Abstract
Experimental studies demonstrated that maternal exposure to certain environmental and dietary factors during early embryonic development can influence the phenotype of offspring as well as the risk of disease development at the later life. DNA methylation, an epigenetic phenomenon, has been suggested as a mechanism by which maternal nutrients affect the phenotype of their offspring in both honeybee and agouti mouse models. Phenotypic changes through DNA methylation can be linked to folate metabolism by the knowledge that folate, a coenzyme of one-carbon metabolism, is directly involved in methyl group transfer for DNA methylation. During the fetal period, organ-specific DNA methylation patterns are established through epigenetic reprogramming. However, established DNA methylation patterns are not immutable and can be modified during our life time by the environment. Aberrant changes in DNA methylation with diet may lead to the development of age-associated diseases including cancer. It is also known that the aging process by itself is accompanied by alterations in DNA methylation. Diminished activity of DNA methyltransferases (Dnmts) can be a potential mechanism for the decreased genomic DNA methylation during aging, along with reduced folate intake and altered folate metabolism. Progressive hypermethylation in promoter regions of certain genes is observed throughout aging and repression of tumor suppressors induced by this epigenetic mechanism appears to be associated with cancer development. In this review we address the effect of folate on early development and aging through an epigenetic mechanism, DNA methylation.
Keywords: DNA methylation, folate, embryonic development, aging, carcinogenesis
1. Introduction
Epidemiologic studies indicate that impaired intrauterine growth and development are associated with higher risk of cardiovascular disease, diabetes mellitus, obesity, and osteoporosis in the adult life of offspring [1-6]. Among many candidate mechanisms by which in utero and early-life conditions affect adult health and diseases, epigenetic machinery including DNA methylation has been highlighted [7]. Epigenetics is a phenomenon that affects gene expression without altering the genomic sequence. DNA methylation is a major epigenetic phenomenon that modifies DNA by methylating cytosine bases at the carbon-5′ position in CpG dinucleotide residues and regulates gene expression and integrity. Appropriate DNA methylation is essential for embryogenesis and early fetal development [8] as inadequate establishment of DNA methylation patterns by insufficient maternal diet such as low folate diet may induce pediatric developmental diseases and even affect health in later life [9, 10].
Established epigenetic patterns during the fetal period can be changed in adult life by environmental factors including nutrition [8]. For example, identical twins possess the same genotype and no distinguishable epigenetic differences in their early life, but they show remarkable differences in genomic DNA methylation and histone acetylation patterns in their later life, so that these epigenetic differences may result in different gene expression and disease susceptibility [11]. This observation emphasizes the concept that the exposure to a particular environment is important during the whole life span. This review summarizes the accumulated body of knowledge for a possible mechanistic explanation on how folate affects both embryonic development and aging.
2. Folate, DNA methylation and early development
2.1. Embryonic development and DNA methylation
During embryonic development each cell, tissue and organ acquires different prototypes of gene expression, which are thought to be mediated by epigenetic modifications such as DNA methylation. In fact, the mammalian genome undergoes profound reprogramming of DNA methylation patterns in the germ cells and the early preimplantation embryos [12]. Upon fertilization gamete methylation patterns from parents are erased by a genome wide demethylation event, and during the implantation methylation patterns are newly established through de novo methylation [13]. These embryonic marks are important for early embryonic development and establishment of toti- or pluripotency [14]. Interestingly, embryonic and fetal exposure to nutrients, which are mainly maternally-derived, can affect this dramatic epigenetic phenomenon, thereby affecting fetal development and even later life health status [15, 16].
DNA methylation plays a crucial role in genomic imprinting as a result of repression of one allele inherited from either parent by DNA methylation. Abnormal derepression of imprinted allele can cause pediatric developmental diseases such as Prader-Willi Syndrome and cancer disease in later life [17-19]. Even though the parental methylation in imprinted genes escapes from the process of demethylation and de novo methylation and a number of imprinted genes remain imprinted throughout the life, it is noteworthy that many genes are imprinted in a tissue-specific manner or during a limited time period, indicating that genomic imprinting by DNA methylation is reversible [20]. Most recently a study using a DNA methyltransferase 1 (Dnmt1) conditional knock-out mouse model demonstrated that a lack of maternal and zygotic Dnmt1 proteins induces demethylation of imprinted loci in blastocysts, indicating that Dnmt1 proteins are a prerequisite for the maintenance of methylation imprints in preimplantation embryo [21]. Thus, alterations in genomic imprinting by maternal diets that are known to affect DNA methylation may also result in pediatric developmental diseases as well as diseases in their life.
2.2. Maternal diet for the development of honeybee
An excellent model demonstrating the effect of maternal diet on the offspring’s phenotype is the honeybee model. Female larvae fed different diets develop to either worker or queen bees, even though they have identical genomic sequences [22]; a royal jelly fed to larvae by young nurse bees let female larvae develop as queens and other larvae fed usual bee bread develop as worker bees. Adult queens are quite different from workers in morphology, reproductive capability, behavioral repertoires and life span. This developmental difference seems to depend on the different expression of an entire collection of genes involved in larval fate [23-26], as microarray analysis demonstrated that 240 genes were differentially expressed between queens and workers [24]. In workers developmental genes were more up-regulated, while in queens physiometabolic genes were more up-regulated, including genes for metabolic enzymes, mass-transforming processes, and the general growth of the organism [24]. This difference in gene activation can be explained by an epigenetic mechanism, DNA methylation. Recently, Kucharski et al. [27] suggested that DNA methylation determined by nutritional input plays a critical role for different developmental fates between fertile queens and sterile workers. Silencing the expression of DNA methyltransferase 3 (Dnmt3) by injection of small interfering RNA (siRNA) in pooled larvae induced the development of large ovary like virgin queens raised on royal jelly. Interestingly, DNA methylation in CpG islands of dynactin p62 gene, which is known to be methylated during development in social insects [28], is significantly decreased in both queen larvae and siRNA treated larvae compared with worker larvae [27]. This study is helpful for the understanding of the potential nutritional effects on reprogramming in early development in humans because honey bees also have three functional DNA methyltransferases (Dnmts) and with similar in vivo properties to human Dnmts [28, 29].
2.3. Methyl donor nutrients for mouse phenotypes
The viable yellow agouti (Avy) mouse is a good animal model to determine the effect of methyl donor nutrients such as folate, methionine and choline on the phenotype of offspring. This model is in fact easy to control and the phenotypes determined by DNA methylation status at the metastable epiallele Avy are clearly definable (e.g., coat color and obesity) [30]. Metastable epialleles are expressed variably in genetically identical individuals by epigenetic modifications during development [16, 31], and the Avy allele is the first metastable epiallele known to be influenced by nutrition [32, 33]. The origin of metastable epiallele is not yet known but its characteristic has been regarded as similar to that of transposable elements (transposons) [10] which are regarded as remnants of ancestral infection and mostly silenced by CpG methylation.
