Abstract
Purpose
Medical images are a significant information source for clinical decision making. Currently available information retrieval and decision support systems rely primarily on the text of scientific publications to find evidence in support of clinical information needs. The images and illustrations are available only within the full text of a scientific publication and do not directly contribute evidence to such systems. Our first goal is to explore whether image features facilitate finding relevant images that appear in publications. Our second goal is to find promising approaches for providing clinical evidence at the point of service, leveraging information contained in the text and images.
Methods
We studied two approaches to finding illustrative evidence: a supervised machine learning approach, in which images are classified as being relevant to an information need or not, and a pipeline information retrieval approach, in which images were retrieved using associated text and then re-ranked using content-based image retrieval (CBIR) techniques.
Results
Our information retrieval approach did not benefit from combining textual and image information. However, given sufficient training data for the machine learning approach, we achieved 56% average precision at 94% recall using textual features, and 27% average precision at 86% recall using image features. Combining these classifiers resulted in improvement up to 81% precision at 96% recall (74% recall at 85% precision, on average) for the requests with over 180 positive training examples.
Conclusions
Our supervised machine learning methods that combine information from image and text are capable of achieving image annotation and retrieval accuracy acceptable for providing clinical evidence, given sufficient training data.
Keywords: Medical Informatics Computing, Information Storage and Retrieval, Image Interpretation, Computer-Assisted, Natural Language Processing
Introduction
Medical images are essential in establishing diagnoses, analyzing and evaluating treatment results, and are useful for educational purposes in many clinical specialties. Despite the fact that in such specialties as dermatology, trauma surgery, and radiology, images are routinely generated for these purposes and subsequently used in publications, few attempts have been made to identify and retrieve images suitable for clinical decision support.
Our long-term goal is to automatically find images from scientific publications and image repositories needed to provide clinical ecidence at the point of service. This goal is attainable only through reliable image annotation and retrieval techniques. Images potentially useful for decision support are found both in large online databases as well as in electronically published biomedical journals. Since approaches to finding evidence support in scientific publications are well studied within the paradigm of Evidence Based Practice (EBP) [1] and we already leverage EBP principles and framework for providing textual decision support [2], our current focus is on semantic annotation and retrieval of EBP-relevant images from scientific publications [3,4]1. Automatic image annotation and subsequent retrieval can be based solely on image analysis [5,6], on indexing of the text associated with images [7], or on a combination of image and text analysis [8,9].
Images in scientific publications elucidate the text and can usually be easily understood in context. For example, Figure 1 and its caption are fairly informative in the context of the paper “Eosinophilic cellulitislike reaction to subcutaneous etanercept injection” [10]. Taken out of context, however, the caption provides little information about the image, and neither does the image provide enough information about the nature of the skin reaction. This example illustrates both the problem of finding text that provides sufficient information about the image without introducing irrelevant information, and the potential benefits of combining information provided by both the text and image. An even greater problem is determining what information about and in an image could be used as evidence in clinical decision making.
Figure 1.
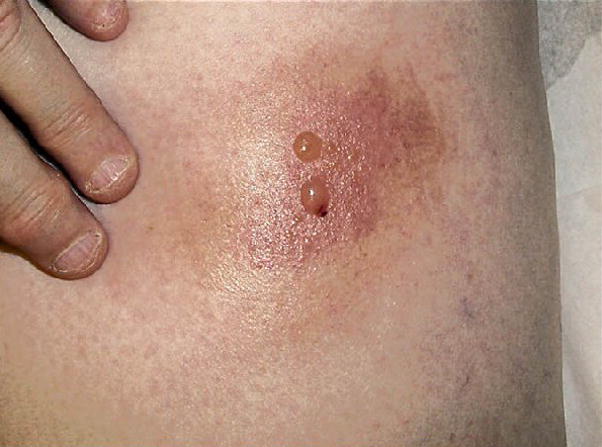
Reaction to intradermal adalimumab 1 to 2 days after the fourth dose [10]
Annotation of an image for clinical decision support involves describing the subject matter of the image, the clinical process it might support, and other aspects that will help find images relevant to a specific clinical information need. We studied available image classifications, such as RadLex2 and IRMA3, and solicited opinions of four physicians experienced in building and using medical ontologies to create facets for image annotation, such as meta-information about the image (image modality, clinical tasks the image will support, and its teaching value) and image content (body location, pathology type, etc.). This information may be at three levels of granularity: 1) coarse, that describes the image and its content at a relatively high level (for example, Microscopy, Eye, and Diagnostic); 2) medium, that uses more specific terms found in controlled vocabularies (for example, anulus fibrosus of mitral orifice or Confocal scanning laser ophthalmoscopy); and 3) specific, that provides detailed descriptions of images and their content (for example, untreated port-wine stain birthmark or nerves stained with panneuronal marker, protein gene product 9.5 (green)) [4,11]. These granularity levels reflect the different types of information needs to be satisfied, and the sophistication of methods required for automatic annotation.
