Abstract
An abasic site called dSpacer has been introduced into duplex regions of the 8–17 DNAzyme and adenosine aptamer for label-free fluorescent detection of Pb2+ and adenosine, respectively. The dSpacer can bind an extrinsic fluorescent compound, 2-amino-5,6,7-trimethyl-1,8-naphthyridine (ATMND), and quench its fluorescence. Addition of Pb2+ enables the DNAzyme to cleave its substrate and release ATMND from DNA duplex, recovering the fluorescence of ATMND. Similarly, the presence of adenosine induces structural switching of the aptamer, resulting in the release of ATMND from the DNA duplex and a subsequent fluorescence enhancement. Under optimized conditions, this label-free method exhibits detection limits of 4 nM for Pb2+ and 3.4 µM for adenosine, which are even lower than those of the corresponding labeled-DNAzyme and aptamer sensors. These low detection limits have been obtained without compromising any of the selectivity of the sensors. Finally, the dynamic range of the adenosine sensor has been tuned by varying the number of hybridized base-pairs in the aptamer duplex. The method demonstrated here can be applied for label-free detection and quantification of a broad range of analytes using other DNAzymes and aptamers.
Introduction
Detection and quantification of metal ions and organic molecules found in biological systems and in the environment remains an active area of research, as these molecules are either quite beneficial or toxic to human health. Instrumental analyses, while highly sensitive, are often limited by costs and sophisticated operation, making it difficult for on-site and real-time detection. To overcome the limitation, a number of sensors that exhibit rapid responses and are both selective and sensitive toward a specific target have been developed.1–9 However, a major challenge in designing sensors for metal ions and organic molecules is the lack of a general method to obtain sensors that can selectively bind their targets, transform the binding event into detectable signals and further tune the sensing dynamic range to match the concentrations of the targets in a given sample of choice. To meet such a challenge, DNAzymes and aptamers have been isolated and developed into sensors using general applicable methodologies to sense a wide range of metal ions and organic molecules.10–21
DNAzymes (also referred to as catalytic DNAs or deoxyribozymes) and aptamers are functional nucleic acids that either display catalytic activity or binding affinity toward a specific target, respectively. Both DNAzymes and aptamers have been obtained through in vitro selection22 or systematic evolution of ligands by exponential enrichment (SELEX),23 which can be tailored to allow these DNA molecules to function in the presence of a specific target of choice. From these selection processes, numerous DNAzymes have been isolated to display high specificity toward various metal ions such as Pb2+,22,24 Cu2+,25,26 Zn2+,27,28 Co2+,29,30 Mn2+,31 and UO2 2+,32 while aptamers have been selected to exhibit specific binding towards a broad range of molecular targets ranging from organic molecules, proteins, to intact bacteria, viruses and cancer cells.10,33–35
The high selectivity of DNAzymes and aptamers toward specific targets make them ideal recognition components for sensing applications.11,34–37 More importantly, general methods have been developed to modify these functional DNA molecules 38–41 in order to transform them into fluorescent,42–45 colorimetric,46–53 and electrochemical54,55 sensors. Amongst these, fluorescent sensors have been a focus of study because of facile operation, high sensitivity, and easily detectable signals.
Most reported fluorescent sensors based either on DNAzymes or aptamers require the covalent attachment of fluorophores to either an internal site or the 3′ or 5′ ends of the DNA, and generally two or more fluorophore and quencher labels are needed to produce an efficient fluorescence switch upon DNA-target interaction.42–44 This results in not only high cost of operation, but also potentially weaker activity of DNAzymes or aptamers in comparison with their non-modified analogues, because linkers used for labeling as well as DNA modifications might interfere with the DNA-target interaction.56,57
Alternatively, label-free fluorescent methods may be cheaper, easier to operate and would not interfere with the activity of the DNAzyme or aptamer used, potentially increasing the overall sensitivity of the sensor. To demonstrate this potential, several label-free fluorescent sensors have been reported using either fluorescent intercalating dyes,58–60 or by coupling the aptamer used with another RNA aptamer for malachite green,61,62 the fluorescence of which changes upon binding. Despite the promise offered by existing label-free functional DNA-based sensors, few can match or exceed the sensitivity of labeled sensors. In addition, while a general method has been developed to tune the dynamic range of DNAzyme sensors,46,48 no method has been reported to tune the dynamic range of aptamer sensors. Herein, we present a general method for the design of label-free, fluorescent DNAzyme and aptamer sensors that match or exceed the sensitivity of labeled sensors and exhibit a tunable dynamic range through the introduction of an abasic site (dSpacer) into the functional DNA duplexes.
Materials and Methods
The fluorophore 2-amino-5,6,7-trimethyl-1,8-naphthyridine (ATMND) was purchased from Ryan Scientific Inc. and was used as received. All oligonucleotides used in this work were purchased from Integrated DNA Technology Inc.(Coralville, IA), with the following sequences (X represents dSpacer):
| Pb2+-dependent DNAzyme: | |
| 17Eab | 5′-ACAGACATCTCTTCTCCGAGCCGGTCGAAATAGXGAG-3′ |
| 17EG | 5′-ACAGACATCTCTTCTCCGAGCCGGTCGAAATAGGGAG-3′ |
| Substrate (a DNA/RNA chimer with a single RNA nucleotide linkage): | |
| 17S | 3′-TGTCTGTAGAGAAGGrATATCCCTC-5′ |
| Adenosine aptamer: | |
| AAP | 5′-TGTCGTTGACCTGGGGGAGTATTGCGGAGGAAGGT-3′ |
| ssDNAs for hybridization with aptamer: | |
| L1ab | 3′-AGCAACTGXACC-5′ |
| L2ab | 3′-AGCAACTGXACCC-5′ |
| L3ab | 3′-AGCAACTGXACCCC-5′ |
| L4ab | 3′-ACAGCAACTGXACCCC-5′ |
| L2G | 3′-AGCAACTGGACCC-5′ |
Fluorescence measurements
For a standard Pb2+ measurement using modified 8–17 DNAzymes, 480 µL buffer A (25 mM HEPES pH 7.0 and 100 mM NaCl), 5 µL ATMND solution (100 µM), 5 µL substrate 17S (102 µM), and 10 µL DNAzyme 17Eab or 17EG (107 µM) were added sequentially into a 1.5 mL microcentrifuge tube and upon vortexing, the tube was allowed to stand at room temperature for 2 min. Since no obvious difference was obtained in the results whether the mixture was annealed (by heating to 80 °C and then cooled to room temperature in 30 min.) or not, the annealing step was skipped in all the fluorescent measurements described in this study for further simplification. The solution was then transferred to a cuvette in a FluoroMax-P fluorimeter (HORIBA Jobin Yvon Inc., USA) with a constant temperature control at 5 °C. After 6 min to allow the temperature to reach equilibrium, 5 µL of Pb2+ stock solution in buffer A was added to the cuvette and followed by vortexing, and time-dependent fluorescent measurement at ex/em = 358/405 nm was immediately started. Typically, the rate of fluorescence enhancement within 3~5 min after Pb2+ addition was calculated for all the measurements. However, when Pb2+ concentration was high (at micromolar levels), the cleavage reaction was extremely fastand therefore, only the initial rates for the first 30 s were calculated.
