Abstract
In order to analyze the structure-function of multi-subunit RNA polymerases (RNAPs), it is necessary to make site-directed mutations in key residues. Because Saccharomyces cerevisiae RNAP II is isolated as a 12 subunit enzyme that has not been amenable to in vitro reconstitution, making site-directed mutations in a particular subunit presents technical issues. In this work, we demonstrate a method to generate and purify site-directed mutants in the second largest (Rpb2) RNAP II subunit from yeast, using a tandem affinity purification tag. Mutants are analyzed for growth defects in vivo and for defects in transcriptional elongation in vitro. We show that Rpb2 R512A/C located just C-terminal to fork loop 2 (Rpb2 500 to 511) has transcriptional defects that are distinct from surrounding fork loop 2 region mutants. Rpb2 E529A/D replacements are faster and E529Q is slower than wild type RNAP II in elongation. E529 appears to form an ion pair with K987, an essential active site residue. Mutations are also analyzed within the active site region indicating key residues for catalysis and the importance of a Rpb2 R983-E1028 ion pair. Rpb2 R983Q and E1028Q are defective in escape from a transcriptional stall.
Introduction
Multi-subunit RNAPs are conserved in structure and function among eubacteria, archaea, eukarya and some viruses. Human RNAP II is a highly regulated enzyme during growth and development and a mis-regulated enzyme during viral infection and cancer. Yeast RNAP II is a close homologue of the human enzyme, and the yeast system offers favorable genetic and genomic strategies to understand aspects of transcriptional control that cannot easily be accessed using other systems.
Recently, high through-put mutagenesis strategies were developed for archaeal RNAPs (1,2). An advantage of the archaeal system is that the catalytic subunits (Rpb1 and Rpb2 in S. cerevisiae (Sc)) are in some archaea divided into smaller subunits (i.e. subunits A' and A”), and the enzyme can be reconstituted in vitro from subunits produced in bacterial expression systems. For an initial screening of many conserved amino acid replacements, therefore, the archaeal system might be favored. However, after initial screening of mutants, it is useful to reproduce the substitution in a powerful genetic system such as yeast, and it is important to analyze the enzyme using the most advanced in vitro techniques. Furthermore, some substitutions, even in conserved residues, may have different activities in divergent systems (i.e. Rpb2 R504; see below).
The active site of Sc RNAP II is comprised of the largest and second largest subunits, Rpb1 and Rpb2 (3,4). In order to generate site-directed mutants in the Rpb2 subunit, we attached a C-terminal tandem affinity purification (TAP) tag to Rpb2 (5). In this way, the mutant protein can be maintained in the untagged wild type Sc RNAP II background. The mutant protein can be selectively purified via the TAP-tag, which allows two rounds of gentle affinity purification: first with IgG binding and TEV protease elution and then with calmodulin-Ca+ binding and EGTA elution (5). In principle, even recessive lethal mutations in RNAP II with extreme defects in transcription might be recovered using this strategy. In practice, enzymes that fail to maintain viability in the absence of the wild type RNAP II appear to be quickly degraded in cells and have not been successfully purified. It is possible, that these enzymes might be recovered using mutant strains of yeast that are defective, for instance, in ubiquitinylation or degradation of RNAP II (6).
In this paper, we show production and purification of a number of site-directed mutants of Sc RNAP II. We analyze a number of amino acid replacements in the extended fork loop region (Rpb2 R504 to E529) and the active site region. We show that Rpb2 R512 is a somewhat unique position in the fork loop 2 region (Rpb2 R500 to 511) compared to surrounding residues. Rpb2 E529 appears to have an important function in regulating the active site. Results with a number of replacements in the vicinity of the active site indicate key residues for catalysis and the function of a Rpb2 R983-E1028 ion pair. Structural models indicate the functions of substituted residues.
Materials and Methods
Strategy
The idea behind the generation of RNAP II mutant proteins was to maintain yeast cells containing both an untagged wild type RNAP II (plasmid pYC2/CT (URA3) (7) borne) and TAP-tagged mutant RNAP II (plasmid pFL39 (TRP1) (8) borne) and to utilize the TAP-tag to purify the mutant protein (9). The wild type RPB2 allele was maintained on plasmid pYC2/CT under control of the galactose promoter, which is induced in the presence of galactose and repressed in the presence of glucose (see Table I for a list of plasmids used in this study). Mutant rpb2 alleles are maintained on plasmid pFL39 under RPB2 promoter control. The pYC2/CT plasmid contains the URA3 gene as a selectable marker and so can be counter-selected by addition of 5-fluoro-orotic acid to the growth medium, which causes cells maintaining the pYC2/CT plasmid to die, eliminating the wild type RPB2 allele (7,8).
Table I.