In earlier work, we explored the feasibility of a machine learning approach to image annotation at the coarse level using the open-source data mining system RapidMiner4. We evaluated text- and image-based classifiers separately, and compared them to an approach which combined image and text features. These features were input to a multi-class Support Vector Machine (SVM) classifier, which classified images by modality (radiological, photo, histology, drawings, charts and graphs, flowcharts, forms, tables, and mixed) and usefulness as evidence (containing characteristic diagnostic clues, instruments and artifacts, procedures, and treatment outcomes.) Text extracted from image captions and discussions in papers was represented as a bag-of-words and/or as a bag of biomedical concepts (which did not yield better results than the bag-of-words representation). Images were represented by texture and color features computed on the entire unsegmented image. Texture features were computed as a 3-level discrete 2-D Daubechies’ Wavelet Transform. Several color features were evaluated, and the four dominant colors and their extent computed in the perceptually uniform CIE LUV color space were found to be most effective. This preliminary work indicated that combining image and text features improves image annotation for EBP-usefulness [11]. For example, the best classification into the Procedures and Outcomes categories was achieved using the bag-of-words model of captions combined with texture and dominant color models.
In this study we extend our previous supervised machine learning image annotation approach from coarse- to medium-level annotation, and compare it to an information retrieval approach in which images are retrieved using their textual descriptions and then re-ranked using image features.
Methods
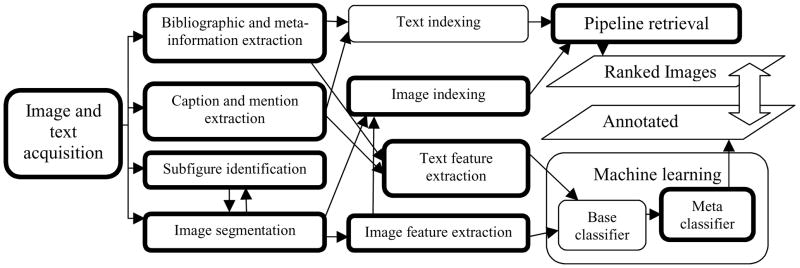
As shown in Figure 2, we combine our tools and those publicly available in a pipeline that starts with text and image pre-processing and ends with retrieving images that are ranked by relevance to a given information need or annotated as relevant (Tools developed by the authors are shown in double-bordered boxes). In this work, we compare the efficacy of these two approaches in finding clinically relevant images.
Figure 2.
Image annotation and retrieval processes
Evaluation of image retrieval approaches requires collections of clinical questions, images, and judgments on relevance of the images to the questions. The collections are available either through individual research efforts or from large-scale community-wide evaluations such as ImageCLEFmed [9].
We evaluated our approaches to image retrieval using the ImageCLEFmed 2008 Medical Image Retrieval task collection5, which consists of over 65,000 images extracted from the Radiology and RadioGraphics journals6. In addition to images, this collection contains image captions, URLs to the full-text scientific publications from which the images were extracted, their PubMed identifiers from the GoldMiner collection7, 30 information need requests expressed as image retrieval questions (topics containing text and images), and judgments about the relevance of images retrieved by teams participating in the evaluation [12, 13]. With the exception of topic Show me images of PowerPoint slides, the 30 information requests can be mapped to our coarse and medium-level annotation. For example, the topic Show me MRI images of the brain with a blood clot, contains coarse-level image modality (MRI) and body part (brain) information and medium-level information describing a clinical problem (blood clot) that can be identified using a controlled vocabulary. Our information retrieval approach used the entire collection of information requests, images and associated text while our supervised machine learning approach used fourteen information requests, for which 50 or more images were judged relevant, and 12,056 images and associated text judged relevant or not to those requests.
Obtaining text for image annotation and retrieval
Extracting image-related text from full-text scientific publications involves finding all text needed for annotation, while introducing as little extraneous information as possible. Text most closely associated with an image can be found in its caption. However, in many cases the captions are uninformative (for example, “Figure 12” merely identifies the image, and does not explain it) or insufficient for image annotation (see Figure 1). Our current approach is to augment an image caption with a discussion of the image (“image mention”), in a paragraph that refers to the image. In the absence of paragraph markers we extract the sentence that mentions the figure. For example, Figure 1 is discussed in the article [10] as follows:
Within 24 to 48 hours of her fourth dose of adalimumab, she developed an ISR initially composed of faint, erythematous macules, which rapidly evolved into an erythematous, indurated plaque surmounted by a bulla (Figure 1).
