Abstract
The major risk factor for developing systemic lupus erythematosus (SLE) is being female. The present study utilized gene profiles of activated T cells from females with SLE and healthy controls to identify signaling pathways uniquely regulated by estradiol that could contribute to SLE pathogenesis. Selected downstream pathway genes (+/− estradiol) were measured by real time polymerase chain amplification. Estradiol uniquely upregulated six pathways in SLE T cells that control T cell function including interferon-α signaling. Measurement of interferon-α pathway target gene expression revealed significant differences (p = 0.043) in DRIP150 (+/− estradiol) in SLE T cell samples while IFIT1 expression was bimodal and correlated moderately (r = 0.55) with disease activity. The results indicate that estradiol alters signaling pathways in activated SLE T cells that control T cell function. Differential expression of transcriptional coactivators could influence estrogen-dependent gene regulation in T cell signaling and contribute to SLE onset and disease pathogenesis.
Keywords: SLE, estradiol, interferon-α, T cell signaling, microarray
Systemic lupus erythematosus (SLE) is an autoimmune disease that affects several organ systems in the body [1]. SLE is characterized by the production of pathogenic autoantibodies [2] resulting in part from abnormal interactions between T and B cell signaling [3]. Genetic susceptibility and environmental triggers contribute to SLE disease onset [4]. However, the greatest risk factor for developing SLE is female gender [5]. Sex hormones contribute to this gender bias in SLE but the mechanisms involved have not been fully elucidated [6–8].
Estradiol is a female sex hormone that binds to specific estrogen receptors (ER) and alters the transcription rates of target genes [9]. ERs function as transcription factors through subdomains that serve as docking surfaces for other essential transcriptional regulatory proteins known as coactivators and corepressors [10]. Nuclear receptor coactivators enhance transcription [11] while nuclear corepressors suppress transcription [12]. The ligand activated ER interacts with the p160 coactivators to open chromatin at the promoter region of target genes [13]. The DRIP coactivators, a second class of coactivator proteins, share subunits with the Mediator complex, suggesting these coactivators operate by recruiting RNA polymerase II to the promoter of estradiol regulated genes [14].
The molecular basis underlying the deregulation of T and B cells in SLE appears to be global in nature since multiple genes are inappropriately regulated [4, 15]. For example, measurement of 160 variants in the sera of SLE patients using a high-throughput protein microarray revealed 30 abnormally regulated proteins including cytokines, chemokines, growth factors and soluble receptors in SLE patients compared with control samples [16]. Gene profiling of heterogeneous populations of peripheral blood mononuclear cells revealed genes within molecular pathways that are consistently altered in SLE and may contribute to the development of autoimmunity [17–19]. The overexpression of the type I interferons (IFNs) and, in particular the IFN-α subtype, is implicated in the initiation and development of SLE. Secretion of IFN-α normally declines within hours after initial viral infection, but in susceptible individuals it can be sustained because genes that control INF-α or downstream targets such as IFN regulatory factor (IRF5) and signal transducer and activator of transcription (STAT4)remain elevated [20, 21]. The IFN signature refers to upregulated genes in the interferon pathway that correlate with disease activity in SLE. The IFN-α signature may also be altered in other autoimmune disorders including Sjögren’s syndrome, dematomyosisits, psoriasis and in a small number of RA patients [22].
Recent completion of a clinical trial showed that monthly administration of the ER antagonist, Faslodex, for one year improved disease activity in SLE patients [23]. Calcineurin and CD154 expression were downregulated in the Faslodex arm of the study. This finding is consistent with our previous reports showing calcineurin and CD154 genes were upregulated by estradiol in SLE but not in normal T cells [24, 25]. To gain additional insight into the molecular basis of estradiol action in SLE T cells, the present study utilized gene profiling and compared estradiol effects in SLE and normal T cells. The results suggest that estradiol alters signaling pathways in SLE T cells that are associated with disease onset and progression. One of the pathways significantly altered by estradiol in SLE T cells is interferon-α. While several genes within the interferon-α pathway appeared to be upregulated, expression of vitamin D receptor interacting protein (DRIP150) is of particular interest because it provides preliminary evidence that deregulated cofactor expression could modulate the response to estradiol in target cells and may underlie the sensitivity to estradiol in some SLE T cells.
PATIENTS AND METHODS
Study subjects
This study was approved by the St. Luke’s Hospital Institutional Review Board and all subjects provided written informed consent prior to participation. Thirteen female SLE patients between the ages of twenty-four and forty-six years with a median age of forty-two years were enrolled in the study. The SLE patients fulfilled the American College of Rheumatology criteria for SLE [26] and were diagnosed with moderate disease activity at the time of blood draw (SLEDAI score 2–18, median 6). Patients enrolled in the study exhibited symmetrical polyarticular non-erosive arthritis (n = 8), lupus malar or discoid rash (n = 8), Raynaud’s syndrome (n =1), serositis (n =3), and renal disease (n = 6). 1 Nine of the patients were taking prednisone, three patients were taking mycophenolate mofetil, three patients were taking azathioprine and seven patients were taking hydroxychloroquine. Duration of lupus in the patients ranged from three to twenty years (median duration, eight years). Eight of the patients were Caucasian, four were African American and one was of Asian descent. Ten healthy control females were enrolled in the study. The age of the control females ranged from twenty-one to fifty-one years with a median age of thirty-seven. To control for the effects of medications, five female patients with rheumatoid arthritis (RA) were enrolled in the study. The age of the RA patients ranged from 28 to 48 years with a median age of 42. Two of the RA patients were taking prednisone, one was taking methotrexate and one was taking azathioprine. Two of the RA patients were not taking medications. Participants had regular menstrual cycles and none were taking hormone replacement therapy, oral contraceptives, or had a history of other collagen vascular diseases.
Collection of T cell enriched peripheral blood mononuclear cells
T cell enriched mononuclear cells were separated from blood samples (~ 90 ml) by density gradient (Histopaque, Sigma, St. Louis, MO, USA). The lymphocytes were removed and washed twice in serum-free medium (RPMI 1640, Fisher Scientific, Hanover Park, IL, USA) and residual red blood cells were lysed (H-Lyse buffer, R&D Systems, Minneapolis, MN, USA). T cells were purified by negative selection through T cell isolation columns (Human T Cell Enrichment Columns, R&D Systems). T cells were cultured overnight (18 h) at 37 °C under 5% CO2 in serum-free medium (Hybridoma, Sigma, St. Louis, MO, USA) supplemented with L-glutamine (200 mM). T cells were activated after 18 h of culture for 4 h with phorbol 12 myristate 13-acetate (PMA, Sigma, 10 ng/ml) and ionomycin (Sigma, 0.5 μg/ml). To test the effect of estradiol on activated T cells, estradiol-17β (10−7 M) was added (or not) to half of the replicate cultures for the entire culture period. We have previously shown that calcineurin and CD154 expression are upregulated in SLE T cells in response to estradiol at this dose and time as described in detail elsewhere [24, 25].