In a standard adenosine measurement, 450 µL buffer B (10 mM HEPES pH 7.0, 100 mM NaCl and 1mM EDTA), 5 µL aptamer AAP (91.8 µM), 6.25 µL ssDNA L1ab ~L4ab or L2G (93.7 µM), and 50 µL ATMND solution (5 µM) were added sequentially into a 1.5 mL microcentrifuge tube. After vortexing, the tube was allowed to stand at room temperature for 1 min. Like in the Pb2+ measurement using DNAzyme described above, we found no obvious difference in the results whether the mixture was annealed or not and therefore all measurements were carried out without annealing. A 5 µL of adenosine stock solution in buffer B was added to the above mixture followed by vortexing and then was allowed to stand at room temperature for 1 min. The solution was then transferred to a cuvette in the fluorimeter with a constant temperature control at 5 °C. After 10 min, the fluorescence intensity at ex/em = 358/405 nm was recorded. The sample showed a stable intensity signal from 8~30 min upon transferring it to the fluorimeter. A lower ATMND concentration (500 nM) was used in the adenosine experiment than in the Pb2+ experiment (1 µM) in order to achieve lower background fluorescence, because the binding affinity of ATMND to the aptamer duplex is weaker than that with the DNAzyme duplex.
Results and Discussion
Design of Pb2+-dependent DNAzyme containing a dSpacer abasic site
The sensor design used in the current study is based on findings from Teramae and co-workers, who reported the use of DNA strands containing an abasic site called dSpacer for nucleobase recognition and target molecule binding.63–65 Given the success of dSpacer-containing DNA for nucleobase recognition and target molecule binding, we propose to create a dSpacer in functional DNA molecules such as DNAzymes and aptamers for label-free sensing applications. Through complementary hydrogen bonding toward the opposite nucleobase and stacking effects from flanking nucleobases, a fluorophore could bind to the abasic site in dsDNA, and lead to its fluorescence quenching. In the presence of the specific target, the DNAzyme-target or the aptamer-target interactions would result in the dehybridization of the DNA duplex region containing an abasic site, thereby releasing the fluorophore into the solution and recovering its quenched fluorescence. By fine tuning of the position of the abasic site for binding different fluorophores we envision that these fluorescent sensors can be used for detecting and quantifying a broad range of targets using this method.
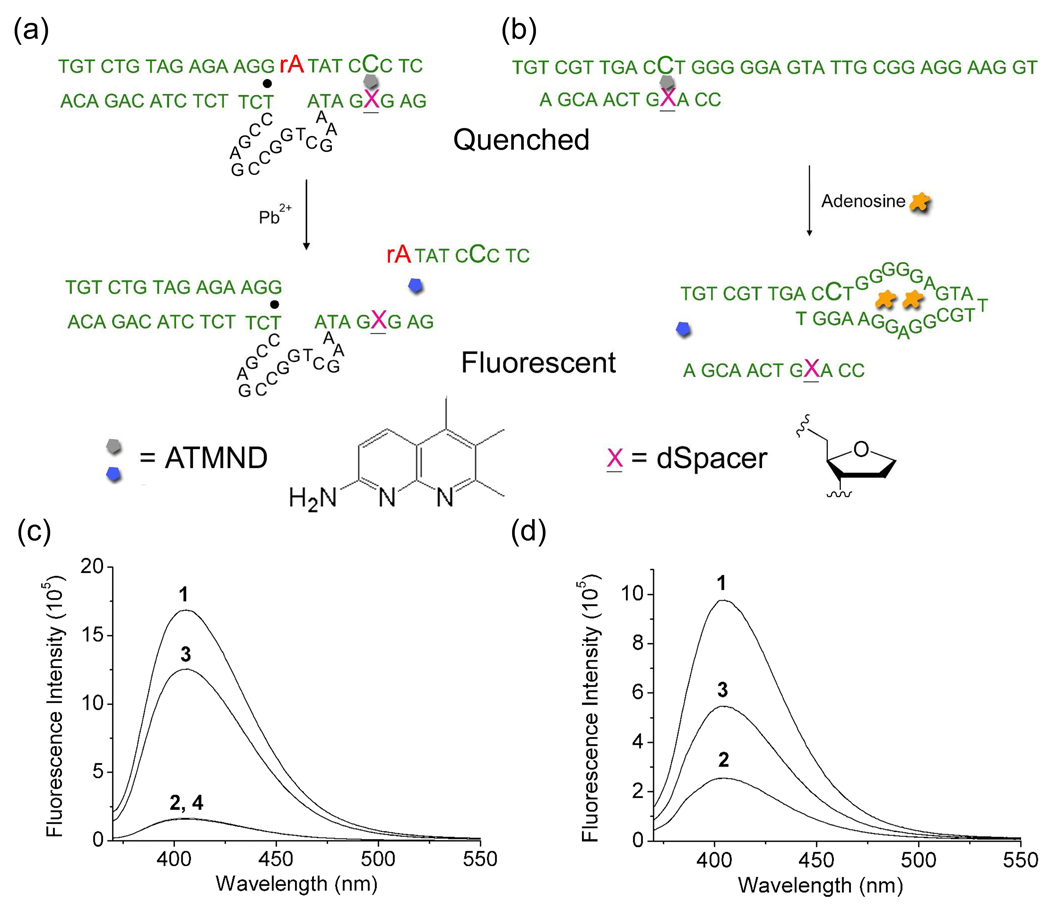
Since fluorophore-labeled 8–17 DNAzymes were successful as highly sensitive and selective catalytic beacon sensors for Pb2+,24,46,66–69 we first chose this DNAzyme system to demonstrate label-free Pb2+ detection using an abasic site and to compare the performance of the two systems. Based on the 8–17 DNAzyme, two mutations at the 3′-end of the enzyme strand, 17E, were introduced (Figure 1a): a T base was mutated to an abasic site for label-free fluorophore binding, and another T base at the 3′-end was eliminated for efficient dehybridization of the abasic site-containing duplex region after the substrate, 17S, was cleaved. This modified enzyme strand containing the abasic site was called 17Eab in this work. In the absence of Pb2+, the fluorophore ATMND would bind to the abasic site in the duplex region of the 17Eab and 17S (Figure 1a), effectively quenching the fluorescence of ATMND. In the presence of Pb2+, on the other hand, the substrate 17S would be cleaved, resulting in release of the cleavage products and formation of ssDNA. In such a case, the binding affinity of ATMND to the ssDNA region would decrease significantly, causing ATMND released and thus enhancing the fluorescent signal.
