santa Cruz) at RT for 2 hr. Between each step, the cells were extensively rinsed in PBS. To stain the nuclei, the cells were incubated in 500 ng/ml 4′,6′-diamidino-2-phenylindole (DAPI) at 37°C for 25 min. Images were captured using a digital video imaging microscope system consisting of an Axiovert 200 M microscope with a 100 × 1.4 UVF objective and three different excitation/emission filters (D360/40-D460/50, D470/40-D535/40, D546/10-D590), and an Axiovision video camera (Carl Zeiss, Thornwood, NY). For quantification, Random 10 fields were counted for a total number of 100 transfected cells in each well for each transfection and this was performed in triplicate, the number of cells exhibiting GFP+G3BP+ SG was used to calculate the percentage of the GFP+ SG-containing cells. Control staining without primary antibodies was performed for all staining to ensure no non-specific or cross reactivity among antibodies.
Data analysis
All values expressed as mean ± SEM. Comparisons among groups were made using two-tailed Student’s t-test. A p value of < 0.05 was considered statistically significant (*), and p < 0.01 highly significant (**).
RESULTS
Recruitment of GFP-tagged Sam68 into SG in response to oxidative stress
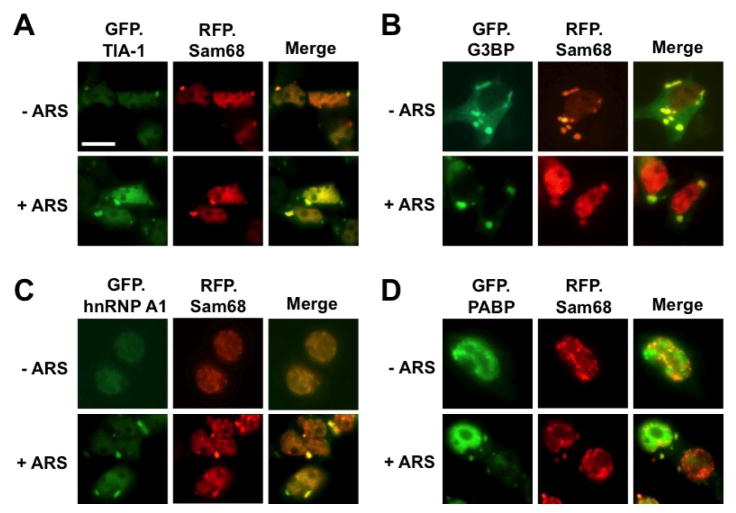
Sam68 is predominantly localized in the nucleus. We have recently shown that over-expression of Sam68 cytoplasmic mutants lacking the NLS results in SG formation [1]. This led us to investigate the possibility that nuclear Sam68 protein is capable of being recruited to cytoplasmic SG under stress conditions. To address this possibility, we took advantage of several well-characterized GFP-tagged SG markers GFP.TIA-1, GFP.G3BP, GFP.PABP, or GFP.hnRNP A1. Over-expression of G3BP and TIA-1 alone is sufficient to induce SG assembly [20, 26], while hnRNP A1 and PABP are recruited into SG only under stress conditions [44]. Thus, we transfected 293T cells with RFP-tagged Sam68 and each of these SG markers and exposed the cells to sodium arsenite (ARS), a widely used oxidative stress inducer. We then determined Sam68 localization in these cells by immunofluorescence microscopic imaging. As expected, expression of GFP.TIA-1 and GFP.G3BP induced SG formation and localized in SG regardless of ARS treatment (Fig. 1A & B). In contrast, without ARS treatment, GFP.hnRNP A1 was detected in the nucleus (Fig. 1C) and GFP.PABP in the nucleus with some also in the cytoplasm [Fig. 1D, [47, 48], and there was no SG formation. With ARS treatment, both GFP.hnRNP A1 and GFP.PABP were partially localized into the cytoplasm, specifically in SG. On the other hand, in GFP.TIA-1- and GFP.G3BP-expressing cells, RFP.Sam68 was co-localized with GFP.TIA-1 or GFP.G3BP in the cytoplasm, although Sam68 was still detected in the nucleus (Fig. 1A & B). Importantly, Sam68 was localized in cytoplasmic SG and in the nucleus in ARS-treated cells and untreated cells. In comparison, in GFP.hnRNP A1- and GFP.PABP-expressing cells, Sam68 was localized in the nucleus in the absence of ARS treatment (Fig. 1C & D). But, in the presence of ARS treatment, Sam68 was not only detected in the cytoplasm, but also co-localized with GFP.hnRNPA 1 and GFP.PABP. These results suggest that Sam68 is recruited into SG in response to oxidative stress.
Figure 1. Localization of exogenous Sam68 into SG upon oxidative stress.
293T cells were transfected with RFP-Sam68 and GFP.TIA-1 (A), GFP.G3BP (B), GFP.hnRNP A1 (C), GFP.PABP (D), GFP.hdcp1α (E). Cells were cultured for 48 hr and then treated with 0.5 mM ARS (+ ARS) or without ARS (− ARS) for 1 hr prior to fixation and microscopic imaging. The images were representative of each transfection from at least three independent experiments. Co-localization of each marker with exogenous Sam68 was shown in the column marked as “Merge”.
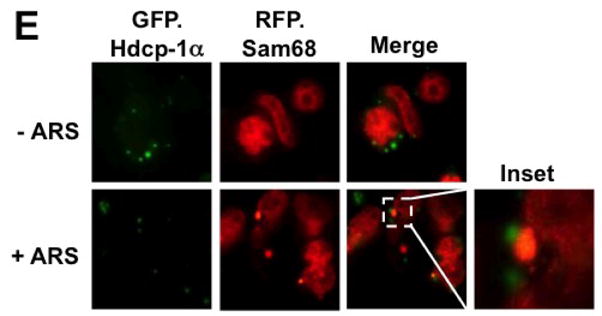
We next determined whether Sam68 was also localized to other cytoplasmic RNA granules, such as processing body (PB), as SG and PB share many common components [49]. Similarly, we transfected 293T cells with RFP.Sam68 and GFP-tagged hcdp1α, a well-characterized PB marker [50] and determined their respective localization. In the absence of ARS treatment, RFP.Sam68 remained in the nucleus, while hdcp1α was localized to PB (Fig. 1E). In the presence of ARS treatment, RFP.Sam68 was partially re-localized to large cytoplasmic foci SG, which was distinct from GFP.hdcp1α-labeled PB. In concordance with the notion that SG and PB are dynamically connected [49], we found that RFP.Sam68-labeled SG were in close proximity to GFP.hdcp1α-labeled PB (Fig. 1E, inset). These results provide additional evidence to support the early finding that Sam68 is conditionally recruited to SG.
Recruitment of endogenous Sam68 to SG in response to oxidative stress
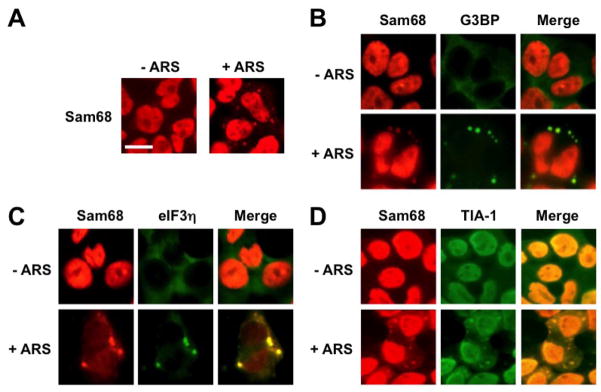
To ascertain that Sam68 possesses the capability of being recruited into SG under oxidative stress, we next compared intracellular localization of endogenous Sam68 in untreated cells with that in ARS-treated cells using immunofluorescence staining. As expected, Sam68 was predominantly localized in the nucleus of untreated cells (Fig. 1E). But in the ARS-treated cells, Sam68 was found to localize in both the nucleus and the cytoplasm in the form of cytoplasmic granules (Fig. 2A). We then characterized these Sam68-containing cytoplasmic granules by double immunofluorescence staining for Sam68 and endogenous SG markers G3BP, eIF3η, or TIA-1. Similarly, we treated 293T cells with ARS prior to the staining. As expected, in the absence of ARS treatment, both endogenous G3BP and eIF3η were detected in the cytoplasm and endogenous TIA-1 and Sam68 in the nucleus, while ARS treatment resulted in enrichment of endogenous TIA-1, G3BP and eIF3η in SG (Fig. 2B-D). Meanwhile, in the presence of ARS treatment, some endogenous Sam68 was co-localized with endogenous G3BP, eIF3η or TIA-1-labeled SG in the cytoplasm. Taken together, these results strongly suggest that Sam68 is recruited to SG under oxidative stress.