RNA isolation
T cell RNA was isolated using the TRIzol reagent (Invitrogen, Carlsbad, CA) and Phase Lock Heavy Gels (Eppendorf, Fisher Scientific). Total RNA was purified from T cells and treated with DNase I according to the manufacturer’s protocol (DNA-free, Ambion, Austin, TX).
Microarray analysis
Gene profiling was carried out at the Kansas University School of Medicine Microarray Facility. The concentration and purity of total RNA was assessed with an Agilent Bioanalyzer and samples with RIN scores above 7.0 were used for complimentary (cRNA) RNA synthesis. Biotinlated cRNA was hybridized to high density Affymetrix human GeneChips HG-U133_Plus_2, which contained 54,675 probe sets. The chips were scanned and analyzed using MAS5 type of data analysis with Affymetrix and Gene spring GX 7.3.1 (Agilent Technologies) software, where the data from the microarray chips were subsequently normalized per gene and chip. The absence, presence, or marginal status of specific gene probes was based on the detection p-value (Student’s t test). Detection p-value under 5% (< 0.05) determines the probe as present. In cases where the detection p-value was between 5 to 6.5% (0.05–0.065) the detection interpretation was marginal lowering the significance level to less than 95%. If the detection value exceeded 6.5% (> 0.065), the probes were designated as absent, since the significance of the detection was less than 94%. Signal intensities of genes present in estradiol-treated samples were compared to the non-treated samples in order to generate a value for fold-change.
Pathway analysis
Cell signaling pathways were identified using the Ingenuity Pathways Analysis (IPA, Ingenuity Systems, Redwood City, CA) library of canonical pathways that were most significant to the data sets. Genes from the data sets that met the fold change cutoff in all SLE patients but not in control females and, in all control females but not in SLE patients were positioned in the appropriate canonical pathways using the IPA Knowledge Base. Fischer’s exact test was used to calculate a p-value determining the probability that the association between the genes in the data set and the canonical pathway were explained by chance alone.
Real-time polymerase chain amplification
Selected target genes within upregulated pathways were independently investigated by examining expression levels using real-time PCR. Total T cell RNA was digested using DNase 1 and cDNA was synthesized from 4 μg of the resulting RNA using a High Capacity cDNA kit (Applied Biosystems, Foster City, CA). Real-time PCR (Step-one, Applied Biosystems) was carried out according to the manufacturer’s protocol. The templates were quantified using a Taqman probe and specific gene primers for vitamin D receptor interacting protein [DRIP150, (also known as mediator complex subunit 14), Hs00188481, Applied Biosystems] and interferon induced with tetratricopeptide repeats 1 (IFIT1, Hs01911452, Applied Biosystems). A Taqman probe and glyceraldehyde 3-phosphate dehydrogenase (GAPDH, Hs99999905, Applied Biosystems) specific gene primers were used for the internal control. In each cycle, fluorescent signals for the target gene and GAPDH were collected from triplicate samples. The value of Ct (target Ct minus GAPDH Ct), which was inversely correlated with the number of target mRNA copies, was calculated and compared with a pooled T cell sample that was used as a positive control for all of the assays. Samples without template were included on each plate as a negative control. The fold change in expression levels was calculated by dividing the sample Ct values obtained from T cells cultured with and without estradiol from the same individual.
Statistical Analysis
Because the entire content of a blood draw was required of an individual assay, samples from each subject enrolled in this study were not tested in all assays. After median normalization for each chip, the Student’s t test was used to compare differences in T cell gene expression without and with estradiol. The significance of the association between a data set and the canonical pathway (IPA, www.ingenuity.com) was measured by: (i) a ratio of the number of genes from the data set that mapped to the pathway divided by the total number of genes that map to the canonical pathway; (ii) Fisher’s exact test to calculate a p-value determining the probability that the association between the genes in the dataset and the canonical pathway was explained by chance alone. A nonparametric Mann-Whitney U test was used to compare differences in DRIP150 and IFIT1 expression. Linear regression analysis correlated SLEDAI scores with IFIT1 and DRIP150 response to estradiol. A p value < 0.05 was considered statistically significant.
RESULTS
Gene profiling reveals differential expression of estrogen responsive genes between SLE and normal T cells
Microarray analysis was used to identify specific genes that were differentially regulated in samples of peripheral T cells purified and activated from SLE patients (n=6) and control females (n=5). Of the six patients used for the microarray study, four were Caucasians and two were African American. The median SLEDAI at the time of blood collection was 6.8. Normalized probe values (see METHODS) were obtained from profiled T cells and the data were used to generate fold changes in gene expression. Since we expected some heterogeneity among human samples we arbitrarily chose to include fold change values ≥ 1 for all upregulated genes and ≤1 for all downregulated genes. Four gene lists were assembled using GeneSpring software (data not shown). The first list contained genes that were upregulated by estradiol in all SLE T cell samples but not in all control T cell samples. The second list contained genes that were downregulated by estradiol in all SLE T cell samples but not in all control T cell samples. The third list contained genes that were upregulated by estradiol in all control T cell samples but not in all SLE T cell samples. The fourth list contained genes that were downregulated by estradiol in all control T cell samples but not in all SLE T cell samples.
Signal transduction pathways are altered by estradiol in SLE T cells
As expected, the expression of a large number of genes was altered in SLE and normal T cells by estradiol (data can be accessed http://bioinformatics.kumc.edu/mdms/login.php). In order to obtain a more focused view of how these genes might alter SLE T cell function, we used the IPA library to investigate whether estradiol altered signaling pathways in all of the T cell samples from SLE patients versus pathways in all of the control T cell samples. In general, estradiol uniquely downregulated more signaling pathways in both SLE and control T cells (20 down versus 16 up in SLE T cell samples; 28 down versus 13 up in control T cell samples, Tables 1–4). Three pathways, including B cell receptor signaling, cardiac β2-adrenergic signaling and IL-4 signaling, were upregulated in both SLE and control T cell samples but the genes involved were different between SLE and control T cells. These pathways are therefore not included as uniquely regulated in SLE or normal T cells.
Table 1.