Figure 1.
Schematic illustration of label-free fluorescent detection of Pb2+ (a) and adenosine (b), and the fluorescence emission spectra of ATMND in the absence and presence of 17Eab-17S (c) and APP-L1ab (d) for Pb2+ and adenosine detection, respectively. The fluorescent spectra in (c) were collected in buffer A (25 mM HEPES pH 7.0 and 100 mM NaCl). 1: ATMND (1 µM); 2: ATMND (1 µM), 17Eab (2.14 µM) and 17S (1.02 µM); 3: ATMND (1 µM), 17Eab (2.14 µM), 17S (1.02 µM) and Pb2+ (1 µM) after 15 min reaction; 4: ATMND (1 µM), 17Eab (2.14 µM), 17S (1.02 µM), Pb2+ (1 µM) and EDTA (1 mM) after 15 min reaction. The fluorescent spectra in (d) were collected in buffer B (10 mM HEPES pH 7.0, 100 mM NaCl, 1 mM EDTA). 1: ATMND (500 nM); 2: ATMND (500 nM), AAP (918 nM) and L1ab (1.17 µM); 3: ATMND (500 nM), AAP (918 nM), S1 (1.17 µM) and adenosine (100 µM). The excitation wavelength was 358 nm. All spectra were recorded at 5 °C.
Formation of duplex DNA between 17Eab and 17S
To check whether 17Eab and 17S could form a duplex that was essential for both ATMND binding into the abasic site as well as Pb2+-dependent catalytic activity, the fluorescence spectra of ATMND in the absence and in the presence of 17Eab and 17S were collected. When alone in solution at a concentration of 1 µM, ATMND exhibited strong blue fluorescence emission with a band centered around 405 nm (Figure 1c, plot 1). Further addition of either 2.14 µM 17Eab or 1.02 µM 17S resulted in little change in the fluorescence spectra of ATMND, suggesting that the interaction between ATMND and ssDNA of either 17Eab or 17S was negligible. In contrast, in the presence of both 17Eab and 17S, the fluorescence emission of ATMND was significantly quenched by more than 90 % (Figure 1c, plot 2), which is attributable to the effective binding of ATMND to the abasic site in DNA duplex formed by 17Eab and 17S.63 Therefore, the hybridization of 17Eab and 17S was efficient and the duplex DNA was conducive for the Pb2+-catalyzed reaction and sensing applications.
Pb2+-catalyzed cleavage of 17S in the 17Eab-17S duplex
To find out the effect of the abasic site on the Pb2+-dependent activity, we first performed enzymatic activity assay using PAGE and found that, in the absence of ATMND, the abasic site within the DNAzyme binding arm weakened the substrate/DNAzyme binding interaction, resulting in reduced activity. However, addition of ATMND restored the substrate/DNAzyme binding affinity and rendered similar activity compared to unmodified enzyme (Figure S1, Supporting Information). To confirm addition of Pb2+ could result in the cleavage of the 17S and release of ATMND, 1 µM of Pb2+ was added to a solution containing 1 µM ATMND, 2.17 µM 17Eab and 1.02 µM 17S and more than a 7-fold fluorescence enhancement was observed after 15 min (Figure 1c, plot 3). To ensure that the fluorescence increase was due to the presence of Pb2+, 1 mM EDTA was added to the above solution as a control before Pb2+ addition. Little change in fluorescence spectra could be observed under the same conditions (Figure 1c, plot 4), suggesting the essential role of Pb2+ for the catalytic reaction. When the essential G•T wobble pair in the 8–17 DNAzyme (Figure 1a) was replaced with a G-C Watson-Crick base pair, the DNAzyme exhibited no Pb2+-dependent activity (Figure S2, Supporting Information), similar to that of original 8–17 DNAzyme. These results suggested that the new label-free DNAzyme sensor construct did not perturb the original activity of the 8–17 DNAzyme, which was crucial for the fluorescence enhancement.
