Abstract
We describe a novel experiment that we conducted with the Drug Interaction Knowledge-base (DIKB) to determine which combinations of evidence enable a rule-based theory of metabolic drug-drug interactions to make the most optimal set of predictions. The focus of the experiment was a group of 16 drugs including six members of the HMG-CoA-reductase inhibitor family (statins). The experiment helped identify evidence-use strategies that enabled the DIKB to predict significantly more interactions present in a validation set than the most rigorous strategy developed by drug experts with no loss of accuracy. The best-performing strategies included evidence types that would normally be of lesser predictive value but that are often more accessible than more rigorous types. Our experimental methods represent a new approach to leveraging the available scientific evidence within a domain where important evidence is often missing or of questionable value for supporting important assertions.
Keywords: knowledge representation, drug safety, drug-drug interactions, HMG-CoA reductase inhibitors, statins, evidentialism, evidence modeling
1 Introduction
Our research focuses on how to best utilize drug-mechanism knowledge to help drug-interaction knowledge-bases expand their coverage beyond what has been tested in clinical trials while avoiding prediction errors that occur when individual drug differences are not recognized. Drug mechanism knowledge presents difficult informatics challenges because it is often dynamic, frequently uncertain, and sometimes missing. A previous publication by our group describes a novel evidential knowledge-representation approach that we designed to address these issues [1]. We have implemented the approach in a system called the Drug Interaction Knowledge Base (DIKB) and have used the system to represent drug-mechanism evidence for 16 drugs including six members of the HMG-CoA-reductase inhibitor family (statins) [2].
An intriguing feature of the DIKB’s knowledge-representation approach is that it enables exploration of the empirical prediction accuracy of a rule-based theory using many sub-sets of a given body of evidence. We tested the usefulness of this feature for integrating information from basic science, clinical research, and authoritative statements for the purpose of making drug-drug interaction (DDI) predictions. This paper briefly reviews the knowledge-representation approach implemented in the DIKB, details the methods and results of our novel experiment, and then discusses its implications for informatics and drug safety.
1.1 The DIKB’s Evidential Knowledge-representation Approach
The DIKB is a knowledge representation system designed to predict DDIs using drug mechanisms. A key component of the system is a rule-based theory of how drugs interact by metabolic inhibition. The system’s knowledge-base contains assertions about specific entities such as drugs, drug metabolites, and enzymes whose relationships with each other are more generally modeled in the rule-based theory. DIKB maintainers place evidence for, or against, each assertion in the system’s knowledge-base in an evidence-base that is kept current by an editorial board. Before each evidence item is entered into the system, maintainers identify its type from a biomedical evidence taxonomy designed to have sufficient coverage of the kinds of evidence relevant for supporting or refuting drug mechanism assertions [2]. They then confirm that the evidence item meets explicitly defined inclusion criteria that apply to all evidence of its type.
The DIKB distinguishes between assertion instances and assertion types. An assertion instance is a clear statement of some property or relationship belonging to one or more entities while an assertion type is the general form of all such instances. For example, the generic (X substrate-of Y) is an assertion type describing a general relationship between an abstract drug and an abstract enzyme. Instances of this assertion type might include assertions such as (carbamazepine substrate-of CYP3A4) and (s-warfarin substrate-of CYP2C9). Expert users map their confidence in drug-mechanism assertions by first defining combinations of evidence types from the system’s evidence taxonomy that might support or refute instances of each assertion type. They then rank the evidence-type combinations by the relative amount of confidence that they would have in an assertion instance of the given assertion type if it were supported by the types of evidence present in the definition. We call such rank-ordered combinations of evidence types levels-of-evidence (LOEs).
For each assertion type in the system, expert users define two, possibly identical, sets of LOEs; one for the types of evidence that can support an assertion type, the other for the types of evidence that refute it. They then select one LOE from each set of LOEs as a belief criterion. Evidence against an assertion dominates evidence for it hence, a query of the DIKB’s knowledge-base for valid drug-mechanism assertions will return only those assertions whose body of evidence for satisfies the belief criterion for supporting evidence and whose body of evidence against does not satisfy the belief criterion for refuting evidence. In this way, the system makes DDI predictions using only those assertions considered current by the system’s maintainers and believable by users.
1.2 The Objective of an Experiment with Belief Criteria
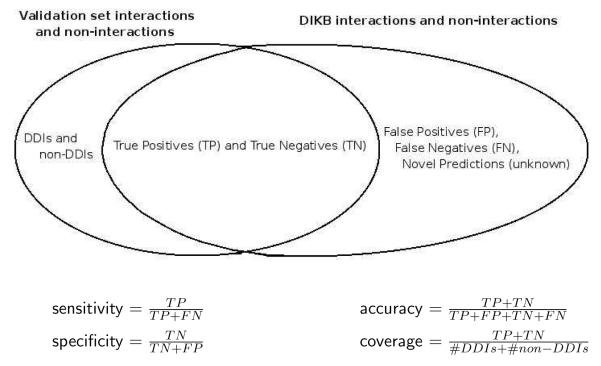
When drug experts define and rank LOEs or choose belief criteria, they are making subjective judgements about the inferential force of an abstract body of evidence. An important question is whether the experts’ choices have any relationship to empirical measures of the system’s performance. Figure 1 defines four metrics relevant to evaluating the DIKB — sensitivity, specificity, coverage, and accuracy. Our hypothesis was that the system would have poorer coverage but greater accuracy when using belief criteria that inspire complete confidence in a drug expert than when using criteria that the expert believes to be less trustworthy. We based this hypothesis on our intuition that the evidence types that experts consider less trustworthy are more readily available in the literature, but often contain less generalizable scientific findings, than types that the same experts view as more trustworthy.
Fig. 1.
The formulas in this figure define four of the metrics that were used to characterize the performance of the DIKB using various belief criteria.
We conducted an experiment to characterize the effect of varying belief criteria on the system’s accuracy and coverage of DDIs present in a reference set of interactions and non-interactions. The experiment was designed to answer the following research questions:
What is the DIKB’s accuracy and coverage of reference set interactions and non-interactions when using expert-defined belief criteria?
Does changing the LOEs that are selected as belief criteria alter the system’s prediction accuracy or coverage?
Do computational experiments imply a particular belief criteria strategy that optimizes the system’s prediction performance?
2 Methods
2.1 Creation of the Drug Mechanism Knowledge-base
We collected sufficient evidence on the pharmacokinetic drug properties of 16 drugs and 19 drug metabolites to perform this experiment. Figure 2 lists the specific drugs and drug metabolites that we chose to represent in the DIKB. The methods used to collect, classify, and enter evidence into the DIKB are described in part I of this two part series [2].
Fig. 2.
The 16 drugs and 19 drug metabolites chosen for DIKB experiments
2.2 Creation of the Experiment’s Validation Set
Once the evidence-base was complete except for minor revisions, we attempted to identify all known pairwise metabolic inhibition interactions and non-interactions between the 35 pharmaceutical entities in the DIKB’s evidence-base. Our intent was to use the interactions and non-interactions that we found as a validation set for determining the accuracy and coverage of the DIKB’s DDI predictions. The DIKB predicts interactions using knowledge of drug mechanisms and a rule-based theory of how interactions occur by metabolic inhibition. The method that we used to confirm interactions and non-interactions approximated an independent reference standard because it relied on quantitative observations and did not consider underlying mechanisms.
We considered a metabolic inhibition interaction to be independently confirmed if any one of following criteria was satisfied:
A pharmacokinetic DDI study provided data showing a statistically significant (defined below) increase in the Area Under the concentration-time Curve (AUC) of the study’s object drug or drug metabolite in the presence of the precipitant drug or drug metabolite.
An observation-based case report provided data showing a measurable increase in the systemic concentration of a drug or drug metabolite in the presence of another drug or drug metabolite and the evidence board could find no viable alternate explanation for the observed increase.
We considered a metabolic inhibition non-interaction to be independently confirmed if all the following criteria were satisfied:
A pharmacokinetic DDI study provided data showing no statistically significant (defined below) increase in the AUC of the study’s object drug or drug metabolite in the presence of the study’s precipitant drug or drug metabolite.
None of the criteria listed above as independently confirming a metabolic inhibition interaction was met.
If neither a metabolic inhibition interaction or non-interaction could be confirmed for any pair of pharmaceutical entities in the DIKB then, we labeled the pair as having no known interaction or non-interaction.
2.2.1 AUC Ratios and Statistical Significance
We defined a statistically significant increase in AUC to be:
| (1) |
Where AUC is the baseline AUC for a DDI study’s object drug or drug metabolite and AUCi is the AUC for the object drug in the presence of the study’s precipitant drug or drug metabolite. Often studies do not provide p-values, in such cases we judged an AUC increase statistically significant if the study provided 95% confidence intervals for the AUC ratio () that did not include 1.0. If the study’s results did not satisfy Equation 1, and/or the 95% confidence intervals for the AUC ratio (if available) included 1.0 then, we defined the metabolic inhibition interaction to not be statistically significant.
2.2.2 Inclusion Criteria for Validation Set Data
We sought evidence for confirming interactions and non-interactions from three sources of pharmacokinetic data — published research articles, drug product labeling, and published observation-based case reports. If the data came from a research article then, the study must have satisfied the definition and inclusion criteria of the evidence type A DDI clinical trial or any of its sub-types in the DIKB evidence taxonomy (Appendices A and B, supplementary material). If we found the data in drug product labeling then it must have met the inclusion criteria for the DIKB evidence type A non-traceable drug-label statement (Appendix B, supplementary material). Finally, case reports needed to meet the inclusion criteria for evidence of their type listed in Appendix B (supplementary material) and provide quantitative measurements of the systemic concentration of the purported object drug before and after administration of the purported precipitant drug.
2.2.3 The Collection of Pharmacokinetic Data
We designed criteria for confirming or refuting a metabolic-inhibition DDI before we began collecting evidence. Once the DIKB’s evidence-base was complete, we began building the validation set. We started by enumerating all ordered pairwise combinations of the 35 drugs and drug metabolites in the DIKB’s final evidence-base. We used a single label in the validation set to represent both possible ways that an interaction might occur between a drug or drug metabolite pair. For example, assuming two drugs X and Y, we used a single label X - Y to represent two possible interactions — that X effects a change in the systemic concentration of Y and vice versa. This resulted in a total of 595 labels (Equation 2) excluding same-compound combinations (e.g. simvastatin-simvastatin). Appendix C (supplementary material) lists 595 labels representing all 1190 potential pairwise interaction and non-interactions between the drugs and drug metabolites in the DIKB.
| (2) |
2.2.4 Avoiding Biased Measures of the DIKB’s Accuracy
Throughout the evidence collection process we often found clinical trials providing data that were relevant to both establishing the validity of an assertion about some metabolic property and confirming a metabolic interaction or non-interaction. To avoid biasing the validation set, we first entered evidence items that could be applied to both the DIKB’s evidence-base and the validation set in both places. Then, before assessing the system’s accuracy, we identified any evidence item that supported a claim made by the validation set and was also used to support a DIKB assertion that could lead to the same conclusion. We dropped from further analysis the interaction or non-interaction that such evidence supported. In total, seven pairs were dropped from further analysis for this reason. These seven drug/drug or drug/drug-metabolite pairs are shown in Table 1 (supplementary material) along with two other pairs that were accidentally excluded from the experiment described in this paper due to a transcription error. Excluding these nine pairs brought the total number of drug/drug and drug/drug-metabolite pairs used for characterizing the DIKB’s accuracy down to 586.