Estradiol uniquely upregulates signaling pathways in activated SLE T cells but not in normal T cells. Gene lists that were generated from microarray data were analyzed for pathways uniquely upregulated in all SLE but not in control T cells. List shown is of pathways in the SLE T cells which met the criteria for significance (p ≤ 0.05).
| Pathway | SLE T cells p-value | Control T cells p-value |
|---|---|---|
| Pentose Phosphate Pathway | 0.0002 | NS |
| Inositol Phosphate Metabolism | 0.0030 | NS |
| Sulfur Metabolism | 0.0037 | NS |
| PI3K/AKT Signaling | 0.0041 | NS |
| Interferon Signaling | 0.0047 | NS |
| T Cell Receptor Signaling | 0.0062 | NS |
| Glucocorticoid Receptor Signaling | 0.0069 | NS |
| GM-CSF Signaling | 0.0071 | NS |
| Insulin Receptor Signaling | 0.0141 | NS |
| β-alanine Metabolism | 0.0155 | NS |
| Propanoate Metabolism | 0.0170 | NS |
| Valine, Leucine and Isoleucine Degradation | 0.0170 | NS |
| Pantothenate and CoA Biosynthesis | 0.0191 | NS |
| Calcium Signaling | 0.0275 | NS |
| Synaptic Long Term Potentiation | 0.0380 | NS |
| SAPK/JNK Signaling | 0.0407 | NS |
| Huntington’s Disease Signaling | 0.0407 | NS |
Table 4.
Estradiol uniquely downregulates signaling pathways in activated control T cells but not in SLE T cells. Gene lists that were generated from microarray data were analyzed for pathways uniquely downregulated in all normal and not in the control T cells. List shown is of pathways which met the criteria for significance (p ≤ 0.05) in control T cells.
| Pathway | Control T cells p-value | SLE T cells p-value |
|---|---|---|
| IGF-1 Signaling | 0.0002 | NS |
| Insulin Receptor Signaling | 0.0002 | NS |
| PI3K/AKT Signaling | 0.0009 | NS |
| Natural Killer Cell Signaling | 0.0016 | NS |
| Fc Epsilon RI Signaling | 0.0017 | NS |
| Mitochondrial Dysfunction | 0.0025 | NS |
| Ceramide Signaling | 0.0030 | NS |
| Estrogen Receptor Signaling | 0.0039 | NS |
| NRF2-mediated Oxidative Stress Response | 0.0044 | NS |
| Synaptic Long Term Depression | 0.0056 | NS |
| Glucocorticoid Receptor Signaling | 0.0058 | NS |
| T Cell Receptor Signaling | 0.0091 | NS |
| Xenobiotic Metabolism Signaling | 0.0100 | NS |
| JAK/Stat Signaling | 0.0117 | NS |
| Oxidative Phosphorylation | 0.0126 | NS |
| VDR/RXR Activation | 0.0138 | NS |
| Integrin Signaling | 0.0145 | NS |
| Protein Ubiquitination Pathway | 0.0195 | NS |
| Neuregulin Signaling | 0.0200 | NS |
| Amyotrophic Lateral Sclerosis Signaling | 0.0234 | NS |
| Death Receptor Signaling | 0.0251 | NS |
| SAPK/JNK Signaling | 0.0282 | NS |
| Ubiquinone Biosynthesis | 0.0347 | NS |
| ERK/MAPK Signaling | 0.0398 | NS |
| FXR/RXR Activation | 0.0407 | NS |
| IL-4 Signaling | 0.0417 | NS |
| NF-κB Signaling | 0.0427 | NS |
| Apoptosis Signaling | 0.0437 | NS |
Sixteen signaling pathways were uniquely upregulated in all of the SLE T cell samples (Table 1). Eight of these pathways are involved in cellular metabolism while six pathways control various aspects of T cell function. Five of the six pathways involved with T cell function are known to be altered in autoimmune diseases. Removed references and discussion of pathways to Discussion as suggested by reviewer 1. Twenty pathways were uniquely downregulated by estradiol in all of the SLE T cell samples (Table 2). The downregulated responses included metabolic pathways, cell signaling pathways and pathways that are crucial for T cell function including PPARα Activation, calcium signaling, cAMP-mediated signaling, and IL-2 signaling. Removed references and discussion of pathways to Discussion as suggested by reviewer 1.
Table 2.
Estradiol uniquely downregulates signaling pathways in activated SLE T cells but not in normal T cells. Gene lists that were generated from microarray data were analyzed for pathways uniquely downregulated in all SLE T cells and not in the control T cells. List shown is of pathways which met the criteria for significance (p ≤ 0.05) in SLE T cells.
| Pathway | SLE T cells p-value | Control T cells p-value |
|---|---|---|
| PPARα/RXRγ Activation | 0.0006 | NS |
| G-Protein Coupled Receptor Signaling | 0.0017 | NS |
| Hepatic Fibrosis/Hepatic Stellate Cell Activation | 0.0019 | NS |
| cAMP-mediated Signaling | 0.0024 | NS |
| Nitrogen Metabolism | 0.0058 | NS |
| Propanoate Metabolism | 0.0102 | NS |
| Circadian Rhythm Signaling | 0.0120 | NS |
| Axonal Guidance Signaling | 0.0174 | NS |
| Keratan Sulfate Biosynthesis | 0.0234 | NS |
| Calcium Signaling | 0.0275 | NS |
| Fatty Acid Metabolism | 0.0282 | NS |
| Valine, Leucine and Isoleucine Degradation | 0.0309 | NS |
| Cardiac β2-adrenergic Signaling | 0.0324 | NS |
| β-alanine Metabolism | 0.0417 | NS |
| Phospholipid Degradation | 0.0417 | NS |
| Synaptic Long Term Potentiation | 0.0437 | NS |
| LPS/IL-1 Mediated Inhibition of RXR Function | 0.0437 | NS |
| TGF-β Signaling | 0.0447 | NS |
| IL-2 Signaling | 0.0457 | NS |
| Amyloid Processing | 0.0457 | NS |
Estradiol increased gene expression of thirteen pathways in all of the control but not the SLE T cell samples (Table 3). The antigen presentation pathway was the only pathway identified that is known to play a role in T cell function. The other types of signaling pathways regulated by estradiol in the control T cell samples involved cell signaling, motility, and metabolism. Twenty eight pathways were significantly downregulated by estradiol in the control T cell samples (Table 4). It is interesting to note that five of those pathways downregulated in the control but not in the SLE T cell samples, regulate processes that are defective in SLE T cells. Removed references and discussion of pathways to Discussion as suggested by reviewer 1.
Table 3.