Figure 2. Localization of endogenous Sam68 into stress granules upon oxidative stress.
293T cells were plated and cultured for 24 hr to reach 60–70% confluence. The cells were treated with 0.5 mM ARS (+ ARS) or without ARS (− ARS) for 1 hr prior to fixation, and then processed for double immunofluorescence staining using pairs of primary and secondary antibodies as specified below: A. rabbit α-Sam68/Rh-conjugated goat α-rabbit; B. rabbit α-Sam68/Rh-conjugated goat α-rabbit, mouse α-G3BP/FITC-conjugated goat α-mouse; C. rabbit α-Sam68/Rh-conjugated bovine α-rabbit, goat α-eIF3η/FITC-conjugated donkey α-goat; D. rabbit α-Sam68/Rh-conjugated bovine α-rabbit, goat α-TIA-1/FITC-conjugated donkey α-goat. The images were representative of each staining from at least three independent experiments. Co-localization of each SG marker with endogenous Sam68 was shown in the column marked as “Merge”.
Requirement of Sam68 proline-/RG-rich domain for its SG recruitment and eIF2α phosphorylation
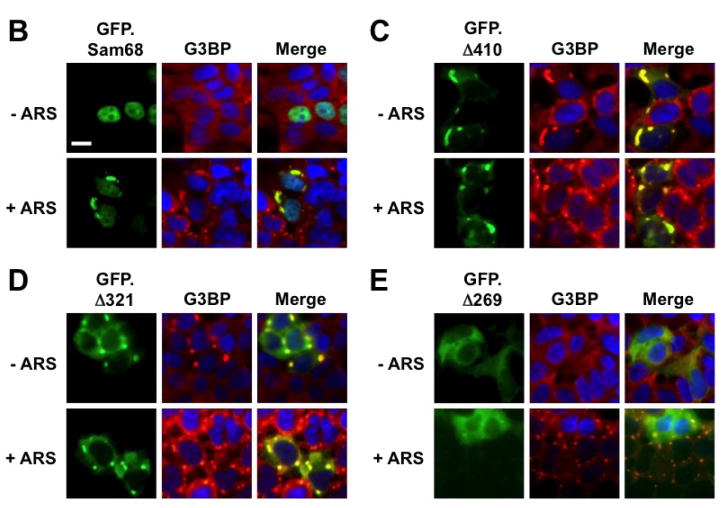
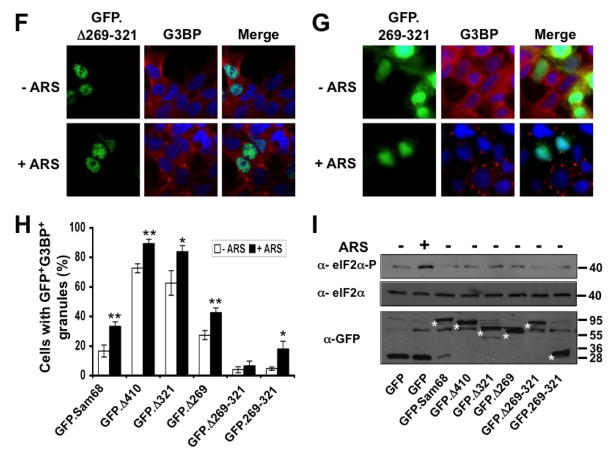
To define the molecular mechanisms whereby Sam68 is recruited to SG, we took advantage of a panel of GFP-tagged Sam68 mutants containing various lengths of the C-terminal deletion [1, 43] and determined Sam68 structural requirement for its SG recruitment. We transfected 293T cells with GFP.Sam68 or each of these mutants (Fig. 3A). We then treated the cells with ARS and performed immunofluorescence staining for endogenous G3BP as the SG marker. We also quantitated the percentage of SG-containing GFP+G3BP+ cells in relation to the total of GFP+ cells in each transfection. As shown previously [1], in the absence of ARS treatment, over-expression of only GFP.Δ410 lacking the NLS and GFP.Δ321 lacking NLS and P4–5 were capable of inducing SG formation, while GFP.Sam68, GFP.Δ269 lacking NLS and P3–5, GFP.Δ260–321 lacking P3, and GFP.269–321 expressing only the P3 showed little SG induction (Fig. 3B–G). Compared to the untreated cells, ARS treatment significantly increased the SG localization of GFP.Sam68 and its mutant GFP.269–321 but not the nuclear mutant GFP.Δ269–321 (Fig. 3H). However, ARS treatment significantly increased the SG localization of all cytoplasmic mutants GFP.Δ410, GFP.Δ321, and GFP.Δ269. These results demonstrated that proline (RG)-rich domain P3 between aa269 and aa321 was essential but not sufficient for SG induction and recruitment and suggest that nuclear localization is a limiting factor for Sam68 recruitment to SG.
Figure 3. Requirement of specific Sam68 domain for its SG recruitment.
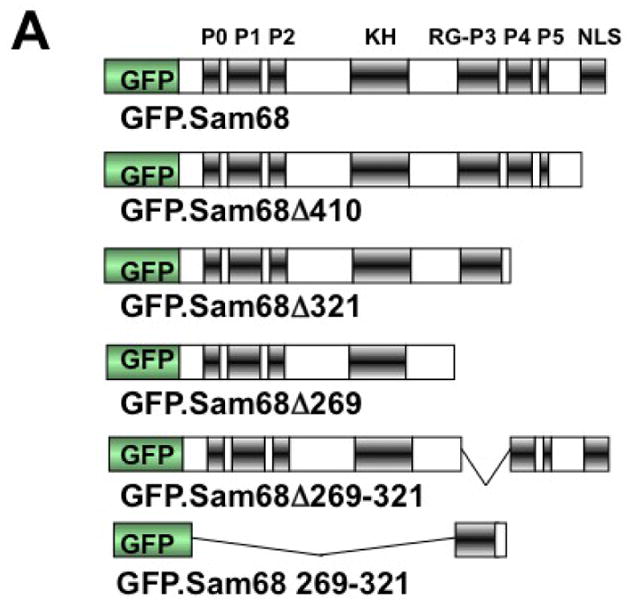
A. P: proline-rich domain; KH: KH domain; RG: arginine-glycine rich domain; and NLS: nuclear localization signal. The single lines in GFP. 269–321 and GFP.269–321 represent deleted regions. B–G. 293T cells were transfected with GFP-tagged Sam68 (B), deletion mutants containing various lengths of C-terminal deletion, i.e. Δ410 (C), Δ321 (D) or Δ 269 (E), Δ269–321 lacking the domain of aa269–321 (F), or 269–321 expressing the domain of aa269–321 (G). Cells were cultured for 48 hr and then treated with 0.5 mM ARS (+ ARS) or without ARS (− ARS) for 1 hr prior to fixation. The cells were then stained using a mouse α-G3BP antibody followed by PE-conjugated goat α-mouse secondary antibody. The images were representative of each transfection. Co-localization of G3BP with Sam68 or each mutant was shown in the column marked as “Merge”. H. Quantitation of GFP+G3BP+ cells in each transfection in B–G, which was expressed as a percentage of the total number of GFP+ cells. The data were mean ± SEM of at least three independent experiments. The comparison was made between ARS-treated and untreated cells. *: p < 0.05; **: p < 0.05. I. 293T cells were transfected with GFP-tagged Sam68 or each of its mutants as indicated, followed by Western blot analysis using α-eIF2α, α-phosphorylated eIF2α (α-eIF2α-P), or α-GFP antibodies. Cells that were transfected with GFP and treated with 0.5 mM ARS or without ARS were included as positive and negative controls for eIF2α phosphorylation, respectively, in the Western blot analysis. *: GFP-tagged proteins.
Over-expression of mutant proteins often overwhelms the protein-folding capacity in the cells and subsequently activates the cellular unfolded protein response and leads to eIF2α phosphorylation and SG assembly [51]. Thus, we next determined if SG induction by Sam68 cytoplasmic mutants was mediated by eIF2α phosphorylation. We transfected 293T cells with GFP.Sam68 or each of its mutants and determined the phosphorylation status of eIF2α. We also included GFP transfection with and without ARS treatment as eIF2α phosphorylation positive and negative controls, respectively. As expected, GFP transfected cells treated with ARS resulted in the phophorylation of eIF2α (Fig. 3I). However, over-expression of Sam68 or each of its mutants did not induce eIF2α phosphorylation. These results suggest that the capacity of Sam68 cytoplasmic mutants to induce SG is eIF2α phosphorylation-independent, reminiscent of other SG-inducing proteins [20, 25, 26], which over-expression may lead to alteration of the equilibrium between mRNPs and polysomes and compete for 48S complexes before initiation is completed [17].