2.2.5 Searching for Validation-set DDIs in the primary literature and Drug-product Labeling
We searched the primary literature for clinical trials involving any of the 586 drug/drug and drug/metabolite pairs in Appendix C (supplementary material) by querying PubMed 1 and the proprietary University of Washington Metabolism and Transport Drug Interaction Database 2 After completing an intensive search for all relevant clinical trials, we conducted a search in drug product labeling for statements mentioning a pharmacokinetic interaction or non-interaction involving any of the 586 pairs. We searched all labels written for each drug product whose only active pharmaceutical ingredient was a drug in the DIKB. The number of qualifying product labels for each drug ranged from one (atorvastatin, fluvastatin, and rosuvastatin) to 18 (diltiazem) but a large proportion of the statements in one product label were repeated in all of the other available labels. All identified statements were noted and then filtered so that only statements providing quantitative data were used to support validation-set interactions. All searches were done using product labeling in the NLM’s DailyMed database. 3
In total, we found 65 drug-product labeling statements that mentioned a pharmacokinetic interaction or non-interaction between the members of some drug or drug-metabolite pair included in our study. A small sample of these 65 statements is shown in Table 2 (supplementary material). Only 21 statements (31%) reported the quantitative results of a pharmacokinetic clinical trial. We approved these 21 for use in the validation set; the remaining 44 statements were retained so that drug-product labeling, clinical trial literature, and case reports could be compared for their agreement on validation set interactions and non-interactions at a later time.
2.2.6 Searching for Validation-set DDIs in Case Report Literature
Having completed searches within clinical trial literature and drug-product labeling, we then did an intensive search of the University of Washington Metabolism and Transport Drug Interaction Database and PubMed for published case reports claiming the occurrence of a DDI between any pair of the active ingredients or metabolites in our study. This search resulted in 35 relevant case reports. We evaluated the full-text article for each of the 35 reports to see if it met the pre-defined inclusion criteria. Unfortunately, none of the 35 reports was accepted for use in the validation set. We rejected most case reports because they did not provide quantitative measurements of the systemic concentration of the purported object drug before and after administration of the purported precipitant drug. Three case reports provided adequate measurements of systemic concentration but failed to meet inclusion criteria by not receiving a causation rating of at least “probable” when co-investigator JH assessed the report using Drug Interaction Probability Scale [3]. Table 3 (supplementary material) cites the reports and provides an explanation for their low causation score.
2.2.7 The Final Validation Set
The interactions and non-interactions in the final validation set are shown in Tables 4 and 5. The validation set claims that some DDI will occur by metabolic inhibition for 41 drug/drug and drug/drug-metabolite pairs and that no DDI will occur by metabolic inhibition for seven pairs. No interaction or non-interaction could be identified for 538 pairs in the validation set (Appendix C, supplementary material). It is important to stress that many of these pairs might have clinically-relevant DDIs that were missed by our evidence collection process or that have not been reported in the sources we searched.
Table 4.
The 41 validation set interactions that were used to characterize the DIKB’s prediction performance.
| Pair | Source |
|---|---|
| alprazolam* - erythromycin | [29] |
| alprazolam* - itraconazole | [30] |
| alprazolam* - ketoconazole | [31], [32] |
| alprazolam* - nefazodone | [33], [34] |
| atorvastatin* - erythromycin | [35] |
| atorvastatin* - nefazodone | [36] |
| clarithromycin - atorvastatin* | [37], [38] |
| clarithromycin* - fluconazole | [39] |
| clarithromycin - pravastatin* | [38] |
| diltiazem - beta-OH-lovastatin* | [40] |
| diltiazem - lovastatin* | [41] |
| diltiazem - midazolam* | [42] |
| diltiazem - simvastatin* | [43] |
| diltiazem - triazolam* | [44] |
| erythromycin - beta-OH-simvastatin* | [45] |
| erythromycin - simvastatin* | [45] |
| fluconazole - 1′-OH-midazolam*† | [46] |
| fluconazole - fluvastatin* | [47] |
| itraconazole - atorvastatin* | [48] |
| itraconazole - beta-OH-lovastatin* | [10] |
| itraconazole* - erythromycin | [49] |
| itraconazole - lovastatin* | [10] |
| itraconazole - ortho-OH-atorvastatin* † | [48] |
| itraconazole - pravastatin* | [48] |
| itraconazole - rosuvastatin* | [14] |
| ketoconazole - simvastatin* | [50] |
| midazolam* - clarithromycin | [51], [52] |
| midazolam* - erythromycin | [53] |
| midazolam* - fluconazole | [46], [54] |
| midazolam* - itraconazole | [55] |
| midazolam* - ketoconazole | [55] |
| midazolam* - nefazodone | [56] |
| nefazodone - 4-OH-alprazolam* † | [34] |
| nefazodone - beta-OH-simvastatin* | [36] |
| nefazodone - simvastatin* | [36] |
| triazolam* - clarithromycin | [17] |
| triazolam* - erythromycin | [57] |
| triazolam* - fluconazole | [58] |
| triazolam* - itraconazole | [59], [60] |
| triazolam* - ketoconazole | [60], [61] |
| triazolam* - nefazodone | [62] |
Asterisks indicate the object of a metabolic inhibition interaction occurring between the pair.
The noted interaction occurs by inhibition of the metabolic clearance of a parent compound.
Table 5.
The seven validation set non-interactions that we used to characterize the DIKB’s prediction performance.
| Pair | Source |
|---|---|
| diltiazem - pravastatin* | [41] |
| erythromycin - rosuvastatin* | [8] |
| fluconazole - 14-OH-clarithromycin* | [39] |
| fluconazole - pravastatin* | [47] |
| fluconazole - rosuvastatin* | [13] |
| itraconazole - fluvastatin* | [10] |
| nefazodone - pravastatin* | [36] |
Asterisks indicate which drug or drug metabolite should not be affected by a metabolic inhibition interaction involving the other drug in the pair.
2.3 The Creation of Prediction Sets for the Experiment
2.3.1 Expert-defined Belief Criteria
Once work on the evidence-base and validation set was complete, the two drug experts in our group (CC and JH) then defined one or more LOEs for each assertion type. This was a two step process; co-investigator RB (an informaticist) first identified all evidence types from the DIKB’s evidence taxonomy (Appendix A, supplementary material) that were applicable to each assertion type. He then helped the two drug experts decide the degree of certainty they would have in an assertion instance whose evidence support consisted of the evidence items represented by each evidence type or combination of evidence types. 4
2.3.2 The Drug Experts’ Levels-of-evidence and Ranking Categories
Table 6 shows the set of labels, or ranking categories, that the drug experts used define LOEs. Some labels in Table 6 represent multiple evidence types that the drug experts believed would confer roughly the same degree of justification for drug mechanism assertions. The experts felt that several evidence types would not be useful for supporting or refuting particular assertion types. These evidence types were grouped into a special “not applicable” ranking category and inserted as the lowest-ranking level-of-evidence for the assertion type’s LOE.
Table 6.
This table shows the set of labels, or ranking categories, that the drug experts used define LOEs.
| Ranking category | Evidence types |
|---|---|
| in vitro categories | |
| iv-met-enz-id-Cyp-450-with-inh |
A CYP450, recombinant, drug metabolism identification experiment using chemical inhibitors A CYP450, human microsome, drug metabolism identifi- cation experiment using chemical inhibitors A CYP450, recombinant, drug metabolism identification experiment using antibody inhibitors A CYP450, human microsome, drug metabolism identifi- cation experiment using antibody inhibitors |
| iv-met-enz-id-Cyp-450-recombinant |
A CYP450, recombinant, drug metabolism identification experiment with possibly NO probe enzyme inhibitor(s) |
| iv-met-enz-id-Cyp-450-microsomal |
A CYP450, human microsome, drug metabolism identifi- cation experiment |
| iv-met-inh-recombinant |
A CYP450, recombinant, metabolic enzyme inhibition ex- periment |
| iv-met-inh-microsomal |
A CYP450, human microsome, metabolic enzyme inhibi- tion experiment |
| Clinical Trial categories | |
| pk-ct-pk |
A randomized DDI clinical trial A genotyped pharmacokinetic clinical trial A phenotyped pharmacokinetic clinical trial |
| pk-ct-pk-genotype | A genotyped pharmacokinetic clinical trial |
| pk-ct-pk-phenotype | A phenotyped pharmacokinetic clinical trial |
| pk-ddi-non-rndm |
A non-randomized DDI clinical trial: A parallel groups DDI clinical trial |
| pk-ddi-rndm | A randomized DDI clinical trial |
| Statement categories | |
| label-statement | A non-traceable drug-label statement |
| nt-statement | A non-traceable, but possibly authoritative, statement |
| Observation-based categories | |
| obs-eval | A published and evaluated observation-based ADE report |
| “not applicable” categories | |
| na-primary-total-clearance-enz † | A non-traceable, but possibly authoritative, statement |
| na-primary-metabolic-clearance-enzyme † |
A CYP450, human microsome, drug metabolism identifi- cation experiment using chemical inhibitors |
| na-substrate-of † |
A CYP450, recombinant, drug metabolism identification experiment with possibly NO probe enzyme inhibitor(s) |
Note: multiple evidence types that the drug experts thought would confer roughly the same degree of justification for drug mechanism assertions are represented by a single label.
A ranking category created to represent evidence types that the drug experts felt would not be applicable to supporting or refuting particular assertions.
While LOEs were designed separately for each assertion type, many of the resulting LOEs were alike. As a result, only 15 LOEs were defined for the 22 assertion types in the DIKB’s rule-based theory of metabolic drug-drug interactions. Table 7 enumerates the assertion types that share the same LOEs. The symbols for each LOE in Table 7 are used in Table 8 to show a concise overview of the ranking categories used for all LOEs. An explicit definition of each LOE is given in Table 9 (supplementary material).
Table 7.
While LOEs were designed separately for each assertion type, many of the resulting LOEs were alike. As a result, only 15 LOEs were defined for the 22 assertion types in the DIKB’s rule-based theory of metabolic drug-drug interactions. This table enumerates the assertion types that share the same LOEs.
| LOE ID | Applies to |
|---|---|
| A | bioavailability maximum-concentration |
| B | first-pass-effect fraction-absorbed |
| C | primary-total-clearance-mechanism |
| D | controls-formation-of substrate-of is-not-substrate-of |
| E | primary-total-clearance-enzyme |
| F | primary-metabolic-clearance-enzyme |
| G | has-metabolite |
| H | inhibition-constant |
| I | inhibits |
| J | does-not-inhibit |
| K | does-not-permanently-deactivate-catalytic-function permanently-deactivates-catalytic-function in-vitro-probe-substrate-of-enzyme |
| L | in-vitro-selective-inhibitor-of-enzyme |
| M | in-viVo-selective-inhibitor-of-enzyme sole-PK-effect-alter-metabolic-clearance |
| N | pceut-entity-of-concern |
| O | polymorphic-enzyme |
Table 8.