Estradiol uniquely upregulates signaling pathways in activated normal T cells but not in SLE T cells. Gene lists that were generated from microarray data were analyzed for pathways uniquely upregulated in all control but not in SLE T cells. List shown is of pathways which met the criteria for significance (p ≤ 0.05) in control T cells.
| Pathway | Control T cells p-value | SLE T cells p-value |
|---|---|---|
| Axonal Guidance Signaling | 0.0001 | NS |
| Ephrin Receptor Signaling | 0.0011 | NS |
| Regulation of Actin-based Motility by Rho | 0.0021 | NS |
| Valine, Leucine and Isoleucine Degradation | 0.0043 | NS |
| Antigen Presentation Pathway | 0.0102 | NS |
| Sonic Hedgehog Signaling | 0.0102 | NS |
| Bile Acid Biosynthesis | 0.0110 | NS |
| Serotonin Receptor Signaling | 0.0123 | NS |
| N-Glycan Biosynthesis | 0.0141 | NS |
| Xenobiotic Metabolism Signaling | 0.0191 | NS |
| FXR/RXR Activation | 0.0372 | NS |
| β-alanine Metabolism | 0.0407 | NS |
| Aryl Hydrocarbon Receptor Signaling | 0.0427 | NS |
The interferon-α pathway is upregulated in response to estradiol in SLE T cells
Type I interferons are suggested to be key modulators in the pathogenesis of SLE [27]. The microarray data showed estradiol upregulated the interferon-α pathway in SLE (p = 0.005) and not in control T cell samples (Table 1). We further investigated interferon-α signaling in order to identify those genes that were contributing to the upregulated response. Normalized values from the microarray data showed seven of the genes including the Janus kinase 2 (JAK2), protein tyrosine phosphatase, non-receptor type 11 (PTPN11), vitamin D receptor interacting protein (DRIP150), 2′, 5′-oligoadenylate synthetase 1 (OAS1), the SEC14-like 2 (S. cerevisiae) (sec14L2), the suppressor of cytokine signaling 1 (SOCS1), and interferon induced with tetratricopeptide repeats 1 (IFIT1) showed mean fold-values greater than 1.5 in response to estradiol across the SLE T cell samples (Figure 1).
Figure 1.
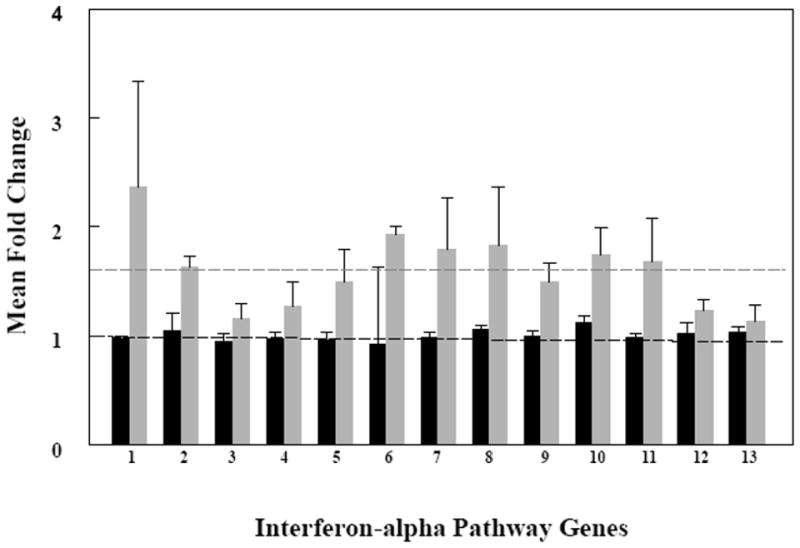
Expression of interferon-related genes in the activated T cells from SLE patients (n=6) increased in response to estradiol but not in the activated T cells from healthy donors (n=5). Activated T cells were cultured without and with estradiol. Fold change (FC) values were calculated from gene profiling data as a ratio of expression levels with estradiol compared to expression levels without estradiol from the same individuals. The mean FC for thirteen genes representing those associated with the interferon-α pathway are displayed (1–13). Dark bars (black) are mean FC values from all T cell samples obtained from control females. Light bars (gray) are mean FC values from all T cell samples obtained from female SLE patients. The horizontal lines represent mean FC values for each group, 1.0 for the control T cells and 1.6 for the SLE T cells. 1 = DRIP150; 2 = IFIT1; 3 = IFNAR1; 4 = INFAR2; 5 = JAK1; 6 = JAK2; 7 = OAS1; 8 = PTPN11; 9 = PTPN2; 10 = SEC14; 11 = SOCS1; 12 = STAT1; 13 = STAT2.
Real-time PCR allowed us to investigate pathway genes in T cells from additional SLE patients as well as T cells from patients with RA to control for the effects of medications. We measured expression levels of two genes in the interferon-α pathway in SLE T cell samples from five of the six SLE patients and five controls who donated blood for microarray study. In addition, we added five additional SLE T cell samples, five additional control T cell samples and T cells from 5 patients with RA. First, we measured the IFIT1 gene product that showed a modest mean-fold increase in response to estradiol in the SLE T cell samples (see Figure 1). The IFIT1 gene was chosen in spite of this modest increase because it codes for a protein that has been suggested to function in the pathogenesis of SLE [28]. Second, was the DRIP150 gene since this gene product functions as a cofactor for ER mediated transcription [29] and could be responsible for the increased sensitivity of SLE T cells to estradiol.
As measured by real-time PCR, the estradiol-induced change in IFIT1 expression was not significantly different (p = 0.63, Mann-Whitney test) between the SLE and control T cell samples. The mean fold change between SLE T cell samples cultured without and with estradiol from the same person was 2.1 (n = 10), while the mean fold change for the control T cell samples was 1.18 (n = 10). The estradiol response in the SLE T cells was bimodal in that five of the T cell samples responded strongly to estradiol (Table 5). The lowest and highest responders were African American suggesting that the bimodal response was not based on race. The five high responders had renal involvement in their lupus. IFIT1 expression was lower in five of the SLE patients in response to estradiol. Two of those patients, both African American, had renal involvement in their disease. Changes in IFIT1 expression showed a moderate correlation (r = 0.55) with the patient’s SLEDAI scores.
Table 5.