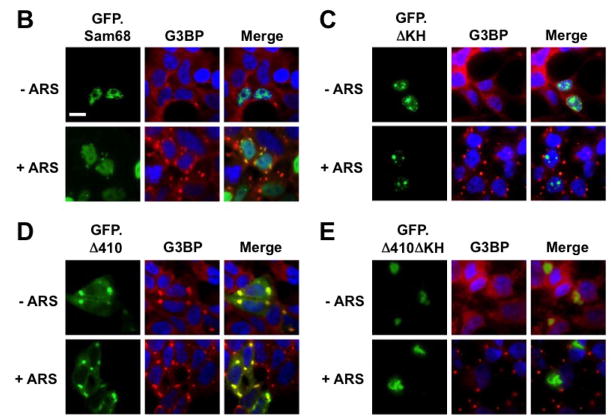
Role of Sam68 KH domain for its SG recruitment and aggregation
The RNA binding property is critical for SG localization of several RBP [20, 25, 26]. Moreover, a recent study has shown that removal of the RNA recognition motifs (RRM) from TIA-1 leads to large insoluble aggregates and as a result, abrogates the ability of TIA-1 to induce SG assembly [20]. Sam68 KH domain is involved in its RNA binding [52], self association [53], and translocation to the cytoplasm [54]. Thus, we further determined the role of RNA binding, i.e. KH domain on Sam68 SG recruitment. To this end, we constructed two KH domain deletion mutants GFP.ΔKH and GFP.Δ410ΔKH using GFP.Sam68 and GFP.Δ410 as respective backbones (Fig. 4A). We transfected 293T cells with GFP.ΔKH or GFP.Δ410ΔKH, treated the cells with ARS, and performed immunofluorescence staining for G3BP as a SG marker. We also included GFP.Sam68 and GFP.Δ410 as controls in these experiments. As shown above, GFP.Δ410 co-localized with G3BP in SG in the absence and presence of ARS treatment, whereas Sam68 localized to SG only in the cells treated with ARS (Fig. 4B & D). Nevertheless, both GFP.ΔKH and GFP.Δ410ΔKH failed to co-localize with G3BP in SG in the presence or absence of ARS treatment (Fig. 4C & E). Of note, GFP.ΔKH expression led to its localization in and formation of nuclear microaggregates (Fig. 4C), while GFP.Δ410.ΔKH was enriched in a single large perinuclear aggregate (Fig. 4E). These findings are reminiscent of the insoluble aggregates induced by over-expression of TIA-1 ΔRRM mutants [20], with the exception that Δ410ΔKH-expressing cells displayed normal assembled SG under the ARS treatment.
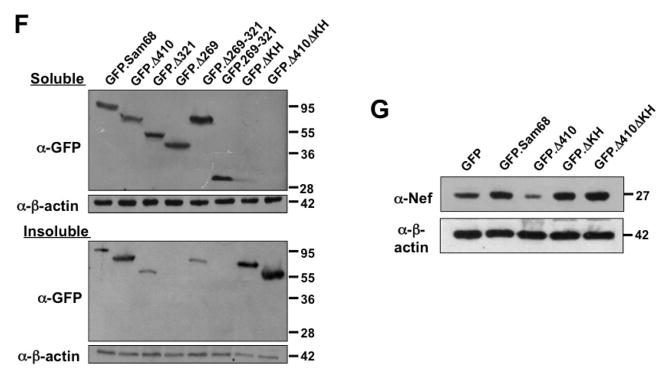
Figure 4. Role of Sam68 KH domain in SG recruitment, Sam68 solubility and HIV-1 Nef mRNA translation.
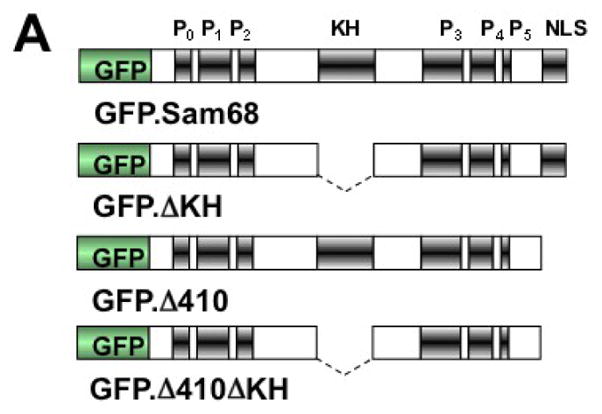
A. KH domain deletion mutants: P: proline-rich domain; KH: KH domain; and NLS: nuclear localization signal. The dotted lines represent deleted regions. B–E. 293T cells were transfected with GFP-tagged GFPSam68 (B), GFP.ΔKH (C), GFP.Δ410 (D), and GFP.Δ410ΔKH (E). Cells were cultured for 48 hr and prior to fixation, the cells were treated with 0.5 mM ARS (+ ARS) or without ARS (− ARS) for 1 hr. The cells were then stained using a mouse α-G3BP antibody followed by phycoerythrin (PE)-conjugated goat anti-mouse antibody. The images were representative of each transfection from at least three independent experiments. Co-localization of G3BP and GFP-tagged Sam68 or its mutants was shown in the column marked as “Merge”. F. 293T cells were transfected with GFP-tagged Sam68 or each of its mutants as indicated, then cell lysates were separated into soluble and insoluble fractions as described in Materials and Methods, followed by Western blot analysis using α-GFP or α-β-actin antibody. G. 293T cells were transfected with HIV-1 isolate NL4-3 and GFP-tagged Sam68 or each of its mutants as indicated, followed by Western blot analysis using α-Nef or α-β-actin antibody.
To further characterize the aggregation nature of these mutants, we transfected 293T cells with GFP.Sam68 or each of its mutants including these two KH mutants, separated the cellular proteins into 1% Triton X-soluble or -insoluble fractions as previously described [46], and performed Western blot analysis to compared the extent of their aggregation. GFP.Sam68, GFP.Δ410, and GFP.Δ321 that were all localized to SG were detected in soluble and insoluble fractions (Fig. 4F). In contrast, GFP.Δ269 and GFP.269–321 which were rarely co-localized into SG were found mainly in the soluble fraction. In agreement with immunofluorescence microscopic imaging (Fig. 4C & E), both GFP.ΔKH and GFP.Δ410ΔKH were exclusively detected in the insoluble fractions (Fig. 4F). To rule out possible effects of GFP on the protein solubility, we also constructed a panel of HA-tagged mutants and obtained similar results (data not shown). Furthermore, we also determined the role of Sam68 KH domain in suppression of HIV-1 Nef translation [1]. We transfected 293T cells with HIV-1 provirus pNL4-3 and these KH deletion mutants as well as their respective backbones and performed Western blot analysis for Nef expression. As previously shown [1], GFP.Δ410 significantly suppressed and GFP.Sam68 slightly increased Nef expression (Fig. 4G). In agreement with its inability to induce or be recruited to SG, GFP.ΔKH and GFP.Δ410ΔKH not only failed to suppress Nef translation, but to the contrary increased Nef expression (Fig. 4G). The mechanisms underlying increased Nef expression by Sam68 and these two KH deletion mutants deserves further investigation. Taken together, these results suggest that Sam68 KH domain is involved in SG induction by Sam68 cytoplasmic mutants or SG recruitment of Sam68 and required for maintaining native and functional structure of Sam68 protein in the cells.
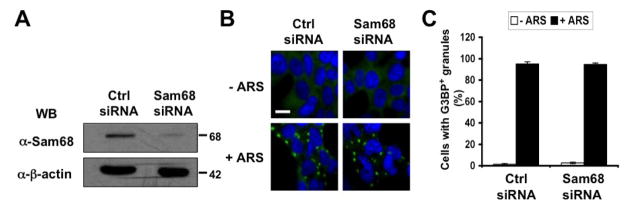
No effects of Sam68 knockdown on SG assembly
Several cellular proteins are absolutely required for SG assembly, and their expression knockout is associated with a muted SG response to multiple stress stimuli [20, 26, 55]. The findings that Sam68 was recruited to SG while some of its mutants induced SG assembly raised the possibility that Sam68 was a component of SG assembly. To address this possibility, we utilized the small interference RNA (siRNA) strategy to knock down endogenous Sam68 expression and determined its effects on SG formation under oxidative stress. Compared to control siRNA, Sam68 siRNA was very effective in knocking down endogenous Sam68 expression (Fig 5A). We then transfected 293T cells with control siRNA or Sam68 siRNA, treated the cells with ARS, and performed immunofluorescence staining for G3BP as the SG marker. There were no apparent changes in the size or morphology of SG (Fig. 5B) and in the percentage of SG-containing cells transfected with control siRNA and Sam68 siRNA (Fig. 5C). These results suggest that Sam68 is not an essential cellular cofactor for SG assembly, at least under the oxidative stress.