The symbols for each LOE in Table 7 are used in this table to show a concise overview of the ranking categories used for all LOEs. While an explicit definition of each each LOE is given in Table 9 (supplementary material), it is possible to derive the same definitions from the columns in this table. For example, the column for LOE-1 shows that the LOE for group “A” (Table 7) consists of the evidence types represented by two ranking categories (Table 6): pk-ct-pk and label-statement.
| Ranking category | LOE-1 | LOE-2 | LOE-3 | LOE-4 | LOE-5 |
|---|---|---|---|---|---|
| pk-ct-pk | A,B,C,G | ||||
| pk-ct-pk-genotype | D,E,F,O | ||||
| pk-ct-pk-phenotype | D,E,F,O | ||||
| pk-ddi-rndm | I,M,N | D,E,F,J | |||
| pk-ddi-non-rndm | I,N | D,E,F,J,M | |||
| iv-met-enz-id-Cyp-450-with-inh | G | D | |||
| iv-met-enz-id-Cyp-450-recombinant | K | ||||
| iv-met-enz-id-Cyp-450-microsomal | K | ||||
| iv-met-inh-recombinant | H,J,L | I | |||
| iv-met-inh-microsomal | H,J | I,L | |||
| label-statement | A,N | B,C,H,I,J,K,O | E,F,G,L,M | D | |
| nt-statement | N | K, | O | L,M | |
| obs-eval | N | ||||
| na-primary-total-clearance-enz | C | ||||
| na-primary-metabolic-clearance-enzyme | F | ||||
| na-substrate-of | G | D |
2.3.3 The Drug Experts’ Belief Criteria Strategy
Once they had defined a complete set of LOEs, the drug experts selected two LOEs for each assertion type as its belief criteria; one for evidence supporting an assertion and the other for evidence refuting an assertion (see Table 10). The experts’ belief criteria strategy used the top level-of-evidence from every LOE definition and, therefore, was the most stringent one tested.
Table 10.
This table shows the evidence board’s belief criteria strategy. The columns next to each assertion type indicate the LOE group and specific LOE that the evidence-board chose as the type’s belief criteria. There are two columns because the DIKB allows the belief criterion for supporting evidence to be different than the belief criterion for refuting evidence. Note that the belief criterion for supporting evidence is the same as the belief criterion for refuting evidence for all but the inhibits and does-not-inhibit assertion types. Assertion types indicated as default assumptions are noted separately because the system uses them for inference even if their evidence does not meet belief criteria
| Assertion types not used as default assumptions | ||
|---|---|---|
| Assertion type | Evidence for | Evidence against |
| bioavailability | A, LOE-1 | A, LOE-1 |
| maximum-concentration | A, LOE-1 | A, LOE-1 |
| first-pass-effect | B, LOE-1 | B, LOE-1 |
| fraction-absorbed | B, LOE-1 | B, LOE-1 |
| primary-total-clearance-mechanism | C, LOE-1 | C, LOE-1 |
| controls-formation-of | D, LOE-1 | D, LOE-1 |
| substrate-of | D, LOE-1 | D, LOE-1 |
| is-not-substrate-of | D, LOE-1 | D, LOE-1 |
| primary-total-clearance-enzyme | E, LOE-1 | E, LOE-1 |
| primary-metabolic-clearance-enzyme | F, LOE-1 | F, LOE-1 |
| has-metabolite | G, LOE-1 | G, LOE-1 |
| inhibition-constant | H, LOE-1 | H, LOE-1 |
| inhibits | I, LOE-1 | J, LOE-1 |
| does-not-inhibit | J, LOE-1 | I, LOE-1 |
| Assertion types used as default assumptions | ||
|---|---|---|
| permanently-deactivates-catalytic-function | K, LOE-1 | K, LOE-1 |
| does-not-permanently-deactivate-catalytic-function | K, LOE-1 | K, LOE-1 |
| in-vitro-probe-substrate-of-enzyme | K, LOE-1 | K, LOE-1 |
| in-vitro-selective-inhibitor-of-enzyme | L, LOE-1 | L, LOE-1 |
| in-viVo-selective-inhibitor-of-enzyme | M, LOE-1 | M, LOE-1 |
| sole-PK-effect-alter-metabolic-clearance | M, LOE-1 | M, LOE-1 |
| pceut-entity-of-concern | N, LOE-1 | N, LOE-1 |
| polymorphic-enzyme | O, LOE-1 | O, LOE-1 |
2.3.4 Automatically-generated Belief Criteria Strategies
We expanded the DIKB so that it could make interaction and non-interaction predictions using every belief criteria strategy possible with the expert-defined LOEs. We were now able to characterize the effect of varying belief criteria on the system’s accuracy and coverage.
A default assumption is a special kind of assertion that is considered justified with no evidence support and that can be asserted or retracted either manually by curators or automatically by the system as it proceeds with inference. Initially, we were going to generate all belief criteria strategies by varying the LOEs chosen as belief criteria for every assertion type that was not labeled as a default assumption. We excluded default assumptions because varying their belief criteria has no effect on the DIKB’s predictions. Table 11 lists the set of assertion types not labeled as default assumptions along with the number of LOEs that the evidence-board defined for each of them. As Equation 3 shows, the total number of belief criteria strategies that the DIKB would have generated for these assertion types is 576,000.
Table 11.
The set of assertion types whose validity in the DIKB’s knowledge-base is based on evidence assessment. The assertion types are shown in decreasing order based on the number of LOEs that we defined for each of them.
| Assertion type | LOE count |
|---|---|
| controls-formation-of | 5 |
| substrate-of | 5 |
| is-not-substrate-of | 5 |
| has-metabolite | 4 |
| primary-metabolic-clearance-enzyme | 4 |
| primary-total-clearance-enzyme | 3 |
| primary-total-clearance-mechanism | 3 |
| does-not-inhibit † | 2 |
| first-pass-effect † | 2 |
| fraction-absorbed † | 2 |
| inhibition-constant † | 2 |
| inhibits | 2 |
| maximum-concentration † | 1 |
| bioavailability † | 1 |
An assertion type for which varying the LOE chosen as a belief criterion would have no effect on the DIKB’s prediction performance because all of the associated evidence was of a type belonging to the highest ranking LOE.
| (3) |
However, inspection of the DIKB’s evidence-base revealed that there were six assertion types for which all of the evidence items, for or against, belonged to the highest-ranked LOE for the type. This meant that varying the LOE chosen as a belief criterion for any of these types (see Table 11) would have no effect on the DIKB’s prediction performance. Excluding these six assertion types and the eight assertion types labeled as default assumptions meant that there were eight assertion types for which varying the LOE chosen as a belief criterion would have an effect on prediction performance. The DIKB generated all 36,000 different belief criteria strategies possible for the remaining eight assertion types by altering the LOEs chosen as belief criteria. The 14 remaining assertion types used the highest-ranking LOEs defined for their types as belief criteria.
2.4 Statistical Analysis and Programming
We used the R statistical language [4] to calculate all descriptive and performance statistics. Bruno Falissard’s psy package [5] was used to calculate a three-valued Cohen’s kappa score as a measure of the degree over random chance to which the DIKB and validation set agreed on interactions, non-interactions, and unknowns. Both R and the Python programming language 5 were used extensively to write various programs that aided our analysis.
3 Results
3.1 Predictions Made Using the Drug-expert’s Belief Criteria Strategy
We began the experiment by testing the accuracy and coverage of the DIKB using the drug experts’ belief criteria strategy (Section 2.3.3). Using this strategy, the DIKB predicted that 15 drug/drug or drug/drug-metabolite pairs would interact by metabolic inhibition and that two would not (see Table 12). Fourteen interaction predictions were present in the validation set and so were considered true positives. The remaining interaction prediction and the two non-interaction predictions were classified as “unknown” because they were neither confirmed nor refuted by the validation set.
Table 12.
Seventeen interaction and non-interaction predictions made by the DIKB using the evidence-board’s belief criteria strategy.
| Pair | Prediction | AUCi/AUC‡ |
|---|---|---|
| diltiazem - midazolam* | ≥ 3 | |
| diltiazem - triazolam* | 2.83 | |
| midazolam* - clarithromycin | ≥ 7 | |
| midazolam* - erythromycin | 4.4 | |
| midazolam* - fluconazole | ≥ 2.6 | |
| midazolam* - itraconazole | 10.8 | |
| midazolam* - ketoconazole | 15.9 | |
| midazolam* - nefazodone | 4.6 | |
| triazolam* - clarithromycin | 5.3 | |
| triazolam* - erythromycin | 2.1 | |
| triazolam* - fluconazole | 2.5 | |
| triazolam* - itraconazole | ≥ 3.1 | |
| triazolam* - ketoconazole | ≥ 9.2 | |
| triazolam* - nefazodone | ≥ 3.9 | |
| triazolam* - atorvastatin † | n/a | |
| triazolam - simvastatin † | no interaction | n/a |
| triazolam - β-OH-simvastatin † | no interaction | n/a |
Asterisks indicate the drug or drug metabolite that the system predicts will be the object of the interaction. The DIKB makes interaction predictions at three levels representing the anticipated AUC ratio of the DDI’s object drug: , , and .
a pair classified as “unknown” in the validation set.
AUC ratios present in at least one evidence item in the validation set that supports the interaction.
The predicted pharmacokinetic magnitude of all 14 confirmed predictions corresponded with levels observed in clinical trial data. While the system’s predictions and magnitude estimates using the drug experts’ strategy had perfect accuracy, its coverage of known interactions was poor. Only 14 (34%) of the 41 pairs known in the validation set to interact by metabolic inhibition were predicted to interact by the DIKB. Also, the system failed to predict any of the seven pairs known in the validation set to not interact by the same mechanism. The system’s poor coverage using the experts’ belief criteria strategy was expected since it was the most stringent strategy tested.
3.2 Predictions Made Using the Computer-generated Belief Criteria Strategies
We then analyzed the computer-generated belief criteria strategies to see what influence they had on the accuracy and coverage DIKB’s predictions. The DIKB failed to make any predictions using one computer-generated strategy due to some unknown error that occurred during the experiment. The results from this strategy were not used in further analysis. We analyzed the remaining 35,999 different strategies for accuracy, coverage, and agreement with the validation set. Table 13 shows summary statistics for each performance parameter we analyzed over all prediction sets.
Table 13.