In the agouti mouse model Cooney et al. [34] demonstrated that maternal methyl supplements increase DNA methylation in the Avy allele and determine the change of coat color which then represents an indicative pattern of future health and particularly of future development of obesity and insulin resistance. The percentage of phenotypes with more agouti coat (combined black and yellow pigment in the hair) are higher as increasing levels of methyl supplement are added to the diet. This shift in the distribution of mouse coat colors was correlated with DNA methylation of the Avy allele [34]. Waterland et al [35] further demonstrated that the dietary methyl supplementation of a/a dams with extra folic acid, vitamin B-12, choline, and betaine alters the phenotype of their Avy/a offspring via increasing CpG methylation at each of seven Avy pseudoexon 1A (PS1A) CpG sites. Dolinoy et al. [36] also demonstrated that maternal exposure to bisphenol A (BPA), an endocrine-active compound that is known to cause adverse effects on the reproduction and the development of animals [37], shifted the coat color distribution of Avy mice offspring toward yellow by decreasing CpG methylation in the intracisternal A particle (IAP) of the agouti gene. IAP sequences are known as endogenous retrovirus-like mobile elements, present at 1000 copies in the mouse genome [38]. Maternal nutritional supplementation such as folic acid or the phytoestrogen genistein counteracts BPA-induced DNA hypomethylation and shifts coat color distribution to black [39].
Another murine metastable epiallele, axin fused (AxinFu), was also examined by Waterland et al. [40]. Intron 6 of the AxinFu gene has a spontaneous IAP and the severity of the tail kinking is negatively associated with the degree of DNA methylation at the IAP in AxinFu [41]. Methyl donor supplementation of female mice during gestation period increased DNA methylation at AxinFu, silenced the expression from the cryptic promoter, and decreased the incidence of tail kinking in AxinFu /+ offspring [40]. This observation indicates that maternal diets may affect epigenetically more than one locus and change the various phenotypes of offspring. More interestingly, maternal methyl donor supplementation during mid-gestation also prevents the tail-specific loss of AxinFu methylation, indicating that nutritional influences on epigenetic regulation of metastable epialleles are not limited to early embryonic development.
There is still some debate over whether maternal diet-induced epigenetic change can be inherited to the next generation. Cropley at al. [42, 43] demonstrated that methyl donor supplementation of the pregnant dam shifts Avy phenotypes not only in exposed fetuses, but also in the fetus’s offspring, implicating that maternal diet may influence succeeding generations, independently of their later changes in diet. However, a recent study reported that diet-induced hypermethylation at Avy is not inherited in the female germ line [44].
2.4. Maternal folate status, DNA methylation and later life outcomes
Folate metabolism is closely associated with fetal development and growth including neural tube defects [45, 46]. It has been hypothesized that inhibition of methyl transfer or reduced folate intake could increase the risk of human neural tube defects by reducing DNA methylation, based on animal studies that demonstrated the importance of adequate methyl availability during cranial neural tube closure [9, 47]. During pregnancy folate status is positively associated with DNA methylation [48]. In a recent study using a hyperhomocysteinemia rat model, Kim et al. [49] demonstrated that folate-supplemented diet increased and folate-deplete diet decreased placental DNA methylation. Placenta DNA methylation was also positively correlated with hepatic folate and hepatic S-adenosylmethionine, the unique methyl donor for DNA methylation [49].
The protein restricted diet model also indicates the association among maternal folate status, DNA methylation and offspring’s health. Rats fed a protein restricted diet during pregnancy, which can induce high blood pressure and alter cardiovascular events in offspring [50, 51], showed decreased promoter methylation and increased expression of the glucocorticoid receptor gene (GR) and peroxisome proliferator-activated receptor gene (PPARa) in the liver of the adult offspring along with reduced Dnmt1 expression. However, folic acid supplementation reversed DNA hypomethylation and reduced the expression of those genes [52, 53]. Sinclair et al. demonstrated that methyl-deficient diet during early development in female mature sheep results in alterations of promoter DNA methylation and leads offspring to obesity, altered immune responses, insulin-resistance, and elevated blood pressure; these effects were most obvious in male offspring [54].
Two possible mechanisms to explain the epigenetic effect of maternal nutrients on phenotypes in their offspring were proposed by Waterland et al [55]; 1) reduced methyl availability may alter the establishment of DNA methylation at metastable epialleles by affecting either one-carbon metabolism or the activity of Dnmt1, and 2) repression of critical genes may occur during de novo DNA methylation in early fetal development. Moreover, this early nutritional effect can result in a permanent defect of epigenetic regulation through DNA methylation, suggesting a possible mechanism for the development of later life diseases [56].
3. Folate, DNA methylation and aging
3.1. DNA methylation and aging
Studies for the relationship between aging and DNA methylation have consistently demonstrated that aging is associated with genomic DNA hypomethylation and gene-specific promoter DNA hypermethylation in a tissue-specific manner [57-61]. Total methylcytosine contents are prone to decrease by aging, leading to genomic hypomethylation in most vertebrate tissues [62, 63], whereas promoter regions tend to undergo hypermethylation in many genes (Table 1). Recently, Bjornsson et al [64] demonstrated that global DNA methylation changes occurred over time in two different cohorts and this alteration may be under genetic control. This study demonstrated that 8-10% of individuals in both populations showed greater than 20% methylation changes over 11-16 years and further methylation changes of familial clustering.
Table 1. Age-associated promoter DNA methylation.
| Gene | Tissues, species | Description | References |
|---|---|---|---|
| ER α | Colon, human | Hypermethylation is associated with gene inactivation | [65] |
| Heart/ vessels, human | Hypermethylation with aging | [66, 67] | |
| Breast, rat | Hypomethylation with aging and tumor | [70] | |
| Prostate, human | Methylation increases with aging | [68, 69] | |
| IGFII | Colon, human | Methylation increases with aging | [71] |
| Brain, rat | Altered methylation in aged brain DNA | [72] | |
| Fibroblast, human | Hypermethylation in SV40 infected aged fibroblast cell | [73] | |
| p16INK4a | Colon, mouse | Hypermethylation with aging | [75] |
| Liver, human | Multiple genes are hypermethylated in aged liver | [76] | |
| Stomach, human | Multiple genes are progressively methylated with aging | [77] | |
| Msh2 | Multiple organ, mouse | Hypermethylation in old breeder mice | [128] |
| hTERT | Peripheral, blood, human | Hypermethylated in Alzheimer disease | [78] |
| N-cadherin | Kidney, rat | Highly methylation in kidney but not liver | [129] |
| E-cadherin | Bladder, human | Hypermethylation with aging and tumor | [130] |
| Stomach, human | Hypermethylation in promoter of 5 genes with aging | [131] | |
| COL1A1 | Periodontal ligament | Methylation decreases the gene expression | [132, 133] |
| C-fos | Liver, human | Methylation increases with aging | [134] |
| RASSF1A | Kidney, human | Age is associated with methylation in kidney | [135] |
| Breast, human | Methylation increases linearly between ages 32-55 | [136] | |
| Lung, human | Methylation is associated with age in lung cancer | [137] |
Estrogen receptor (ER) gene was first studied to show an association between age and promoter DNA methylation [62]. Issa et al. [65] demonstrated that the ER gene is repressed by promoter hypermethylation in the aged human colonic mucosa, which is similar to the earliest epigenetic event that predispose to colonic tumorigenesis. However, methylation status in this gene is tissue-specific; hypermethylated in aged heart, arteries, vessels [66, 67] and prostate [68, 69] and hypomethylated in aged breast tissue [70]. Another gene studied by the same group is the insulin-like growth factor II gene (IGF2) [71]. During aging, this promoter methylation also becomes more extensive and involves the originally unmethylated allele in human colon. Similar methylation patterns are reported in other tissues such as rat brain and human fibroblast [72, 73].