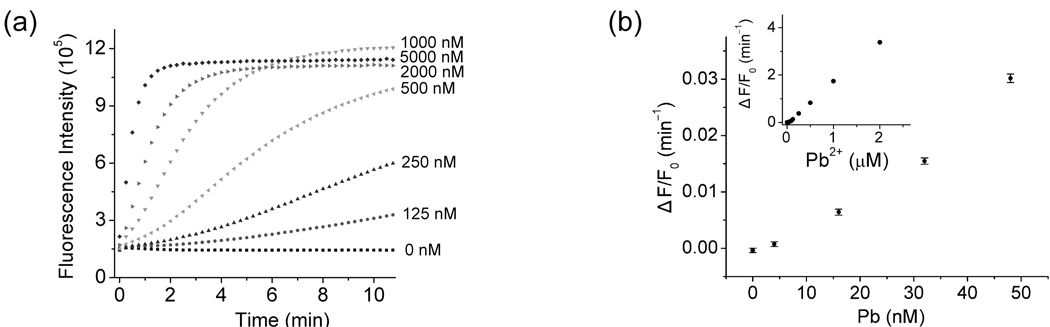
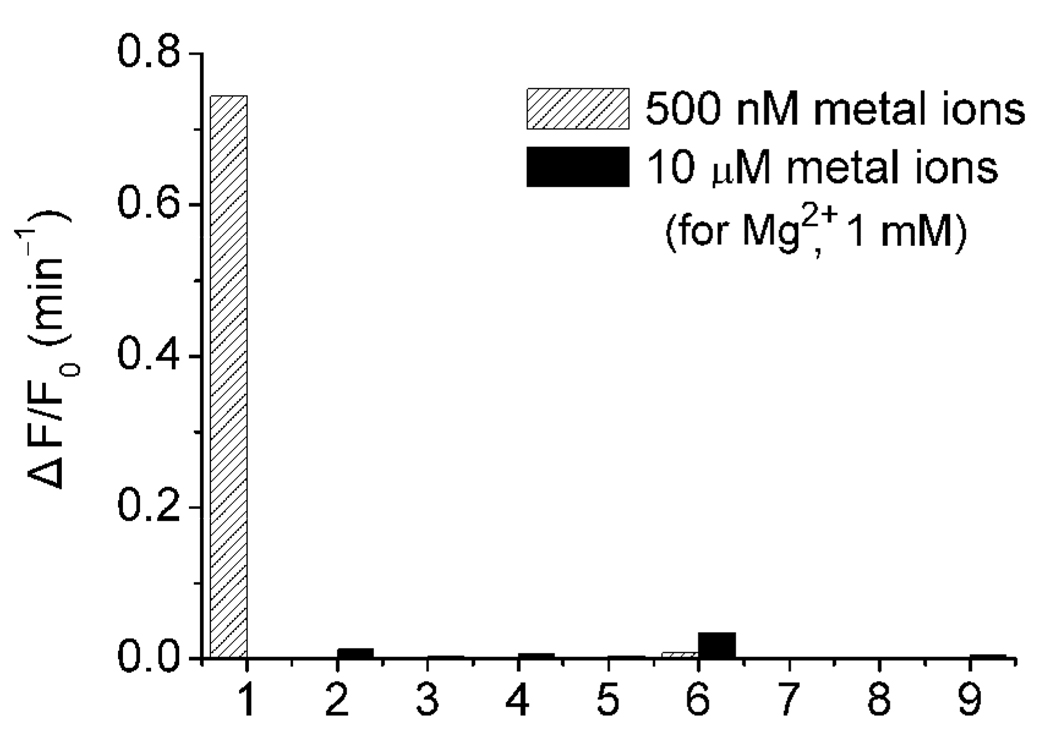
A kinetics study was carried out to monitor the time-dependent emission of ATMND at 405 nm after the reaction was initiated through the addition of various amounts of Pb2+ (Figure 2a; for lower Pb2+ concentration range, Figure S3, Supporting Information). The rate of fluorescence enhancement ratio (ΔF/F0 per minute) showed an approximately linear relationship with Pb2+ concentration (CPb 2+) at least in 0~1 µM range as ΔF/F0 (min−1) = 1.697×CPb 2+ (µM) (Figure 2b). For Pb2+ measurements, the rate (ΔF/F0 per minute) within 3~5 min after Pb2+ addition was recorded rather than fluorescence intensity at a specific time point, because the rate measurement was much less vulnerable to fluctuations in the background fluorescence of the samples. This new method was very sensitive to the concentration of Pb2+, with a detection limit measured by 3σb/slope (σb, standard deviation of the blank samples) of 4 nM, which was even lower than previously reported labeled fluorescent methods.24,46,70 In addition, this method maintained excellent selectivity over other divalent metal ions, such as Mg2+, Ca2+, Sr2+, Ba2+, Mn2+, Fe2+, Co2+, Zn2+, Ni2+, Hg2+, Cd2+ (Figure 3).
Figure 2.
Kinetics of fluorescence enhancement by Pb2+-catalyzed cleavage (a), and relationship between fluorescence enhancement rate and Pb2+ concentrations (b). Fluorescence enhancement rate was calculated within 3~5 min after Pb2+ addition. λex/λem = 358/405 nm. Condition: 1 µM ATMND, 2.14 µM 17Eab, 1.02 µM 17S in 25 mM HEPES pH 7.0, 100 mM NaCl at 5 °C.
Figure 3.
Selectivity of ATMND-17Eab-17S system for Pb2+ detection over other divalent metal ions. 1: 500 nM Pb; 2: Mg2+; 3: Mn2+; 4: Fe2+; 5: Co2+; 6: Zn2+; 7: Ni2+; 8: Hg2+; 9: Cd2+.
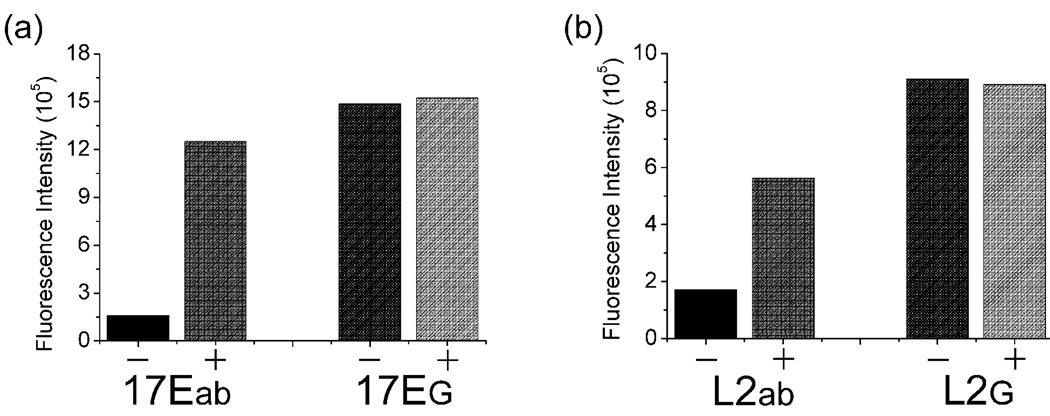
To confirm the essential role of the abasic site in this method, a control DNAzyme 17EG of the same DNA sequence with 17Eab, except that the abasic site was replaced by a G, was also investigated under the same conditions. Little fluorescence quenching of ATMND was observed when the fluorophore was added to a solution containing 17EG and 17S, and subsequent addition of Pb2+ to the solution yielded negligible fluorescence enhancement (Figure 4a). These results suggested that there was barely any binding of ATMND to the completely complementary 17EG-17S duplex, and the Pb2+-dependent catalytic cleavage of 17S by 17EG could not be transformed into a fluorescence signal change in the absence of the abasic site.