In addition to image captions and mentions, more image-related information may appear in the title, abstract, and MeSH terms (assigned by expert indexers to describe the publication and provided in MEDLINE citations) [4].
As a further complication, the extracted captions often pertain to multi-panel figures composed of several individual images or to a series of images. This requires a preprocessing step of segmenting the multi-panel images and multi-part captions. We developed several rule-based algorithms for caption segmentation. Segmented captions were most beneficial to our supervised machine learning approach to coarse-level annotation [11], and in our current evaluation of the information retrieval approach, we study the benefits of using segmented captions alone, as well as combining them with other text excerpts.
Supervised machine learning approach to image retrieval
The RapidMiner implementation of the SVM learner that was found most effective in our earlier coarse-level annotation was used for text-based and image-based annotation. Each retrieved image was classified as to whether or not it was an answer to an information request. The predictions of the text-based and image-based learners were combined using our own implementation of stacking [2] that combines predictions from lower-level models into a higher-level model using a least squares linear regression adapted for classification [14].
Based on our previous findings [11], textual features were extracted from the segmented captions and represented as a bag of words using the RapidMiner text plugin with Stopword, PorterStemmer and TokenLength Filters.
Image features for supervised machine learning
Our image features consisted of several described below measurements of texture and color information obtained using methods we developed with MATLAB8. Texture features included Gabor Filters, which are used to derive a low level representation of the image using spectral and coarsely localized information, and Daubechies Discrete Wavelet Transform that treats the image as a 2D non-stationary signal and computes principal horizontal, vertical, and diagonal frequency components at various image resolutions [15]. Color features included color moments computed in the perceptually linear L*a*b* color space, and dominant colors in the image in the standard RGB (Red, Green, Blue) color space. All of these features are “global” (they are computed over the entire image), and are computationally intensive.
The images in the ImageCLEFmed 2008 Medical Retrieval task collection vary in size, (approximately 2000 to 3000 pixels by 1000 to 1500 pixels). In order to obtain a uniform measure and for computational efficiency the features were computed on images of reduced size measuring 256×256 pixels. Not all images are equal in both dimensions and resizing images to a fixed square size resulted in skewed images. However, this does not have a significant effect on the global image features that were selected (rather than local features) in line with the coarse- and medium-level image classification described previously. To extract meaningful local features, it would be necessary to process the images at a significantly higher resolution, and we reserve this task for our future work.
We used Gabor filters to capture image “gist” (coarse texture and spatial layout) without processing it through an object detector [16]. The gist computation is resistant to image degradation and has been shown to be very effective for natural scene images [16]. The texture and layout features were computed in 4 scales with 8 orientations per scale, resulting in a 512-element feature vector.
The Discrete Wavelet Tranform (DWT) has been successfully used for multi-resolution image analysis [17]. That is, since an image can be considered a 2-dimensional non-stationary signal comprising many frequency components, the DWT can be used as an effective feature to capture these frequency components at varying scales. The mean and standard deviation of the magnitude of the vertical, horizontal and diagonal frequencies were computed at 5 levels, resulting in a 32-element feature vector.
Color plays an important role in the human visual system and its distribution can provide valuable discriminating data in the image. The color was measured using the three central color moment features: mean, the standard deviation, and skewness [18], resulting in a 9-element feature vector. Additionally, 4 dominant colors and their frequencies of occurence are computed using the k-means clustering algorithm, resulting in a 16-element vector. This results in a combined 25-element color feature vector for each image.
Images used for training were annotated with an additional feature: relevant or not to a specific information request. This feature was withheld in the automatic annotation of images used for testing, which were thus annotated as unknown. After applying machine learning, each test image was automatically annotated as relevant or not to each of the fourteen information requests used for testing.
The total number of extracted features for each image is quite large and it would be desirable to identify a smaller number of features that contribute most effectively to classification [19]. This step deserves a thorough investigation and is part of our ongoing research (the parallel effort on selection of textual features is presented in [20].) It was not performed in the current experiments which focused on understanding contributions to image classification of each image feature individually and in combination with text features as shown in the Results section.
Information retrieval approach
In this method, we employed a pipeline approach to image retrieval. In the first step, we used (and compared) two open-source search engines, Lucene9 and Terrier10, for indexing the set of the extracted text fields: captions, segmented captions, image mentions, article titles, abstracts and MeSH terms. Each ImageCLEFmed 2008 information request (topic) consisted of a text component and an image component. In the first step, we used the text component of the topic to retrieve images based on their associated text. For this we formed two types of queries: 1) information requests as provided, and 2) expanded queries, in which information requests were mapped to the Unified Medical Language System (UMLS) [21] Metathesaurus using MetaMap [22] and represented supplementing image modality, findings, and anatomy terms identified in the original requests, with their preferred UMLS names and synonyms. For example, the expanded query for the topic Show me MRI images of the brain with a blood clot, included terms Magnetic Resonance Imaging, MR Tomography and other synonyms of the query term MRI, as well as Thrombus and other synonyms of the query term blood clot.