Other genes for tumor suppression, cell cycle, apoptosis, detoxification, lipid metabolism and housekeeping [74] are also associated with altered promoter DNA methylation in aging. In a young and old mice study, aged mice colon demonstrated p16 promoter hypermethylation compared to young mice, and the p16 promoter methylation increase paralleled with the increased dietary folate levels [75]. Recently, multiple tumor suppressor genes including lysil oxidase (LOX), runt-related transcription factor 3 (RUNX3) and tazarotene-induced gene 1 (TIG1) showed aberrant methylation in their promoter regions in normal aged liver [76] and gastric epithelium [77]. Many other genes have been studied in a variety of tissues to demonstrate associations between aging and DNA methylation (Table 1). However, longevity associated genes such as SIRT3, SMARCA5, and CDH1 in peripheral blood did not show any significant differences in promoters methylation between young and elderly groups [78]. Evidence also indicates that the aging process induces gene silencing by other epigenetic phenomena such as histone modifications and chromatin remodeling in combination with DNA methylation [74, 79].
3.2. Mechanisms for altered DNA methylation in aging
Several potential mechanisms are suggested for age-associated changes in genomic DNA methylation [62]. 1) The progressive decrement of genomic DNA methylation during aging results mainly from passive demethylation by the progressively diminished activity of Dnmt1 that maintains the hypermethylated status of heterochromatic DNA [80]. Expression of Dnmt1 declines significantly from birth to age and decreased Dnmt1 activity may lead to reduced replication of methylation patterns during mitosis [73]. 2) Aging per se increases homocysteine, a sulfur containing amino acid formed in folate-mediated one-carbon metabolism [81]. A major cause of hyperhomocysteinemia is the disturbance of methyl transfer in one-carbon metabolism, which is known to decrease genomic DNA methylation status by increasing the cellular S-adenosylhomocysteine that inhibits DNA methyltransferases [82]. 3) Friso et al. demonstrated that estrogen replacement therapy in menopause women reduces total plasma homocysteine concentration and increases genomic DNA methylation of peripheral mononuclear cells [83]. This study indicates that the decline of sex hormone during aging may also reduce genomic DNA methylation. 4) Nutritional factors associated with aging are also involved in DNA hypomethylation [62]. Folate status declines with aging due to both decreased folate intake and altered folate availability [84], and folate depletion results in DNA hypomethylation in elderly women [85, 86]. Both a folate deficient diet and a methyl donor deficient diet are known to decrease methyl-CpG-binding protein and Dnmts, resulting in genomic DNA hypomethylation in the rat liver [87, 88]. Deficiency of trace elements such as zinc and selenium also causes genomic DNA hypomethylation by altering one-carbon metabolism in the elderly [89].
The mechanism for aging-associated promoter hypermethylation is not yet clear. In contrast to reduced genome-wide methylation in aged cells by reduced Dnmt1 activity, gene specific promoter hypermethylation observed during aging may be linked to increased de novo methylation by other Dnmts such as Dnmt3b, which showed increased activity in aged and immortalized cells [90]. It is also suggested that heterochromatin, which is highly methylated and has the ability to propagate and influence gene expression in a region-specific and sequence-independent manner, may spread to euchromatin, which is less methylated and easily transcribed, over the boundary elements by aging. This encroachment of heterochromatin may progressively methylate the promoter regions of adjacent genes by aging [91] .
3.3. DNA methylation, folate and life span
Lin et al [92] demonstrated that overexpression of Drosophila DNA methyltransferase 2 (dDnmt2) could extend the life span of Drosophila melanogaster. The upstream-activating sequences-dDnmt2 transgene (UAS-dDnmt2) inserted fly, which over-expresses dDnmt2, shows a greater mean life span compared to controls. Although the functional role of the Dnmt2 proteins is still unclear and Dnmt2 is more known to mediate RNA methylation [93], this experiment implies that DNA methylation might affect aging or longevity of eukaryotes. Interestingly, mice with disruption of proliferation associated SNF-2-like gene (PASG), which displayed a loss of 33 to 43% of total methylcytosine, demonstrated cellular senescence and an age-associated phenotype, indicating that altered DNA methylation can result in cell senescence and reduced longevity [94, 95].
Telomere shortening is associated with reduced human life span and has been proposed as a mechanism of age-associated disorders such as cancer [96]. Increasing amounts of evidence suggest that telomere length is regulated by neighboring histone modifications [97-99] and DNA methylation of the subtelomeric region adjacent to the telomere, which is comprised of repeated (TTAGGG)n sequences lacking CpG dinucleotides necessary for Dnmts [100]. During aging, subtelomeric regions become hypomethylated due to decreased Dnmt activity, resulting in telomere shortening [100].
Homocysteine is known to accelerate the onset of endothelial progenitor cell senescence by the suppression of telomerase activity which results in shortening of telomere [101]. A recent population-based cohort study demonstrated that plasma homocysteine concentrations are negatively correlated with leukocytic telomere length (LTL) [102]. Based on these observations, it can be hypothesized that a low folate diet or methyl deficient diet, both of which can induce hyperhomocysteinemia, might also be involved in the shortening of telomere length. However, it is not known whether the effects of hyperhomocysteinemia on telomerase activity or the length of telomere are conveyed through genomic DNA hypomethylation or promoter DNA hypermethylation [103].
Low dietary folate can alter, genetically, the sequence and length of telomere [104]. Under low folate condition, thymidine rich telomere sequences are prone to DNA strand breaks, which may lead to shortening of telomere length. It is known that folate deficiency increases uracil misincorporation into DNA by reducing the synthesis of thymidylate, thereby producing DNA strand breaks by simultaneously increased activity of uracil glycosylases and reduced DNA repair [103].
3.4. DNA methylation, an epigenetic transition between aging and cancer
A possible epigenetic link between aging and cancer has been suggested because genomic DNA hypomethylation, promoter DNA hypermethylation and altered Dnmt expression can occur in both aging and cancer [105]. Toyota et al. [106] demonstrated a similar pattern of promoter methylation in colon cancer clones can occur in normal colonic cells during the aging process. Since epigenetic silencing of tumor suppressor genes by promoter DNA hypermethylation can initiate tumorigenesis [107, 108], it appears that progressively accumulated promoter hypermethylation by aging could induce cellular transformation to malignancy [80].