Figure 5. Effects of Sam68 knockdown on SG assembly.
A. 293T cells were plated at a density of 1.75×105 per well and transfected with 75 nM of control siRNA or Sam68 siRNA. The cells were harvested 24 hr after transfection for Western Blot analysis using anti-Sam68 or anti-β-actin antibodies. B. 293T cells were plated and transfected with control siRNA or Sam68 siRNA as above. The cells were treated with 0.5 mM ARS (+ ARS) or without ARS (− ARS) for 1 hr prior to fixation. The cells were then stained using a mouse α-G3BP antibody followed by FITC-conjugated α-mouse antibody. Transfected cells were also stained with 500 ng/ml of DAPI for nuclei. The images were representative of each transfection from at least three independent experiments. C. Quantitation of G3BP+ SG-containing cells in B, which was expressed as percentage of the total number of G3BP+ cells. The data were mean ± SEM of at least three independent experiments.
Interaction of Sam68 with major SG proteins and its SG recruitment
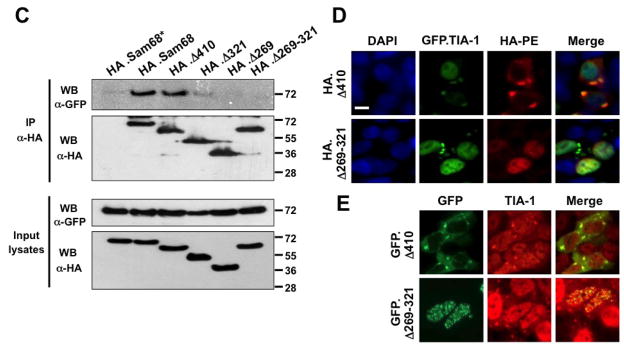
There are several possible ways by which a protein can be recruited into SG, and one of the common pathways is through interaction with constitutive core components of SG [17]. To investigate whether this was the case for Sam68 SG recruitment, we initially performed a BLAST search in a database, which predicts human protein-protein interactions [56], for possible Sam68-interacting proteins. We obtained a list of 108 proteins as potential candidates, among which was TIA-1. TIA-1 has been shown to recruit proteins to SG through direct binding [57–60]. One recent example is FAST, a protein that binds Sam68 [39]. Thus, we focused on the possibility of Sam68/TIA-1 interaction. We first transfected 293T cells with HA-tagged Sam68 (HA.Sam68), GFP.TIA-1, or both and performed immunoprecipitation (IP) followed by Western blotting analysis (WB). We also performed Western blot analysis for the input lysates to ensure comparable levels of protein expression among transfections (top panel, Fig. 6A), as well as IP with control IgG antibody followed by WB with α-HA or α-GFP to ensure no non-specific background binding (2nd panel, Fig. 6A). IP with α-HA antibody followed by WB with α-GFP antibody showed complex formation between Sam68 and TIA-1 in cells co-expressing Sam68 and TIA-1, and with α-HA antibody showed similar IP efficiency between cells expressing Sam68 alone and cells expressing both Sam68 and TIA-1 (3rd panel, Fig. 6A). Conversely, IP with α-TIA-1 antibody followed by WB with α-HA antibody also showed that Sam68 complexed with TIA-1 in cells expressing both Sam68 and TIA-1, and with α-GFP antibody showed similar IP efficiency between cells expressing Sam68 alone and cells expressing both Sam68 and TIA-1 (4th panel, Fig. 6A). Next, we determined whether there was complex formation between endogenous Sam68 and endogenous TIA-1 using the similar IP/WB strategy, we obtained similar results (Fig. 6B).
Figure 6. Complex formation between Sam68 and TIA-1 and Sam68 SG recruitment.
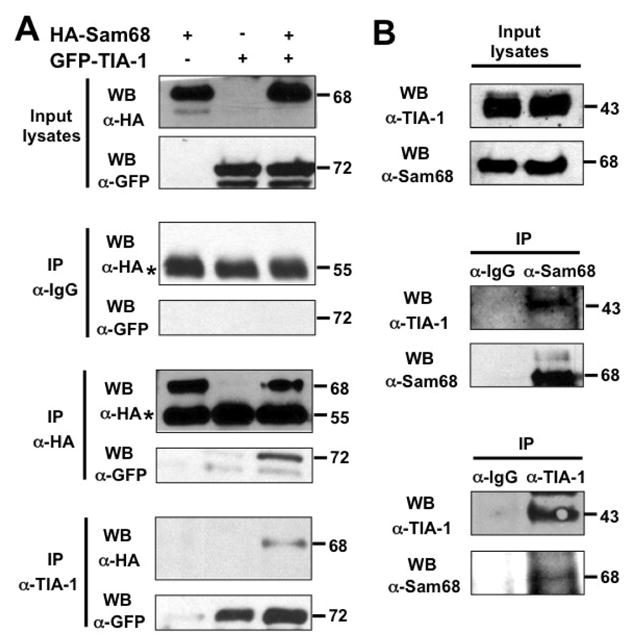
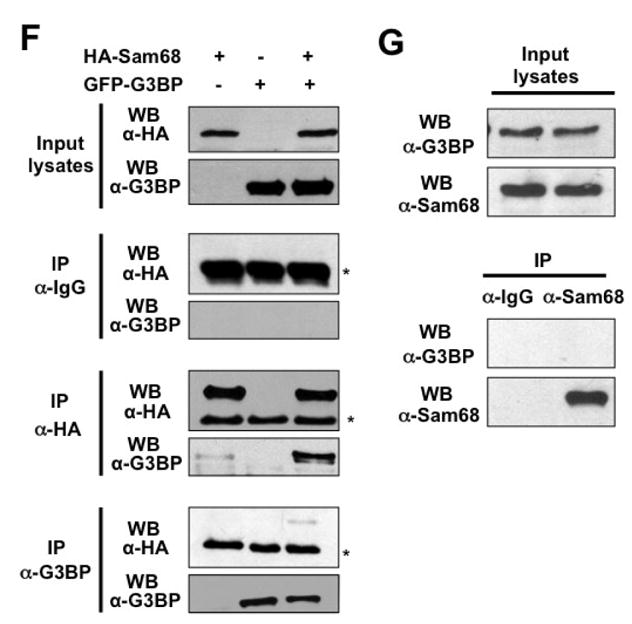
A. 293T cells were transfected with HA.Sam68, GFP-TIA-1, or both and harvested 48 hr after transfection for cell lysates. Cell lysates were then immunoprecipitated with normal IgG, anti-HA, or anti-TIA-1 antibody, followed by Western blotting using α-HA or α-GFP antibody. Input lysates were directly blotted to ensure comparable Sam68 and TIA-1 expression among transfections. *: reactive IgG bands. B. Cell lysates were prepared from 293T cells and blotted for endogenous Sam68 and TIA-1 expression (Input lysates), or immunoprecipitated with normal IgG, α-Sam68, or α-TIA-1, followed by Western blot analysis using α-TIA-1 or α-Sam68 antibody. C. 293T cells were transfected with GFP-TIA-1, in combination with HA.Sam68, or each of its mutants. Input lysates were directly blotted with α-GFP or α-HA antibody to ensure comparable protein expression, or immunoprecipitated with α-HA antibody, followed by Western blot analysis using α-GFP or α-HA antibody. D. 293T cells were transfected with GFP-TIA-1 in combination with HA.Δ410 or HA.Δ269–321. The cells were then stained using an anti-HA antibody followed by PE-conjugated goat anti-mouse antibody. Transfected cells were also stained with 500 ng/ml of DAPI for nuclei. E. 293T cells were transfected with GFP.Δ410 or GFP.Δ269–321, cultured for 48 hr and then treated with 0.5 mM ARS for 1 hr prior to fixation. The cells were then stained using a mouse α-TIA-1 antibody followed by PE-conjugated donkey α-goat secondary antibody. The images were representative of each transfection from at least three independent experiments. F. 293T cells were transfected with HA.Sam68, GFP-G3BP, or both and harvested 48 hr after transfection for cell lysates. Cell lysates were then immunoprecipitated with normal IgG, anti-HA, or anti-G3BP antibody, followed by Western blotting using α-HA or α-G3BP antibody. Input lysates were directly blotted to ensure comparable Sam68 and G3BP expression among transfections. G. Cell lysates were prepared from 293T cells and blotted for endogenous Sam68 and G3BP expression (Input lysates), or immunoprecipitated with normal IgG or α-Sam68, followed by Western blot analysis using α-G3BP or α-Sam68 antibody.*: reactive IgG bands.