Yeast strains and plasmids used in this work.
| Strain/Plasmid | Description | Source |
|---|---|---|
| Yeast strains | ||
| YBC-1 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 | Langelier et al., 2005 |
| YBC-9 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 | Langelier et al., 2005 |
| YBC-10 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-RPB2 | This study |
| YBC-15 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-R504A | This study |
| YBC-16 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-D505A | This study |
| YBC-17 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-R512A | This study |
| YBC-18 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-Q513A | This study |
| YBC-19 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-R983Q | This study |
| YBC-20 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-E1028Q | This study |
| YBC-21 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-K510A | This study |
| YBC-23 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-R1020Q | This study |
| YBC-24 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39 | This study |
| YBC-25 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pFL39-RPB2 | This study |
| YBC-26 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pFL39-rpb2-R504A | This study |
| YBC-27 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pFL39-rpb2-D505A | This study |
| YBC-28 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pFL39-rpb2-K510A | This study |
| YBC-29 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pFL39-rpb2-Q513A | This study |
| YBC-30 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pFL39-rpb2-R983Q | This study |
| YBC-31 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pFL39-rpb2-E1028Q | This study |
| YBC-32 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-RPB2 | This study |
| YBC-33 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-R504A | This study |
| YBC-34 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-D505A | This study |
| YBC-35 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-K510A | This study |
| YBC-36 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-R512A | This study |
| YBC-37 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-Q513A | This study |
| YBC-39 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-R983Q | This study |
| YBC-40 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-R1020Q | This study |
| YBC-41 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-E1028Q | This study |
| YBC-42 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39 | This study |
| YBC-44 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-R512C | This study |
| YBC-45 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-E529A | This study |
| YBC-46 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-K987R | This study |
| YBC-47 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-K987Q | This study |
| YBC-48 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-E529D | This study |
| YBC-49 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-E529Q | This study |
| YBC-50 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-R1020K | This study |
| YBC-51 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-S1019F | This study |
| YBC-52 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-K979R | This study |
| YBC-53 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-K979Q | This study |
| YBC-54 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-P528DG530A | This study |
| YBC-55 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pYC2/CT-RPB2 pFL39-rpb2-G985AG988A | This study |
| YBC-56 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pFL39-rpb2-E529A | This study |
| YBC-57 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pFL39-rpb2-E529D | This study |
| YBC-58 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pFL39-rpb2-S1019F | This study |
| YBC-59 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pFL39-rpb2-P528DG530A | This study |
| YBC-60 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pFL39-rpb2-E529Q | This study |
| YBC-61 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pFL39-rpb2-G985AG988A | This study |
| YBC-64 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-R512C | This study |
| YBC-65 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-E529A | This study |
| YBC-66 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-E529D | This study |
| YBC-67 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-E529Q | This study |
| YBC-68 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-P528DG530A | This study |
| YBC-69 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-K979Q | This study |
| YBC-70 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-K979R | This study |
| YBC-71 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-K987Q | This study |
| YBC-72 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-K987R | This study |
| YBC-73 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-G985AG988A | This study |
| YBC-74 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-S1019F | This study |
| YBC-75 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 dst1::kanMX pYC2/CT-RPB2 pFL39-rpb2-R1020K | This study |
| YBC-76 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pFL39-rpb2-R512A | This study |
| YBC-77 | MATa can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 ade2–1 rpb2::LEU2 pFL39-rpb2-R512C | This study |
| Plasmids | ||
| pYC2/CT | URA3, ampR, CEN6, ARSH4 | Sikorski and Hieter, 1989 |
| pFL39 | TRP1, ampR, CEN6, ARS | Bonneaud etal., 1991 |
Description of expression plasmid
Cloning steps were done in E. coli XL1 blue (Stratagene). Plasmids used and constructed in this work were confirmed by DNA sequencing. The cDNA encoding yeast RPB2 was cloned with its endogenous promoter in the vector pFL39 (TRP1). Then this vector was modified to insert a copy of the TAP-tag at the C-terminus of RPB2. Creation of rpb2 mutants was done by site-directed mutagenesis with inverse PCR on the plasmid containing RPB2 TAP-tagged at the C-terminus. The primers used at this step were phosphorylated at the 5'end and held specific mutation sites.
Construction of yeast strains
The Sc strain yBC-10 (see Table I for a list of yeast strains used in this study), which conditionally expresses Rpb2 under the control of the Gal1 promoter and Rpb2 TAP-tagged under control of the Rpb2 promoter, was constructed in several steps. The strain yBC-10 was constructed from yBC-1 (10). This strain was transformed with the plasmid pFL39-RPB2-TAP (TRP1). All the other strains with mutations of rpb2 were constructed in the same way.
The strain yBC-25 was constructed from yBC-10 using a plasmid shuffling procedure based on the 5-FOAR selection. Growth on 5-fluoro-orotic acid selects for yeast lacking plasmid pYC2/CT -RPB2 (URA3). The resulting strain contains only one copy of RPB2 encoded in pFL39 with the TAP-tag. All the other strains with mutations of rpb2 were constructed in the same way.