Genomic DNA hypomethylation, which frequently occurs during aging, is also observed in many malignant tissues, but the role of aging-associated genomic hypomethylation in carcinogenesis is not yet fully understood [105]. Decreased DNA methylation is thought to promote chromosomal instability eventually leading to carcinogenesis [109, 110]. Genome-wide DNA hypomethylation also affects transcription through loss of imprinting [111], ectopic onco-fetal gene expression, and up-regulation of silent genes, all of which might induce tumor development [8, 112].
3.5. Interactions between folate and aging in cancer
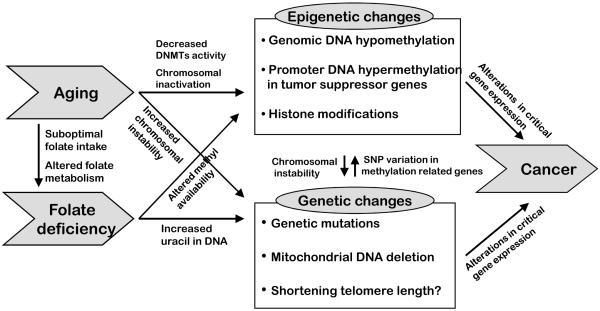
As a coenzyme in one-carbon metabolism, folate participates in both nucleotide synthesis (genetic pathway) and DNA methylation (epigenetic pathway) [84]. Figure 1 shows a complex mechanism through which folate and aging interact genetically and epigenetically, thereby promoting cancer development.
Figure 1.
Genetic and epigenetic interactions between aging and folate deficiency in carcinogenesis
Inadequate folate intake reduces the synthesis of thymidylate from deoxyuridylate, resulting in excessive uracil misincorporation into DNA, consequently leading to mutagenesis [113, 114]. Choi et al [115] demonstrated that elder rats increased uracil content in colonic DNA compared with young rats and dietary folate depletion further increased uracil misincorporation. Aging and folate also affect the integrity of mitochondrial DNA, the instability of which is thought to play an important role in tumorigenesis [116-118].
In one-carbon metabolism folate depletion decreases S-adenosylmethionine, the universal methyl donor, and increases S-adenosylhomocysteine, an inhibitor of methyltransferases. Aberrant DNA methylation mediated by folate depletion has been regarded as one candidate mechanism that explains the association between folate and cancer [84]. Since aging by itself affects DNA methylation, a synergistic effect between folate status and aging on DNA methylation has been investigated. In the elder rat liver folate supplementation increases genomic DNA methylation in a dose dependent manner [119]. Keyes et al. [75] demonstrated that genomic DNA methylation and promoter methylation of p16 increased in parallel with an increment of dietary folate levels in old mice colon but not in the young. Interestingly, DNA methylation may interact with genetic mechanisms, because 1) genomic DNA hypomethylation is associated with instability of chromosome and DNA [120, 121] and 2) specific allelic sequence variants such as single nucleotide polymorphisms (SNPs) may act in cis or trans to influence DNA methylation [122]. Genetic variations in genes that regulate epigenetic phenomena such as Dnmt genes or polymorphisms of genes involved in one-carbon metabolism such as 5,10-methylenetetrahydrofolate reductase gene (MTHFR) can affect DNA methylation [123-126]. In another example, a G>C SNP in the LRP1B promoter creates an additional CpG site and increases the probability that this allele becomes methylated [127].
4. Conclusions
As seen in the experimental models, maternal diet can affect the offspring’s phenotype as well as the risk of disease development at the later life. The amount of methyl donors in the maternal diet is especially critical for embryonic development by affecting DNA methylation in animal models. In this regard, folate, which ultimately transfers methyl groups for DNA methylation, is expected to significantly influence the extent of DNA methylation reprogramming during the embryonic development period that is particularly vulnerable to the low methyl availability.
During aging, genomic and gene specific DNA methylation can be altered in a tissue-specific manner. Folate status further modifies DNA methylation in the elderly. It appears that aging and folate deficiency synergistically provide an epigenetic milieu toward cancer development.
For healthy embryonic growth and aging, as well as for the prevention of aging-associated chronic diseases, proper folate status is needed to secure optimal DNA methylation status. Further studies to investigate the effect of folate on fetal epigenetic programming and aging focused on the role of DNA methylation and other major epigenetic phenomena will provide us with a better understanding of maintaining health and preventing chronic diseases.
Acknowledgments
This material is based upon work supported by the U.S. Department of Agriculture, under agreement No. 581950-9-001. Any opinions, findings, conclusion, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the U.S. Dept of Agriculture. Authors do not have any competing interest. This project has been supported in part by the National Institute of Health Grants R21 AA016681 and R01 AG025834 (SWC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- [1].Barker DJ, Osmond C. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet. 1986;1:1077–1081. doi: 10.1016/s0140-6736(86)91340-1. [DOI] [PubMed] [Google Scholar]
- [2].Kensara OA, Wootton SA, Phillips DI, Patel M, Jackson AA, Elia M. Fetal programming of body composition: relation between birth weight and body composition measured with dual-energy X-ray absorptiometry and anthropometric methods in older Englishmen. The American journal of clinical nutrition. 2005;82:980–987. doi: 10.1093/ajcn/82.5.980. [DOI] [PubMed] [Google Scholar]
- [3].Osmond C, Barker DJ, Winter PD, Fall CH, Simmonds SJ. Early growth and death from cardiovascular disease in women. BMJ (Clinical research ed. 1993;307:1519–1524. doi: 10.1136/bmj.307.6918.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hales CN, Barker DJ. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia. 1992;35:595–601. doi: 10.1007/BF00400248. [DOI] [PubMed] [Google Scholar]
- [5].Cooper C, Fall C, Egger P, Hobbs R, Eastell R, Barker D. Growth in infancy and bone mass in later life. Annals of the rheumatic diseases. 1997;56:17–21. doi: 10.1136/ard.56.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rich-Edwards JW, Kleinman K, Michels KB, et al. Longitudinal study of birth weight and adult body mass index in predicting risk of coronary heart disease and stroke in women. BMJ (Clinical research ed. 2005;330:1115. doi: 10.1136/bmj.38434.629630.E0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. The New England journal of medicine. 2008;359:61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Shames DS, Minna JD, Gazdar AF. DNA methylation in health, disease, and cancer. Current molecular medicine. 2007;7:85–102. doi: 10.2174/156652407779940413. [DOI] [PubMed] [Google Scholar]