In the second step, the images that were retrieved using various search strategies applied to the text of the above fields were re-ranked using image features that were extracted from example query images provided for each of the 30 information requests. (The same image features were also used for the machine learning approach.) Using these features, the images were automatically assigned to one of three broad categories: grayscale images (e.g., x-rays, CT, MRI, ultrasound images), color images (e.g., histopathology images, photographs), and figure images (e.g. Graphs, charts, tables). This classification was done using a simple color histogram analysis based on the following intuition. Grayscale images tend to have a simple histogram with few to no pixels with different values for the Red, Green, and Blue channels; figure images tend to be bimodal with a greater number of white pixels than others; and the remainder, which are classified as color images, also tend to have a mixed histogram. The extracted query features and broad categories were compared to those computed for images retrieved in the first step (text-based retrieval) using the L2-norm. Retrieved images were then re-ranked according to their proximity to query images.
Retrieval results were evaluated using the trec_eval package11 which computes Mean Average Precision, precision at different retrieval levels, and other metrics widely accepted in information retrieval evaluations. We specifically focused on precision @ 5 (precision for five images retrieved or classified with highest confidence as answers to a specific topic P@5). Precision @ 5 measures practical improvements in image retrieval that might be seen at the point of care. Supervised machine learning results were evaluated using recall, precision, P@5, and F-score. Precision was computed as the number of images correctly annotated as relevant to the question divided by the total number of images automatically annotated as relevant. Recall was computed as the number of images correctly annotated as relevant by the classifier divided by the total number of images judged to be relevant to the question. P@5 was computed by sorting images in descending order of the classifier confidence scores, and then dividing the number of images correctly annotated as relevant to the question within the five highest ranking by 5. F-score was computed as the weighted harmonic mean of precision and recall (Fβ = (1+β2)·(precision·recall)/β2·precision + recall, where β=1).
Results
We used all 30 of the ImageCLEFmed 2008 information requests in the evaluation of our information retrieval approach. As our goal was to evaluate machine learning methods on a reserved set of images, we could use only 14 requests that had 50 or more relevant images (which could be divided into the training and test sets, and still provide enough positive training examples).
Evaluation of the information retrieval approach
To evaluate the overall performance of our retrieval methods we use Mean Average Precision (MAP) that averages the precision of each of the 30 individual requests. Average precision for each request is the mean of the precision scores after each relevant image is retrieved. This measure shows both how well the system finds all known relevant images and how it orders those images. For clinical decision support, it is crucial to present the most relevant images at the top of the ranked list; therefore we also consider average precision for the first five images retrieved by our systems (P@5).
Table I presents MAP and P@5 for image retrieval based on various combinations of segments of text pertaining to images. Our pipeline approach to image retrieval invariably resulted in MAP around 0.04 and P@5 around 0.12, which is within the range of visual retrieval results reported for the ImageCLEFmed 2008 medical image retrieval task, and is consistent with the observation that visual retrieval techniques can degrade the overall performance [12].
Table I.
Mean average precision and precision at 5 (P@5) for 30 image retrieval requests
| Indexed text and query type ( the request was used as supplied if query expansion is not indicated) | MAP | Precision at 5 | ||
|---|---|---|---|---|
| Lucene | Terrier | Lucene | Terrier | |
| Short captions provided by ImageCLEFmed | 0.151 | 0.045 | 0.347 | 0.200 |
| Full captions | 0.142 | 0.079 | 0.347 | 0.160 |
| Segmented captions | 0.149 | 0.081 | 0.353 | 0.167 |
| Mentions | 0.026 | 0.036 | 0.166 | 0.000 |
| Captions and mentions | 0.122 | 0.160 | 0287 | 0.386 |
| Captions + query expansion | 0.153 | 0.082 | 0.420 | 0.200 |
| Captions and mentions + query expansion | 0.131 | 0.169 | 0406 | 0.387 |
Evaluation of the supervised machine learning approach
The images and associated text judged as relevant or not to 14 requests each of which having 50 or more relevant images, contained on average 159 positive training examples, 616 negative examples, and 85 images randomly withheld for testing while still preserving the proportion of the positive and negative examples for each request. Table II presents average recall, precision, precision for five images classified with highest confidence as answers to a specific topic, and F-scores obtained for text-based and image-based classifiers and their combinations. We explored all possible combinations of the base classifiers. The representative stacking results are also shown in Table II.