The strain yBC-9 expresses Rpb2 under the control of the Gal1 promoter and is deleted for the chromosomal copy of the DST1 gene, encoding transcription factor IIS (TFIIS) (10). From the strain yBC-9, the strain yBC-32 was constructed by transformation with pFL39-RPB2-TAP (TRP1). Strains containing mutant rpb2-TAP alleles and Δdst1 were constructed in the same way.
All strains constructed for this work are listed in Table I. Yeast transformations were performed as previously described (11).
Isolation of mutant RNAP II
The protocol for TAP purifications was adapted from Rigaud et al. (5), with some minor modifications. The buffers used to purify TAP-tagged RNAP II contained 500 mM NaCl. After purification, proteins are dialyzed and concentrated in buffer F (10 mM Hepes pH 7.9, 100 mM NaCl, 0.1 mM EDTA pH 8.0, 20% PEG 8000, 0.5 mM DTT, 20% glycerol). For 10 grams of cells, we obtained between 15 and 35 μg of RNAP II, depending on the replacement.
Genetic analyses
Mutant rpb2 alleles were tested in vivo for the ability to maintain yeast cell viability in the absence of the wild type RPB2 allele. This was done in two different ways: 1) by curing the pYC2/CT -RPB2 plasmid by addition of 5-fluoro-orotic acid; and 2) by repressing transcription of RPB2 from the plasmid by addition of glucose to the growth medium and repression of transcription from the galactose promoter. In the first case, no wild type Rpb2 is expected to be present. In the second case, leaky expression from the galactose promoter may allow low level Rpb2 to be produced.
To indicate possible defects of mutants in RNAP II transcription, two challenges were made to the transcriptional elongation system. First, growth was compared to wild type RPB2 in the presence of glucose and 6-azauracil, a treatment that depletes cellular GTP and UTP pools (12). Second, RNAP II mutant strains were combined with a ΔTFIIS (Δdst1) strain in the presence of glucose. Many RNAP II mutant proteins are synthetic lethal in combination with the Δdst1 allele (12–14).
Transcription assays
RNAP II mutant proteins were tested for defects in transcriptional elongation in vitro. Briefly, RNA9 (5'-AUCGAGAGG-3') (also referred to as G9 TEC for 9 nucleotide TEC ending in 3'-GMP) was annealed to template strand DNA, mixed with RNAP II and combined with non-template DNA (15,16). NTP substrates were added as indicated in Figure 3. Transcriptional activities of RNAP II mutants were compared to wild type Rpb2, and are summarized in Table I. RNAP II mutant proteins were analyzed for: 1) RNA synthesis rate; and 2) the ability to recover from stalling at A20. The non-template DNA strand is designated NDS82D111G34: 5' CCT ATA GGA TAC TTA CAG CCA TCG AGA GGG ACA CGG CGA ATA GCC ATC CCA ATG CAC ACG TCC AAC GGG GCA ACC GTA TGT A 3' (RNA9 sequence is italic; DNA sequence represented in RNA is underlined). The DNA template strand is designated
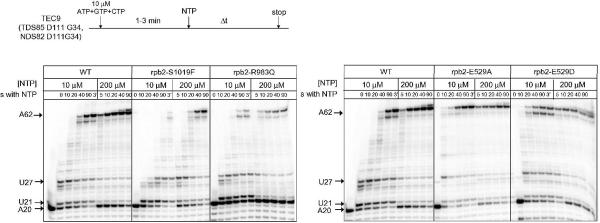
Figure 3.
RNAP II proteins were assembled into RNA9 (G9) TECs. ATP, GTP and CTP were added for 1–3 min to stall the TEC at A20, and then all 4 NTPs were supplied at 10 or 200 μM for the times shown. Persistence of the A20 TEC during the chase for Rpb2 R983Q, indicates long-term stalling or arrest at A20.
TDS85D111G34: 5' TAC ATA CGG TTG CCC CGT TGG ACG TGT GCA TTG GCA TGG CTA TTC GCC GTG TCC CTC TCG ATG GCT GTA AGT ATC CTA TAG GTG T 3' (RNA9 complement is italic; DNA sequence transcribed into RNA is underlined).