- [9].Dunlevy LP, Burren KA, Mills K, Chitty LS, Copp AJ, Greene ND. Integrity of the methylation cycle is essential for mammalian neural tube closure. Birth Defects Res A Clin Mol Teratol. 2006;76:544–552. doi: 10.1002/bdra.20286. [DOI] [PubMed] [Google Scholar]
- [10].Waterland RA, Jirtle RL. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20:63–68. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- [11].Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science (New York, NY. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- [13].Tang WY, Ho SM. Epigenetic reprogramming and imprinting in origins of disease. Rev Endocr Metab Disord. 2007;8:173–182. doi: 10.1007/s11154-007-9042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(1):R47–58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- [15].Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- [16].Dolinoy DC, Das R, Weidman JR, Jirtle RL. Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatr Res. 2007;61:30R–37R. doi: 10.1203/pdr.0b013e31804575f7. [DOI] [PubMed] [Google Scholar]
- [17].Murphy SK, Jirtle RL. Imprinting evolution and the price of silence. Bioessays. 2003;25:577–588. doi: 10.1002/bies.10277. [DOI] [PubMed] [Google Scholar]
- [18].Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- [19].Feinberg AP. The epigenetics of cancer etiology. Seminars in cancer biology. 2004;14:427–432. doi: 10.1016/j.semcancer.2004.06.005. [DOI] [PubMed] [Google Scholar]
- [20].Ideraabdullah FY, Vigneau S, Bartolomei MS. Genomic imprinting mechanisms in mammals. Mutat Res. 2008;647:77–85. doi: 10.1016/j.mrfmmm.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hirasawa R, Chiba H, Kaneda M, et al. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22:1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lue PF, Dizon SE. Studies in the mode of action of royal jelly in honeybee development. VII. The free amino acids in the haemolymph of developing larvae. Can J Zool. 1967;45:205–214. doi: 10.1139/z67-027. [DOI] [PubMed] [Google Scholar]
- [23].Grozinger CM, Fan Y, Hoover SE, Winston ML. Genome-wide analysis reveals differences in brain gene expression patterns associated with caste and reproductive status in honey bees (Apis mellifera) Mol Ecol. 2007;16:4837–4848. doi: 10.1111/j.1365-294X.2007.03545.x. [DOI] [PubMed] [Google Scholar]
- [24].Barchuk AR, Cristino AS, Kucharski R, Costa LF, Simoes ZL, Maleszka R. Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Dev Biol. 2007;7:70. doi: 10.1186/1471-213X-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Evans JD, Wheeler DE. Expression profiles during honeybee caste determination. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0001. RESEARCH0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Evans JD, Wheeler DE. Differential gene expression between developing queens and workers in the honey bee, Apis mellifera. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:5575–5580. doi: 10.1073/pnas.96.10.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science (New York, NY. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- [28].Wang Y, Jorda M, Jones PL, et al. Functional CpG methylation system in a social insect. Science (New York, NY. 2006;314:645–647. doi: 10.1126/science.1135213. [DOI] [PubMed] [Google Scholar]
- [29].Schaefer M, Lyko F. DNA methylation with a sting: an active DNA methylation system in the honeybee. Bioessays. 2007;29:208–211. doi: 10.1002/bies.20548. [DOI] [PubMed] [Google Scholar]
- [30].Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nature genetics. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- [31].Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18:348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- [32].Waterland RA. Do maternal methyl supplements in mice affect DNA methylation of offspring? The Journal of nutrition. 2003;133:238. doi: 10.1093/jn/133.1.238. author reply 239. [DOI] [PubMed] [Google Scholar]
- [33].Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. Faseb J. 1998;12:949–957. [PubMed] [Google Scholar]
- [34].Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. The Journal of nutrition. 2002;132:2393S–2400S. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- [35].Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yaoi T, Itoh K, Nakamura K, Ogi H, Fujiwara Y, Fushiki S. Genome-wide analysis of epigenomic alterations in fetal mouse forebrain after exposure to low doses of bisphenol A. Biochem Biophys Res Commun. 2008;376:563–567. doi: 10.1016/j.bbrc.2008.09.028. [DOI] [PubMed] [Google Scholar]
- [38].Dupressoir A, Heidmann T. Expression of intracisternal A-particle retrotransposons in primary tumors of oncogene-expressing transgenic mice. Oncogene. 1997;14:2951–2958. doi: 10.1038/sj.onc.1201148. [DOI] [PubMed] [Google Scholar]
- [39].Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Waterland RA, Dolinoy DC, Lin JR, Smith CA, Shi X, Tahiliani KG. Maternal methyl supplements increase offspring DNA methylation at Axin Fused. Genesis. 2006;44:401–406. doi: 10.1002/dvg.20230. [DOI] [PubMed] [Google Scholar]
- [41].Rakyan VK, Chong S, Champ ME, et al. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cropley JE, Suter CM, Beckman KB, Martin DI. Germ-line epigenetic modification of the murine A vy allele by nutritional supplementation. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17308–17312. doi: 10.1073/pnas.0607090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Cooney CA. Germ cells carry the epigenetic benefits of grandmother’s diet. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17071–17072. doi: 10.1073/pnas.0608653103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Waterland RA, Travisano M, Tahiliani KG. Diet-induced hypermethylation at agouti viable yellow is not inherited transgenerationally through the female. Faseb J. 2007;21:3380–3385. doi: 10.1096/fj.07-8229com. [DOI] [PubMed] [Google Scholar]
- [45].Antony AC. In utero physiology: role of folic acid in nutrient delivery and fetal development. The American journal of clinical nutrition. 2007;85:598S–603S. doi: 10.1093/ajcn/85.2.598S. [DOI] [PubMed] [Google Scholar]
- [46].Rondo PH, Tomkins AM. Folate and intrauterine growth retardation. Ann Trop Paediatr. 2000;20:253–258. doi: 10.1080/02724936.2000.11748144. [DOI] [PubMed] [Google Scholar]
- [47].Weingartner J, Lotz K, Fanghanel J, Gedrange T, Bienengraber V, Proff P. Induction and prevention of cleft lip, alveolus and palate and neural tube defects with special consideration of B vitamins and the methylation cycle. J Orofac Orthop. 2007;68:266–277. doi: 10.1007/s00056-007-0701-6. [DOI] [PubMed] [Google Scholar]
- [48].Park BH, Kim YJ, Park JS, et al. Folate and homocysteine levels during pregnancy affect DNA methylation in human placenta. J Prev Med Pub Health. 2005;38:437–442. [PubMed] [Google Scholar]