Estradiol differentially regulates expression of genes involved in the interferon-α pathway in SLE T cells. Data shown are the fold change (FC) in target gene expression from the same T cell sample cultured without and with estradiol.
| SLE | DRIP150 FC | IFIT1 FC | Controls | DRIP150 FC | IFIT1 FC | Rheumatoid Arthritis | DRIP150 FC | IFIT1 FC |
|---|---|---|---|---|---|---|---|---|
| SLE 1 | 1.11 | 0.80 | CTRL 1 | 1.06 | 1.19 | RA 1 | 1.18 | 0.93 |
| SLE 2 | 0.97 | CTRL 2 | 1.02 | 1.60 | RA 2 | 0.90 | 1.74 | |
| SLE 3 | 2.07 | 2.78 | CTRL 3 | 1.71 | 1.09 | RA 3 | 1.22 | 2.92 |
| SLE 4 | 16.57 | 0.01 | CTRL 4 | 0.62 | 0.05 | RA 4 | 0.46 | 0.93 |
| SLE 5 | 18.89 | 0.20 | CTRL 5 | 1.25 | 0.72 | RA 5 | 0.49 | 0.89 |
| SLE 6 | 1.08 | 2.22 | CTRL 6 | 0.63 | 0.74 | |||
| SLE 7 | 15.27 | ND | CTRL 7 | 0.74 | 1.25 | |||
| SLE 8 | 0.34 | 0.59 | CTRL 8 | 1.80 | 2.34 | |||
| SLE 9 | 3.22 | 1.93 | CTRL 9 | 0.97 | 2.08 | |||
| SLE 10 | 8.03 | 0.88 | CTRL 10 | 1.06 | 0.74 | |||
| SLE 11 | ND | 5.15 | ||||||
| SLE 12 | ND | 6.60 | ||||||
| Mean FC: | 6.75 | 2.12 | Mean FC: | 1.09 | 1.18 | Mean FC: | 0.85 | 1.48 |
| Median FC: | 2.65 | 1.41 | Median FC: | 1.04 | 1.14 | Median FC: | 0.90 | 0.93 |
| Std. Dev.: | 7.38 | 2.20 | Std. Dev.: | 0.41 | 0.68 | Std. Dev.: | 0.36 | 0.88 |
ND = not done.
Relative DRIP150 expression (+/− estradiol) was significantly different in SLE T cells compared with the control T cells (p = 0.043, 2-sided Mann-Whitney test). The mean fold change in DRIP150 expression for the SLE T cells was 6.8 (n=10). The median fold change in DRIP150 expression among the SLE patient’s T cells was 2.6. Among the top five responders, the mean fold change for DRIP150 expression was 8.4 while in the five lower responders the mean fold increase in expression was 1.1. T cell samples from six of the ten SLE patients showed a greater than two fold increase in DRIP150 and those six patients had renal involvement in their lupus. The T cell samples from two SLE patients showed a decline in DRIP150 expression compared with the same T cell samples cultured without estradiol. Both of those patients initially presented with arthritis and one exhibited Raynaud’s syndrome. Neither of those patients exhibited renal involvement in their disease. DRIP150 expression in the control and RA T cell samples showed a mean fold difference of 1 (n=10) and 0.85 (n=5), respectively. The values for the RA T cell samples were closer to the controls than to the SLE T cell samples indicating that altered DRIP150 expression is a disease effect rather than a response to medications.
DISCUSSION
Previous results from our laboratory showed SLE T cells exhibit an increased sensitivity to estradiol [6, 7]. Administration of the ER antagonist Faslodex reduced disease activity in some SLE patients [23]. We have extended those findings and used gene profiling to gain greater insight into signaling pathways that are stimulated by estradiol and may alter T cell function. We selected pathways that were uniquely controlled by estradiol in all six female SLE and all five control T cell samples obtained from healthy females. It is important to note that such an approach will not detect altered pathways in subsets of patients. Our rationale was to select those pathways differentially regulated by estradiol in all T cell samples in order to identify signaling pathways that were targets in the majority of SLE patient’s T cells. Confirming the validity of this approach was the identification of the T cell receptor signaling pathway as an estrogen target shown in earlier studies [24] and known to be altered in SLE T cells [15].
Estradiol increases signaling through PI3K/AKT, T Cell Receptor, Interferon, GM-CSF, Calcium and SAPK/JNK pathways (Table 1) that are normally associated with regulating T cell function. Estradiol increases expression of the PI3K/AKT pathway in SLE and not in control T cells. In normal T cells, activation of the PI3K/AKT pathway stimulates cell survival, glucose metabolism and inducible transcription [30]. It is important to note that alteration in the PI3K/AKT pathway is sufficient to promote autoimmunity [15]. Estradiol enhances the T cell receptor signaling pathway which we [24, 25], and others [3, 4, 31] have shown is altered in human SLE T cells. Estradiol increases GM-CSF signaling uniquely in activated SLE T cells (Table 1). GM-CSF is a critical hematopoietic growth factor that affects circulating leukocytes and is produced by activated T cells [32]. GM-CSF can increase antigen-induced immune responses and alter the Th1/Th2 cytokine balance in T cells. Estradiol also augmented the stress-activated protein kinase (SAPK/JNK) pathway in SLE T cells. JNK1 and JNK2 stimulate the release of proinflammatory cytokines in activated T cells and modulate the differentiation of Th1 and Th2 cells [33].
Calcium signaling is dysregulated in SLE T cells by several mechanisms (34–39). Engagement of the T cell receptor leads to a transient release of calcium that occurs earlier and is of greater magnitude in SLE compared with normal T cells [36]. By contrast, long-term changes in calcium signaling is diminished in SLE T cells and may underlie abnormal T cell activation and/or the inability of SLE T cells to produce IL-2 [37, 38]. It is interesting to note that estradiol both stimulates and reduces calcium signaling in SLE T cells (compare Tables 1 and 2). This finding suggests that estradiol plays a role in differentially affecting short term versus sustained calcium signaling in SLE T cells. Some clues regarding the molecular mechanisms involved in this apparent paradox are suggested by increased signaling through the pentose phosphate pathway in SLE T cells, which is increased by estradiol (Table 1). Previous studies have shown that the mitochondrial membrane in SLE T cells is hyperpolarized owing to reductions in ATP and glutathione [38, 39]. Mitochondrial hyperpolarization and persistent ATP depletion is a critical checkpoint that predispose SLE T cells to undergo cell death by necrosis rather than apoptosis. The mitochondrial transmembrane potential is regulated in part by the supply of NADPH produced by the pentose phosphate pathway [39]. Since abnormal T cell activation and cell death underpin the pathology of SLE it is important to clarify the estrogen-dependent steps that contribute to alterations in this metabolic pathway and may change calcium signaling in SLE T cells.