Figure 4.
Effect of abasic site on the fluorescence response of DNAzyme to Pb2+ (a) and aptamer to adenosine (b) in this label-free method. For (a), in the absence (−) and presence (+) of 1 µM Pb2+; for (b), in the absence (−) and presence (+) of 1 mM adenosine.
Design of adenosine-dependent aptamer containing a dSpacer abasic site
Encouraged by the above DNAzyme-based sensing results, we would like to explore whether the same concept could be extended to aptamers so that the same method could be used to sense even a wider range of analytes beyond metal ions. To achieve this goal, we decided to use the adenosine aptamer53,71 as a model system for demonstration. The challenge was to transform the structural switching43,72,73 of the aptamer upon target-binding into fluorescence enhancement. Because the location of ATMND on the DNA was controllable in this label-free method, an abasic site-containing ssDNA linker (called L1ab) was designed to hybridize to the adenosine aptamer DNA (called AAP, see Figure 1b). In the absence of the target adenosine, ATMND bound strongly to the abasic site in the duplex region, resulting in suppression of its fluorescent signal. In the presence of adenosine, the aptamer bound adenosine strongly, thereby releasing the abasic-site containing the ssDNA and ATMND from the duplex DNA into solution and recovering the quenched fluorescence of ATMND. Consequently, adenosine could be effectively detected through the fluorescence enhancement response.
Performance of label-free fluorescent adenosine detection
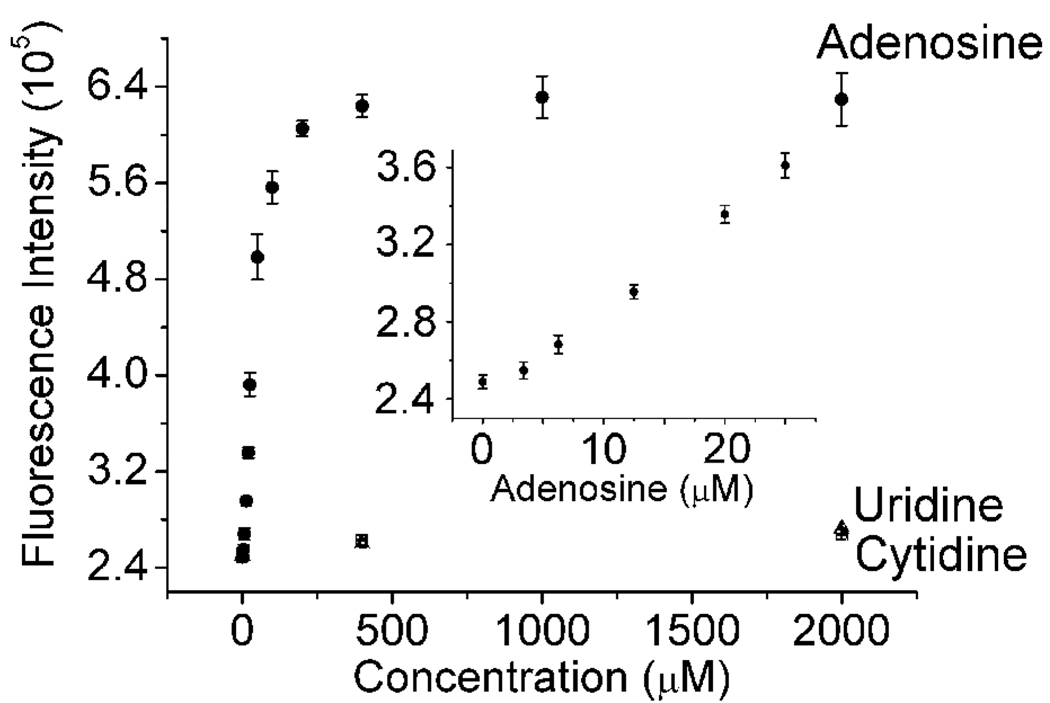
To check the binding of the fluorophore to the AAP-L1ab duplex, ATMND was added to a solution containing AAP and L1ab. ATMND underwent fluorescence quenching upon binding to the AAP-L1ab duplex (Figure 1d, plot 1 and 2), similar to that of the 17Eab-17S system. Next, addition of adenosine to the solution resulted in recovering some of the quenched fluorescence of ATMND (Figure 1d, plot 3), probably through structural switching of AAP that allows single strand DNA formation and release of ATMND from DNA duplex into solution. The kinetics of the binding and subsequent switching was fast, and a stable signal over at least 30 min could be obtained, making it possible to record fluorescence spectra 10 min after adenosine addition, at which point the equilibrium was established. The emission intensity at 405 nm was found to rise with increasing concentration of adenosine in solution, with a linear response in the range of 0~25 µM and a detection limit (3Sb/slope, σb, standard deviation of the blank samples) of 3.4 µM (Figure 5), which is lower than some reported sensors for adenosine.53,71,74 This relatively low detection limit may be ascribed to the short duplex region of AAP-L1ab, which facilitated the binding of adenosine and release of L1ab. The selectivity of this method toward adenosine over two other nucleotides, uridine and cytidine, was very high, with little fluorescence enhancement observed for them even in concentrations at the millimolar level (Figure 5). Guanosine was not tested because it is not soluble in aqueous solutions at the millimolar concentration range. In addition, five mutants of AAP were also investigated under the same conditions. These mutants could only induce little or no fluorescence enhancement response toward adenosine compared to APP (Figure S4, Supporting Information), suggesting that the activity of the aptamer was crucial for the fluorescence enhancement.
Figure 5.
Fluorescence intensity of ATMND-AAP-L1ab in the presence of different concentrations of adenosine, uridine and cytidine. Inset: adenosine in 0~25 µM range. λex/λem = 358/405 nm. Condition: 500 nM ATMND, 918 nM AAP, 1.17 µM L1ab in 10 mM HEPES pH 7.0, 100 mM NaCl, 1 mM EDTA at 5 °C.