Summary statistics for each performance parameter that we analyzed for all 35,599 belief criteria strategies. The columns labeled “n” show the number of strategies whose predictions shared each minimum and maximum value.
| statistic | Min. | n | Median | Mean | Max. | n |
|---|---|---|---|---|---|---|
| true positives | 14.0 | 1,440 | 33.0 | 30.8 | 34.0 | 17,280 |
| false positives | 0.0 | 17,279 | 1.0 | 0.6 | 2.0 | 2,880 |
| true negatives | 0.0 | 21,599 | 0.0 | 0.7 | 2.0 | 11,520 |
| false negatives | 0.0 | 19,583 | 0.0 | 0.6 | 2.0 | 4,032 |
| sensitivity | 0.88 | 576 | 1.00 | 0.98 | 1.00 | 19,583 |
| specificity | 0.00 | 11,232 | 0.50 | 0.44 | 1.00 | 6,912 |
| positive predictive value | 0.94 | 240 | 0.97 | 0.98 | 1.00 | 17,279 |
| DIKB-only unknown | 10.0 | 5,760 | 13.0 | 15.3 | 34.0 | 576 |
| validation-set-only unknown | 3.0 | 864 | 40.0 | 42.4 | 62.0 | 2,880 |
| kappa | 0.41 | 576 | 0.47 | 0.48 | 0.52 | 768 |
The DIKB’s sensitivity ranged from 0.88 to 1.0 with 19,583 (54%) of the belief criteria strategies causing the system to operate at maximum sensitivity. The systems specificity ranged from 0.0 to 1.0 with 6,912 (19%) of the belief criteria strategies causing the system to operate at maximum specificity. The system had excellent positive predictive value (range: 0.94 to 1) across all belief criteria strategies. However, we could not characterize the system’s negative predictive value in a meaningful way because the DIKB never predicted more than two of the seven validation set non-interactions using any belief criteria strategy. We also calculated three-valued kappa scores for every prediction set using Cohen’s kappa to see how the agreement between the DIKB and the validation set compared with agreement expected by random chance. The DIKB’s predictions across all prediction sets had moderate agreement (0.4 to 0.5) with the validation set and never reached levels typically considered indicative of significant agreement (≥ 0.7).
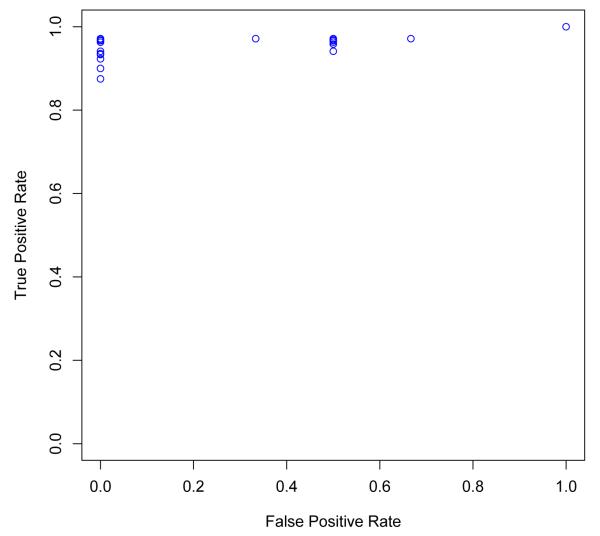
It would be informative to show the trade-off in the true-positive and false-positive rate of the DIKB over all belief criteria strategies using a receiver operating characteristic (ROC) curve. However, there is no way to create an ROC curve for the DIKB’s prediction sets because LOEs are rank-ordered. Another issue is that the false-positive rate of the system can not be calculated for 28,511 strategies for which the system made no true negative or false positive predictions. Figure 3 shows a scatter-plot of the DIKB’s true-positive rate versus its false-positive rate over the 7,488 belief criteria strategies for which a false-positive rate can be calculated. The plot shows that a number of strategies cause the system to make no false-positives predictions with high, or nearly perfect, sensitivity.
Fig. 3.
The set of predictions that the DIKB makes is influenced by varying the LOEs chosen as belief criteria for each assertion type. Since LOEs are rank-ordered there is no way to create a continuous receiver operating characteristic (ROC) curve. This figure shows a scatter plot of the true-positive rate (sensitivity) versus the false-positive rate (1.0 — specificity) over the 7,488 belief criteria strategies for which both values could be calculated. The system made no “true negative” or “false positive” predictions using the strategy chosen by the drug experts so, its performance using that strategy cannot be shown in the scatter-plot.
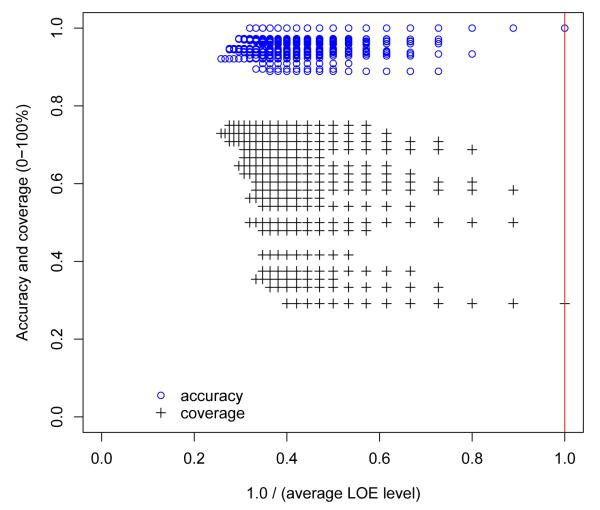
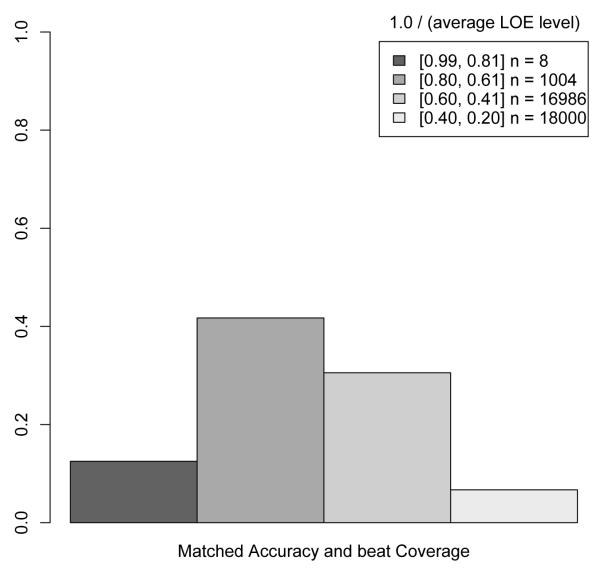
Figure 4 shows the accuracy and coverage of the DIKB as the proportion of higher-ranking LOEs present in a belief criteria strategy increases. To create the x axis for this plot, we first assigned an integer rank to each level-of-evidence in the LOEs for the eight assertion types for which varying belief criteria had an effect on prediction performance (see Section 2.3.4). The integer ranks were assigned such that the highest-ranking level-of-evidence was assigned a one, the next a two, and so-on. Then, for all 35,999 belief criteria strategies possible with the selected LOEs, the average integer-rank of each level-of-evidence used in the strategy was divided into 1.0. The resulting metric has a range of 0.26 (least stringent) to 1.0 (most stringent) for the set of strategies used in the current study. Figure 4 indicates that the most stringent belief criteria strategy resulted in very accurate predictions but that a large number of less-stringent strategies produced the same accuracy while covering a greater proportion of validation set interactions. Figure 5 shows this finding more clearly by plotting, at four different stringency levels, the proportion of belief criteria strategies that matched the most stringent strategy’s accuracy but had better coverage.
Fig. 4.
A scatterplot of the accuracy and coverage of the DIKB as the proportion of higher-ranking LOEs present in a belief criteria strategy increases. The vertical line intersecting 1.0 on the x axis indicates the accuracy and coverage of the most stringent belief criteria strategy that was defined by the drug experts. The numerical metric for stringency used in this plot (1.0 / average LOE level) was developed as follows: 1) An integer rank was assigned to each level-of-evidence in the LOEs for the eight assertion types for which varying belief criteria had an effect on prediction performance. This was done such that the highest-ranking level-of-evidence was assigned a one, the next a two, and so-on. 2) For all 35,999 belief criteria strategies, the average integer-rank of each level-of-evidence used in the strategy was divided into 1.0. The resulting metric has a range of 0.26 (least stringent) to 1.0 (most stringent) for the set of strategies used in the current study.
Fig. 5.
A barplot of the proportion of belief criteria strategies at four different stringency levels that matched the accuracy but had better coverage than the most stringent strategy. Figure 4 explains the numerical metric for stringency that is used in this plot. The plot makes it clear that less stringent strategies often result in a greater coverage of known interactions with no loss of accuracy.
A notable result of this experiment is that 8,351 (23%) of the 35,599 tested strategies caused the DIKB to have equal or better performance in terms of sensitivity, positive predictive value, and agreement with the validation set than the most stringent strategy. Table 14 shows the performance characteristics for 1,152 (3%) strategies that performed at the top level in these three performance categories. It is important to note that neither the most stringent strategy nor any of these “best-performing” strategies predicted non-interactions that were confirmed or refuted by the validation set. This meant that we could not calculate the specificity of the DIKB using either expert or computer-generated strategies.
Table 14.
1,152 (3%) belief criteria strategies caused the DIKB to perform optimally in terms of sensitivity, positive predictive value, and agreement with the validation set as measured by Cohen’s kappa. This table shows all measured performance characteristics for these “best-performing” strategies.
| statistic | value | statistic | value |
|---|---|---|---|
| true positives | 34 | false positives | 0 |
| true negatives | 0 | false negatives | 0 |
| sensitivity | 1 | specificity | n/a |
| positive predictive value | 1 | kappa | 0.52 |
| DIKB-only unknown | 14 | validation-set-only unknown | 40 |
All of the “best-performing” strategies caused the DIKB to make the same set of 65 interaction and non-interaction predictions. These strategies predicted a metabolic inhibition interaction for 34 (83%) of the 41 interacting pairs in the validation set while making no false positive and no false negative predictions. The system’s coverage of the validation set using any of these “best-performing” strategies was incomplete — it made no predictions for seven interactions and seven non-interactions listed in the validation set. As Table 15 shows, the pharmacokinetic magnitude of 30 of the 34 confirmed (88%) predictions made using the best performing belief criteria strategies matched levels observed in clinical trial data.
Table 15.
Each of the 1,152 strategies that caused the DIKB to perform optimally in terms of sensitivity, positive predictive value, and agreement with the validation set caused the system to make the same 34 interaction predictions that were confirmed by the validation set.
| pair | AUCi/AUC | |
|---|---|---|
| alprazolam* - erythromycin | (0, 1.3) | 1.61 |
| alprazolam* - itraconazole | (0, 1.3) † | 2.7 |
| alprazolam* - ketoconazole | (0, 1.3) † | 3.98 |
| alprazolam* - nefazodone | (0, 1.3) | 1.98 |
| atorvastatin* - erythromycin | > 2† | 1.4 |
| atorvastatin* - nefazodone | > 2 | 3-4 |
| clarithromycin - atorvastatin* ‡ | > 2 | ≥ 1.8, max 5.4 |
| clarithromycin* - fluconazole | > 0 | ≥ 1.18, max 1.33 |
| diltiazem - lovastatin* | > 2 | ≥ 3 |
| diltiazem - beta-OH-lovastatin* | > 2 | 3.57 |
| diltiazem - midazolam* | > 2 | 3.75 |
| diltiazem - simvastatin* | > 2 | 4.8 |
| diltiazem - triazolam* | > 2 | 2.83 |
| erythromycin - simvastatin* | > 2 | 6.3 |
| erythromycin - beta-OH-simvastatin* | > 2 | 3.9 |
| fluconazole - fluvastatin* | > 2 † | 1.83 |
| itraconazole - atorvastatin* | > 2 | 2.5 |
| itraconazole - lovastatin* | > 2 | 14.8 |
| itraconazole - beta-OH-lovastatin* | > 2 | 8.56 |
| ketoconazole - simvastatin* | > 2 | 12.55 |
| midazolam* - clarithromycin | > 2 | ≥ 7 |
| midazolam* - erythromycin | > 2 | 4.4 |
| midazolam* - fluconazole | > 2 | ≥ 2.6 |
| midazolam* - itraconazole | > 2 | 10.8 |
| midazolam* - ketoconazole | > 2 | 15.9 |
| midazolam* - nefazodone | > 2 | 4.6 |
| nefazodone - simvastatin* | > 2 | 20 |
| nefazodone - beta-OH-simvastatin* | > 2 | 20 |
| triazolam* - clarithromycin | > 2 | 5.3 |
| triazolam* - erythromycin | > 2 | 2.1 |
| triazolam* - fluconazole | > 2 | 2.5 |
| triazolam* - itraconazole | > 2 | ≥ 3.1 |
| triazolam* - ketoconazole | > 2 | ≥ 9.2 |
| triazolam* - nefazodone | > 2 | ≥ 3.9 |
Asterisks indicate the drug or drug metabolite that the system predicts will be the object of the interaction.