Of the twenty signaling pathways that were uniquely downregulated by estradiol (Table 2), the decrease in IL-2 signaling is a central component of defective SLE T cell signaling [40, 41]. Local increases in nitric oxide at the site of T cell receptor engagement reduce IL-2 production [42]. Nitric oxide is regulated by NADPH and, as revealed by our study, the pentose phosphate pathway is altered by estradiol in SLE T cells. Alternatively, IL-2 is regulated by a number of transcription factors including the cAMP response element-binding protein (CREB). Of relevance for IL-2 regulation in SLE T cells is the report from Katsiari et al. (43) showing that protein phosphatase 2A expression is higher in SLE than normal T cells leading to reduced transcription of IL-2. Protein kinase A (PKA) is a major target of cAMP activation. PKA signaling is maintained by a balance between kinase and phosphatase activity and the pathway is deficient in SLE T cells [44]. Although additional studies are required to identify the targets for reduced cAMP signaling in response to estradiol, our data indicate that estradiol targets pathways known to be markers for abnormal signaling in SLE T cells.
Estradiol uniquely downregulates 28 signal transduction pathways in activated control T cells (Table 4). Several of these pathways are known to be defective in SLE T cells including cell receptor signaling [45], ERK/MAPK signaling [46, 47], IL-4 [48], NF-κB [49] and apoptosis signaling [50]. Although nothing is known about estradiol-dependent signaling in normal T cells, our results suggest that estradiol downregulates key signaling pathways in activated normal T cells. Estrogen is considered a promoter of the immune response (51). Our results indicate that estradiol downregulates numerous pathways in activated T cells and suggests that failure to downregulate these same pathways in SLE T cells could contribute to SLE T cell hyperactivation. The concept now requires further investigation because it suggests downregulation is an important mechanism that is defective in the SLE T cell response to estradiol.
In this report, we chose to further study the interferon-α pathway owing to its importance in SLE and other autoimmune diseases [17, 18, 22]. Seven of thirteen genes identified in interferon-α signaling showed greater than a 1.5 mean-fold increase in the SLE T cell samples (Figure 1). If regulation of this pathway is typical for estradiol targets, it suggests that estrogen-dependent upregulation of signaling will occur by increased expression of multiple genes within a pathway rather than a large change in the magnitude of expression of a single gene. We found that expression of IFIT1 in SLE T cell samples was bimodal in that some patient’s T cells responded strongly to estradiol while others were less responsive to the hormone. Changes in IFIT1 expression correlated moderately with SLE disease activity. Interestingly, T cells from patients with renal disease showed the greatest sensitivity to estradiol. Other studies have found significant differences in IFIT1 expression in SLE compared with control T cells [28, 52] and increased expression correlated with renal disease [53]. It is likely that our study underestimates estradiol effects on IFIT1 expression since the T cells were cultured for 16 h in serum free medium without added interferon-α.
The interferon stimulated gene factor 3 (ISGF3) is a heterotrimeric complex central to the regulation of IFN gene transcription [54, 55]. Binding of IFN to cell surface receptors activates STAT1 and STAT2 proteins which complex with interferon regulatory factor 9 (IRF9) forming the ISGF3 activation complex. The ISGF3 complex enters the nucleus where it binds to specific DNA sequences and increases the rates of transcription of interferon gene targets. While many of the signaling events downstream of interferon signaling are known [22, 27, 54], the mechanisms involved in regulating IFN-dependent gene transcription are less well understood.
Transcriptional coactivators are important modulators of gene regulation. Coactivators regulate gene transcription by modifying chromatin structure and enhancing promoter accessibility. In addition, coactivators can recruit other factors to the promoter of responsive genes that enhance the transcriptional response. DRIP150 is a member of a multi-protein complex that shares several subunits with the mammalian Mediator complex [29, 54]. Mediator complexes function as a bridge between distal transcription activators and RNA polymerase II. The Mediator complex is essential for the regulation of NF-κB, SP1 and most nuclear hormone receptor signaling pathways including the ER [13]. DRIP150 interacts with both ER-α and ER-β [56]. ISGF3-mediated transcription is dependent on STAT2 interactions with DRIP150 [55]. Interestingly, one of the DRIP coactivators, TRAP220 showed preference for interacting with ER-β over ER-α [56].
Our data indicate that DRIP150 expression is altered in SLE compared with normal T cells in response to estradiol. These results are the first to suggest that abnormal cofactor recruitment to the promoter of genes involved in cell signaling could lead to enhanced pathway activation and disrupt normal T cell function. Over and or under expression of key signaling pathways is expected to contribute to the development of autoimmune disease. It may be that abnormal coactivator expression recruits ERs to the promoters of genes that are not normally regulated by estradiol. Alternatively, inappropriate cofactor recruitment could occur by differential expression of ER subtypes such that ratio of ER-α/ER-β is altered in SLE T cells [57] leading to abnormal signal transduction. It is now clear that the time has come to explore the role of coactivators and corepressors as possible mechanisms underlying the deregulation of numerous genes in SLE T cells.
Footnotes
The research reported in this manuscript is funded by the National Institutes of Health (AI49272 and P20RR0-16475 for the INBRE Program of the National Center for Research Resources). We are grateful to the patients who donated blood for this study. We thank Phyllis Smotherman and Clark Bloomer for technical assistance. We are grateful to our colleagues at the KUMC Microarray Facility for their support and guidance.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Boumpas DT, Fessler BJ, Austin HA, 3rd, Balow JE, Klippel JH, Lockshin MD. Systemic lupus erythematosus: emerging concepts. Part 2: Dermatologic and joint disease, the antiphospholipid antibody syndrome, pregnancy and hormonal therapy, morbidity and mortality, and pathogenesis. Ann Intern Med. 1995;123:42–53. doi: 10.7326/0003-4819-123-1-199507010-00007. [DOI] [PubMed] [Google Scholar]