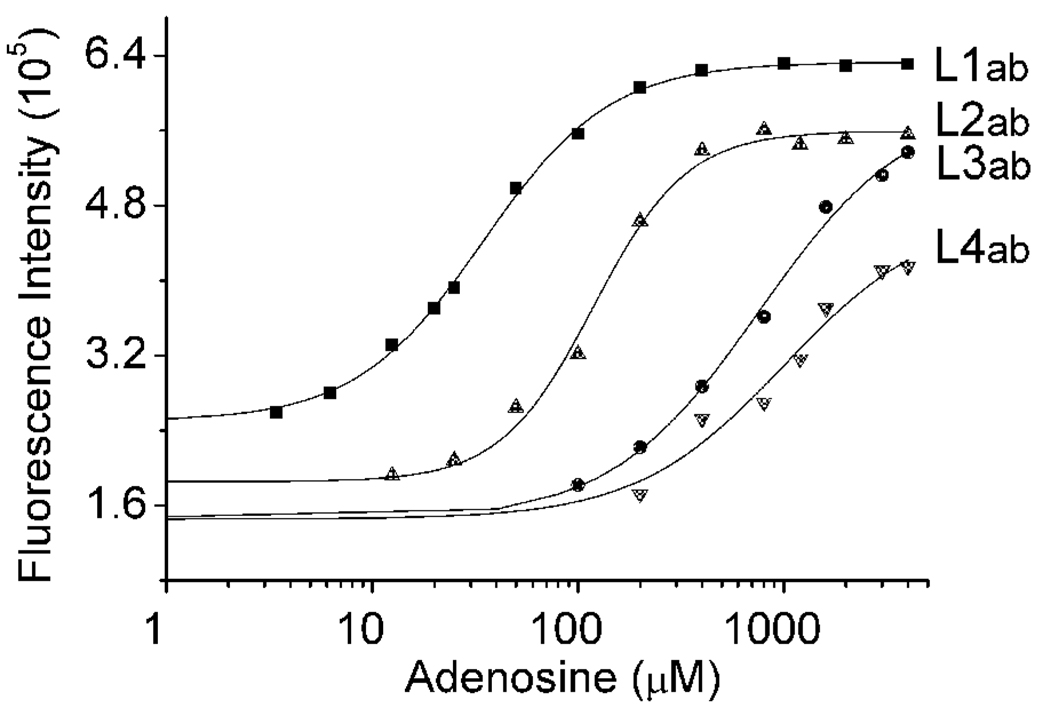
Most analytes of interest have varied concentration ranges in different compartments of cells or at different locations in the environment. A practical sensor needs to have a tunable dynamic range that matches the concentration ranges for the different locations; too strong a binding by the sensor can perturb the equilibrium of the sensing environment while too weak a binding by the sensor may not allow detection. Previously we have shown that DNAzyme-based sensors can be designed to have a tunable dynamic range.46,48 However, the same strategy cannot be applied to aptamers because catalytic cleavage and turnovers were required in tuning the dynamic range of the DNAzyme sensors. Here we explore a new strategy of tuning the dynamic range of aptamers by varying the length of the abasic site-containing ssDNA, L1ab~L4ab, which hybridize to AAP. As shown in Figure 6, with increasing length of L1ab to L2ab~L4ab by more complementary base pairs to AAP, the dynamic range of the sensor can be tuned to be 3.4–200 µM, 25–400 µM, 100 µM-4 mM, and 200 µM-4 mM, respectively. These results can be attributed to the fact that, with increasing number of base pairs in the duplex region formed by AAP with ssDNA from L1ab to L4ab, the duplex becomes more stable, making it more difficult to carry out the structural switching that leads to fluorescence enhancement under the same experimental conditions. To achieve the same effects, more adenosine is needed. Therefore, by designing appropriate number of base pairs in the ssDNA/aptamer duplex region that is critical to the adenosine-induced APP structural switching, the dynamic range of this label-free fluorescent method can be controlled, facilitating adenosine detection over broad concentration ranges.
Figure 6.
Fine-tuning of the dynamic range of adenosine detection using different lengths of abasic site-containing ssDNA.
To make sure that the effects observed were due to the abasic site, a control ssDNA L2G, which is the same with L2ab except that the abasic site is replaced by a G base, was used in the experiment under identical conditions. Such a control showed neither fluorescence quenching of ATMND with L2G-AAP in the absence of adenosine nor subsequent recovery of quenching in the presence of adenosine (Figure 4b), suggesting the essential role of the abasic site.
Conclusion
By incorporating an abasic site of dSpacer into the duplexes of either DNAzymes or aptamers, the label-free fluorescent detection of Pb2+ and adenosine with a fluorescent signal enhancement response was achieved using ATMND as an extrinsic fluorophore. Detection limits as low as 4 nM for Pb2+ and 3.4 µM for adenosine were obtained for this label-free method, which also showed good selectivity toward Pb2+ and adenosine over other divalent metal ions and nucleotides, respectively. The effect of the hybridized base-pair number on the efficiency of structural switching upon target binding with the adenosine aptamer provided the opportunity to tune the dynamic range of adenosine detection from the micromolar to the millimolar range. Since DNAzymes and aptamers specific for many other metal ions and organic molecules can be obtained through in vitro selection, and the described method is independent of exact sequences used, and the location of fluorophore binding in the DNA can be controlled through the creation of abasic sites, this label-free approach can be further extended to other DNA-based systems for detecting and quantifying a broad range of analytes.
Supplementary Material
Acknowledgement
We wish to thank the US Department of Energy (DE- FG02-08ER64568), and the National Science Foundation (Grant No. CTS-0120978 and DMI-0328162) for financial supports.
