Abstract
Kruppel-like factor 5 (KLF5) is implicated in human breast cancer by frequent genomic deletion and expressional deregulation, but the molecular mechanisms by which KLF5 affects breast tumorigenesis are still unknown. The present study was conducted to examine whether and how KLF5 affects the function of ER in breast cancer cells. Using different cell lines, we found that restored expression of KLF5 inhibited estrogen-promoted cell proliferation in ER-positive MCF-7 and T-47D cell lines but had no effect on ER-negative SK-BR-3 cells. Transcriptional activity of ER was also suppressed by KLF5, as detected by using estrogen-stimulated ER responsive element-mediated reporter assay and expression analysis of ER target genes including c-MYC and cathepsin D (CSTD). Chromatin immunoprecipitation (ChIP) assays showed that KLF5 inhibited ERα binding to the promoter of c-myc and CSTD. Furthermore, estrogen induced an interaction between KLF5 and ERα. These results suggest that KLF5 inhibits the function of ERα in gene regulation and cell proliferation through protein interaction that interrupts the binding of ERα to target gene promoters to prevent target gene induction.
Keywords: KLF5, ER, Estrogen, breast cancer
INTRODUCTION
Human Kruppel-like factor 5 (KLF5/IKLF/BTEB2) has important roles in various aspects of carcinogenesis including cell proliferation, differentiation, cell cycle regulation, and angiogenesis 1–8. It belongs to the Sp/KLF zinc finger transcription factor family, which is comprised of over 15 mammalian members that share three C2H2-type zinc fingers at the carboxyl terminus forming the DNA-binding domain (DBD) 2, 9. Interaction between KLF5 and other transcription factors such as CBP/p300 10, retinoid acid receptor α 6 and NF-κB 11 is important in the transcriptional regulatory function of KLF5.
The role of KLF5 in carcinogenesis appears to be context-dependent. KLF5 is a positive regulator of proliferation in untransformed cells 7, 12 and even transforms normal fibroblasts 3, but it suppresses the proliferation of cancer cells from the prostate 5, breast 4 and colon 7. Moreover, KLF5 undergoes frequent genomic deletion in human cancers 4, 5, which results in a loss of function for KLF5 during carcinogenesis because KLF5 is haplo-insufficient 6. In breast cancer, while the expression of KLF5 is reduced in most cell lines, expression of KLF5 mRNA in primary tumors was reported to correlate significantly with shorter patient survival and increased expression of oncogene HER2 and proliferation marker MKI67 13. While it is clear that KLF5 plays a role in breast cancer, the molecular mechanisms remain to be elucidated.
Approximately 70% of human breast cancers express estrogen receptors (ERs) including ERα and ERβ 14. Many ERα-positive breast cancers require estrogen for cell proliferation, and they undergo apoptotic cell death when deprived of estrogen 15. Estrogen 17β-estradiol (E2) plays pivotal roles in normal breast and in the genesis and progression of breast cancer 16. The biological actions of estrogen are mediated through its binding to ERs, which belong to the nuclear receptor superfamily of transcription factors. Interestingly, KLF9 (BTEB1), which is high homologous to KLF5 (BTEB2) 9, is a negative regulator of legend-dependent ERα signaling in human Ishikawa endometrial epithelial cells 17. On the other hand, KLF5 physically interacts with nuclear receptors such as retinoic acid receptor (RAR) α to regulate the transcription of target genes 18, 19. Taken together, a relationship between KLF5 and ERα could be related to the function of KLF5 in breast cancer.
In the present study, we investigated whether and how KLF5 could function as a coregulator of ERα in ER positive breast cancer cells. We demonstrated that KLF5 inhibited ER function in cell proliferation and gene regulation, likely through estrogen-induced protein interaction with ERα.
MATERIAL AND METHODS
Cell Lines and Other Materials
The MCF 10A mammary epithelial cell line, HEK-293 human kidney epithelial cell line, and breast cancer cell line MCF-7, T47D and SK-BR-3 were purchased from the American Type Culture Collection (ATCC) (Manassas, VA) and propagated following ATCC’s instructions. MCF-7 cells and T47D cells were grown in phenol red-free medium supplemented with 10% charcoal-dextran-stripped fetal bovine serum (FBS) for at least 3 days before cells were treated with 17β-estradiol (E2).