Results
Overview of mutagenesis
Many amino acid replacements have been generated in multi-subunit RNAPs using both genetic and recombinant strategies (reviewed in (17)). In the current work, a number of site-directed amino acid replacements were generated by mutation of the RPB2 gene (Table II), encoding the Rpb2 subunit of yeast RNAP II, which combines with the Rpb1 subunit to form the RNAP II active site. RNAP II is isolated as a 12 subunit enzyme (Rpb1-Rpb12). In every case, the entire rpb2 gene was sequenced to confirm that the desired mutation was made and that no secondary mutations were introduced. Mutations were constructed in an extended fork region (Rpb2 R504 to E529) and the active site region because, among other reasons, we had a particular interest in the rpb2 R512C and R983G replacements originally obtained as allele-specific suppressors of ssu72–2 (18,19). SSU72 encodes the RNAP II CTD phosphatase that removes serine 5 phosphorylation marks, placed during initiation by the TFIIH CTD kinase Kin28, from the CTD repeat YSPTSPS (26 repeats in Sc). Modeling from x-ray crystal structures was also utilized in planning mutagenesis. The main conclusions of this work are that Rpb2 R512A/C replacements are very slow in transcriptional elongation. R512 appears to have a role that is distinct from many neighboring residues in the fork loop 2 region, including R504, D505, K510, and Q513. Rpb2 E529, which extends from the fork loop 2 region toward the active site, appears to have a function in regulating elongation rate and possibly fidelity. E529A/D replacements are faster than wild type in elongation. Within the active site region, K979, K987, and R1020 appear to be important for transcription and essential for viability. An ion pair between R983 and E1028 plays a role in transcription, and S1019 adjacent to R1020 is important. Specific amino acid substitutions are discussed in more detail below.
Table II.
Sc RNAP II mutant proteins. Abbreviations: Gal (galactose), Glu (glucose), cs (cold sensitive), 6AU (6-azauracil), 5FOA (5-fluoro-orotic acid; counter-selection against URA3 plasmid carrying RPB2 wild type), KO (knockout), Purif (successful purification), Tx (transcription elongation; +++ represents wild type activity; efs (efs) indicates a strong (weak) defect in escape from a transcriptional stall).
| Sc Pol II | Tt RNAP | Region | Rationale | Gal | Glu | 5FOA | 6AU + Glu | KO TFIIS + Glu | Purif | Tx |
|---|---|---|---|---|---|---|---|---|---|---|
| Rpb2 R504A | β R420 | Fork Loop 2 | i+2 NTP interaction? | + | + | + | + | + | + | +++ |
| Rpb2 D505A | β E421 | Fork Loop 2 | i+2 NTP interaction? | + | + | +/− | + | + | + | ++ (efs) |
| Rpb2 K510A | β D426 | Fork Loop 2 | i+2 NTP interaction? | + | + | + | + | + | + | ++ |
| Rpb2 R512A | β R428 | Fork | Rpb2 R512C (forward genetics) | + | +/− cs | − cs | − | +/− | + | + |
| Rpb2 R512C* | β R428 | Fork | Rpb2 R512C (forward genetics) | + | + cs | − cs | − | +/− | + | + |
| Rpb2 Q513A | β D429 | Fork | i+2 NTP interaction? | + | + | + | + | + | + | +++ (efs) |
| Rpb2 E529A | β E445 | Fork/ Active Site | interaction with i+1 NTP? | + | + | +/− | +/− | +/− | + | ++++ |
| Rpb2 E529Q | β E445 | Fork/ Active Site | interaction with i+1 NTP? | + | + | +/− | +/− | +/− | + | ++ |
| Rpb2 E529D | β E445 | Fork/ Active Site | interaction with i+1 NTP? | + | + | + | + | + | + | ++++ (efs) |
| Rpb2 P528D G530A | β P444 G446 | Fork/ Active Site | alter flexibility of Fork | + | + | + | + | + | + | +++ |
| Rpb2 K979Q | β K838 | Active Site | i-site RNA phosphate interaction | + | − | − | − | − | n.d | n.d. |
| Rpb2 K979R | β K838 | Active Site | i-site RNA phosphate interaction | + | +/− | − | − | − | n.d | n.d. |
| Rpb2 G985A G988A | β G844 G847 | Active Site | alter flexibility of Active Site loop | + | + | + | + | + | + | +++ efs |
| Rpb2 R983Q | β R842 | Active Site | Rpb2 R983G (forward genetics) | + | + | + | +/− | + | + | ++ efs |
| Rpb2 K987Q | β K846 | Active Site | i-site RNA phosphate interaction | + | − | − | − | − | n.d | n.d. |
| Rpb2 K987R | β K846 | Active Site | i-site RNA phosphate interaction | + | − | − | − | − | n.d. | n.d. |
| Rpb2 S1019F | β S878 | Active Site | adjacent to Rpb2R1020 | + | + | + | − | + | + | + |
| Rpb2 R1020K | β R879 | Active Site | i+1 NTP γ-phosphate interaction | + | − | − | − | − | n.d. | n.d. |
| Rpb2 R1020Q | β R879 | Active Site | i+1 NTP γ-phosphate interaction | + | − | − | − | − | n.d. | n.d. |
| Rpb2 E1028Q | β E887 | Active Site | ion pair to Rpb2 R983 | + | + | +/− | + | +/− | + | +++ efs |
| wt | positive Control | + | + | + | + | + | + | +++ |
In vivo tests
A number of analyses were done to determine whether rpb2 mutant alleles would support cell viability and, therefore, RNAP II function in vivo. An example of these analyses is shown in Figure 1 and the results are summarized in Table II. Growth of a strain on galactose, in which transcription of wild type RNAP II is induced, indicates that the mutant RNAP II is not dominant lethal to wild type RNAP II, and all strains grew on galactose. Growth on glucose indicates strain viability when wild type RNAP II is limiting because of repression of the pYC2/CT plasmid borne galactose promoter driving wild type RNAP II expression. Rpb2 K979Q, K987Q/R, and R1020K are inviable in glucose medium. K979R shows very limited potential for growth. Rpb2 R512C/A substitutions do not grow well at 25° C because of the cold sensitivity of these strains. A more stringent viability test for mutant RNAP II strains is to cure the URA3 plasmid carrying the wild type RPB2 allele by counter selection with 5-fluoro-orotic acid. In addition to the strains that show defects in growth on glucose media, Rpb2 E529Q/A and E1028Q demonstrate growth defects with selection on 5-fluoro-orotic acid. Rpb2 E529D, a more conservative substitution than E529Q/A, does not show a defect with 5-fluoro-orotic acid.