Table II.
Results of machine-learning approach to image annotation and retrieval averaged over 14 information requests (A) and 7 requests with the training set containing over 180 positive examples (S)
| Classifier: features | Precision | P@5 | Recall | F-score | ||||
|---|---|---|---|---|---|---|---|---|
| A | S | A | S | A | S | A | S | |
| SVM: Segmented caption text (bag-of-words) TEXT BASELINE | 0.341 | 0.588 | 0.443 | 0.714 | 0.853 | 0.939 | 0.488 | 0.723 |
| SVM: DWT (Image) | 0.135 | 0.270 | 0.057 | 0.057 | 0.429 | 0.856 | 0.205 | 0.410 |
| SVM: Gabor filters (Image) | 0.199 | 0.307 | 0.129 | 0,171 | 0.789 | 0.706 | 0.317 | 0.428 |
| SVM: Color (Image) | 0.202 | 0.315 | 0.171 | 0.343 | 0.817 | 0.778 | 0.324 | 0.449 |
| Stacking: Text + DWT | 0.372 | 0.744 | 0.457 | 0.771 | 0.424 | 0.848 | 0.396 | 0.793 |
| Stacking: Text + Gabor filters | 0.314 | 0.628 | 0.357 | 0.571 | 0.382 | 0.765 | 0.345 | 0.690 |
| Stacking: Text + Color | 0.344 | 0.688 | 0.457 | 0.714 | 0.426 | 0.852 | 0.380 | 0.761 |
| Stacking: Color + Gabor filters | 0.177 | 0.345 | 0.186 | 0.371 | 0.310 | 0.604 | 0.226 | 0.439 |
| Stacking: all classifiers | 0.310 | 0.618 | 0.329 | 0.571 | 0.394 | 0.788 | 0.346 | 0.692 |
Table III shows differences in precision at five retrieved images between the information retrieval and machine learning approaches, as well as differences in information retrieval results for topics included and excluded in machine learning experiments.
Table III.
Lucene retrieval results (IR) for topics included and excluded from machine learning (ML) experiments. The IR results for caption retrieval with query expansion (text) are shown.
| Topics | Features | IR | ML | ||
|---|---|---|---|---|---|
| MAP | P@5 | Precision | P@5 | ||
| 14 included in ML | Text | 0.198 | 0.471 | 0.341 | 0.443 |
| Text + image | 0.041 | 0.129 | 0.372 | 0.457 | |
| 7 best in ML | Text | 0.202 | 0.400 | 0.588 | 0.714 |
| Text + image | 0.043 | 0.200 | 0.744 | 0.771 | |
| 7 worst in ML | Text | 0.098 | 0.271 | 0.095 | 0.171 |
| Text + image | 0.019 | 0.029 | 0 | 0.143 | |
| 16 excluded from ML | Text | 0.112 | 0.375 | Topics excluded from machine learning experiments due to lacking or insufficient positive examples | |
| Text + image | 0.033 | 0.100 | |||
| All topics | Text | 0.153 | 0.420 | ||
| Text + image | 0.039 | 0.119 | |||
The quality and small differences in our information retrieval results do not warrant evaluation of statistical significance. The small sample sizes in machine learning experiments are best addressed by the two-tailed Kolmogorov-Smirnov test and the Wilcoxon two-sample test, the most powerful non-parametric statistics for determining whether two samples are from the same population when the underlying distributions are unknown [23]. The results were obtained using SAS 9.1 npar1way procedure. The difference in Mean Average Precision between the topics included and excluded in machine learning experiments is not statistically significant, which indicates there is no difference in the difficulty of the topics between the groups. The improvement in machine learning precision results for seven best topics is significant at the 0.05 level.
Discussion
The results of the supervised machine learning approach were consistent with our previous findings for the base classifiers, indicating that methods developed for coarse-level annotation scale up to the medium-level and do not depend on the biomedical subdomain. On the other hand, the stacking results were somewhat surprising. Contrary to our previous experience [2] and results reported in [14], on average, stacking did not show improvement over the base classification results. Further analysis revealed a clear separation between the questions and classifiers. The text-based, Gabor-filter, and color-based classifiers had fairly high recall for all questions. For seven questions, these classifiers yielded much higher precision than for the other seven (0.588 and 0.095 respectively). The DWT classifier assigned the same confidence of being an answer to all images (with confidence close to our threshold of 0.5 for classifying an image as an answer for the same seven questions on which other classifiers achieved higher precision.)