SiRNA Transfection
The siRNA for KLF5 (5’-AAGCTCACCTGAGGACTCA-3’), as described previously 11, was used in transfection. Luciferase siRNA was used as a negative control. MCF 10A cells were transfected with 200 nM chemically synthesized siRNA (Dharmacon, Chicago, IL) using SiPORT Amine (Ambion, Austin, TX). Cells were collected 48 hours after transfection for analysis.
Cell Proliferation Assay
Cells were seeded at 1 × 105 cells/well onto 24-well tissue culture plates and grown for 24 hours. Using the Lipofectamine reagent (Invitrogen, Carlsbad, CA) (for MCF-7 and SK-BR-3 cells) or FuGENE6 reagent (Roche, Indianapolis, IN) (for T47D cells), pcDNA3 or pcDNA3-KLF5 4 was transfected into cells following the manufacturer's instructions. After 30 hours, 1 nM E2 was added to treat cells for 18 hours, at which point 3H-thymidine (MP Biomedicals, Irvine, CA) was added at 0.5 µCi/well. Four hours later, cells were washed with PBS, fixed in 5% ice-cold trichloroacetic acid for 2 hours, lysed with 1 M NaOH, transferred onto glass fiber filters, and dried. Radioactivity was measured using a scintillation counter.
Colony Formation Assay
MCF-7 cells were seeded onto six well plates at a density of 5 × 105 per well. On the following day, 2 µg of pcDNA3 or pcDNA3-KLF5 were transfected in triplicate. Thirty hours later, culture medium was replaced with medium containing G418 (800 µg/ml, Invitrogen), and thereafter the selection medium was replaced every 3 days. After 2 weeks of selection in G418, cells were fixed with 10% trichloroacetic acid and stained with sulforhodamine B. Absorbance, which indicated cell numbers, were measured following a previously described protocol 20.
Promoter-Luciferase Reporter Assay
ERE-TK-Luc 21, pcDNA3 and pcDNA3-KLF5 were transfected into MCF-7 cells using Lipofectamine reagent (Invitrogen, Carlsbad, CA), following the manufacturer's instructions. Forty hours later, 1 nM E2 was added and incubated with cells for 20 hours. Luciferase assay was carried out using the Promega luciferase assay kit as previously described 22. Three wells of cells were used for each data point.
Real-time PCR
The first-strand cDNA was synthesized from total RNA using the iScript DNA synthesis Kit (Bio-Rad Laboratories, Hercules, CA). Real-time PCR was conducted following the procedures described in our previous study 23.
Chromatin Immunoprecipitation (ChIP) Assay
MCF-7 cells were transfected with pcDNA3-KLF5 or pcDNA3.1 (Invitrogen) plasmid with Lipofectamine 2000 reagent (Invitrogen). Forty hours after transfection, cells were incubated in the presence or absence of 25 nM of E2 for 1 hour. ChIP was performed according to the protocol from Upstate (Lake Placid, NY). Antibody against ERα (Estrogen Receptor Ab-10, Thermo Fishier Scientific, Fremont, CA) was used to precipitate protein/DNA complex. Precipitated DNA was analyzed by PCR with c-Myc promoter-specific primers 5’-AGGCGCGCGTAGTTAATTCAT- 3’ (forward) and 5’-CGCCCTCTGCTTTGGGA-3’ (reverse) 24 and CSTD promoter-specific primers 5’-GGTTTCTCTGGAAGCCCTGTAG-3’ (forward) and 5’-TCCTGCACCTGCTCCTCC-3’ (reverse) 24. The procedure for the real-time PCR was described previously 25, and the same primers as those for regular PCR were used. The value for each sample, as shown in Figures 3D and 3G, was normalized by the value of respective input and that for KLF5 with E2 treatment was defined to 1.