The DIKB’s magnitude estimate did not correspond with validation set data
The DIKB also predicted a metabolic inhibition interaction at the level with clarithromycin as the object drug.
3.3 Evaluation of the DIKB’s Novel DDI Predictions Made Using the Best-performing Belief Criteria Strategies
The system also predicted 31 metabolic inhibition interactions and nine non-interactions using the 1,152 “best-performing” belief criteria strategies whose validity was unknown by the validation set. These novel interaction predictions (Table 16, supplementary material) represent potentially interacting drug combinations that our review of the literature indicate have not been studied. We searched PubMed for clinical trials involving these pairs and could only find one clinical trial that was not already included in the validation set [6] unfortunately, this study did not meet the inclusion criteria for its evidence type and so could not be used as evidence for, or against, any interactions.
Fifteen of the published case reports we had collected while constructing the validation set claimed the occurrence of a DDI that matched one of the 31 novel predictions. Each report was reviewed using the Drug Interaction Probability Scale (DIPS) [3] by clinician co-investigator JH. The DIPS defines four qualitative levels (Doubtful, Possible, Probable, and Highly Probable) representing the degree to which the information provided by the report supports the proposition that a specific drug combination effected an adverse event or events. Six novel predictions were matched with case reports that met the DIPS Probable level; meaning that the predicted interactions were the likely cause of an adverse event occurring in a patient. Seven novel predictions were matched with reports that met the DIPS Possible level; meaning that the predicted interactions could not be excluded from consideration as the cause of an adverse event in a patient.
3.4 Comparing the DIKB Predictions to Labeling Statements
We could not do a quantitative comparison of the system’s predictions with drug-drug interaction statements from product labeling because a significant proportion of the validation set was constructed from labeling statements. We examined the 44 statements that were not used in the validation set and found that two of the seven novel predictions that were matched with reports meeting the DIPS Possible level could not be inferred from any of the product labeling statements (see Table 17). We believe that the combination of evidence from the case report literature supporting these interactions and the lack of discussion in drug product labeling makes these interactions especially important to investigate further.
Table 17.
The two novel interaction predictions shown in this table were supported by case reports but were not present in any of the 65 drug-drug interaction statements from product labeling that we had collected while building the validation set.
| Predicted interaction | report and DIPS score | |
|---|---|---|
| diltiazem - atorvastatin* | > 2 | [63]:possible |
| fluconazole - simvastatin* | > 2 | [64]:possible |
Asterisks indicate the interaction’s predicted object drug.
We also found one product label statement declaring an interaction that is refuted by clinical trial data present in the validation set. This statement extrapolated an interaction observed between erythromycin and one or more HMG-CoA reductase inhibitors to all drugs in that class:
Erythromycin has been reported to increase concentrations of HMG-CoA reductase inhibitors (e.g., lovastatin and simvastatin). Rare reports of rhabdomyolysis have been reported in patients taking these drugs concomitantly [7].
The active ingredient rosuvastatin is among the HMG-CoA reductase inhibitors included in our study. The labeling statement indirectly declares a potential pharmacokinetic interaction between erythromycin and rosuvastatin that is refuted by a randomized clinical trial in the validation set [8]. None of the interaction predictions made by the DIKB using the evidence board or the best performing belief criteria strategies were refuted by the validation set (i.e. false positives or false negatives). While the system made no prediction involving erythromycin and rosuvastatin with these strategies, it correctly predicted a non-interaction between erythromycin and rosuvastatin using other, lower specificity, belief criteria strategies. These results indicate that, depending on belief criteria strategies, DDI prediction using drug-mechanism knowledge can be accurate while avoiding the kinds of false predictions that occur when individual drug differences are not recognized.
4 Discussion
Changing the LOEs selected as belief criteria altered the system’s prediction accuracy and coverage in the way that we had hypothesized. We found for this data set that, as the criteria for including assertions were relaxed, the DIKB predicted a larger number of true interactions; sometimes at the expense of also making more false predictions. By having the computer iterate through a large set of possible belief criteria strategies we found that a significant proportion (23%) of them caused the DIKB to have equal or better performance in terms of sensitivity, positive predictive value, and agreement with the validation set than the most stringent strategy designed by drug experts.
4.1 Further Analysis of the “Best-performing” Strategies
The experiment also found a particular family of belief criteria strategies that optimized the system’s prediction accuracy and coverage. Table 18 shows the range of LOEs used by the 1,152 “best performing” belief criteria strategies. To better understand the information in Table 18, it is useful to note the situations where a level-of-evidence can be selected as a belief criterion for an assertion type without having any effect on the system’s predictions.
Table 18.
This table shows the range of LOEs used by 1,152 belief criteria strategies that had perfect sensitivity and positive predictive value and optimized the DIKB’s coverage and agreement with the validation set. The LOEs that were chosen as belief criteria for supporting and refuting evidence were always the same for the assertions shown in the table. The columns Evidence for and Evidence against show the distribution of evidence items across the LOEs. Not shown are the assertion types for which varying the LOEs chosen as belief criteria would have no effect on the DIKB’s prediction performance (Section 2.3.4).
| Assertion type | Belief criteria | Evidence for | Evidence against |
|---|---|---|---|
| inhibits | I: LOE-1, LOE-2 | n=11 ∼ LOE-1(91%), LOE-2(9%) | n=4 ∼ LOE-1(50%), LOE-2(50%) |
| substrate-of | D: LOE-3 | n=29 ∼ LOE-1(10%), LOE-2(21%) LOE-3(17%), LOE-4(21%) LOE-5(31%) |
n=11 ∼ LOE-1(0%), LOE-2(0%) LOE-3(36%), LOE-4(9%) LOE-5(55%) |
| is-not-substrate-of | D: LOE-1 → LOE-3 | n=11 ∼ LOE-1(0%), LOE-2(0%) LOE-3(36%), LOE-4(9%) LOE-5(55%) |
|
| primary-total-clearance-mechanism | C: LOE-1 → LOE-3 | n=14 ∼ LOE-1(21%), LOE-2(64%), LOE-3(14%) |
|
| primary-total-clearance-enzyme | E: LOE-3 | n=8 ∼ LOE-1(25%), LOE-2(62%) LOE-3(13%) |
|
| primary-metabolic-clearance-enzyme | F: LOE-1 → LOE-4 | n=3 ∼ LOE-1(0%), LOE-2(0%) LOE-3(67%), LOE-4(33%) |
n=1 ∼ LOE-1(100%), LOE-2(0%) LOE-3(0%), LOE-4(0%) |
| controls-formation-of 6 | D: LOE-1 → LOE-3, LOE-4, LOE-5 |
n=17 ∼ LOE-1(0%), LOE-2(0%), LOE-3(12%), LOE-4(6%), LOE-5(82%) |
|
| has-metabolite6 | G: LOE-1 → LOE-2 LOE-3, LOE-4 |
n=27 ∼ LOE-1(19%), LOE-2(0%) LOE-3(44%), LOE-4(37%) |
No belief criteria strategy in the set of 1,152 used LOE-4 or any lower-ranking LOE for the controls-formation-of assertion type and LOE-3 or any lower-ranking LOE for the has-metabolite assertion type.
Any level-of-evidence can be chosen as a belief criterion for an assertion type that is not relevant for the predictions-of-interest
In the current study, the types substrate-of and inhibits were key to making all interaction predictions. In contrast, one assertion type, primary-metabolic-clearance-enzyme, was used exclusively to predict the magnitude of an interaction. Varying this assertion’s belief criteria could not affect the system’s non-magnitude predictions and, therefore, had no effect on the study’s performance metrics. Indeed, Table 18 shows that all four of this assertion type’s LOEs were used by the “best-performing” strategies.
Any level-of-evidence can be chosen as a belief criterion for an assertion type when there is no evidence that supports or refutes its instances
Table 18 shows that there were multiple cases where no evidence items were mapped to a particular level-of-evidence for an assertion type. For example, since none of the 17 evidence items that were linked to controls-formation instances map to the assertion type’s top two ranking LOEs (D: LOE-1 and D: LOE-2), the system’s predictions were not affected when either was chosen as a belief criterion. Similar situations held for the is-not-substrate-of, and primary-metabolic-clearance-enzyme assertion types.
Choosing a lower-ranking level-of-evidence as a belief criterion has no effect on predictions if all instances of an assertion type are justified by a higher-ranking level-of-evidence
While the inhibits assertion type was relevant for making DDI predictions, Table 18 shows that both of its LOEs (I: LOE-1 and I: LOE-2) were used by the “best-performing” belief criteria strategies. This was because there was only one evidence item that mapped to LOE-2; an item supporting the assertion (ketoconazole inhibits CYP3A4). The same assertion had two other evidence items that mapped to LOE-1. As a result, the assertion was justified no matter which LOE the system chose as a belief criterion for the inhibits assertion type.
These observations suggest that the experiment’s “best-performing” belief criteria strategies are quite likely unique to the set of drugs and evidence items used in the current experiment. For example, further work on the evidence-base might identify new evidence items that map to LOE-2 for the inhibits assertion and that cause the system to make false predictions. Similarly, the effect of evidence items that map to LOE-1 or LOE-2 for the controls--formation assertion type are unknown because such evidence was not present in the current evidence base. We intend to explore if the “best-performing” strategies perform as well for a different set of drugs and evidence items in future work.
4.2 Prediction Sets that Produced Invalid Predictions
So far in our discussion we have focused only on the performance of the DIKB using the “best-performing” strategies. However, 27,648 (77%) of the strategies that we tested caused the DIKB to predict at least one interaction or non-interaction considered invalid by the validation set. The maximum number of interaction or non-interaction predictions refuted by the validation set for any single strategy was three and included either two invalid interactions and one invalid non-interaction or vice versa. Table 19 shows the four invalid predictions that appeared in various combinations among the predictions made using a wide range of strategies. We now briefly discuss the conditions under which each of these predictions were made.
Table 19.