To determine whether the complex formation between Sam68 and TIA-1 was responsible for Sam68 recruitment into SG, we further characterized Sam68 interaction with TIA-1 using a similar panel of HA-tagged Sam68 and mutants. We transfected 293T cells with HA.Sam68 or each of its mutants, in combination with GFP-TIA-1, and analyzed their binding to TIA-1 using the similar IP/WB strategy. We first performed WB with α-GFP or α-HA antibody to ensure a comparable level of Sam68 and each of its mutants and a comparable level of TIA-1 expression among transfections (Input lysates, Fig. 6C). We then performed IP with α-HA antibody followed by WB with α-GFP antibody as well as WB with α-HA antibody. Similar to Sam68, Δ410 showed complexed formation with TIA-1, so did Δ321 but to a lesser extent (top panel, Fig. 6C). On the other hand, Δ269 or Δ269–321 did not show any complex formation with TIA-1, despite that Sam68 and all mutants showed no difference in the IP efficiency with α-HA antibody. These results showed that Sam68 domain 269–321 was involved in interaction with TIA-1.
To further corroborate the role of domain aa269–321 in TIA-1 binding and its relationship to Sam68 SG recruitment, we transfected 293T cells with GFP-TIA-1 in combination with HA.Δ410 or HA.Δ269–321, followed by immunofluorescence staining using α-HA antibody. As shown above, Δ410 was co-localized with TIA-1 in SG in Δ410/TIA-1 co-expressing cells (Fig. 6D). In contrast, in Δ269–321/TIA-1-co-expressing cells, Δ269–321 remained in nucleus, while TIA-1 was detected in SG in the cytoplasm. Similar results were obtained with SG containing endogenous TIA-1 (Fig. 6E).
Our previous results showed that Sam68 deletion mutant Δ321 is strongly recruited to SG (Fig. 6); however, it binds weakly to TIA-1 (Fig. 6C), suggesting that additional factors could be involved in Sam68 recruitment to SG. In fact, several studies have shown that Sam68 also interacts with other SG components such as FAST, PABP, and hnRNP A1 [11, 38, 39]. G3BP, which is absolutely required for SG assembly [26], has been suggested to cooperate with Sam68 in a pathway that links RAS-GAP and RNA [61]. Thus, we decided to determine if Sam68 and G3BP interacted in vivo. We transfected 293T cells with HA.Sam68, GFP.G3BP, or both and performed IP followed by WB analysis. IP with a-HA antibody followed by WB with a-G3BP antibody showed complex formation between Sam68 and G3BP in cells co-expressing Sam68 and G3BP (6th panel; Fig. 6F), and a-HA antibody showed similar IP efficiency between cells expressing Sam68 alone and cells expressing both Sam68 and G3BP (5th panel; Fig. 6F). Conversely, IP with a-G3BP antibody followed by WB with a-HA antibody also showed similar results (7th panel; Fig. 6F). However, we failed to detect any complex formation between endogenous Sam68 and endogenous G3BP in the absence (Fig. 6G) or presence of ARS (data not shown), suggesting a possible interference of the antibodies with complex formation, or the low sensitivity or affinity of the antibodies. Taken together, these results show that Sam68 domain aa269–321 (P3/RG-rich) is directly involved in Sam68 binding to TIA-1 and strongly suggest that Sam68/TIA-1 complex formation is, at least in part, responsible for Sam68 recruitment into SG and provide additional evidence that Sam68 is recruited into the SG in a complex that contains TIA-1 and other SG proteins.
Sam68 phosphorylation during oxidative stress
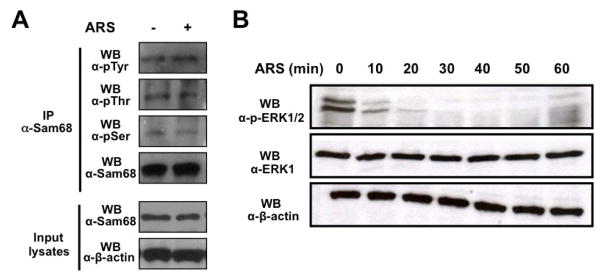
The activation of signaling pathways has been shown to control the activity and subcellular distribution of RNA-binding proteins, including Sam68. For instance, Sam68 translocates to the cytoplasm upon serine/threonine phosphorylation by ERK1/2 [36]. Furthermore, phosphorylation of tyrosine 440 is required for Sam68 nuclear localization [62]. Therefore, an intriguing possibility is that ARS treatment leads to changes in Sam68 phophorylation status, which in turn results in its translocation to the cytoplasm and SG localization. To test this possibility, we treated 293T cells with ARS and then analyzed Sam68 serine, threonine, and tyrosine phosphorylation levels using an IP/WB strategy. We first performed WB with α-Sam68 and α-β-actin antibodies to ensure a comparable level of Sam68 among immunoprecipitation (Input lysates, Fig. 7A). We then performed IP with α-Sam68 antibody followed by WB with α-pSer, α-pThr, or α-pTyr antibodies as well as WB with α-Sam68 antibody. We did not observe a change on the threonine or tyrosine phosphorylation levels of Sam68 upon ARS treatment despite Sam68 was immunoprecipitated equally in untreated and treated cells (1st and 2nd panels; Fig 7A). However, we found that serine phosphorylation levels of Sam68 were slightly decreased in ARS treated cells (3rd panel; Fig. 7A). In concordance with this finding, ERK1/2 were potently deposphorylated upon ARS treatment in a time-dependent manner (Fig. 7B). Taken together, these results suggest that Sam68 posphorylation may not play a role in its recruitment to SG during oxidative stress.
Figure 7. Effects of oxidative stress on Sam68 phosphorylation.
A. 293T cells were treated with 0.5 mM ARS (+ ARS) or without ARS (− ARS) for 1 hr prior to harvesting the cells. Cell lysates were then immunoprecipitated with normal anti-Sam68 antibody, followed by Western blotting using α-pTyr, α-pThr, α-pSer or α-Sam68 antibody. Input lysates were directly blotted to ensure comparable Sam68 and actin expression among samples. B. 293T cells were treated with 0.5 mM ARS for 0, 10, 20, 30, 40, 50, and 60 min, followed by Western blot analysis using anti-ERK1, anti-phosphorylated ERK1/2 (p-ERK1/2), or anti-β-actin.
DISCUSSION
In the current study, we demonstrated that both exogenous and endogenous Sam68 was recruited into SG upon oxidative stress (Fig. 1 & 2). Removal of the P3 (RG-rich) domain aa269–321, or the KH domain aa171–211, abrogated Sam68 ability to be recruited into SG (Fig. 3 & 4). Knockdown of Sam68 expression had no effects on SG assembly (Fig. 5). Importantly, Sam68 formed a complex with TIA-1 through the P3 (RG-rich) domain aa269–321, deletion of which prevented Sam68 recruitment into SG (Fig. 6). In addition, Sam68 interacted in vivo with G3BP (Fig. 6). Moreover, oxidative stress has minimal effects on the phosphorylation status of Sam68 (Fig. 7). Thus, we concluded that Sam68 was recruited to SG under oxidative stress, likely through a mechanism involving direct/indirect interactions with SG core protein component.
Nuclear envelope is a limiting factor for Sam68 relocalization to SG
Despite the fact that Sam68 possesses a NLS, it has been found in the cytoplasm under various conditions, including poliovirus infection [27], ischemia [28], urate crystals treatment [31], HIV infection [63], sperm cell meiosis [36], mitosis [64], neuronal depolarization [32], signaling transduction [8], and in some mouse cells [15]. The findings that Sam68 can be rapidly accumulated in the cytoplasm during acute conditions such as depolarization and ischemia suggest a shuttling mechanism; however, classical heterokaryon assays have failed to detect any apparent nuclear-cytoplasmic shuttling of Sam68 [65]. On the other hand, it is possible that Sam68 localization in the cytoplasm results from alterations in the nuclear importation machinery such as poliovirus infection [66], disruption of the nuclear membrane and nuclear protein leaking such as meiosis and mitosis [67], or to high concentration of sam68 binding proteins that prevent nuclear importation just after translation in T cells and adipocytes [68, 69]. Meanwhile, multiple nuclear RBP proteins have been described to localize to SG upon oxidative stress. Some of them including TIA-1, HuR, FMRP constantly shuttle between the nucleus and the cytoplasm with mRNA and are retained in the cytoplasm during SG assembly [70–72]; others including hnRNP A1 undergo post-translational modifications during stress, which alters the relative export/import rates and SG localization [44]. The current study shows that oxidative stress significantly enhanced Sam68 recruitment into SG, although not occurring in all cells (Fig. 1–3), and deletion of its NLS directly induces SG in an overwhelmingly majority of the cells (Fig. 3 & 6D). Moreover, oxidative stress does not impact significantly the phosphorylation status of Sam68 (Fig. 7A) suggesting that this post-translational modification is not required for this phenomenon. Therefore, it is conceivable that the nuclear envelope is a limiting factor for Sam68 recruitment to SG. While it is not likely that oxidative stress lead to Sam68 shuttling from the nucleus to cytoplasm, it seems plausible that Sam68 recruitment to SG only occurs in cells that happen to have Sam68 in the cytoplasm. One of the likely scenarios is possible variations on Sam68 localization in somatic cells during cell cycle progression, and this scenario clearly deserves further investigation.