- [49].Kim JM, Hong K, Lee JH, Lee S, Chang N. Effect of folate deficiency on placental DNA methylation in hyperhomocysteinemic rats. J Nutr Biochem. 2008 doi: 10.1016/j.jnutbio.2008.01.010. [DOI] [PubMed] [Google Scholar]
- [50].Watkins AJ, Wilkins A, Cunningham C, et al. Low protein diet fed exclusively during mouse oocyte maturation leads to behavioural and cardiovascular abnormalities in offspring. J Physiol. 2008;586:2231–2244. doi: 10.1113/jphysiol.2007.149229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kappen C. Folate supplementation in three genetic models: implications for understanding folate-dependent developmental pathways. Am J Med Genet C Semin Med Genet. 2005;135C:24–30. doi: 10.1002/ajmg.c.30050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. The Journal of nutrition. 2005;135:1382–1386. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- [53].Lillycrop KA, Slater-Jefferies JL, Hanson MA, Godfrey KM, Jackson AA, Burdge GC. Induction of altered epigenetic regulation of the hepatic glucocorticoid receptor in the offspring of rats fed a protein-restricted diet during pregnancy suggests that reduced DNA methyltransferase-1 expression is involved in impaired DNA methylation and changes in histone modifications. The British journal of nutrition. 2007;97:1064–1073. doi: 10.1017/S000711450769196X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sinclair KD, Allegrucci C, Singh R, et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19351–19356. doi: 10.1073/pnas.0707258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr. 2007;27:363–388. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- [56].McKay JA, Williams EA, Mathers JC. Folate and DNA methylation during in utero development and aging. Biochem Soc Trans. 2004;32:1006–1007. doi: 10.1042/BST0321006. [DOI] [PubMed] [Google Scholar]
- [57].Hamatani T, Falco G, Carter MG, et al. Age-associated alteration of gene expression patterns in mouse oocytes. Human molecular genetics. 2004;13:2263–2278. doi: 10.1093/hmg/ddh241. [DOI] [PubMed] [Google Scholar]
- [58].Ono T, Takahashi N, Okada S. Age-associated changes in DNA methylation and mRNA level of the c-myc gene in spleen and liver of mice. Mutation research. 1989;219:39–50. doi: 10.1016/0921-8734(89)90039-8. [DOI] [PubMed] [Google Scholar]
- [59].Xu J. Age-related changes in Usp9x protein expression and DNA methylation in mouse brain. Brain research. 2005;140:17–24. doi: 10.1016/j.molbrainres.2005.06.009. [DOI] [PubMed] [Google Scholar]
- [60].Richardson BC. Role of DNA methylation in the regulation of cell function: autoimmunity, aging and cancer. The Journal of nutrition. 2002;132:2401S–2405S. doi: 10.1093/jn/132.8.2401S. [DOI] [PubMed] [Google Scholar]
- [61].Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science (New York, NY. 1983;220:1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- [62].Richardson B. Impact of aging on DNA methylation. Ageing research reviews. 2003;2:245–261. doi: 10.1016/s1568-1637(03)00010-2. [DOI] [PubMed] [Google Scholar]
- [63].Golbus J, Palella TD, Richardson BC. Quantitative changes in T cell DNA methylation occur during differentiation and ageing. European journal of immunology. 1990;20:1869–1872. doi: 10.1002/eji.1830200836. [DOI] [PubMed] [Google Scholar]
- [64].Bjornsson HT, Sigurdsson MI, Fallin MD, et al. Intra-individual change over time in DNA methylation with familial clustering. Jama. 2008;299:2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nature genetics. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- [66].Post WS, Goldschmidt-Clermont PJ, Wilhide CC, et al. Methylation of the estrogen receptor gene is associated with aging and atherosclerosis in the cardiovascular system. Cardiovascular research. 1999;43:985–991. doi: 10.1016/s0008-6363(99)00153-4. [DOI] [PubMed] [Google Scholar]
- [67].Kim J, Kim JY, Song KS, et al. Epigenetic changes in estrogen receptor beta gene in atherosclerotic cardiovascular tissues and in-vitro vascular senescence. Biochimica et biophysica acta. 2007;1772:72–80. doi: 10.1016/j.bbadis.2006.10.004. [DOI] [PubMed] [Google Scholar]
- [68].Kwabi-Addo B, Chung W, Shen L, et al. Age-related DNA methylation changes in normal human prostate tissues. Clin Cancer Res. 2007;13:3796–3802. doi: 10.1158/1078-0432.CCR-07-0085. [DOI] [PubMed] [Google Scholar]
- [69].Li LC, Shiina H, Deguchi M, et al. Age-dependent methylation of ESR1 gene in prostate cancer. Biochemical and biophysical research communications. 2004;321:455–461. doi: 10.1016/j.bbrc.2004.06.164. [DOI] [PubMed] [Google Scholar]
- [70].Yenbutr P, Hilakivi-Clarke L, Passaniti A. Hypomethylation of an exon I estrogen receptor CpG island in spontaneous and carcinogen-induced mammary tumorigenesis in the rat. Mechanisms of ageing and development. 1998;106:93–102. doi: 10.1016/s0047-6374(98)00093-1. [DOI] [PubMed] [Google Scholar]
- [71].Issa JP, Vertino PM, Boehm CD, Newsham IF, Baylin SB. Switch from monoallelic to biallelic human IGF2 promoter methylation during aging and carcinogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:11757–11762. doi: 10.1073/pnas.93.21.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kitraki E, Bozas E, Philippidis H, Stylianopoulou F. Aging-related changes in IGF-II and c-fos gene expression in the rat brain. Int J Dev Neurosci. 1993;11:1–9. doi: 10.1016/0736-5748(93)90029-d. [DOI] [PubMed] [Google Scholar]
- [73].Vertino PM, Issa JP, Pereira-Smith OM, Baylin SB. Stabilization of DNA methyltransferase levels and CpG island hypermethylation precede SV40-induced immortalization of human fibroblasts. Cell Growth Differ. 1994;5:1395–1402. [PubMed] [Google Scholar]
- [74].Burzynski SR. Aging: gene silencing or gene activation? Medical hypotheses. 2005;64:201–208. doi: 10.1016/j.mehy.2004.06.010. [DOI] [PubMed] [Google Scholar]
- [75].Keyes MK, Jang H, Mason JB, et al. Older age and dietary folate are determinants of genomic and p16-specific DNA methylation in mouse colon. The Journal of nutrition. 2007;137:1713–1717. doi: 10.1093/jn/137.7.1713. [DOI] [PubMed] [Google Scholar]
- [76].Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology (Baltimore, Md. 2008;47:908–918. doi: 10.1002/hep.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].So K, Tamura G, Honda T, et al. Multiple tumor suppressor genes are increasingly methylated with age in non-neoplastic gastric epithelia. Cancer science. 2006;97:1155–1158. doi: 10.1111/j.1349-7006.2006.00302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Silva PN, Gigek CO, Leal MF, et al. Promoter methylation analysis of SIRT3, SMARCA5, HTERT and CDH1 genes in aging and Alzheimer’s disease. J Alzheimers Dis. 2008;13:173–176. doi: 10.3233/jad-2008-13207. [DOI] [PubMed] [Google Scholar]