27,648 strategies led the DIKB to predict one or more of the interactions or non-interactions shown in this table that were refuted by the validation set. For refuted interactions, asterisks indicate the drug that the DIKB considers the object of a metabolic inhibition interaction via the enzyme shown in parentheses. For refuted non-interactions, they indicate the drug that should not be affected by inhibition of the enzyme shown in parentheses.
| Refuted interactions |
|---|
| itraconazole - fluvastatin* (CYP3A4) |
| fluconazole - rosuvastatin* (CYP2C9) |
| Refuted non-interactions |
|---|
| itraconazole - rosuvastatin* (CYP3A4) |
| clarithromycin* - fluconazole (CYP2C9) |
4.2.1 Invalid Predictions Caused by Low-ranking Evidence
itraconazole-fluvastatin
The itraconazole-fluvastatin interaction prediction occurred when the system used strategies that accept drug labeling statements as belief criteria because 1) the assertion (itraconazole inhibits CYP3A4) was a default assumption and 2) the evidence-base recorded one labeling statement (based on a non-cited in vitro study) proposing that CYP3A4 is a minor elimination pathway for fluvastatin (<20% of total clearance) [9]. This prediction was refuted in the validation set by a randomized controlled trial involving 10 healthy volunteers that showed no significant increase in the systemic concentration of fluvastatin in the presence of itraconazole [10].
fluconazole-rosuvastatin
The DIKB predicted the fluconazole-rosuvastatin interaction using strategies that allow statements in product labeling to justify the controls-formation and has-metabolite assertion types and non-randomized clinical trial data to justify the inhibits assertion type. In this case, the system inferred that rosuvastatin is a substrate of CYP2C9 because the two assertions (rosuvastatin has-metabolite N-desmethylrosuvastatin) and (CYP2C9 controls-formation-of N-desmethylrosuvastatin) were each supported by evidence items based on labeling information [11] and the assertion (fluconazole inhibits CYP2C9) was supported by a non-randomized clinical trial [12]. This prediction was refuted in the validation set by a randomized controlled trial involving 14 healthy volunteers that found no statistically significant increase in the systemic concentration of rosuvastatin in the presence of fluconazole [13].
itraconazole-rosuvastatin
The DIKB predicted a non-interaction between itraconazole and rosuvastatin via CYP3A4 inhibition using strategies that allow statements in product labeling to justify the is-not-substrate-of assertion type. This was because the evidence-base contained one evidence item, based on a labeling statement, declaring CYP3A4 to not have a role in the metabolic clearance of rosuvastatin [11]. The validation set contained a randomized clinical trial that found a small, but statistically significant, increase in the systemic concentration of rosuvastatin in the presence of itraconazole during [14].
4.2.2 An Invalid Prediction Caused by Using More Stringent LOEs
fluconazole-clarithromycin
The system predicted a non-interaction between fluconazole and clarithromycin using strategies that considered 1) inhibits type justified by non-randomized clinical trial data, 2) the is-not-substrate-of assertion type justified by in vitro metabolism identification studies using human microsomes and chemical inhibitors, and 3) the substrate-of assertion type justified by any of the clinical trial types. These strategies caused the system to apply one evidence item [12] to justify the assertion (fluconazole inhibits CYP2C9) and another item [15] to justify the assertion (clarithromycin is-not--substrate-of CYP2C9).
This case is interesting because the invalid prediction was overruled when using a less stringent levels-of-evidence for the substrate-of assertion type. The first two LOEs for the substrate-of assertion type are defined by various clinical trial types. Strategies that used the assertion type’s third-ranking LOE, in vitro metabolism identification experiments, as belief criterion predicted a validated interaction between fluconazole and clarithromycin via CYP3A4 inhibition. We think that this case shows the value of examining the performance of the system at all possible belief criteria strategies.
4.3 Why was the DIKB’s Coverage of Validation-set Interactions Always Incomplete?
Table 20 shows six interactions and four non-interactions present in the validation set that were never predicted by the DIKB using any belief criteria strategy. We now briefly examine why the system missed these interactions and non-interactions.
Table 20.
Pairs in the validation set (Tables 4 and 5) for which the DIKB made no prediction using any the 35,599 belief criteria strategies.
| missing interactions | missing non-interactions |
|---|---|
| clarithromycin - pravastatin | diltiazem - pravastatin |
| fluconazole - 1′-OH-midazolam | fluconazole - pravastatin |
| itraconazole - pravastatin | nefazodone - pravastatin |
| itraconazole - erythromycin | fluconazole - 14-OH-clarithromycin |
| itraconazole - ortho-OH-atorvastatin | |
| nefazodone - 4-OH-alprazolam |
Missing non-interaction involving pravastatin
The system missed the non-interactions involving pravastatin because its evidence-base was incomplete in regards to the pravastatin’s metabolic properties. Although pravastatin’s drug product label states that it is not metabolized by CYP3A4 “to any significant extent” [16], this evidence was never entered into the DIKB. Doing so would have caused the system to predict the three missing non-interactions involving pravastatin using some belief criteria strategies. This case shows that missing interactions and non-interaction can help identify errors made while building the DIKB’s evidence-base. We think that an analysis of missing interactions and non-interactions should be done as quality assurance every time a set of “best-performing” strategies is created for a new set of drugs. Once errors are corrected, the process of identifying “best-performing” strategies can be repeated.
Missing interactions involving pravastatin
It is important to note that entering the aforementioned product label evidence would have caused the system to predict non-interactions for the two missing interactions involving pravastatin (Table 20). If the non-interaction predictions are incorrect then, the interactions likely occur by mechanisms other than those modeled by the DIKB’s current DDI theory. If these mechanisms are known then, the DDI theory and evidence-base should be expanded to include them. Another possible explanation is that non-interaction predictions are correct, but that the studies that support the interactions contain some limitation. In this situation, it would be important to identify the limitation and integrate it into the inclusion criteria used to collect evidence.
Missing interactions involving three drug metabolites
Three missing interactions and one non-interaction are accounted for by the fact that were no assertions in the system indicating which enzymes do or do not metabolize 1′-OH-midazolam, ortho-OH-atorvastatin, 4-OH-alprazolam, 14-OH-clarithromycin. Neither are there assertions indicating that these metabolites inhibit a drug-metabolizing enzyme present in the system. The DIKB’s rule-based theory is capable of predicting when inhibition of the parent compound will or will not affect the formation of these metabolites made but we did not include these kinds of predictions in the study
The missing interaction between itraconazole and erythromycin
There were two evidence items in the system that supported the assertion (erythromycin inhibits CYP3A4) [17,18] but no assertion or evidence in the system claiming that itraconazole is a substrate of that enzyme. Hence, The system did not predict an interaction between erythromycin and itraconazole with itraconazole as the effected drug.
Conversely, the system had three default assumptions that separately established itraconazole to be both an in vivo and in vitro selective inhibitor of CYP3A4 and erythromycin to be an in vitro probe substrate. However, the system did not predict that itraconazole would interact with erythromycin because it does not assume that all properties established in vitro will hold in vivo. We think that the decision to accept in vitro knowledge as sufficient for inferring an in vivo result should occur within the evidence-model, not the rule-base.
4.4 Limitations
It’s important to note that our experiment only looked at binary performance criteria — predictions were classified as “true” or “false” according to the validation set and the goal was to maximize “true” predictions and minimize “false” predictions. It is likely that a different set of best-performing strategies would be found if our goal was to optimize the accuracy of the system’s magnitude predictions. This could be a worthwhile experiment because the system is capable of order-of-magnitude predictions for some drug combinations. As is shown in Section 3, the DIKB’s magnitude estimates for 14 interactions using the drug experts’ belief criteria strategy corresponded with levels reported in clinical trial data present in the validation set. A set of belief criteria strategies that focused on optimizing magnitude would seek to expand the DIKB’s coverage of known interactions past these 14 while still making correct magnitude predictions. However, this kind of analysis should also examine if there are some features of the rule-based theory of metabolic inhibition interactions used by the DIKB that could limit its ability to make accurate magnitude predictions. The DIKB’s current method for magnitude estimation makes a number of assumptions about many of the factors that can sometimes contribute to an increase in the AUC of an object drug in pharmacokinetic DDI study. It might be desirable to apply a more sophisticated approach that makes fewer assumptions such as that described by Ohno et al [19].
If a clinical trial is applied as support for an interaction or non-interaction in the validation set while a labeling statement echoing, but not citing, the study supports an assertion that the DIKB uses to predict the same interaction, an assessment of the system’s accuracy will be somewhat biased in favor of the system. The same bias will exist if a validation set interaction or non-interaction rests on only a single non-traceable statement and there are assertions in the DIKB used to predict the interaction or non-interaction that depend on the study that inspired the statement Both situations have the same remedy — exclude the “dual-use” validation set interaction or non-interaction from calculations of the systems accuracy. Unfortunately, we did not implement any strategy to avoid this kind of bias so it is possible that some labeling data was used to support mechanistic assertions that led to predictions validated by the same data but appearing in a different source. Future work will examine if this bias was present and, if so, what effect removing the affected interactions or non-interactions has on the calculations of DIKB accuracy.
Another limitation is that all case report evaluations were conducted using a causality assessment tool (DIPS [3]) whose validity and reproducibility has not yet been formally evaluated. Furthermore, the reviewer (co-investigator JH) was both the developer of DIPS and a member of the DIKB evidence board. Future work should involve a more rigorous evaluation of the DIKB’s novel interactions designed to overcome any potential bias not addressed in this study.
5 Related Work
Mechanism-based DDI prediction is currently part of pre-clinical drug development where it used to identify potential interactions between a new drug candidate and drugs currently on the market [20]. These early-phase modeling efforts are geared towards identifying interactions between a new drug and drugs with which it might be co-administered early on, before much time and money is invested [21,22]. The predictions made using drug-mechanisms in this context are generally qualitative; they indicate that two drugs might interact via a mechanism but offer no estimate of the magnitude of the interaction. Scientists can use qualitative predictions to select the set of clinical trials necessary to establish a new drug’s safety profile [23].
Considerable effort has been invested in both industry and academia researching how to make quantitative estimates of in vivo DDIs from in vitro evidence. Unfortunately, there is currently no general method for making accurate quantitative estimates of the magnitude of a metabolic inhibition DDI using in vitro data [23]. In spite of its difficulties, software tools have been developed that combine in vitro and in vivo findings to help clinicians reason about potential metabolic DDIs [24,25]. These systems rely heavily on mathematical models to make quantitative estimates of the magnitude of their DDI predictions. In contrast, the DIKB’s theory of metabolic DDIs makes simple magnitude estimates based on qualitative assertions about the degree to which inhibition of an enzyme would affect the clearance of some drug. A comparison of the DIKB’s ability to accurately predict magnitude with that of other systems would be worthwhile and should be the topic of future work.
To the best of our knowledge the DIKB’s computational model of evidence provides features that are unique among all current system’s that represent drug-mechanism knowledge. While other drug knowledge-bases and knowledge-based systems have linked evidence to their drug facts (e.g. DRUGDEX ® 6, Q-DIPS [24], PharmGKB [26], and BioCyc [27]), only the DIKB classifies all evidence entered into the system using a biomedical evidence ontology oriented toward confidence assignment. This enables the system to provide customized views of a body of drug-mechanism knowledge to users who do not agree about the inferential value of particular evidence types. This feature also allowed us to develop the novel approach to identifying the optimal use of available evidence that we have presented in this paper.