Role of the KH and RG rich domains in the process of Sam68 re-localization to SG
Several proteins are recruited to SG via RNA-binding KH or RG rich domains. Deletion of RG-rich sequences from proteins such as FMRP [73], Caprin 1 [25], G3BP [26], or RHAU [74] prevents their accumulation on SG; similarly, KH domains target PCBP2 [75], ZBP1 [76], or hMex 3B [77] proteins to SG. Sam68 binds RNA through a single KH domain (aa171–211). Sam68 also contains a methylated P3/RG-rich domain aa269–321 that is involved in non-specific RNA binding [78], homotypic interactions [43], HIV replication inhibition [41, 43], and cellular localization [79]. Here, we demonstrated that deletion of each of these two domains abrogated Sam68 recruitment to SG (Fig. 3 & 4). However, it should be noted that these deletions differentially altered the localization of Sam68 or its mutant. Deletion of the P3/RG-rich domain resulted in a diffuse cytoplasmic (Δ269) or nuclear (Δ269–321) localization in the majority of the cells, whereas deletion of the KH domain led to formation of insoluble protein aggregates that are different from SG (Fig. 4). Therefore, our results and the fact that Δ410 selectively targets mRNAs to free mRNP/monosomes fractions [41] suggest that cytoplasmic Sam68 binds to pre-initiation complexes through its KH domain, and that the P3/RG-rich region promotes nucleation of SG through homo-heterotypic interactions.
Role of heterotypic protein interactions on Sam68 recruitment to SG
Recently, SG protein components have been classified into four categories: stalled initiation complexes (universal markers), RBP linked to translational regulation (reliable markers), RBP that regulate aspects of mRNA metabolism other than translation or decay, and proteins recruited to SG by core components through piggy-bag interactions [17]. SRC3, KSRP, PMR1, and FAST are proteins recruited to SG through interactions with core component TIA-1 [57–60]. Interestingly, Sam68 directly interacts with FAST [39], and forms a unique RNP complex with KSRP and intron 6 of the β-tropomyosin gene [40]. Here, we showed that Sam68 was not a core component of SG, since its knockdown did not affect SG assembly (Fig. 5). In addition, using the bioinformatics and immunoprecipitation/Western blot analysis, we demonstrated that Sam68 bound to TIA-1 through its domain aa269–321 (Fig. 6C). Furthermore, we also identified G3BP as an additional Sam68 interacting SG core protein (Fig. 6F). Therefore, it is plausible to speculate that when present in the cytoplasm, Sam68 is targeted to SG through its interaction with TIA. However, whether this is a direct interaction, indirectly mediated by RNA or FAST, or the only interaction with SG core components required for its recruitment to these structures remains to be determined. The later point will be particularly difficult to elucidate since the depletion of SG core components results in impaired SG assembly during stressful conditions [20, 26] (data not shown).
STAR proteins as new components of RNA granules
The extended family of STAR proteins is involved in multiple process of mRNA metabolism. Two best characterized members, GLD-1 and Quakin 6, are bona fide translational repressors and involved in sexual identity [80], tumor suppression [81], and cell differentiation [82]. GLD-1 is targeted to cytoplasmic particles that resemble PB and SG in C. elegans [37]. The mouse qk gene produces three major isoforms with different cellular localization. QK-5 is predominantly nuclear, while QK-6 and QK-7 are localized in the cytoplasm. Interestingly, over-expression of QKI-6 and QKI-7 but not QKI-5 leads to their localization in cytoplasmic foci similar to SG. All of the above studies indicate that STAR family proteins including Sam68, when present in the cytoplasm, can regulate mRNA translation through targeting mRNA to cytoplasmic granules. A number of RNA targets have been identified for Sam68, including poly (U), poly (A), A/U-rich sequences such as UAAA and UUUA, and several other cellular mRNA [6, 35, 53, 83–85]. Further characterization of Sam68 interaction with these RNA, identification of additional cellular RNA targets, and elucidation of translational control of these RNA under oxidative stress will certainly add to our understanding of Sam68 biological functions.
Acknowledgments
We thank Dr. Janice Blum, Dr. Ann Roman, Dr. Jeremy Sanford and Dr. Ronald Wek for advices. We also thank Dr. Sonia Guil and Dr. Javier Caceres for GFP.TIA-1, GFP.hnRNP A1, GFP.PABP, and GFP.G3BP plasmids. This work was supported by the grants R01NS039804 and R01NS065785 (to JJH) from the National Institutes of Health.
ABBREVIATIONS
- eIF2α
eukaryotic elongation factor 2α
- G3BP
Ras-GTPase-activating protein SH3 domain-binding protein 1
- hnRNP A1
heteronuclear ribonucleoprotein A1
- KH
heteronuclear ribonucleoprotein particle K homology domain
- PABP
poly(A)-binding protein
- PB
processing body
- RBP
RNA-binding proteins
- Sam68
Src-associated protein in mitosis
- SG
stress granules
- TIA-1
T cell intracellular antigen-1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Henao-Mejia J, Liu Y, Park IW, Zhang J, Sanford J, He JJ. Suppression of HIV-1 Nef Translation by Sam68 Mutant-Induced Stress Granules and nef mRNA Sequestration. Mol Cell. 2009;33:87–96. doi: 10.1016/j.molcel.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellis C, Moran M, McCormick F, Pawson T. Phosphorylation of GAP and GAP-associated proteins by transforming and mitogenic tyrosine kinases. Nature. 1990;343:377–381. doi: 10.1038/343377a0. [DOI] [PubMed] [Google Scholar]
- 3.Moran MF, Koch CA, Anderson D, Ellis C, England L, Martin GS, Pawson T. Src homology region 2 domains direct protein-protein interactions in signal transduction. Proc Natl Acad Sci U S A. 1990;87:8622–8626. doi: 10.1073/pnas.87.21.8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong G, Muller O, Clark R, Conroy L, Moran MF, Polakis P, McCormick F. Molecular cloning and nucleic acid binding properties of the GAP-associated tyrosine phosphoprotein p62. Cell. 1992;69:551–558. doi: 10.1016/0092-8674(92)90455-l. [DOI] [PubMed] [Google Scholar]
- 5.Fumagalli S, Totty NF, Hsuan JJ, Courtneidge SA. A target for Src in mitosis. Nature. 1994;368:871–874. doi: 10.1038/368871a0. [DOI] [PubMed] [Google Scholar]
- 6.Taylor SJ, Shalloway D. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature. 1994;368:867–871. doi: 10.1038/368867a0. [DOI] [PubMed] [Google Scholar]
- 7.Lukong KE, Richard S. Sam68, the KH domain-containing superSTAR. Biochim Biophys Acta. 2003;1653:73–86. doi: 10.1016/j.bbcan.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Najib S, Martin-Romero C, Gonzalez-Yanes C, Sanchez-Margalet V. Role of Sam68 as an adaptor protein in signal transduction. Cell Mol Life Sci. 2005;62:36–43. doi: 10.1007/s00018-004-4309-3. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Liu Y, Kim BO, He JJ. Direct participation of Sam68, the 68-kilodalton Src-associated protein in mitosis, in the CRM1-mediated Rev nuclear export pathway. J Virol. 2002;76:8374–8382. doi: 10.1128/JVI.76.16.8374-8382.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLaren M, Asai K, Cochrane A. A novel function for Sam68: enhancement of HIV-1 RNA 3′ end processing. RNA. 2004;10:1119–1129. doi: 10.1261/rna.5263904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol. 2007;176:929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Babic I, Cherry E, Fujita DJ. SUMO modification of Sam68 enhances its ability to repress cyclin D1 expression and inhibits its ability to induce apoptosis. Oncogene. 2006;25:4955–4964. doi: 10.1038/sj.onc.1209504. [DOI] [PubMed] [Google Scholar]