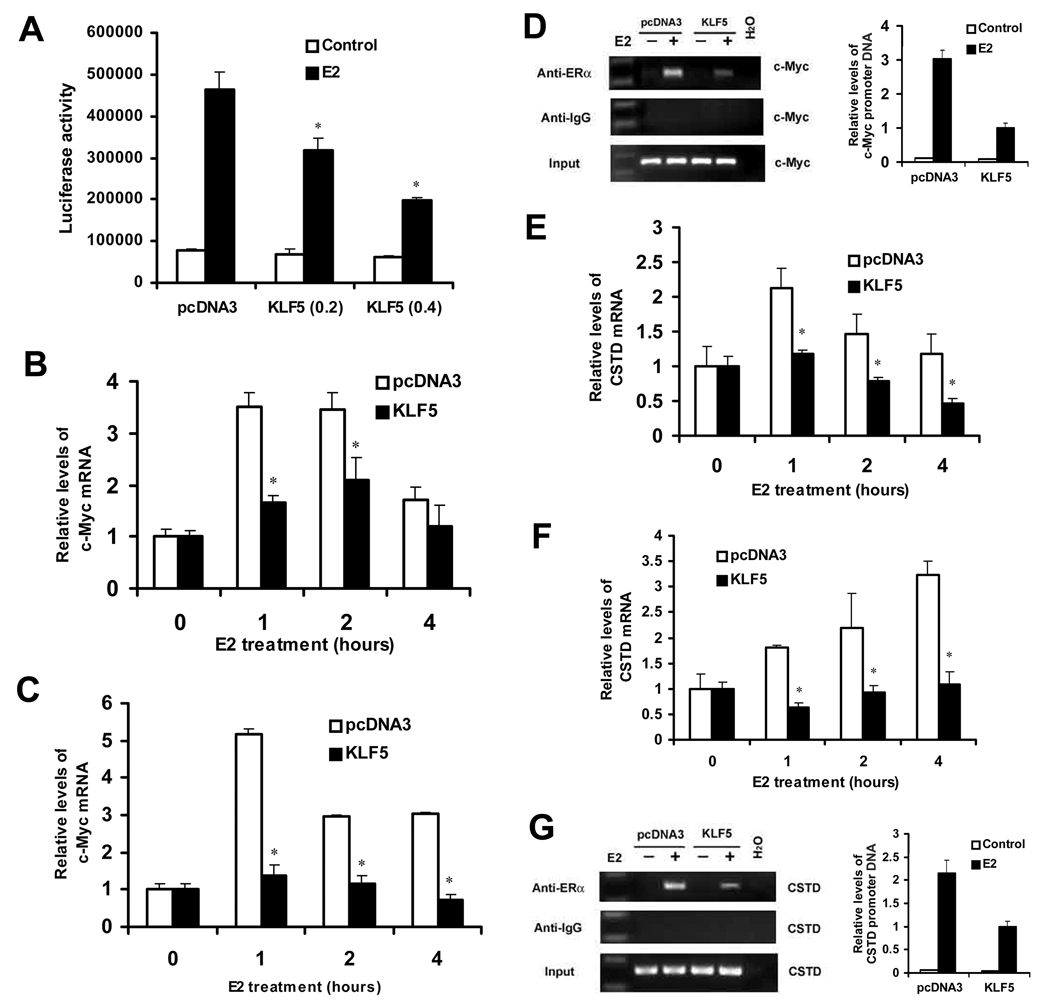
Figure 3. KLF5 inhibits E2/ERα-induced promoter activity and expression of both c-Myc and Cathepsin D (CSTD) in ER-positive breast cancer cells, apparently by reducing the binding of ER to c-Myc and CSTD promoters.