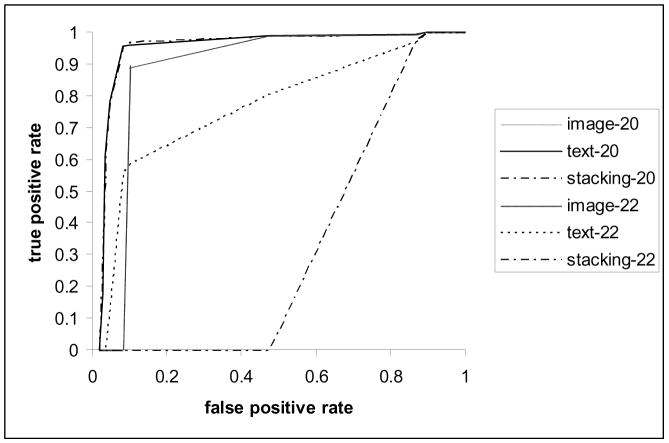
The difference in classification precision cannot be explained by the nature of the questions, as the better and worse performing questions were distributed evenly over question categories, complexity levels, and difficulty for retrieval measured by the average Mean Average Precision obtained for these topics in the ImageCLEFmed 2008 evaluation [12]. The number of positive training examples could have influenced the machine learning results: all poorly performing questions had 100 or less positive training examples, whereas all better performing questions had between 189 and 288 positive examples. Figure 3 presents performance of the text-based and DWT-based classifiers for topics 2012 (MRI or CT of colonoscopy) and 22 (Show me images of muscle cells). The combination of these classifiers achieved the best performance (81% precision and 96% recall at the 0.5 threshold level) for topic 20. Topic 22 is representative of the poorly performing topics.
Figure 3.
Comparative ROC curves of the text-based and image-based classifiers and their combination for the best and worst performing questions.
The benefits of combining the text and image features are illustrated in the following example: Based on its caption, the image presented in Figure 413 was classified with high probability as not relevant to the topic MRI or CT of colonoscopy, however, combining the low probability of relevance based on the textual features (0.268) with the higher probability of relevance based on the image features (0.453), the meta-classifier annotated the image as relevant with 0.891 probability. The error in text-based annotation as well as text-based retrieval for this image can be explained by the vocabulary mismatch: none of the query terms can be found in the caption text. Even query expansion in the information retrieval approach was not helpful in this case because in the UMLS MR is not synonymous with MRI, and colonoscopy cannot be mapped to colonography.
Figure 4.
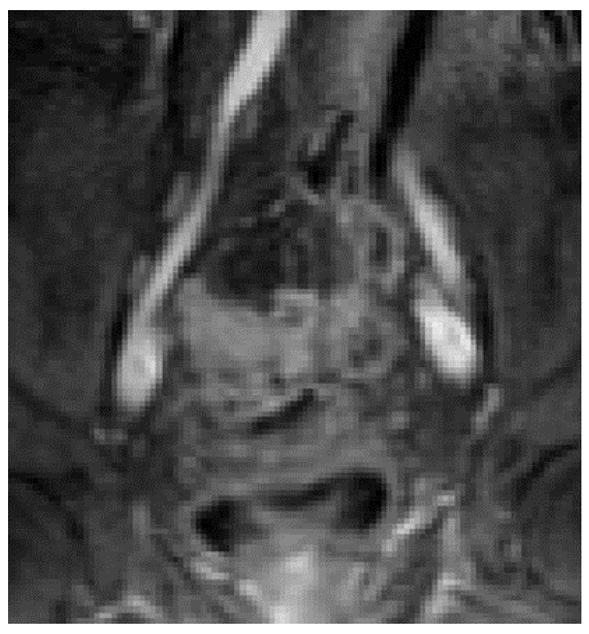
An image and its caption relevant to topic “MRI or CT of colonoscopy”
Our results demonstrate that the benefits of combining text and image features for the classification of images found in biomedical articles—which were observed in classifying images into six modality categories on a set of 554 images (73.66% average F-score) [25]—can be extended to medium level annotation using different fusion methods applied in this study and in other work [25].
The results of the information retrieval approach provide interesting insights into the nature and amount of text needed for a comparable performance of different information retrieval methods. Whereas the vector space model implemented in Lucene performed best on segmented captions, all extracted text was needed for comparable performance of the Terrier Inverse Document Frequency model with Laplace after-effect and normalization 2 (InL2), which we selected to gain early precision (boost mean precision at five retrieved documents). Although the Terrier L2 model was found to be less sensitive to the variation of document length in several text collections [26], our results demonstrate that the model behaves differently for document collections with average document length of 66 words in the segmented captions, and for expanded documents with average length of 149 words. To our knowledge, this is the first report of the differences between the vector space and the InL2 models caused by the document length. Notably, information contained in the descriptions of images in the body of the text is not sufficient for image retrieval and does not add value to captions when using the vector space model. The image retrieval component of our approach tends to be sensitive to the variety of features available in the image queries. Consequently, the results degraded even further when it was observed that example query images provided with the questions were not sufficiently represented in the image collection.