Figure 1.
In vivo tests of RNAP II strains. Growth of strains on galactose, glucose, 5-fluoro-orotic acid (5FOA), 6-azauracil (6AU) and growth of strains in a Δdst1 (TFIIS knockout; KO DST1_glucose) strain.
Many RNAP II mutants with defects in transcription elongation fail to support growth in the presence of 6-azauracil, which has the effect of depleting cellular GTP and UTP pools (12). Rpb2 D505A, R512C/A, E529Q/A, R983Q, S1019F and E1028Q have growth defects when challenged with 6-azauracil. Because this test involves addition of glucose, Rpb2 K979Q/R, K987Q/R, and R1020Q/K are inviable in the presence of glucose + 6-azauracil. Because 6-azauracil sensitivity can be caused by effects that are not related to transcription, these results must be interpreted with some caution, but, because amino acid replacements are in RPB2, most likely these mutations represent transcriptional defects.
Another genetic test that may relate to transcriptional defects is to determine whether strains are viable in the absence of the gene DST1 encoding the transcriptional elongation factor TFIIS (13). TFIIS rescues arrested elongation complexes by inducing endonuclease cleavage of the nascent RNA (20–22). Domain III of TFIIS (a Zn2+ ribbon) projects through the secondary pore, a solvent accessible channel, to the RNAP II active site. A DE motif at the end of the Zn+2 ribbon is thought to hold a Mg2+ close to the RNAP II active site to participate in the cleavage reaction. Many RNAP II mutant strains with defects in elongation are inviable in the Δdst1 strain. Rpb2 R512C/A, E529A, and E1028Q have reduced viability in the Δdst1 strain. Because this test is done in glucose to suppress wild type RPB2, strains requiring RPB2 for growth are inviable in this test.
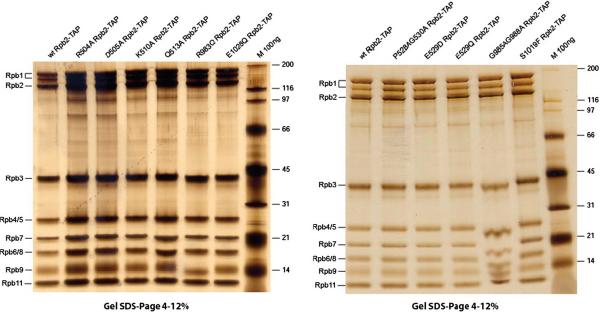
Purification of RNAP II
In Figure 2, we show the purification of a number of RNAP II mutant proteins. Using a 4–12% gradient SDS-PAGE gel, 8 of 12 RNAP II subunits are separately resolved in what appears to be approximately stoichiometric recovery. There is some evidence of proteolysis of the largest (Rpb1) subunit, because a band is detected between the Rpb1 and Rpb2-TAP bands. Probably, this band is generated by proteolysis of the carboxy terminal domain (CTD) of Rpb1, a degenerate repeat of the sequence YSPTSPS (26 repeats in Sc). The TAP-tag was engineered on the Rpb2 subunit, which carries amino acid replacements, allowing the selective purification of mutant RNAP II. From SDS-PAGE, we conclude that the proteins isolated through the TAP-tag are of high quality and should be suitable for varied in vitro analyses. The high quality of RNAP II is due to TAP purification strategy utilizing two rounds of selective affinity purification and the use of buffers with a high salt concentration.
Figure 2.
TAP-purification of RNAP II proteins. Proteins are stained with silver.