- 2.Zhang J, Jacobi AM, Wang T, Berlin R, Volpe BT, Diamond B. Polyreactive autoantibodies in systemic lupus erythematosus have pathogenic potential. J Autoimmun. 2009 doi: 10.1016/j.jaut.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tenbrock K, Juang YT, Kyttaris VC, Tsokos GC. Altered signal transduction in SLE T cells. Rheumatology. 2007;46:1525–30. doi: 10.1093/rheumatology/kem154. [DOI] [PubMed] [Google Scholar]
- 4.Kyttaris VC, Krishnan S, Tsokos GC. Systems biology in systemic lupus erythematosus: integrating genes, biology and immune function. Autoimmunity. 2006;39:705–9. doi: 10.1080/08916930601061363. [DOI] [PubMed] [Google Scholar]
- 5.Cervera R, Khamashta MA, Font J, Sebastiani GD, Gil A, Lavilla P, et al. Systemic lupus erythematosus: clinical and immunologic patterns of disease expression in a cohort of 1,000 patients. The European Working Party on Systemic Lupus Erythematosus. Medicine (Baltimore) 1993;72:113–24. [PubMed] [Google Scholar]
- 6.Rider V, Abdou NI. Gender differences in autoimmunity: molecular basis for estrogen effects in systemic lupus erythematosus. Int Immunopharmacol. 2001;1:1009–24. doi: 10.1016/s1567-5769(01)00046-7. [DOI] [PubMed] [Google Scholar]
- 7.Rider V, Li X, Abdou NI. Hormonal Influences in the Expression of Systemic Lupus Erythematosus. In: Tsokos GC, Gordon PC, Smolen JS, editors. Systemic Lupus Erythematosus: A Companion to Rheumatology. Elsevier; New York, NY: 2007. pp. 87–94. [Google Scholar]
- 8.Cohen-Solal JF, Jeganathan V, Grimaldi CM, Peeva E, Diamond B. Sex hormones and SLE: influencing the fate of autoreactive B cells. Curr Top Microbiol Immunol. 2006;305:67–88. doi: 10.1007/3-540-29714-6_4. [DOI] [PubMed] [Google Scholar]
- 9.Khorasanizadeh S, Rastinejad F. Nuclear-receptor interactions on DNA-response elements. Trends Biochem Sci. 2001;26:384–90. doi: 10.1016/s0968-0004(01)01800-x. [DOI] [PubMed] [Google Scholar]
- 10.O’Malley BW. Coregulators: from whence came these “master genes”. Mol Endocrinol. 2007;21:1009–13. doi: 10.1210/me.2007-0012. [DOI] [PubMed] [Google Scholar]
- 11.Wolf IM, Heitzer MD, Grubisha M, DeFranco DB. Coactivators and nuclear receptor transactivation. J Cell Biochem. 2008;104:1580–6. doi: 10.1002/jcb.21755. [DOI] [PubMed] [Google Scholar]
- 12.Ordentlich P, Downes M, Evans RM. Corepressors and nuclear hormone receptor function. Curr Top Microbiol Immunol. 2001;254:101–16. doi: 10.1007/978-3-662-10595-5_5. [DOI] [PubMed] [Google Scholar]
- 13.Burakov D, Crofts LA, Chang CP, Freedman LP. Reciprocal recruitment of DRIP/mediator and p160 coactivator complexes in vivo by estrogen receptor. J Biol Chem. 2002;277:14359–62. doi: 10.1074/jbc.C200099200. [DOI] [PubMed] [Google Scholar]
- 14.Chiba N, Suldan Z, Freedman LP, Parvin JD. Binding of liganded vitamin D receptor to the vitamin D receptor interacting protein coactivator complex induces interaction with RNA polymerase II holoenzyme. J Biol Chem. 2000;275:10719–22. doi: 10.1074/jbc.275.15.10719. [DOI] [PubMed] [Google Scholar]
- 15.Ohashi PS. T-cell signalling and autoimmunity: molecular mechanisms of disease. Nat Rev Immunol. 2002;2:427–38. doi: 10.1038/nri822. [DOI] [PubMed] [Google Scholar]
- 16.Bauer JW, Baechler EC, Petri M, Batliwalla FM, Crawford D, Ortmann WA, et al. Elevated serum levels of interferon-regulated chemokines are biomarkers for active human systemic lupus erythematosus. PLoS Med. 2006;3:2274–84. doi: 10.1371/journal.pmed.0030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA. 2003;100:2610–5. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crow MK, Kirou KA, Wohlgemuth J. Microarray analysis of interferon-regulated genes in SLE. Autoimmunity. 2003;36:481–90. doi: 10.1080/08916930310001625952. [DOI] [PubMed] [Google Scholar]
- 19.Crow MK, Kirou KA. Interferon-alpha in systemic lupus erythematosus. Curr Opin Rheumatol. 2004;16:541–7. doi: 10.1097/01.bor.0000135453.70424.1b. [DOI] [PubMed] [Google Scholar]
- 20.Kozyrev SV, Alarcon-Riquelme ME. The genetics and biology of Irf5-mediated signaling in lupus. Autoimmunity. 2007;40:591–601. doi: 10.1080/08916930701510905. [DOI] [PubMed] [Google Scholar]
- 21.Kariuki SN, Kirou KA, MacDermott EJ, Barillas-Arias L, Crow MK, Niewold TB. Cutting edge: autoimmune disease risk variant of STAT4 confers increased sensitivity to IFN-alpha in lupus patients in vivo. J Immunol. 2009;182:34–8. doi: 10.4049/jimmunol.182.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baechler EC, Batliwalla FM, Reed AM, Peterson EJ, Gaffney PM, Moser KL, Gregersen PK, Behrens TW. Gene expression profiling in human autoimmunity. Immunol Rev. 2006;210:120–37. doi: 10.1111/j.0105-2896.2006.00367.x. [DOI] [PubMed] [Google Scholar]
- 23.Abdou NI, Rider V, Greenwell C, Li X, Kimler BF. Fulvestrant (Faslodex), an estrogen selective receptor downregulator, in therapy of women with systemic lupus erythematosus. Clinical, serologic, bone density, and T cell activation marker studies: a double-blind placebo-controlled trial. J Rheumatol. 2008;35:797–803. [PubMed] [Google Scholar]
- 24.Rider V, Foster RT, Evans M, Suenaga R, Abdou NI. Gender differences in autoimmune diseases: estrogen increases calcineurin expression in systemic lupus erythematosus. Clin Immunol Immunopathol. 1998;89:71–80. doi: 10.1006/clin.1998.4604. [DOI] [PubMed] [Google Scholar]
- 25.Rider V, Jones S, Evans M, Bassiri H, Afsar Z, Abdou NI. Estrogen increases CD40 ligand expression in T cells from women with systemic lupus erythematosus. J Rheumatol. 2001;28:2644–9. [PubMed] [Google Scholar]
- 26.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 27.Crow MK. Interferon pathway activation in systemic lupus erythematosus. Curr Rheumatol Rep. 2005;7:463–8. doi: 10.1007/s11926-005-0053-4. [DOI] [PubMed] [Google Scholar]
- 28.Ye S, Pang H, Gu YY, Hua J, Chen XG, Bao CD, et al. Protein interaction for an interferon-inducible systemic lupus associated gene. IFIT1 Rheumatology. 2003;42:1155–63. doi: 10.1093/rheumatology/keg315. [DOI] [PubMed] [Google Scholar]