- 13.Coyle JH, Guzik BW, Bor YC, Jin L, Eisner-Smerage L, Taylor SJ, Rekosh D, Hammarskjold ML. Sam68 enhances the cytoplasmic utilization of intron-containing RNA and is functionally regulated by the nuclear kinase Sik/BRK. Mol Cell Biol. 2003;23:92–103. doi: 10.1128/MCB.23.1.92-103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukong KE, Richard S. Motor coordination defects in mice deficient for the Sam68 RNA-binding protein. Behav Brain Res. 2008;189:357–363. doi: 10.1016/j.bbr.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Richard S, Torabi N, Franco GV, Tremblay GA, Chen T, Vogel G, Morel M, Cleroux P, Forget-Richard A, Komarova S, Tremblay ML, Li W, Li A, Gao YJ, Henderson JE. Ablation of the Sam68 RNA binding protein protects mice from age-related bone loss. PLoS Genet. 2005;1:e74. doi: 10.1371/journal.pgen.0010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richard S, Vogel G, Huot ME, Guo T, Muller WJ, Lukong KE. Sam68 haploinsufficiency delays onset of mammary tumorigenesis and metastasis. Oncogene. 2008;27:548–556. doi: 10.1038/sj.onc.1210652. [DOI] [PubMed] [Google Scholar]
- 17.Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Anderson P, Kedersha N. Stressful initiations. J Cell Sci. 2002;115:3227–3234. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- 19.Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am J Physiol Cell Physiol. 2003;284:C273–284. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- 20.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazroui R, Di Marco S, Kaufman RJ, Gallouzi IE. Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol Biol Cell. 2007;18:2603–2618. doi: 10.1091/mbc.E06-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeGracia DJ, Hu BR. Irreversible translation arrest in the reperfused brain. J Cereb Blood Flow Metab. 2007;27:875–893. doi: 10.1038/sj.jcbfm.9600388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dang Y, Kedersha N, Low WK, Romo D, Gorospe M, Kaufman R, Anderson P, Liu JO. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J Biol Chem. 2006;281:32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- 24.Mazroui R, Sukarieh R, Bordeleau ME, Kaufman RJ, Northcote P, Tanaka J, Gallouzi I, Pelletier J. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol Biol Cell. 2006;17:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solomon S, Xu Y, Wang B, David MD, Schubert P, Kennedy D, Schrader JW. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol Cell Biol. 2007;27:2324–2342. doi: 10.1128/MCB.02300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.McBride AE, Schlegel A, Kirkegaard K. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc Natl Acad Sci U S A. 1996;93:2296–2301. doi: 10.1073/pnas.93.6.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daoud R, Mies G, Smialowska A, Olah L, Hossmann KA, Stamm S. Ischemia induces a translocation of the splicing factor tra2-beta 1 and changes alternative splicing patterns in the brain. J Neurosci. 2002;22:5889–5899. doi: 10.1523/JNEUROSCI.22-14-05889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeGracia DJ, Rudolph J, Roberts GG, Rafols JA, Wang J. Convergence of stress granules and protein aggregates in hippocampal cornu ammonis 1 at later reperfusion following global brain ischemia. Neuroscience. 2007;146:562–572. doi: 10.1016/j.neuroscience.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paronetto MP, Bianchi E, Geremia R, Sette C. Dynamic expression of the RNA-binding protein Sam68 during mouse pre-implantation development. Gene Expr Patterns. 2008;8:311–322. doi: 10.1016/j.gep.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Gilbert C, Barabe F, Rollet-Labelle E, Bourgoin SG, McColl SR, Damaj BB, Naccache PH. Evidence for a role for SAM68 in the responses of human neutrophils to ligation of CD32 and to monosodium urate crystals. J Immunol. 2001;166:4664–4671. doi: 10.4049/jimmunol.166.7.4664. [DOI] [PubMed] [Google Scholar]
- 32.Ben Fredj N, Grange J, Sadoul R, Richard S, Goldberg Y, Boyer V. Depolarization-induced translocation of the RNA-binding protein Sam68 to the dendrites of hippocampal neurons. J Cell Sci. 2004;117:1079–1090. doi: 10.1242/jcs.00927. [DOI] [PubMed] [Google Scholar]
- 33.Grange J, Boyer V, Fabian-Fine R, Fredj NB, Sadoul R, Goldberg Y. Somatodendritic localization and mRNA association of the splicing regulatory protein Sam68 in the hippocampus and cortex. J Neurosci Res. 2004;75:654–666. doi: 10.1002/jnr.20003. [DOI] [PubMed] [Google Scholar]
- 34.Belly A, Moreau-Gachelin F, Sadoul R, Goldberg Y. Delocalization of the multifunctional RNA splicing factor TLS/FUS in hippocampal neurones: exclusion from the nucleus and accumulation in dendritic granules and spine heads. Neurosci Lett. 2005;379:152–157. doi: 10.1016/j.neulet.2004.12.071. [DOI] [PubMed] [Google Scholar]
- 35.Grange J, Belly A, Dupas S, Trembleau A, Sadoul R, Goldberg Y. Specific interaction between Sam68 and neuronal mRNAs: implication for the activity-dependent biosynthesis of elongation factor eEF1A. J Neurosci Res. 2009;87:12–25. doi: 10.1002/jnr.21824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paronetto MP, Zalfa F, Botti F, Geremia R, Bagni C, Sette C. The nuclear RNA-binding protein Sam68 translocates to the cytoplasm and associates with the polysomes in mouse spermatocytes. Mol Biol Cell. 2006;17:14–24. doi: 10.1091/mbc.E05-06-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noble SL, Allen BL, Goh LK, Nordick K, Evans TC. Maternal mRNAs are regulated by diverse P body-related mRNP granules during early Caenorhabditis elegans development. J Cell Biol. 2008;182:559–572. doi: 10.1083/jcb.200802128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paronetto MP, Messina V, Bianchi E, Barchi M, Vogel G, Moretti C, Palombi F, Stefanini M, Geremia R, Richard S, Sette C. Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J Cell Biol. 2009;185:235–249. doi: 10.1083/jcb.200811138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simarro M, Mauger D, Rhee K, Pujana MA, Kedersha NL, Yamasaki S, Cusick ME, Vidal M, Garcia-Blanco MA, Anderson P. Fas-activated serine/threonine phosphoprotein (FAST) is a regulator of alternative splicing. Proc Natl Acad Sci U S A. 2007;104:11370–11375. doi: 10.1073/pnas.0704964104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grossman JS, Meyer MI, Wang YC, Mulligan GJ, Kobayashi R, Helfman DM. The use of antibodies to the polypyrimidine tract binding protein (PTB) to analyze the protein components that assemble on alternatively spliced pre-mRNAs that use distant branch points. RNA. 1998;4:613–625. doi: 10.1017/s1355838298971448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marsh K, Soros V, Cochrane A. Selective translational repression of HIV-1 RNA by Sam68DeltaC occurs by altering PABP1 binding to unspliced viral RNA. Retrovirology. 2008;5:97. doi: 10.1186/1742-4690-5-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Liu Y, Park IW, He JJ. Expression of exogenous Sam68, the 68-kilodalton SRC-associated protein in mitosis, is able to alleviate impaired Rev function in astrocytes. J Virol. 2002;76:4526–4535. doi: 10.1128/JVI.76.9.4526-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Liu Y, Henao J, Rugeles MT, Li J, Chen T, He JJ. Requirement of an additional Sam68 domain for inhibition of human immunodeficiency virus type 1 replication by Sam68 dominant negative mutants lacking the nuclear localization signal. Gene. 2005;363:67–76. doi: 10.1016/j.gene.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 44.Guil S, Long JC, Caceres JF. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol Cell Biol. 2006;26:5744–5758. doi: 10.1128/MCB.00224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng C, Sharp PA. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol Cell Biol. 2006;26:362–370. doi: 10.1128/MCB.26.1.362-370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 47.Afonina E, Stauber R, Pavlakis GN. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J Biol Chem. 1998;273:13015–13021. doi: 10.1074/jbc.273.21.13015. [DOI] [PubMed] [Google Scholar]
- 48.Cande C, Vahsen N, Metivier D, Tourriere H, Chebli K, Garrido C, Tazi J, Kroemer G. Regulation of cytoplasmic stress granules by apoptosis-inducing factor. J Cell Sci. 2004;117:4461–4468. doi: 10.1242/jcs.01356. [DOI] [PubMed] [Google Scholar]