In Vitro Assay
Because many of the rpb2-TAP constructs support yeast viability in the absence of the wild type RPB2 allele, i.e. after 5-fluoro-orotic acid URA3 plasmid counter-selection, these constructs must produce active RNAP II in vivo. Purified RNAP II mutant proteins are also active in supporting transcriptional elongation in vitro, but show a wide spectrum of differences from wild type RNAP II. The assay shown in Figure 3 was done on ternary elongation complexes (TECs) assembled from a DNA template strand, a G9 complementary RNA, RNAP II, and a non-template DNA strand (15,16). For yeast RNAP II, in vitro assembly of TECs is efficient and circumvents some difficulties associated with promoter-dependent transcription. For instance, from a promoter, yeast RNAP II uses a scanning mechanism to locate a transcription start site, which may be located 70–100 nucleotides downstream from the core promoter sequence. Furthermore, a number of accessory transcription factors (i.e. TBP/TFIID, TFIIB, TFIIF, TFIIE, and TFIIH) are required for Sc RNAP II to recognize and initiate from a promoter, and these factors are omitted during TEC assembly.
The experiment in Figure 3 shows examples of transcriptional elongation assays for select mutant RNAP IIs compared to wild type. Transcriptional data are summarized in Table II. In the top panel, Rpb2 S1019F is shown to be very slow in elongation compared to wild type RNAP II. Rpb2 R983Q is slower than wild type RNAP II but more rapid in elongation than S1019F. In addition to its slow elongation in vitro, Rpb2 R983Q fails to elongate as efficiently as wild type from the A20 stall position (Figure 3, top panel). Interestingly, E1028Q and G985A, G987A, show similar defects to R983Q in escape from the A20 stall (data not shown; Table II). E1028 is in an apparent ion pair to R983, and the G985A, G987A double substitution is expected to subtly affect the conformation and/or dynamics of K987, an essential residue (Table II; Figure 1) that is close to R983. Rpb2 D505A, Q513A, and E529D have a somewhat weaker defect in the escape from a transcriptional stall at A20.
In the bottom panel of Figure 3, Rpb2 E529A and E529D are shown to be faster in elongation compared to wild type RNAP II. Other substitutions (data not shown) with notable transcription elongation defects include Rpb2 R512C/A (very slow) and E1028Q (slow). We conclude that we have predicted and produced a number of amino acid substitutions in the Rpb2 subunit of Sc RNAP II with interesting defects in transcription that are detected by both in vivo and in vitro assays.
Discussion
Using a TAP-tag on the C-terminus of the Rpb2 subunit of yeast RNAP II (9), a series of rpb2 site-directed mutant RNAP IIs has been constructed, purified and assayed. These substitutions show a wide spectrum of effects ranging from recessive lethal in vivo to apparently wild type activity in vivo and in vitro. Replacements were obtained that are faster and slower than wild type in elongation. Mutants were made in the vicinity of Rpb2 fork loop 2 region and the active site. A structural summary of substituted and potentially interacting residues is shown in Figure 4.
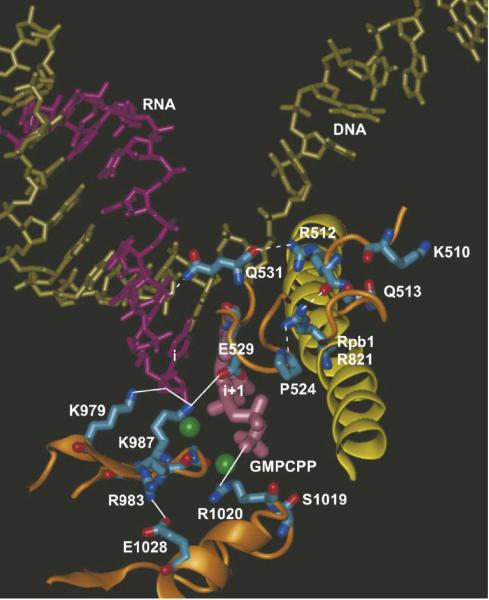
Figure 4.
Structural summary of mutational analysis. The bridge helix (Rpb1 810 to 845) is yellow; Rpb2 segments are orange; RNA is purple; the DNA template strand is tan; the i+1 active site GMPCPP is mauve; Mg2+ is green; apparent ionic interactions are indicated by solid lines; apparent hydrogen bonds are indicated by dashed lines. The image is from the Sc RNAP II TEC structure PDB 2NVT, which has an open trigger loop conformation, but contacts are very similar in 2E2H, which has a closed trigger helix (catalytic) conformation (not shown). Rpb2 R504 and D505 are not visible because they are located within a disordered segment of fork loop 2. The image was generated using Visual Molecular Dynamics (27) software.