- [79].Burzynski SR. Gene silencing--a new theory of aging. Medical hypotheses. 2003;60:578–583. doi: 10.1016/s0306-9877(03)00050-1. [DOI] [PubMed] [Google Scholar]
- [80].Fraga MF, Agrelo R, Esteller M. Cross-talk between aging and cancer: the epigenetic language. Ann N Y Acad Sci. 2007;1100:60–74. doi: 10.1196/annals.1395.005. [DOI] [PubMed] [Google Scholar]
- [81].Jacques PF, Bostom AG, Wilson PW, Rich S, Rosenberg IH, Selhub J. Determinants of plasma total homocysteine concentration in the Framingham Offspring cohort. Am J Clin Nutr. 2001;73:613–621. doi: 10.1093/ajcn/73.3.613. [DOI] [PubMed] [Google Scholar]
- [82].Yi P, Melnyk S, Pogribna M, Pogribny IP, Hine RJ, James SJ. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation [In Process Citation] J Biol Chem. 2000;275:29318–29323. doi: 10.1074/jbc.M002725200. [DOI] [PubMed] [Google Scholar]
- [83].Friso S, Lamon-Fava S, Jang H, Schaefer EJ, Corrocher R, Choi SW. Oestrogen replacement therapy reduces total plasma homocysteine and enhances genomic DNA methylation in postmenopausal women. The British journal of nutrition. 2007;97:617–621. doi: 10.1017/S0007114507433013. [DOI] [PubMed] [Google Scholar]
- [84].Choi SW, Mason JB. Folate and carcinogenesis: an integrated scheme. The Journal of nutrition. 2000;130:129–132. doi: 10.1093/jn/130.2.129. [DOI] [PubMed] [Google Scholar]
- [85].Rampersaud GC, Kauwell GP, Hutson AD, Cerda JJ, Bailey LB. Genomic DNA methylation decreases in response to moderate folate depletion in elderly women. Am J Clin Nutr. 2000;72:998–1003. doi: 10.1093/ajcn/72.4.998. see comments. [DOI] [PubMed] [Google Scholar]
- [86].Jacob RA, Gretz DM, Taylor PC, et al. Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J Nutr. 1998;128:1204–1212. doi: 10.1093/jn/128.7.1204. [DOI] [PubMed] [Google Scholar]
- [87].Esfandiari F, Green R, Cotterman RF, Pogribny IP, James SJ, Miller JW. Methyl deficiency causes reduction of the methyl-CpG-binding protein, MeCP2, in rat liver. Carcinogenesis. 2003;24:1935–1940. doi: 10.1093/carcin/bgg163. [DOI] [PubMed] [Google Scholar]
- [88].Ghoshal K, Li X, Datta J, et al. A folate- and methyl-deficient diet alters the expression of DNA methyltransferases and methyl CpG binding proteins involved in epigenetic gene silencing in livers of F344 rats. The Journal of nutrition. 2006;136:1522–1527. doi: 10.1093/jn/136.6.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Cooney CA. Dietary selenium and arsenic affect DNA methylation. The Journal of nutrition. 2001;131:1871–1872. doi: 10.1093/jn/131.6.1871. [DOI] [PubMed] [Google Scholar]
- [90].Lopatina N, Haskell JF, Andrews LG, Poole JC, Saldanha S, Tollefsbol T. Differential maintenance and de novo methylating activity by three DNA methyltransferases in aging and immortalized fibroblasts. J Cell Biochem. 2002;84:324–334. doi: 10.1002/jcb.10015. [DOI] [PubMed] [Google Scholar]
- [91].Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- [92].Lin MJ, Tang LY, Reddy MN, Shen CK. DNA methyltransferase gene dDnmt2 and longevity of Drosophila. J Biol Chem. 2005;280:861–864. doi: 10.1074/jbc.C400477200. [DOI] [PubMed] [Google Scholar]
- [93].Lyko F, Whittaker AJ, Orr-Weaver TL, Jaenisch R. The putative Drosophila methyltransferase gene dDnmt2 is contained in a transposon-like element and is expressed specifically in ovaries. Mech Dev. 2000;95:215–217. doi: 10.1016/s0925-4773(00)00325-7. [DOI] [PubMed] [Google Scholar]
- [94].Sun LQ, Arceci RJ. Altered epigenetic patterning leading to replicative senescence and reduced longevity. A role of a novel SNF2 factor, PASG. Cell cycle (Georgetown, Tex. 2005;4:3–5. doi: 10.4161/cc.4.1.1341. [DOI] [PubMed] [Google Scholar]
- [95].Sun LQ, Lee DW, Zhang Q, et al. Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev. 2004;18:1035–1046. doi: 10.1101/gad.1176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Aragona M, Maisano R, Panetta S, et al. Telomere length maintenance in aging and carcinogenesis. International journal of oncology. 2000;17:981–989. doi: 10.3892/ijo.17.5.981. [DOI] [PubMed] [Google Scholar]
- [97].Fingerman IM, Li HC, Briggs SD. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes & development. 2007;21:2018–2029. doi: 10.1101/gad.1560607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Garcia-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nature genetics. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- [99].Grafi G, Ben-Meir H, Avivi Y, Moshe M, Dahan Y, Zemach A. Histone methylation controls telomerase-independent telomere lengthening in cells undergoing dedifferentiation. Developmental biology. 2007;306:838–846. doi: 10.1016/j.ydbio.2007.03.023. [DOI] [PubMed] [Google Scholar]
- [100].Gonzalo S, Jaco I, Fraga MF, et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nature cell biology. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- [101].Zhu JH, Chen JZ, Wang XX, Xie XD, Sun J, Zhang FR. Homocysteine accelerates senescence and reduces proliferation of endothelial progenitor cells. Journal of molecular and cellular cardiology. 2006;40:648–652. doi: 10.1016/j.yjmcc.2006.01.011. [DOI] [PubMed] [Google Scholar]
- [102].Richards JB, Valdes AM, Gardner JP, et al. Homocysteine levels and leukocyte telomere length. Atherosclerosis. 2008 doi: 10.1016/j.atherosclerosis.2007.12.035. [DOI] [PubMed] [Google Scholar]
- [103].Bull C, Fenech M. Genome-health nutrigenomics and nutrigenetics: nutritional requirements or ‘nutriomes’ for chromosomal stability and telomere maintenance at the individual level. The Proceedings of the Nutrition Society. 2008;67:146–156. doi: 10.1017/S0029665108006988. [DOI] [PubMed] [Google Scholar]
- [104].Toussaint M, Dionne I, Wellinger RJ. Limited TTP supply affects telomere length regulation in a telomerase-independent fashion. Nucleic acids research. 2005;33:704–713. doi: 10.1093/nar/gki219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Liu L, Wylie RC, Andrews LG, Tollefsbol TO. Aging, cancer and nutrition: the DNA methylation connection. Mechanisms of ageing and development. 2003;124:989–998. doi: 10.1016/j.mad.2003.08.001. [DOI] [PubMed] [Google Scholar]
- [106].Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Issa JP. CpG-island methylation in aging and cancer. Current topics in microbiology and immunology. 2000;249:101–118. doi: 10.1007/978-3-642-59696-4_7. [DOI] [PubMed] [Google Scholar]