- 49.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 51.Lin JH, Walter P, Yen TS. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol. 2008;3:399–425. doi: 10.1146/annurev.pathmechdis.3.121806.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin Q, Taylor SJ, Shalloway D. Specificity and determinants of Sam68 RNA binding. Implications for the biological function of K homology domains. J Biol Chem. 1997;272:27274–27280. doi: 10.1074/jbc.272.43.27274. [DOI] [PubMed] [Google Scholar]
- 53.Chen T, Damaj BB, Herrera C, Lasko P, Richard S. Self-association of the single-KH-domain family members Sam68, GRP33, GLD-1, and Qk1: role of the KH domain. Mol Cell Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McBride AE, Taylor SJ, Shalloway D, Kirkegaard K. KH domain integrity is required for wild-type localization of Sam68. Exp Cell Res. 1998;241:84–95. doi: 10.1006/excr.1998.4047. [DOI] [PubMed] [Google Scholar]
- 55.McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem. 2005;280:16925–16933. doi: 10.1074/jbc.M412882200. [DOI] [PubMed] [Google Scholar]
- 56.McDowall MD, Scott MS, Barton GJ. PIPs: human protein-protein interaction prediction database. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang F, Peng Y, Murray EL, Otsuka Y, Kedersha N, Schoenberg DR. Polysome-bound endonuclease PMR1 is targeted to stress granules via stress-specific binding to TIA-1. Mol Cell Biol. 2006;26:8803–8813. doi: 10.1128/MCB.00090-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu C, York B, Wang S, Feng Q, Xu J, O’Malley BW. An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Mol Cell. 2007;25:765–778. doi: 10.1016/j.molcel.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rothe F, Gueydan C, Bellefroid E, Huez G, Kruys V. Identification of FUSE-binding proteins as interacting partners of TIA proteins. Biochem Biophys Res Commun. 2006;343:57–68. doi: 10.1016/j.bbrc.2006.02.112. [DOI] [PubMed] [Google Scholar]
- 60.Li W, Simarro M, Kedersha N, Anderson P. FAST is a survival protein that senses mitochondrial stress and modulates TIA-1-regulated changes in protein expression. Mol Cell Biol. 2004;24:10718–10732. doi: 10.1128/MCB.24.24.10718-10732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tocque B, Delumeau I, Parker F, Maurier F, Multon MC, Schweighoffer F. Ras-GTPase activating protein (GAP): a putative effector for Ras. Cell Signal. 1997;9:153–158. doi: 10.1016/s0898-6568(96)00135-0. [DOI] [PubMed] [Google Scholar]
- 62.Lukong KE, Larocque D, Tyner AL, Richard S. Tyrosine phosphorylation of sam68 by breast tumor kinase regulates intranuclear localization and cell cycle progression. J Biol Chem. 2005;280:38639–38647. doi: 10.1074/jbc.M505802200. [DOI] [PubMed] [Google Scholar]
- 63.Najib S, Rodriguez-Bano J, Rios MJ, Muniain MA, Goberna R, Sanchez-Margalet V. Sam68 is tyrosine phosphorylated and recruited to signalling in peripheral blood mononuclear cells from HIV infected patients. Clin Exp Immunol. 2005;141:518–525. doi: 10.1111/j.1365-2249.2005.02867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guitard E, Barlat I, Maurier F, Schweighoffer F, Tocque B. Sam68 is a Ras-GAP-associated protein in mitosis. Biochem Biophys Res Commun. 1998;245:562–566. doi: 10.1006/bbrc.1998.8374. [DOI] [PubMed] [Google Scholar]
- 65.Soros VB, Carvajal HV, Richard S, Cochrane AW. Inhibition of human immunodeficiency virus type 1 Rev function by a dominant-negative mutant of Sam68 through sequestration of unspliced RNA at perinuclear bundles. J Virol. 2001;75:8203–8215. doi: 10.1128/JVI.75.17.8203-8215.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gustin KE, Sarnow P. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 2001;20:240–249. doi: 10.1093/emboj/20.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lenart P, Ellenberg J. Monitoring the permeability of the nuclear envelope during the cell cycle. Methods. 2006;38:17–24. doi: 10.1016/j.ymeth.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 68.Sanchez-Margalet V, Gonzalez-Yanes C, Najib S, Fernandez-Santos JM, Martin-Lacave I. The expression of Sam68, a protein involved in insulin signal transduction, is enhanced by insulin stimulation. Cell Mol Life Sci. 2003;60:751–758. doi: 10.1007/s00018-003-2342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smida M, Posevitz-Fejfar A, Horejsi V, Schraven B, Lindquist JA. A novel negative regulatory function of the phosphoprotein associated with glycosphingolipid-enriched microdomains: blocking Ras activation. Blood. 2007;110:596–615. doi: 10.1182/blood-2006-07-038752. [DOI] [PubMed] [Google Scholar]
- 70.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.David PS, Tanveer R, Port JD. FRET-detectable interactions between the ARE binding proteins, HuR and p37AUF1. RNA. 2007;13:1453–1468. doi: 10.1261/rna.501707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim SH, Dong WK, Weiler IJ, Greenough WT. Fragile X mental retardation protein shifts between polyribosomes and stress granules after neuronal injury by arsenite stress or in vivo hippocampal electrode insertion. J Neurosci. 2006;26:2413–2418. doi: 10.1523/JNEUROSCI.3680-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mazroui R, Huot ME, Tremblay S, Filion C, Labelle Y, Khandjian EW. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum Mol Genet. 2002;11:3007–3017. doi: 10.1093/hmg/11.24.3007. [DOI] [PubMed] [Google Scholar]
- 74.Chalupnikova K, Lattmann S, Selak N, Iwamoto F, Fujiki Y, Nagamine Y. Recruitment of the RNA Helicase RHAU to Stress Granules via a Unique RNA-binding Domain. J Biol Chem. 2008;283:35186–35198. doi: 10.1074/jbc.M804857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fujimura K, Kano F, Murata M. Identification of PCBP2, a facilitator of IRES-mediated translation, as a novel constituent of stress granules and processing bodies. RNA. 2008;14:425–431. doi: 10.1261/rna.780708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stohr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, Huttelmaier S. ZBP1 regulates mRNA stability during cellular stress. J Cell Biol. 2006;175:527–534. doi: 10.1083/jcb.200608071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Courchet J, Buchet-Poyau K, Potemski A, Bres A, Jariel-Encontre I, Billaud M. Interaction with 14-3-3 adaptors regulates the sorting of hMex-3B RNA-binding protein to distinct classes of RNA granules. J Biol Chem. 2008;283:32131–32142. doi: 10.1074/jbc.M802927200. [DOI] [PubMed] [Google Scholar]
- 78.Chen T, Cote J, Carvajal HV, Richard S. Identification of Sam68 arginine glycine-rich sequences capable of conferring nonspecific RNA binding to the GSG domain. J Biol Chem. 2001;276:30803–30811. doi: 10.1074/jbc.M102247200. [DOI] [PubMed] [Google Scholar]
- 79.Cote J, Boisvert FM, Boulanger MC, Bedford MT, Richard S. Sam68 RNA binding protein is an in vivo substrate for protein arginine N-methyltransferase 1. Mol Biol Cell. 2003;14:274–287. doi: 10.1091/mbc.E02-08-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jan E, Motzny CK, Graves LE, Goodwin EB. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 1999;18:258–269. doi: 10.1093/emboj/18.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schumacher B, Hanazawa M, Lee MH, Nayak S, Volkmann K, Hofmann ER, Hengartner M, Schedl T, Gartner A. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell. 2005;120:357–368. doi: 10.1016/j.cell.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 82.Larocque D, Galarneau A, Liu HN, Scott M, Almazan G, Richard S. Protection of p27(Kip1) mRNA by quaking RNA binding proteins promotes oligodendrocyte differentiation. Nat Neurosci. 2005;8:27–33. doi: 10.1038/nn1359. [DOI] [PubMed] [Google Scholar]
- 83.Lin Q, Taylor SJ, Shalloway D. Specificity and determinants of Sam68 RNA binding. Implications for the biological function of K homology domains. J Biol Chem. 1997;272:27274–27280. doi: 10.1074/jbc.272.43.27274. [DOI] [PubMed] [Google Scholar]
- 84.Itoh M, Haga I, Li QH, Fujisawa J. Identification of cellular mRNA targets for RNA-binding protein Sam68. Nucleic Acids Res. 2002;30:5452–5464. doi: 10.1093/nar/gkf673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tremblay GA, Richard S. mRNAs associated with the Sam68 RNA binding protein. RNA Biol. 2006;3:1–4. doi: 10.4161/rna.3.2.3204. [DOI] [PubMed] [Google Scholar]