Rpb2 R512C/A replacements, located just C-terminal to fork loop 2 (Rpb2 500 to 511) have a much more severe outcome phenotypically and transcriptionally than replacements in surrounding residues Rpb2 R504A, D505A, K510A, and Q513A. Initially, these substitutions were planned based on a model for interaction of fork loop2 region residues with a pre-loaded NTP at the i+2 position. This model is termed the NTP-driven translocation model (23). Based on differences in the activities of fork loop 2 substitutions and the R512 substitutions, however, a previously proposed structural model for i+2 NTP placement is not likely to be correct (23), whether or not NTPs load to downstream DNA template sites (24,25). Interestingly, in Archaeal RNAP, replacement of the residue homologous to Rpb2 R504 (Pyrococcus furiosus B R445) shows significant defects in a number of transcription assays (1), although no similar effects of the Rpb2 R504 substitution were noted in the current work. This comparison indicates the potential importance of testing substitution of homologous residues in more than one system. Rpb2 R512C/A replacements are very slow in elongation. We suggest that Rpb2 R512 is part of a hydrogen bonding network that may link the extended fork region to active site function. The proposed hydrogen bonding network connects Rpb2 Pro524-Rpb1 R821 (on the bridge α-helix)-Rpb2 R512-Rpb2 Q531 (Figure 4). These residues, with the exception of Q531, which is replaced by alanine in bacterial RNAP, and all of these hydrogen bonds appear to be conserved in bacterial RNAP. Rpb2 Q531 is very close to the RNA chain near the active site (i-2 phosphate oxygen) and is also very close to Rpb2 E529, in which we have constructed the E529A/Q/D substitutions.
Very interestingly, Rpb2 E529A/D replacements are significantly faster than wild type RNAP II in elongation (Figure 3). By contrast, E529Q is slightly slower than wild type in elongation. Because RNAP II mutant proteins that are fast in elongation have a tendency to be error prone (i.e. Rpb1 E1103G/A) (16), we are currently analyzing the fidelity of Rpb2 E529A/Q/D. We speculate that breaking the Rpb2 R512-Rpb2 Q531 main chain hydrogen bond through the R512C/A substitutions affects the interactions of Q531 and/or nearby E529. Another aspect of Rpb2 E529 is that it forms an apparent ion pair to Rpb2 K987, which appears to be an essential residue for catalysis (Table II; Figure 4). Breaking or weakening the Rpb2 E529-K987 connection may cause rapid elongation by the E529A/D substitutions. Additional experiments, molecular dynamics simulations and/or mutagenesis may be required to understand why E529 substitutions are faster than wild type RNAP II in elongation.
Mutations were made in highly conserved residues Rpb2 K979, K987, and R1020 within the active site region of RNAP II, at positions that potentially might be essential for catalytic function (Figure 4). Lethal substitutions included K979Q/R, K987Q/R, and R1020Q/K. As noted above, K987 may form an ion pair to E529, and E529A/D substitutions are fast in elongation (Figure 3). Although an attempt was made to engineer conservative substitutions at K979, K987, and R1020, none of these proteins was recovered, even from the wild type RNAP II background, indicating that RNAP II within inactive and/or arrested TECs or inactive pre-initiation complexes may be subject to degradation. At least one future aim of this project is to track the fate of defective or inactive RNAP II in vivo. Although the R1020Q/K substitutions could not be recovered, the adjacent Rpb2 S1019F substitution was made. This mutant is very slow in elongation and should provide a useful tool to study the mechanism of RNAP II elongation. S1019F may mis-orient R1020, potentially explaining slow elongation by S1019F.
In Escherichia coli (Ec) RNAP, substitutions at homologous positions to Sc Rpb2 K979 (Ec β K1065A/R), R983 (Ec β R1069A), and K987 (Ec β K1073A) were made (26). These substitutions are dominant lethal in vivo, consistent with our failure to recover Sc Rpb2 K979Q/R and K987Q/R proteins. The Ec RNAP mutants support transcription in vitro. Ec β K1065A/R and K1073A are slow in elongation; Ec β R1069A is more active than wild type in some transcription assays. Ec β K1065A, R1069A and K1073A are defective in readthrough of the trpL attenuator.
Because of the identification of the Rpb2 R983G mutation in a forward genetic screen (19), we became interested in an apparent ion pair linking R983 and E1028 (Figure 4). We considered it possible that the R983-E1028 ion pair, which is directed away from the active site, might link essential K979 and K987, which interact with the phosphate oxygen of the i site NMP RNA base, with R1020, an essential residue, which interacts with the γ-phosphate of the i+1 NTP (Figure 4). The R983Q substitution is somewhat slow in elongation, and R983Q and E1028Q have difficulties in escaping from a transcriptional stall at A20 (Figure 3, Table II and data not shown). There may also be evidence for transcriptional arrest and/or stalling by Rpb2 R983Q at the U21 and U27 positions, particularly at 10 μM NTPs (Figure 3). The G985A, G988A double substitution mutation located nearby is also defective in escape from A20 (data not shown; Table II). Rpb2 985-GQKG-988 forms a turn and a segment of β sheet that projects K987 toward the active site. Because the G985A, G988A double substitution, which flanks K987, shows similar effects to R983Q and E1028Q, the suggestion is strengthened that the R983-E1028 ion pair may be coupled to K987 function, as described above. Interestingly, with the exception of clustered R983Q, E1028Q, and G985A, G988A substitutions, no other RNAP II mutants that we have assayed have a strong defect in escape from the A20 stall (Figure 3 and data not shown). Rpb2 R504A, Q513A, and E529D have a much weaker defect in escaping from the A20 stall.