Footnotes
Supporting Information Available: Activity assays and fluorescent measurements of mutant 8–17 DNAzyme and adenosine aptamer. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Nolan EM, Lippard SJ. Chem. Rev. 2008;108:3443–3480. doi: 10.1021/cr068000q. [DOI] [PubMed] [Google Scholar]
- 2.Wong BA, Friedle S, Lippard SJ. J. Am. Chem. Soc. 2009;131:7142–7152. doi: 10.1021/ja900980u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burdette SC, Walkup GK, Spingler B, Tsien RY, Lippard SJ. J. Am. Chem. Soc. 2001;123:7831–7841. doi: 10.1021/ja010059l. [DOI] [PubMed] [Google Scholar]
- 4.Que EL, Domaille DW, Chang CJ. Chem. Rev. 2008;108:1517–1549. doi: 10.1021/cr078203u. [DOI] [PubMed] [Google Scholar]
- 5.Chang CJ, Jaworski J, Nolan EM, Sheng M, Lippard SJ. Proc. Nat. Acad. Sci. U.S.A. 2004;101:1129–1134. doi: 10.1073/pnas.0308079100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon S, Albers AE, Wong AP, Chang CJ. J. Am. Chem. Soc. 2005;127:16030–16031. doi: 10.1021/ja0557987. [DOI] [PubMed] [Google Scholar]
- 7.Chen P, He C. J. Am. Chem. Soc. 2004;126:728–729. doi: 10.1021/ja0383975. [DOI] [PubMed] [Google Scholar]
- 8.Chen PR, He C. Curr. Opin. Chem. Biol. 2008;12:214–221. doi: 10.1016/j.cbpa.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 9.Xue XJ, Wang F, Liu XG. J. Am. Chem. Soc. 2008;130:3244–3245. doi: 10.1021/ja076716c. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Cao Z, Lu Y. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Lu Y. Functional Nucleic Acids for Sensing and Other Analytical Applications. New York: Springer; 2009. [Google Scholar]
- 12.Schlosser K, Li YF. Chem. Biol. 2009;16:311–322. doi: 10.1016/j.chembiol.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Baker BR, Lai RY, Wood MS, Doctor EH, Heeger AJ, Plaxco KW. J. Am.Chem. Soc. 2006;128:3138–3139. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- 14.Swensen JS, Xiao Y, Ferguson BS, Lubin AA, Lai RY, Heeger AJ, Plaxco KW, Soh HT. J. Am. Chem. Soc. 2009;131:4262–4266. doi: 10.1021/ja806531z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao Y, Lubin AA, Heeger AJ, Plaxco KW. Angew. Chem., Int. Ed. 2005;44:5456–5459. doi: 10.1002/anie.200500989. [DOI] [PubMed] [Google Scholar]
- 16.Xiao Y, Piorek BD, Plaxco KW, Heeger AJ. J. Am. Chem. Soc. 2005;127:17990–17991. doi: 10.1021/ja056555h. [DOI] [PubMed] [Google Scholar]
- 17.Zuo XL, Xiao Y, Plaxco KW. J. Am. Chem. Soc. 2009;131:6944–6945. doi: 10.1021/ja901315w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Wang LH, Liu XF, Liang ZQ, Song SP, Li WX, Li GX, Fan CH. Adv. Mater. 2007;19:3943–3946. [Google Scholar]
- 19.Wang LH, Liu XF, Hu XF, Song SP, Fan CH. Chem. Commun. 2006:3780–3782. doi: 10.1039/b607448k. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, Wang LH, Pan D, Song SP, Boey FYC, Zhang H, Fan CH. Small. 2008;4:1196–1200. doi: 10.1002/smll.200800057. [DOI] [PubMed] [Google Scholar]
- 21.Zuo XL, Song SP, Zhang J, Pan D, Wang LH, Fan CH. J. Am. Chem. Soc. 2007;129:1042–1043. doi: 10.1021/ja067024b. [DOI] [PubMed] [Google Scholar]
- 22.Breaker RR, Joyce GF. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 23.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Lu Y. J. Am. Chem. Soc. 2000;122:10466–10467. [Google Scholar]
- 25.Cuenoud B, Szostak JW. Nature. 1995;375:611–614. doi: 10.1038/375611a0. [DOI] [PubMed] [Google Scholar]
- 26.Carmi N, Shultz LA, Breaker RR. Chem. Biol. 1996;3:1039–1046. doi: 10.1016/s1074-5521(96)90170-2. [DOI] [PubMed] [Google Scholar]
- 27.Santoro SW, Joyce GF, Sakthivel K, Gramatikova S, Barbas CF., III J. Am. Chem. Soc. 2000;122:2433–2439. doi: 10.1021/ja993688s. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Zheng WC, Kwon AH, Lu Y. Nucleic Acids Res. 2000;28:481–488. doi: 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mei SHJ, Liu Z, Brennan JD, Li Y. J. Am. Chem. Soc. 2003;125:412–420. doi: 10.1021/ja0281232. [DOI] [PubMed] [Google Scholar]
- 30.Bruesehoff PJ, Li J, Angustine AJ, Lu Y. Comb. Chem. High Throughput Screening. 2002;5:327–335. doi: 10.2174/1386207023330264. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Silverman SK. J. Am. Chem. Soc. 2003;125:6880–6881. doi: 10.1021/ja035150z. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB, Lu Y. Proc. Nat. Acad. Sci. U.S.A. 2007;104:2056–2061. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JF, Hesselberth JR, Meyers LA, Ellington AD. Nucleic Acids Res. 2004;32:D95–D100. doi: 10.1093/nar/gkh094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navani NK, Li Y. Curr. Opin. Chem. Biol. 2006;10:272–281. doi: 10.1016/j.cbpa.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Song S, Wang L, Li J, Fan C, Zhao J. TrAC, Trends Anal. Chem. 2008;27:108–117. [Google Scholar]