Conclusions and future work
This study concludes that combining text and image features is beneficial for finding clinically relevant images. Our image annotation and retrieval results demonstrate that machine learning methods have a potential to achieve retrieval accuracy required for finding evidence for clinical decision making. We defer studying methods of combining image and text features other than in a pipeline information retrieval approach (for example, fusion) until we improve our base retrieval methods. We plan to explore leveraging domain knowledge beyond ontology-based query expansion and MeSH terms for text pre-processing. For improved image annotation and retrieval, the results indicate a need to use image features that are relevant to concepts derived from text analysis. We also plan to study methods that extract and apply features in stages (for example, texture features using grayscale image characteristics before use of color features). Finally, local image characteristics are essential in ranking the most relevant images first. Steps toward this include analysis and extraction of image overlays in addition to segmentation of local image features.
We attribute the high accuracy of the machine learning approach seen on seven of the questions to the relatively large number of annotated images. Since obtaining hundreds of training examples for each clinical information request is not feasible, we plan to explore methods of reducing requests to questions with known answers available in our knowledge base.
We also plan to explore methods for training set size reduction. For example, these methods may include selection of the most informative training samples, feature selection methods, and consideration of other learners whose performance is less sensitive to the small sample size and noisy data.
Summary points
What is already known on the topic:
Medical images are a significant information source for clinical decision making.
Image retrieval for clinical decision making can be based on image analysis, on indexing of the text assigned to images, or on a combination of image and text analysis.
High quality image retrieval is required for clinical decision support.
What the study has added to our knowledge:
This work highlights the challenges in image retrieval.
The information retrieval and classification-based systems for image retrieval have been evaluated and compared.
Information retrieval models known to be stable with respect to document length were found to be sensitive to average document length of 66 words.
The quality of image retrieval has to be improved to reliably provide illustrative evidence.
Given sufficient training data, the supervised machine learning approach based on combining image and text features has a better potential to achieve image annotation and retrieval accuracy acceptable for providing clinical evidence than either method alone.
Acknowledgments
This research was supported by the Lister Hill National Center for Biomedical Communications and intramural R&D division of the National Library of Medicine, at the National Institutes of Health, U.S. Department of Health and Human Services. We thank Emilia Apostolova for implementing the caption segmentation algorithm.
Footnotes
Henceforth we use “image” as equivalent to “images and illustrations found in scientific publications”
Published by the Radiological Society of North America (RSNA)
Topic numbers refer to those in the list of 30 ImageCLEFmed 2008 medical image retrieval task topics
Authors’ contributions
DDF, SA, and GRT conceived and designed the project. DDF and MS developed and coded the text-based systems. SA developed and coded the image-based systems. DDF and SA designed, performed, and analyzed the evaluation. DDF and SA wrote the manuscript. GRT and MS edited the manuscript. All authors read and approved the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sackett DL, Straus SE, Richardson WS, Rosenberg W, Haynes RB. Evidence-Based Medicine: How to Practice and Teach EBM. 2. Churchill Livingstone; Edinburgh, Scotland: 2000. [Google Scholar]
- 2.Demner-Fushman D, Lin J. Answering Clinical Questions with Knowledge-Based and Statistical Techniques. Computational Linguistics. 2007;33(1):63–103. [Google Scholar]
- 3.Antani S, Demner-Fushman D, Li J, Srinivasan B, Thoma GR. Exploring use of images in clinical articles for decision support in evidence-based medicine. Proc. SPIE-IS&T Electronic Imaging; San Jose, CA. January 2008; p. 6815. 68150Q(1–10) [Google Scholar]