The Sc RNAP II system offers significant advantages and some challenges for site directed mutagenesis structure-function studies. Because yeast is an excellent genetic system, many important residues can be identified using genetic screens, and site-directed mutants with useful properties can be tested using well characterized genetic systems. Excellent x-ray crystal structures of Sc RNAP II are available to support mutagenic studies. A great advantage of the yeast system is that it allows rich in vivo and in vitro assessment of mutant proteins. In the future, the relatively small yeast genome should allow the fate of defective RNAP II proteins to be tracked in vivo on active genes. A challenge of mutagenizing RNAP II is the large size of the catalytic Rpb1 and Rpb2 subunits and the difficulties in reconstituting active enzyme from subunits. To circumvent these problems, we have used a TAP-tag on Rpb2 to purify mutant RNAP II.
Acknowledgements
This work was in part supported by the National Institutes of Health Grant GM57461 (to Z.F.B.). Z.F.B. receives support from Michigan State University, the Michigan State University Agricultural Experiment Station, and the Michigan State University College of Osteopathic Medicine. The contents of this publication do not necessarily reveal the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
Abbreviations
- RNAP II
RNA polymerase II
- Sc
Saccharomyces cerevisiae
- Ec
Escherichia coli
- TFIIS
Transcription Factor IIS
References
- 1.Naji S, Bertero MG, Spitalny P, Cramer P, Thomm M. Nucleic Acids Res. 2008;36:676–687. doi: 10.1093/nar/gkm1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan L, Wiesler S, Trzaska D, Carney HC, Weinzierl RO. J Biol. 2008;7:40. doi: 10.1186/jbiol98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westover KD, Bushnell DA, Kornberg RD. Cell. 2004;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 6.Svejstrup JQ. Trends Biochem Sci. 2007;32:165–171. doi: 10.1016/j.tibs.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Sikorski RS, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonneaud N, Ozier-Kalogeropoulos O, Li GY, Labouesse M, Minvielle-Sebastia L, Lacroute F. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 9.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 10.Langelier MF, Baali D, Trinh V, Greenblatt J, Archambault J, Coulombe B. Nucleic Acids Res. 2005;33:2629–2639. doi: 10.1093/nar/gki570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gietz RD, Woods RA. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 12.Malagon F, Kireeva ML, Shafer BK, Lubkowska L, Kashlev M, Strathern JN. Genetics. 2006;172:2201–2209. doi: 10.1534/genetics.105.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Archambault J, Lacroute F, Ruet A, Friesen JD. Molecular and cellular biology. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malagon F, Tong AH, Shafer BK, Strathern JN. Genetics. 2004;166:1215–1227. doi: 10.1534/genetics.166.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kireeva ML, Lubkowska L, Komissarova N, Kashlev M. Methods Enzymol. 2003;370:138–155. doi: 10.1016/S0076-6879(03)70012-3. [DOI] [PubMed] [Google Scholar]
- 16.Kireeva ML, Nedialkov YA, Cremona GH, Purtov YA, Lubkowska L, Malagon F, Burton ZF, Strathern JN, Kashlev M. Mol Cell. 2008;30:557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trinh V, Langelier MF, Archambault J, Coulombe B. Microbiol Mol Biol Rev. 2006;70:12–36. doi: 10.1128/MMBR.70.1.12-36.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pappas DL, Jr., Hampsey M. Molecular and cellular biology. 2000;20:8343–8351. doi: 10.1128/mcb.20.22.8343-8351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reyes-Reyes M, Hampsey M. Molecular and cellular biology. 2007;27:926–936. doi: 10.1128/MCB.01361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeon C, Yoon H, Agarwal K. Proc Natl Acad Sci U S A. 1994;91:9106–9110. doi: 10.1073/pnas.91.19.9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kettenberger H, Armache KJ, Cramer P. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 22.Kim B, Nesvizhskii AI, Rani PG, Hahn S, Aebersold R, Ranish JA. Proc Natl Acad Sci U S A. 2007;104:16068–16073. doi: 10.1073/pnas.0704573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burton ZF, Feig M, Gong XQ, Zhang C, Nedialkov YA, Xiong Y. Biochem Cell Biol. 2005;83:486–496. doi: 10.1139/o05-059. [DOI] [PubMed] [Google Scholar]
- 24.Gong XQ, Zhang C, Feig M, Burton ZF. Mol Cell. 2005;18:461–470. doi: 10.1016/j.molcel.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Xiong Y, Burton ZF. The Journal of biological chemistry. 2007;282:36582–36592. doi: 10.1074/jbc.M707014200. [DOI] [PubMed] [Google Scholar]
- 26.Sagitov V, Nikiforov V, Goldfarb A. The Journal of biological chemistry. 1993;268:2195–2202. [PubMed] [Google Scholar]
- 27.Humphrey W, Dalke A, Schulten K. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]