6 Conclusions
The results of the experiment discussed in this paper suggest that the DIKB’s unique approach to representing evidence provides two advantages over the representation methods used in current drug knowledge-bases. One advantage of the method is that experts can prospectively map their confidence in each assertion type to some arrangement of one or more abstract evidence types rather than having to review every relevant evidence item in the system. The experts can then define belief criteria that will influence the prediction accuracy of the system in a predictable way. Relaxing belief criteria causes the system to predict a larger number of true interactions, sometimes at the expense of making more false predictions.
Another advantage of the method is that researchers can characterize the prediction accuracy of a DDI theory using many different sub-sets of a specific body of evidence. This process can identify which evidence-use strategies optimize the system’s prediction accuracy and coverage of known DDIs. While the resulting evidence-use strategies might be unique to the set of drugs and evidence items used to create them, it is reasonable that the clinical relevance of any novel DDIs predicted using such “best-performing” strategies will be within the range present for the known interactions. This would have important implications for drug safety and is an area we intend to explore in future work.
The experiment has also helped to identify an opportunity for research into new computational methods to help support analysis of belief criteria strategies. Currently, the task is difficult because of the complex interplay between the kinds of evidence present in the knowledge-base, how it is linked to each assertion instance, and the relationship between each assertion type and the variables chosen for scoring the system’s prediction performance. In future work we hope to explore this research area as well as validate the DIKB’s approach to a broader range of drugs and drug-interaction mechanisms. We also hope to explore methods for quantifying the amount of confidence that expert users should have in DDI predictions the system makes using LOEs that don’t meet belief criteria. Progress on this topic might allow DIKB predictions to be interpreted in terms of a normative decision-theoretic frame-work such as expected utility theory. Understanding the relationship between the DIKB’s rank-ordered LOEs and quantitative measures of evidential support such as those reviewed by Tentori et al [28] might be an important first step in this direction.
We envision that, over the next decade, a new generation of highly accurate tools will become available that use pharmacologic theory, drug mechanism knowledge, and patient-specific data to help clinicians assess the combined effect of multiple drugs, the effect of removing a drug from a patients drug regimen, and individual response to therapy due to enzyme polymorphisms. These tools will be a significant advance in medicine and a radical change from the functionality that current prescribing software offers. Our research on how to best represent drug mechanism knowledge for the purpose of making clinically relevant DDI predictions is a small, through important step, toward understanding how to build and deploy the highly accurate tools that we envision.
Supplementary Material
7 Acknowledgments
This project was funded in part by a National Library of Medicine Biomedical and Health Informatics Training Program grant (T15 LM07442) and an award from the Elmer M. Plein Endowment Research Fund (University of Washington School of Pharmacy). The DIKB ontology and evidence taxonomy were developed using the Protégé resource, which is supported by grant LM007885 from the United States National Library of Medicine. The author expresses sincere appreciation to Drs. Tom Hazlet and John Gennari for their comments during the course of this project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The questionnaire shown in Appendix G (supplementary material) was used to help the experts reach consensus on the inductive strength of each evidence-type combination.
References
- [1].Boyce R, Collins C, Horn J, Kalet I. Modeling Drug Mechanism Knowledge Using Evidence and Truth Maintenance. IEEE Transactions on Information Technology in Biomedicine. 2007;11(4):386–397. doi: 10.1109/titb.2007.890842. [DOI] [PubMed] [Google Scholar]
- [2].Boyce R, Collins C, Horn J, Kalet I. Computing with evidence part I: A drug-mechanism evidence taxonomy oriented toward confidence assignment. Journal of Biomedical Informatics. 2009 doi: 10.1016/j.jbi.2009.05.001. DOI:10.1016/j.jbi.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Horn J, Hansten P, Chan L. Proposal for a new tool to evaluate drug interaction cases. Ann Pharmacother. 2007;41(4):674–80. doi: 10.1345/aph.1H423. [DOI] [PubMed] [Google Scholar]
- [4].R Development Core Team, R . A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2006. ISBN 3-900051-07-0. URL http://www.R-project.org. [Google Scholar]
- [5].Falissard B. psy: Various procedures used in psychometry. R package version 0.7 2007.
- [6].Shi J, Chapel S, Montay G, Hardy P, Barrett JS, Sica D, Swan SK, Noveck R, Leroy B, Bhargava VO. Effect of ketoconazole on the pharmacokinetics and safety of telithromycin and clarithromycin in older subjects with renal impairment. Int J Clin Pharmacol Ther. 2005;43(3):123–133. doi: 10.5414/cpp43123. [DOI] [PubMed] [Google Scholar]
- [7].Abbot, pce (erythromycin) tablet, FDA-approved drug product labeling. Feb, 2007. [last accessed on DailyMed 03/03/2008].
- [8].Cooper KJ, Martin PD, Dane AL, Warwick MJ, Raza A, Schneck DW. The effect of erythromycin on the pharmacokinetics of rosuvastatin. Eur J Clin Pharmacol. 2003;59(1):51–56. doi: 10.1007/s00228-003-0573-7. [DOI] [PubMed] [Google Scholar]
- [9].Novartis Pharmaceuticals Corporation lescol (fluvastatin sodium) capsule; lescol XL (fluvastatin sodium) tablet, extended release, FDA-approved drug product labeling. Jan, 2007. [last accessed on DailyMed 07/02/2008].
- [10].Kivisto KT, Kantola T, Neuvonen PJ. Different effects of itraconazole on the pharmacokinetics of fluvastatin and lovastatin. Br J Clin Pharmacol. 1998;46(1):49–53. doi: 10.1046/j.1365-2125.1998.00034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Astrazeneca, crestor (Rosuvastatin calcium) tablet, film coated for oral use, FDA-approved drug product labeling. Nov, 2007. [last accessed on DailyMed 04/17/2008].
- [12].Black DJ, Kunze KL, Wienkers LC, Gidal BE, Seaton TL, McDonnell ND, Evans JS, Bauwens JE, Trager WF. Warfarin-fluconazole. II. A metabolically based drug interaction: in vivo studies. Drug Metab Dispos. 1996;24(4):422–428. [PubMed] [Google Scholar]
- [13].Cooper KJ, Martin PD, Dane AL, Warwick MJ, Schneck DW, Cantarini MV. The effect of fluconazole on the pharmacokinetics of rosuvastatin. Eur J Clin Pharmacol. 2002;58(8):527–531. doi: 10.1007/s00228-002-0508-8. [DOI] [PubMed] [Google Scholar]
- [14].Cooper KJ, Martin PD, Dane AL, Warwick MJ, Schneck DW, Cantarini MV. Effect of itraconazole on the pharmacokinetics of rosuvastatin. Clin Pharmacol Ther. 2003;73(4):322–329. doi: 10.1016/s0009-9236(02)17633-8. [DOI] [PubMed] [Google Scholar]
- [15].Rodrigues AD, Roberts EM, Mulford DJ, Yao Y, Ouellet D. Oxidative metabolism of clarithromycin in the presence of human liver microsomes. Major role for the cytochrome P4503A (CYP3A) subfamily. Drug Metab Dispos. 1997;25(5):623–630. [PubMed] [Google Scholar]
- [16].Squibb B-M. Pravachol (pravastatin sodium) tablet, FDA-approved drug product labeling. Nov, 2007. [last accessed on DailyMed 03/19/2009].
- [17].Greenblatt DJ, von Moltke LL, Harmatz JS, Counihan M, Graf JA, Durol AL, Mertzanis P, Duan SX, Wright CE, Shader RI. Inhibition of triazolam clearance by macrolide antimicrobial agents: in vitro correlates and dynamic consequences. Clin Pharmacol Ther. 1998;64(3):278–285. doi: 10.1016/S0009-9236(98)90176-X. [DOI] [PubMed] [Google Scholar]
- [18].Zimmermann T, Yeates RA, Laufen H, Scharpf F, Leitold M, Wildfeuer A. Influence of the antibiotics erythromycin and azithromycin on the pharmacokinetics and pharmacodynamics of midazolam. Arzneimittelforschung. 1996;46(2):213–217. [PubMed] [Google Scholar]
- [19].Ohno Y, Hisaka A, Suzuki H. General framework for the quantitative prediction of CYP3A4-mediated oral drug interactions based on the AUC increase by coadministration of standard drugs. Clin Pharmacokinet. 2007;46(8):681–696. doi: 10.2165/00003088-200746080-00005. [DOI] [PubMed] [Google Scholar]
- [20].Internal Authorship [Last accessed 09/25/2006];FDA guideline: Drug interaction studies — study design, data analysis, and implications for dosing and labeling. 2006 September; doi: 10.1038/sj.clpt.6100054. Internet, http://www.fda.gov/Cber/gdlns/interactstud.htm. [DOI] [PubMed]
- [21].van de Waterbeemd H, Gifford E. ADMET in silico modeling: towards prediction paradise? Nat Rev Drug Discov. 2003;2(3):192–204. doi: 10.1038/nrd1032. [DOI] [PubMed] [Google Scholar]
- [22].Yamashita F, Hashida M. In silico approaches for predicting ADME properties of drugs. Drug Metab Phamacokin. 2004;19(5):327–338. doi: 10.2133/dmpk.19.327. [DOI] [PubMed] [Google Scholar]
- [23].Obach RS, Walsky RL, Venkatakrishnan K, Houston JB, Tremaine L. In vitro cytochrome P450 inhibition data and the prediction of drug-drug interactions: qualitative relationships, quantitative predictions, and the rank-order approach. Clin Pharmacol Ther. 2005;78:582–92. doi: 10.1016/j.clpt.2005.09.004. [DOI] [PubMed] [Google Scholar]
- [24].Bonnabry P, Sievering J, Leemann T, Dayer P. Quantitative Drug Interactions Prediction System (Q-DIPS): A Dynamic Computer-Based Method to Assist in the Choice of Clinically Relevant In Vivo Studies. Clinical Pharmacokinetics. 2001;40(9):631–640. doi: 10.2165/00003088-200140090-00001. [DOI] [PubMed] [Google Scholar]
- [25].Zvada SP, Chagwedera TE, Chigwanda R, Masimirembwa CM. An experimental pharmacokinetic computer program to predict potential drug-drug interactions. The Open Drug Metabolism Journal. 2009;3:8–16. [Google Scholar]
- [26].Klein T, Chang J, Cho M, Easton K, Fergerson R, Hewett M, Lin Z, Liu Y, Liu S, Oliver D, Rubin D, Shafa F, Stuart J, Altman R. Integrating Genotype and Phenotype Information: An Overview of the PharmGKB Project. The Pharmacogenomics Journal. 2001;1:167–170. doi: 10.1038/sj.tpj.6500035. [DOI] [PubMed] [Google Scholar]
- [27].Karp P, Paley S, Krieger C, Zhang P. Pacific Symposium on Biocomputing 2004. 2004. An evidence ontology for use in pathway/genome databases; pp. 190–201. [DOI] [PubMed] [Google Scholar]
- [28].Tentori K, Crupi V, Bonini N, Osherson D. Comparison of confirmation measures. Cognition. 2007;103(1):107–119. doi: 10.1016/j.cognition.2005.09.006. [DOI] [PubMed] [Google Scholar]
- [29].Yasui N, Otani K, Kaneko S, Ohkubo T, Osanai T, Sugawara K, Chiba K, Ishizaki T. A kinetic and dynamic study of oral alprazolam with and without erythromycin in humans: in vivo evidence for the involvement of CYP3A4 in alprazolam metabolism. Clin Pharmacol Ther. 1996;59(5):514–519. doi: 10.1016/S0009-9236(96)90179-4. [DOI] [PubMed] [Google Scholar]
- [30].Yasui N, Kondo T, Otani K, Furukori H, Kaneko S, Ohkubo T, Nagasaki T, Sugawara K. Effect of itraconazole on the single oral dose pharmacokinetics and pharmacodynamics of alprazolam. Psychopharmacology (Berl) 1998;139(3):269–273. doi: 10.1007/s002130050715. [DOI] [PubMed] [Google Scholar]
- [31].Greenblatt DJ, Wright CE, von Moltke LL, Harmatz JS, Ehrenberg BL, Harrel LM, Corbett K, Counihan M, Tobias S, Shader RI. Ketoconazole inhibition of triazolam and alprazolam clearance: differential kinetic and dynamic consequences. Clin Pharmacol Ther. 1998;64(3):237–247. doi: 10.1016/S0009-9236(98)90172-2. [DOI] [PubMed] [Google Scholar]
- [32].Schmider J, Brockmoller J, Arold G, Bauer S, Roots I. Simultaneous assessment of CYP3A4 and CYP1A2 activity in vivo with alprazolam and caffeine. Pharmacogenetics. 1999;9(6):725–734. doi: 10.1097/01213011-199912000-00007. [DOI] [PubMed] [Google Scholar]
- [33].DeVane CL, Donovan JL, Liston HL, Markowitz JS, Cheng KT, Risch SC, Willard L. Comparative CYP3A4 inhibitory effects of venlafaxine, fluoxetine, sertraline, and nefazodone in healthy volunteers. J Clin Psychopharmacol. 2004;24(1):4–10. doi: 10.1097/01.jcp.0000104908.75206.26. [DOI] [PubMed] [Google Scholar]
- [34].Greene DS, Salazar DE, Dockens RC, Kroboth P, Barbhaiya RH. Coadministration of nefazodone and benzodiazepines: III. A pharmacokinetic interaction study with alprazolam. J Clin Psychopharmacol. 1995;15(6):399–408. doi: 10.1097/00004714-199512000-00003. [DOI] [PubMed] [Google Scholar]
- [35].Siedlik PH, Olson SC, Yang BB, Stern RH. Erythromycin coadministration increases plasma atorvastatin concentrations. J Clin Pharmacol. 1999;39(5):501–504. [PubMed] [Google Scholar]
- [36].Teva, nefazodone hydrochloride (Nefazodone Hydrochloride) tablet, FDA-approved drug product labeling. Nov, 2006. [last accessed on DailyMed 04/21/2009].