- 29.Lee J, Safe S. Coactivation of estrogen receptor alpha (ER alpha)/Sp1 by vitamin D receptor interacting protein 150 (DRIP150) Arch Biochem Biophys. 2007;461:200–10. doi: 10.1016/j.abb.2006.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kane LP, Weiss A. The PI-3 kinase/Akt pathway and T cell activation: pleiotropic pathways downstream of PIP3. Immunol Rev. 2003;192:7–20. doi: 10.1034/j.1600-065x.2003.00008.x. [DOI] [PubMed] [Google Scholar]
- 31.Zikherman J, Weiss A. Antigen receptor signaling in the rheumatic diseases. Arthritis Res Ther. 2009;11:202. doi: 10.1186/ar2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi Y, Liu CH, Roberts AI, Das J, Xu G, Ren G, et al. Granulocyte-macrophage colony-stimulating factor (GM-CSF) and T-cell responses: what we do and don’t know. Cell Res. 2006;16:126–33. doi: 10.1038/sj.cr.7310017. [DOI] [PubMed] [Google Scholar]
- 33.Rincón M, Davis RJ. Regulation of the immune response by stress-activated protein kinases. Immunol Rev. 2009;228:212–24. doi: 10.1111/j.1600-065X.2008.00744.x. [DOI] [PubMed] [Google Scholar]
- 34.Fernandez D, Bonilla E, Mirz N, Niladn B, Perl A. Rapamycin reduces disease activity and normalizes T cell activation-induced calcium fluxing in patients with systemic lupus erythematosus. Arthritis Rheum. 2006;54:2983–8. doi: 10.1002/art.22085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feske S. Calcium signaling in lymphocyte activation and disease. Nature. 2007;7:690–702. doi: 10.1038/nri2152. [DOI] [PubMed] [Google Scholar]
- 36.Tsokos GC. Calcium signaling in systemic lupus erythematosus lymphocytes and its therapeutic exploitation. Arthritis Rheum. 2008;58:1216–9. doi: 10.1002/art.23445. [DOI] [PubMed] [Google Scholar]
- 37.Nagy G, Koncz A, Fernandez D, Perl A. Nitric oxide, mitochondrial hyperpolarization and T-cell activation. Free Radic Biol Med. 2007:1625–31. doi: 10.1016/j.freeradbiomed.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez D, Perl A. Metabolic control of T cell activation and death in SLE. Auotimmunity Rev. 2009:184–9. doi: 10.1016/j.autrev.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perl A, Gergely P, Jr, Koncz A, Banki K. Mitochondrial hyperpolarization: a checkpoint of T cell life, death, and autoimmunity. Trends Immunol. 2004;25:360–7. doi: 10.1016/j.it.2004.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crispín JC, Tsokos GC. Transcriptional regulation of IL-2 in health and autoimmunity. Autoimmunity Rev. 2009;8:190–5. doi: 10.1016/j.autrev.2008.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gómez-Martín D, Díaz-Zamudio M, Crispín JC, Alcocer-Varela J. Interleukin 2 and systemic lupus erythematosus: Beyond the transcriptional regulatory net abnormalities. Autoimmun Rev. 2009 doi: 10.1016/j.autrev.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 42.Ibiza S, Víctor VM, Boscá I, Ortega A, Urzainqui A, O’Connor JE, et al. Endothelial nitric oxide synthase regulates T cell receptor signaling at the immunological synapse. Immunity. 2006;24:753–65. doi: 10.1016/j.immuni.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Katsiari CG, Vasileios VC, Juang Y-T, Tsokos GC. Protein phosphatase 2A is a negative regulator of IL-2 production in patients with systemic lupus erythematosus. J Clin Invest. 2005;115:3193–204. doi: 10.1172/JCI24895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kammer GM. Deficient protein kinase a in systemic lupus erythematosus: a disorder of T lymphocyte signal transduction. Ann NY Acad Sci. 2002;968:96–105. doi: 10.1111/j.1749-6632.2002.tb04329.x. [DOI] [PubMed] [Google Scholar]
- 45.Zikherman J, Weiss A. Antigen receptor signaling in the rheumatic diseases. Arthritis Res Ther. 2009;11:202. doi: 10.1186/ar2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gorjestani S, Rider V, Kimler BF, Greenwell C, Abdou NI. Extracellular signal-regulated kinase 1/2 signalling in SLE T cells is influenced by oestrogen and disease activity. Lupus. 2008;17:548–54. doi: 10.1177/0961203307087982. [DOI] [PubMed] [Google Scholar]
- 47.Sawalha AH, Jeffries M, Webb R, Lu Q, Gorelik G, Ray D, et al. Defective T-cell ERK signaling induces interferon-regulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes Immun. 2008;9:368–78. doi: 10.1038/gene.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh RR. IL-4 and many roads to lupus like autoimmunity. Clin Immunol. 2003;108:73–9. doi: 10.1016/s1521-6616(03)00145-1. [DOI] [PubMed] [Google Scholar]
- 49.Brown KD, Claudio E, Siebenlist U. The roles of the classical and alternative nuclear factor-kappaB pathways: potential implications for autoimmunity and rheumatoid arthritis. Arthritis Res Ther. 2008;10:212. doi: 10.1186/ar2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maniati E, Potter P, Rogers NJ, Morley BJ. Control of apoptosis in autoimmunity. J Pathol. 2008;214:190–8. doi: 10.1002/path.2270. [DOI] [PubMed] [Google Scholar]
- 51.Straub RH. The complex role of estrogens in inflammation. Endocr Rev. 2007;28:521–74. doi: 10.1210/er.2007-0001. [DOI] [PubMed] [Google Scholar]
- 52.Kirou KA, Lee C, George S, Louca K, Papagiannis IG, Peterson MG, et al. Coordinate overexpression of interferon-alpha-induced genes in systemic lupus erythematosus. Arthritis Rheum. 2004;50:3958–67. doi: 10.1002/art.20798. [DOI] [PubMed] [Google Scholar]
- 53.Kirou KA, Lee C, George S, Louca K, Peterson MG, Crow MK. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 2005;52:1491–503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- 54.Lau JF, Nusinzon I, Burakov D, Freedman LP, Horvath CM. Role of metazoan mediator proteins in interferon-responsive transcription. Mol Cell Biol. 2003;23:620–8. doi: 10.1128/MCB.23.2.620-628.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chadick JZ, Asturias FJ. Structure of eukaryotic Mediator complexes. Trends Biochem Sci. 2005;30:264–71. doi: 10.1016/j.tibs.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 56.Wärnmark A, Almlöf T, Leers J, Gustafsson JA, Treuter E. Differential recruitment of the mammalian mediator subunit TRAP220 by estrogen receptors ERalpha and ERbeta. J Biol Chem. 2001;276:23397–404. doi: 10.1074/jbc.M011651200. [DOI] [PubMed] [Google Scholar]
- 57.Rider V, Li X, Peterson G, Dawson J, Kimler BF, Abdou NI. Differential expression of estrogen receptors in women with systemic lupus erythematosus. J Rheumatol. 2006;33:1093–101. [PubMed] [Google Scholar]