- 4.Demner-Fushman D, Antani SK, Simpson M, Thoma GR. Combining Medical Domain Ontological Knowledge and Low-level Image Features for Multimedia Indexing. Proceedings of 2nd International Language Resources for Content-Based Image Retrieval Workshop (OntoImage 2008), 6th Language Resources and Evaluation Conference (LREC 2008); Marrakech, Morrocco. May 2008; Available from: http://archive.nlm.nih.gov/pubs/ceb2008/2008010.pdf and http://www.lrec-conf.org/proceedings/lrec2008/ [Google Scholar]
- 5.Smeulders AWM, Worring M, Santini S, Gupta A, Jain R. Content-based image retrieval at the end of the early years. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2000 Dec;22(12):1349–80. [Google Scholar]
- 6.Antani S, Katuri R, Jain R. A survey on the use of pattern recognition methods for abstraction, indexing and retrieval of images and video. Pattern Recognition. 2002;35(4):945–65. [Google Scholar]
- 7.Hearst M, Divoli A, Wooldridge M, Ye J. Exploring the efficacy of caption search for bioscience journal search interfaces. Proceedings of BioNLP 2007, ACL 2007 Workshop; Prague, Czech Republic. [Google Scholar]
- 8.Chen Z, Wenyin L, Zhang F, Li M, Zhang H. Web mining for Web image retrieval. J Am Soc Information Science & Technology. 2001 Jun;52(10):831–9. [Google Scholar]
- 9.Hersh WR, Müller H, Jensen J, Yang J, Gorman P, Ruch P. Advancing biomedical image retrieval: development and analysis of a test collection. Journal of the American Medical Informatics Association. 2006;13:488–496. doi: 10.1197/jamia.M2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winfield H, Lain E, Horn T, Hoskyn J. Eosinophilic cellulitislike reaction to subcutaneous etanercept injection. Arch Dermatol. 2006 Feb;142(2):218–20. doi: 10.1001/archderm.142.2.218. [DOI] [PubMed] [Google Scholar]
- 11.Demner-Fushman D, Antani SK, Thoma GR. Automatically Finding Images for Clinical Decision Support. Proceedings of IEEE International Workshop on Data Mining in Medicine (DM-Med 2007); Omaha, NE. 2007 Oct; pp. 139–144. Available from http://archive.nlm.nih.gov/pubs/ceb2007/2007022.pdf. [Google Scholar]
- 12.Müller H, Kalpathy-Cramer J, Kahn CE, Jr, Hatt W, Bedrick S, Hersh W. Overview of the ImageCLEFmed 2008 medical image retrieval task. Available from: http://www.clef-campaign.org/2008/working%5Fnotes/
- 13.Kahn CE, Jr, Thao C. GoldMiner: a radiology image search engine. AJR. 2007;188:1475–1478. doi: 10.2214/AJR.06.1740. [DOI] [PubMed] [Google Scholar]
- 14.Ting KM, Witten IH. Issues in stacked generalization. J Artif Intell Res. 1999;10:271–89. [Google Scholar]
- 15.Manjunath BS, Ma WY. Texture features for browsing and retrieval of image data. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1996;18(8):837–42. [Google Scholar]
- 16.Torralba A. Contextual priming for object detection. Intl J Computer Vision. 2003;53(2):153–167. [Google Scholar]
- 17.Rui Y, Huang T, Chang S. Image retrieval: current techniques, promising directions and open issues. J Visual Communication and Image Representation. 1999;10(4):39–62. [Google Scholar]
- 18.Mojsilovic A, Hu J, Soljanin E. Extraction of perceptually important colors and similarity measurement for image matching, retrieval, and analysis. IEEE Transactions on Image Processing. 2002;11(11):1238–48. doi: 10.1109/TIP.2002.804260. [DOI] [PubMed] [Google Scholar]
- 19.Witten IH, Frank E. Data Mining: Practical Machine Learning Tools and Techniques. 2. San Francisco, CA: Morgan Kaufmann Publishers/Elsevier; 2005. pp. 286–296. [Google Scholar]
- 20.Simpson M, Demner-Fushman D, Sneiderman C, Antani SK, Thoma GR. Using Non-lexical Features to Identify Effective Indexing Terms for Biomedical Illustrations. Proceedings of the 12th Conference of the European Chapter of the Association for Computational Linguistics (EACL-09); Athens, Greece. April 2009. [Google Scholar]
- 21.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993 Aug;32(4):281–91. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 23.Siegel S, Castellan N. Nonparametric statistics for the behavioral sciences. 2. New York: McGraw-Hill; 1988. [Google Scholar]
- 24.Ajaj W, Lauenstein TC, Pelster G, Holtmann G, Ruehm SG, Debatin JF, Goehde SC. MR colonography in patients with incomplete conventional colonoscopy. Radiology. 2005 Feb;234(2):452–9. doi: 10.1148/radiol.2342032001. Available from: http://radiology.rsnajnls.org/cgi/content/full/234/2/452. [DOI] [PubMed]
- 25.Rafkind B, Lee M, Chang SF, Yu H. Exploring text and image features to classify images in bioscience literature. Proceedings of the HLT-NAACL BioNLP Workshop on Linking Natural Language and Biology. 2006 Jun;:73–80. [Google Scholar]
- 26.Amati G, Van Rijsbergen CJ. Probabilistic models of information retrieval based on measuring the divergence from randomness. ACM Trans Inf Syst. 2002 Oct;20(4):357–389. [Google Scholar]