- [37].Amsden GW, Kuye O, Wei GC. A study of the interaction potential of azithromycin and clarithromycin with atorvastatin in healthy volunteers. J Clin Pharmacol. 2002;42(4):444–449. [PubMed] [Google Scholar]
- [38].Jacobson TA. Comparative pharmacokinetic interaction profiles of pravastatin, simvastatin, and atorvastatin when coadministered with cytochrome P450 inhibitors. Am J Cardiol. 2004;94(9):1140–1146. doi: 10.1016/j.amjcard.2004.07.080. [DOI] [PubMed] [Google Scholar]
- [39].Abbott, Biaxin filmtab (clarithromycin) tablet, film coated, FDA-approved drug product labeling. Mar, 2007. [last accessed on DailyMed 05/29/2008].
- [40].Azie NE, Brater DC, Becker PA, Jones DR, Hall SD. The interaction of diltiazem with lovastatin and pravastatin. Clin Pharmacol Ther. 1998;64(4):369–377. doi: 10.1016/S0009-9236(98)90067-4. [DOI] [PubMed] [Google Scholar]
- [41].Biovail, cardizem (diltiazem hydrochloride) tablet, coated, FDA-approved drug product labeling. Apr, 2006. [last accessed on DailyMed 04/21/2009].
- [42].Backman JT, Olkkola KT, Aranko K, Himberg JJ, Neuvonen PJ. Dose of midazolam should be reduced during diltiazem and verapamil treatments. Br J Clin Pharmacol. 1994;37(3):221–225. doi: 10.1111/j.1365-2125.1994.tb04266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mousa O, Brater DC, Sunblad KJ, Hall SD. The interaction of diltiazem with simvastatin. Clin Pharmacol Ther. 2000;67(3):267–274. doi: 10.1067/mcp.2000.104609. [DOI] [PubMed] [Google Scholar]
- [44].Varhe A, Olkkola KT, Neuvonen PJ. Diltiazem enhances the effects of triazolam by inhibiting its metabolism. Clin Pharmacol Ther. 1996;59(4):369–375. doi: 10.1016/S0009-9236(96)90103-4. [DOI] [PubMed] [Google Scholar]
- [45].Kantola T, Kivisto KT, Neuvonen PJ. Erythromycin and verapamil considerably increase serum simvastatin and simvastatin acid concentrations. Clin Pharmacol Ther. 1998;64(2):177–182. doi: 10.1016/S0009-9236(98)90151-5. [DOI] [PubMed] [Google Scholar]
- [46].Ahonen J, Olkkola KT, Neuvonen PJ. Effect of route of administration of fluconazole on the interaction between fluconazole and midazolam. Eur J Clin Pharmacol. 1997;51(5):415–419. doi: 10.1007/s002280050223. [DOI] [PubMed] [Google Scholar]
- [47].Kantola T, Backman JT, Niemi M, Kivisto KT, Neuvonen PJ. Effect of fluconazole on plasma fluvastatin and pravastatin concentrations. Eur J Clin Pharmacol. 2000;56(3):225–229. doi: 10.1007/s002280000127. [DOI] [PubMed] [Google Scholar]
- [48].Mazzu AL, Lasseter KC, Shamblen EC, Agarwal V, Lettieri J, Sundaresen P. Itraconazole alters the pharmacokinetics of atorvastatin to a greater extent than either cerivastatin or pravastatin. Clin Pharmacol Ther. 2000;68(4):391–400. doi: 10.1067/mcp.2000.110537. [DOI] [PubMed] [Google Scholar]
- [49].Janssen, sporanox (itraconazole) capsule, FDA-approved drug product labeling. Jan, 2008. [last accessed on DailyMed 05/16/2008].
- [50].Chung E, Nafziger AN, Kazierad DJ, Bertino JSJ. Comparison of midazolam and simvastatin as cytochrome P450 3A probes. Clin Pharmacol Ther. 2006;79(4):350–361. doi: 10.1016/j.clpt.2005.11.016. [DOI] [PubMed] [Google Scholar]
- [51].Gorski JC, Jones DR, Haehner-Daniels BD, Hamman MA, O’Mara EMJ, Hall SD. The contribution of intestinal and hepatic CYP3A to the interaction between midazolam and clarithromycin. Clin Pharmacol Ther. 1998;64(2):133–143. doi: 10.1016/S0009-9236(98)90146-1. [DOI] [PubMed] [Google Scholar]
- [52].Gurley B, Hubbard MA, Williams DK, Thaden J, Tong Y, Gentry WB, Breen P, Carrier DJ, Cheboyina S. Assessing the clinical significance of botanical supplementation on human cytochrome P450 3A activity: comparison of a milk thistle and black cohosh product to rifampin and clarithromycin. J Clin Pharmacol. 2006;46(2):201–213. doi: 10.1177/0091270005284854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Olkkola KT, Aranko K, Luurila H, Hiller A, Saarnivaara L, Himberg JJ, Neuvonen PJ. A potentially hazardous interaction between erythromycin and midazolam. Clin Pharmacol Ther. 1993;53(3):298–305. doi: 10.1038/clpt.1993.25. [DOI] [PubMed] [Google Scholar]
- [54].Olkkola KT, Ahonen J, Neuvonen PJ. The effects of the systemic antimycotics, itraconazole and fluconazole, on the pharmacokinetics and pharmacodynamics of intravenous and oral midazolam. Anesth Analg. 1996;82(3):511–516. doi: 10.1097/00000539-199603000-00015. [DOI] [PubMed] [Google Scholar]
- [55].Olkkola KT, Backman JT, Neuvonen PJ. Midazolam should be avoided in patients receiving the systemic antimycotics ketoconazole or itraconazole. Clin Pharmacol Ther. 1994;55(5):481–485. doi: 10.1038/clpt.1994.60. [DOI] [PubMed] [Google Scholar]
- [56].Lam YW, Alfaro CL, Ereshefsky L, Miller M. Pharmacokinetic and pharmacodynamic interactions of oral midazolam with ketoconazole, fluoxetine, fluvoxamine, and nefazodone. J Clin Pharmacol. 2003;43(11):1274–1282. doi: 10.1177/0091270003259216. [DOI] [PubMed] [Google Scholar]
- [57].Phillips JP, Antal EJ, Smith RB. A pharmacokinetic drug interaction between erythromycin and triazolam. J Clin Psychopharmacol. 1986;6(5):297–299. [PubMed] [Google Scholar]
- [58].Varhe A, Olkkola KT, Neuvonen PJ. Fluconazole, but not terbinafine, enhances the effects of triazolam by inhibiting its metabolism. Br J Clin Pharmacol. 1996;41(4):319–323. doi: 10.1046/j.1365-2125.1996.03189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Neuvonen PJ, Varhe A, Olkkola KT. The effect of ingestion time interval on the interaction between itraconazole and triazolam. Clin Pharmacol Ther. 1996;60(3):326–331. doi: 10.1016/S0009-9236(96)90059-4. [DOI] [PubMed] [Google Scholar]
- [60].Varhe A, Olkkola KT, Neuvonen PJ. Oral triazolam is potentially hazardous to patients receiving systemic antimycotics ketoconazole or itraconazole. Clin Pharmacol Ther. 1994;56(6 Pt 1):601–607. doi: 10.1038/clpt.1994.184. [DOI] [PubMed] [Google Scholar]
- [61].von Moltke LL, Greenblatt DJ, Harmatz JS, Duan SX, Harrel LM, Cotreau-Bibbo MM, Pritchard GA, Wright CE, Shader RI. Triazolam biotransformation by human liver microsomes in vitro: effects of metabolic inhibitors and clinical confirmation of a predicted interaction with ketoconazole. J Pharmacol Exp Ther. 1996;276(2):370–379. [PubMed] [Google Scholar]
- [62].Barbhaiya RH, Shukla UA, Kroboth PD, Greene DS. Coadministration of nefazodone and benzodiazepines: II. A pharmacokinetic interaction study with triazolam. J Clin Psychopharmacol. 1995;15(5):320–326. doi: 10.1097/00004714-199510000-00003. [DOI] [PubMed] [Google Scholar]
- [63].Gladding P, Pilmore H, Edwards C. Potentially fatal interaction between diltiazem and statins. Ann Intern Med. 2004;140(8):W31. doi: 10.7326/0003-4819-140-8-200404200-00037-w1. [DOI] [PubMed] [Google Scholar]
- [64].Shaukat A, Benekli M, Vladutiu GD, Slack JL, Wetzler M, Baer MR. Simvastatin-fluconazole causing rhabdomyolysis. Ann Pharmacother. 2003;37(78):1032–1035. doi: 10.1345/aph.1C467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.