- 36.Lu Y. Chem. Eur. J. 2002;8:4588–4596. doi: 10.1002/1521-3765(20021018)8:20<4588::AID-CHEM4588>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 37.Rajendran M, Ellington AD. Comb. Chem. High Throughput Screening. 2002;5:263–270. doi: 10.2174/1386207023330246. [DOI] [PubMed] [Google Scholar]
- 38.Freeman R, Sharon E, Tel-Vered R, Willner I. J. Am. Chem. Soc. 2009;131:5028–5029. doi: 10.1021/ja809496n. [DOI] [PubMed] [Google Scholar]
- 39.Weizmann Y, Cheglakov Z, Willner I. J. Am. Chem. Soc. 2008;130:17224–17225. doi: 10.1021/ja806222e. [DOI] [PubMed] [Google Scholar]
- 40.Willner I, Shlyahovsky B, Zayats M, Willner B. Chem. Soc. Rev. 2008;37:1153–1165. doi: 10.1039/b718428j. [DOI] [PubMed] [Google Scholar]
- 41.Willner I, Zayats M. Angew. Chem., Int. Ed. 2007;46:6408–6418. doi: 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]
- 42.Cho EJ, Rajendran M, Ellington AD. Top. Fluoresc. Spectrosc. 2005;10:127–155. [Google Scholar]
- 43.Nutiu R, Li Y. Chem. Eur. J. 2004;10:1868–1876. doi: 10.1002/chem.200305470. [DOI] [PubMed] [Google Scholar]
- 44.Cao Z, Suljak SW, Tan W. Curr. Proteomics. 2005;2:31–40. [Google Scholar]
- 45.Liu J, Lu Y. Methods Mol. Biol. 2006;335:275–288. doi: 10.1385/1-59745-069-3:275. [DOI] [PubMed] [Google Scholar]
- 46.Liu J, Lu Y. J. Am. Chem. Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- 47.Lee JH, Wang Z, Liu J, Lu Y. J. Am. Chem. Soc. 2008;130:14217–14226. doi: 10.1021/ja803607z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, Lee JH, Lu Y. Adv. Mater. 2008;20:3263–3267. [Google Scholar]
- 49.Zhao W, Lam JC, Chiuman W, Brook MA, Li Y. Small. 2008;4:810–816. doi: 10.1002/smll.200700757. [DOI] [PubMed] [Google Scholar]
- 50.Pavlov V, Xiao Y, Shlyahovsky B, Willner I. J. Am. Chem. Soc. 2004;126:11768–11769. doi: 10.1021/ja046970u. [DOI] [PubMed] [Google Scholar]
- 51.Huang C-C, Huang Y-F, Cao Z, Tan W, Chang H-T. Anal. Chem. 2005;77:5735–5741. doi: 10.1021/ac050957q. [DOI] [PubMed] [Google Scholar]
- 52.Lee J-S, Han MS, Mirkin CA. Angew. Chem., Int. Ed. 2007;46:4093–4096. doi: 10.1002/anie.200700269. [DOI] [PubMed] [Google Scholar]
- 53.Liu J, Lu Y. Angew. Chem., Int. Ed. 2006;45:90–94. [Google Scholar]
- 54.Xiao Y, Rowe AA, Plaxco KW. J. Am. Chem. Soc. 2007;129:262–263. doi: 10.1021/ja067278x. [DOI] [PubMed] [Google Scholar]
- 55.Willner I, Zayats M. Angew. Chem., Int. Ed. 2007;46:6408–6418. doi: 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]
- 56.Wang J, Jiang Y, Zhou C, Fang X. Anal. Chem. 2005;77:3542–3546. doi: 10.1021/ac050165w. [DOI] [PubMed] [Google Scholar]
- 57.Jiang Y, Fang X, Bai C. Anal. Chem. 2004;76:5230–5235. doi: 10.1021/ac049565u. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Liu B. Analyst. 2008;133:1593–1598. doi: 10.1039/b806908e. [DOI] [PubMed] [Google Scholar]
- 59.Li B, Wei H, Dong S. Chem. Commun. 2007:73–75. doi: 10.1039/b612080f. [DOI] [PubMed] [Google Scholar]
- 60.Joseph MJ, Taylor JC, McGown LB, Pitner B, Linn CP. Biospectroscopy. 1996;2:173–183. [Google Scholar]
- 61.Babendure JR, Adams SR, Tsien RY. J. Am. Chem. Soc. 2003;125:14716–14717. doi: 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- 62.Stojanovic MN, Kolpashchikov DM. J. Am. Chem. Soc. 2004;126:9266–9270. doi: 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]
- 63.Yoshimoto K, Nishizawa S, Minagawa M, Teramae N. J. Am. Chem. Soc. 2003;125:8982–8983. doi: 10.1021/ja029786m. [DOI] [PubMed] [Google Scholar]
- 64.Sankaran NB, Nishizawa S, Seino T, Yoshimoto K, Teramae N. Angew. Chem., Int. Ed. 2006;45:1563–1568. doi: 10.1002/anie.200502979. [DOI] [PubMed] [Google Scholar]
- 65.Ihara T, Uemura A, Futamura A, Shimizu M, Baba N, Nishizawa S, Teramae N, Jyo A. J. Am. Chem. Soc. 2009;131:1386–1387. doi: 10.1021/ja809023n. [DOI] [PubMed] [Google Scholar]
- 66.Liu JW, Lu Y. Chem. Mater. 2004;16:3231–3238. [Google Scholar]
- 67.Swearingen CB, Wernette DP, Cropek DM, Lu Y, Sweedler JV, Bohn PW. Anal. Chem. 2005;77:442–448. doi: 10.1021/ac0401016. [DOI] [PubMed] [Google Scholar]
- 68.Wernette DP, Mead C, Bohn PW, Lu Y. Langmuir. 2007;23:9513–9521. doi: 10.1021/la701303k. [DOI] [PubMed] [Google Scholar]
- 69.Wernette DP, Swearingen CB, Cropek DM, Lu Y, Sweedler JV, Bohn PW. Analyst. 2006;131:41–47. doi: 10.1039/b510071b. [DOI] [PubMed] [Google Scholar]
- 70.Liu J, Lu Y. J. Am. Chem. Soc. 2005;127:12677–12683. doi: 10.1021/ja053567u. [DOI] [PubMed] [Google Scholar]
- 71.Liu J, Lu Y. Nat. Protoc. 2006;1:246–252. doi: 10.1038/nprot.2006.38. [DOI] [PubMed] [Google Scholar]
- 72.Nutiu R, Li Y. Angew. Chem., Int. Ed. 2005;44:1061–1065. doi: 10.1002/anie.200461848. [DOI] [PubMed] [Google Scholar]
- 73.Nutiu R, Li Y. J. Am. Chem. Soc. 2003;125:4771–4778. doi: 10.1021/ja028962o. [DOI] [PubMed] [Google Scholar]
- 74.Liu J, Lu Y. Anal. Chem. 2003;75:6666–6672. doi: 10.1021/ac034924r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.