- [108].Toyota M, Issa JP. The role of DNA hypermethylation in human neoplasia. Electrophoresis. 2000;21:329–333. doi: 10.1002/(SICI)1522-2683(20000101)21:2<329::AID-ELPS329>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- [109].Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science (New York, NY. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- [110].Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science (New York, NY. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- [111].Holm TM, Jackson-Grusby L, Brambrink T, Yamada Y, Rideout WM, 3rd, Jaenisch R. Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer cell. 2005;8:275–285. doi: 10.1016/j.ccr.2005.09.007. [DOI] [PubMed] [Google Scholar]
- [112].Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nature genetics. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- [113].Mason JB, Choi SW. Folate and carcinogenesis: developing a unifying hypothesis. Advances in enzyme regulation. 2000;40:127–141. doi: 10.1016/s0065-2571(99)00037-0. [DOI] [PubMed] [Google Scholar]
- [114].Blount BC, Mack MM, Wehr CM, et al. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:3290–3295. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Choi SW, Friso S, Dolnikowski GG, et al. Biochemical and molecular aberrations in the rat colon due to folate depletion are age-specific. The Journal of nutrition. 2003;133:1206–1212. doi: 10.1093/jn/133.4.1206. [DOI] [PubMed] [Google Scholar]
- [116].Shay JW, Werbin H. New evidence for the insertion of mitochondrial DNA into the human genome: significance for cancer and aging. Mutation research. 1992;275:227–235. doi: 10.1016/0921-8734(92)90026-l. [DOI] [PubMed] [Google Scholar]
- [117].Zhu W, Qin W, Sauter ER. Large-scale mitochondrial DNA deletion mutations and nuclear genome instability in human breast cancer. Cancer detection and prevention. 2004;28:119–126. doi: 10.1016/j.cdp.2003.11.008. [DOI] [PubMed] [Google Scholar]
- [118].Crott JW, Choi SW, Branda RF, Mason JB. Accumulation of mitochondrial DNA deletions is age, tissue and folate-dependent in rats. Mutation research. 2005;570:63–70. doi: 10.1016/j.mrfmmm.2004.09.009. [DOI] [PubMed] [Google Scholar]
- [119].Choi SW, Friso S, Keyes MK, Mason JB. Folate supplementation increases genomic DNA methylation in the liver of elder rats. The British journal of nutrition. 2005;93:31–35. doi: 10.1079/bjn20041283. [DOI] [PubMed] [Google Scholar]
- [120].Toyota M, Issa JP. CpG island methylator phenotypes in aging and cancer. Seminars in cancer biology. 1999;9:349–357. doi: 10.1006/scbi.1999.0135. [DOI] [PubMed] [Google Scholar]
- [121].Shen L, Toyota M, Kondo Y, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18654–18659. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Dobrovic A, Kristensen LS. DNA methylation, epimutations and cancer predisposition. The international journal of biochemistry & cell biology. 2009;41:34–39. doi: 10.1016/j.biocel.2008.09.006. [DOI] [PubMed] [Google Scholar]
- [123].Miremadi A, Oestergaard MZ, Pharoah PD, Caldas C. Cancer genetics of epigenetic genes. Human molecular genetics. 2007;16(1):R28–49. doi: 10.1093/hmg/ddm021. [DOI] [PubMed] [Google Scholar]
- [124].Cebrian A, Pharoah PD, Ahmed S, et al. Genetic variants in epigenetic genes and breast cancer risk. Carcinogenesis. 2006;27:1661–1669. doi: 10.1093/carcin/bgi375. [DOI] [PubMed] [Google Scholar]
- [125].Koushik A, Kraft P, Fuchs CS, et al. Nonsynonymous polymorphisms in genes in the one-carbon metabolism pathway and associations with colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:2408–2417. doi: 10.1158/1055-9965.EPI-06-0624. [DOI] [PubMed] [Google Scholar]
- [126].Siva A, De Lange M, Clayton D, Monteith S, Spector T, Brown MJ. The heritability of plasma homocysteine, and the influence of genetic variation in the homocysteine methylation pathway. Qjm. 2007;100:495–499. doi: 10.1093/qjmed/hcm054. [DOI] [PubMed] [Google Scholar]
- [127].Taylor KH, Kramer RS, Davis JW, et al. Ultradeep bisulfite sequencing analysis of DNA methylation patterns in multiple gene promoters by 454 sequencing. Cancer research. 2007;67:8511–8518. doi: 10.1158/0008-5472.CAN-07-1016. [DOI] [PubMed] [Google Scholar]
- [128].Conde-Perezprina JC, Luna-Lopez A, Lopez-Diazguerrero NE, Damian-Matsumura P, Zentella A, Konigsberg M. Msh2 promoter region hypermethylation as a marker of aging-related deterioration in old retired female breeder mice. Biogerontology. 2008 doi: 10.1007/s10522-008-9144-8. [DOI] [PubMed] [Google Scholar]
- [129].Akintola AD, Crislip ZL, Catania JM, et al. Promoter methylation is associated with the age-dependent loss of N-cadherin in the rat kidney. American journal of physiology. 2008;294:F170–176. doi: 10.1152/ajprenal.00285.2007. [DOI] [PubMed] [Google Scholar]
- [130].Bornman DM, Mathew S, Alsruhe J, Herman JG, Gabrielson E. Methylation of the E-cadherin gene in bladder neoplasia and in normal urothelial epithelium from elderly individuals. The American journal of pathology. 2001;159:831–835. doi: 10.1016/S0002-9440(10)61758-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Kang GH, Lee HJ, Hwang KS, Lee S, Kim JH, Kim JS. Aberrant CpG island hypermethylation of chronic gastritis, in relation to aging, gender, intestinal metaplasia, and chronic inflammation. The American journal of pathology. 2003;163:1551–1556. doi: 10.1016/S0002-9440(10)63511-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Ohi T, Uehara Y, Takatsu M, Watanabe M, Ono T. Hypermethylation of CpGs in the promoter of the COL1A1 gene in the aged periodontal ligament. Journal of dental research. 2006;85:245–250. doi: 10.1177/154405910608500308. [DOI] [PubMed] [Google Scholar]
- [133].Takatsu M, Uyeno S, Komura J, Watanabe M, Ono T. Age-dependent alterations in mRNA level and promoter methylation of collagen alpha1(I) gene in human periodontal ligament. Mechanisms of ageing and development. 1999;110:37–48. doi: 10.1016/s0047-6374(99)00041-x. [DOI] [PubMed] [Google Scholar]
- [134].Choi EK, Uyeno S, Nishida N, et al. Alterations of c-fos gene methylation in the processes of aging and tumorigenesis in human liver. Mutation research. 1996;354:123–128. doi: 10.1016/0027-5107(96)00056-5. [DOI] [PubMed] [Google Scholar]
- [135].Peters I, Vaske B, Albrecht K, Kuczyk MA, Jonas U, Serth J. Adiposity and age are statistically related to enhanced RASSF1A tumor suppressor gene promoter methylation in normal autopsy kidney tissue. Cancer Epidemiol Biomarkers Prev. 2007;16:2526–2532. doi: 10.1158/1055-9965.EPI-07-0203. [DOI] [PubMed] [Google Scholar]
- [136].Euhus DM, Bu D, Milchgrub S, et al. DNA methylation in benign breast epithelium in relation to age and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:1051–1059. doi: 10.1158/1055-9965.EPI-07-2582. [DOI] [PubMed] [Google Scholar]
- [137].Kim DH, Kim JS, Ji YI, et al. Hypermethylation of RASSF1A promoter is associated with the age at starting smoking and a poor prognosis in primary non-small cell lung cancer. Cancer research. 2003;63:3743–3746. [PubMed] [Google Scholar]