(A) Expression of KLF5 inhibits estrogen-promoted ER responsive promoter-mediated luciferase reporter activity in MCF-7 cells. E2 treatment was at 1 nM for 20 hours. Asterisks indicate statistically significant differences in luciferase activities between cells transfected with pcDNA3 and cells transfected with pcDNA3-KLF5 in the presence of estrogen (p < 0.05). (B, C) Stimulation of c-Myc expression by estrogen is suppressed by expression of KLF5 in MCF-7 (B) and T-47D (C) cells, as detected by real-time PCR. Cells were transfected with pcDNA3 or pcDNA3-KLF5, and 30 hours later were treated with 100 nM E2. (D) Binding of ERα to c-Myc promoter is reduced by KLF5, as detected by chromatin immunoprecipitation (ChIP) assay combined with regular PCR (left) and real-time PCR (right) assays in MCF-7 cells. MCF-7 cells transfected with pcDNA3-KLF5 or pcDNA3.1 were incubated in the presence or absence of 25 nM E2 for 1 hour, and antibody against ERα was used to precipitate DNA-protein complexes. Precipitated DNA was analyzed by PCR with primers specific to Myc promoter. (E, F) Measurement of Cathepsin D (CSTD) mRNA levels by real time PCR in MCF-7 (E) and T-47D (F) cells transfected with KLF5 or vector control (pcDNA3). Cells were treated with 100 nM E2. In panels B, C, E and F, asterisks indicate statistically significant differences in mRNA expression between cells transfected with pcDNA3 and cells transfected with pcDNA3-KLF5 in the presence of estrogen (P < 0.05). (G) Detection of ERα binding to CSTD promoter by ChIP assay combined with regular PCR (left) or real-time PCR (right) assays in MCF-7 cells transfected with KLF5 and vector control. The value shown in panels D and G was normalized with that of inputs, with that for the group of KLF5 plus E2 treatment defined to 1. These experiments were repeated twice and consistent results were obtained.
Co-Immunoprecipitation (co-IP) Assay
Co-IP was conducted following a standard protocol. Briefly, MCF-7 cells were transfected with pcDNA3-FLAG-KLF5 26 or pcDNA3-FLAG (Invitrogen) plasmid with Lipofectamine 2000 reagent (Invitrogen). Forty hours after transfection, cells were incubated in the presence of 10 nM MG132 for 4 hours and in the presence or absence of 25 nM of E2 for 1 hour and then lysed in lysis buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100) containing protease and phosphatase inhibitors. Cell extracts were incubated with FLAG antibody-agarose beads (Sigma), and then protein complexes were separated by SDS–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore, Billerica, MA). Specific proteins were detected by western blot analysis using antibodies against ERα (Clone 60C, Millipore, Billerica, MA).
GST Pull-down Assay
Full-length KLF5 was cloned into Hind III and BamHI sites of pGEX vector (GE Healthcare, Piscataway, NJ) to generate GST-KLF5 expression construct by PCR method with pcDNA3-FLAG-KLF5 plasmid as a template. GST-KLF5 or GST expressed in BL21 bacteria was purified with glutathione-Sepharose 4B slurry beads (GE Healthcare) following the manufacturer's instructions. Equal molar amounts of purified GST and GST fusion proteins (3.1 µg of GST or 10 µg of GST-KLF5) were immobilized to 50 µl of 50% glutathione-Sepharose 4B slurry beads (GE Healthcare) in 0.5 ml of GST pull-down buffer (10 mM HEPES, pH 7.6, 3mM MgCl2, 100 mM, KCl, 5 mM EDTA, 5% glycerol, and 0.5% CA630). After incubation for 1 hour at 4°C with rotation, the beads were washed three times with GST pull-down buffer. One nM E2 and 10 µl of 35S-labeled in vitro translated ER protein, which was synthesized as previously described 22, were added and mixed for 2 hours at 4°C with rotation. Bound proteins were eluted by boiling in 30 µl of loading buffer. 35S-labeled proteins were detected by gel electrophoresis and autoradiography. In another set of experiment, 500 pmol of double strand oligonucleotides (NWG-Biotech, High Point, NC) from the promoter of EBAG9, which contains a typical ERE, were added to the mixture of GST protein and 35S-ER. The sequences for the oligonucleotides were as follows, 5’-GGTACTGTTTCCGGGTCAGGGTGACCTCTGGG-3’ (forward) and 5’-CCCAGAGGTCACCCTGACCCGGAAACAGTACC-3’ (reverse). This pair of complementary oligos were annealed following standard protocol.
RESULTS
KLF5 inhibits estrogen-induced cell proliferation in ER-positive breast cancer cells
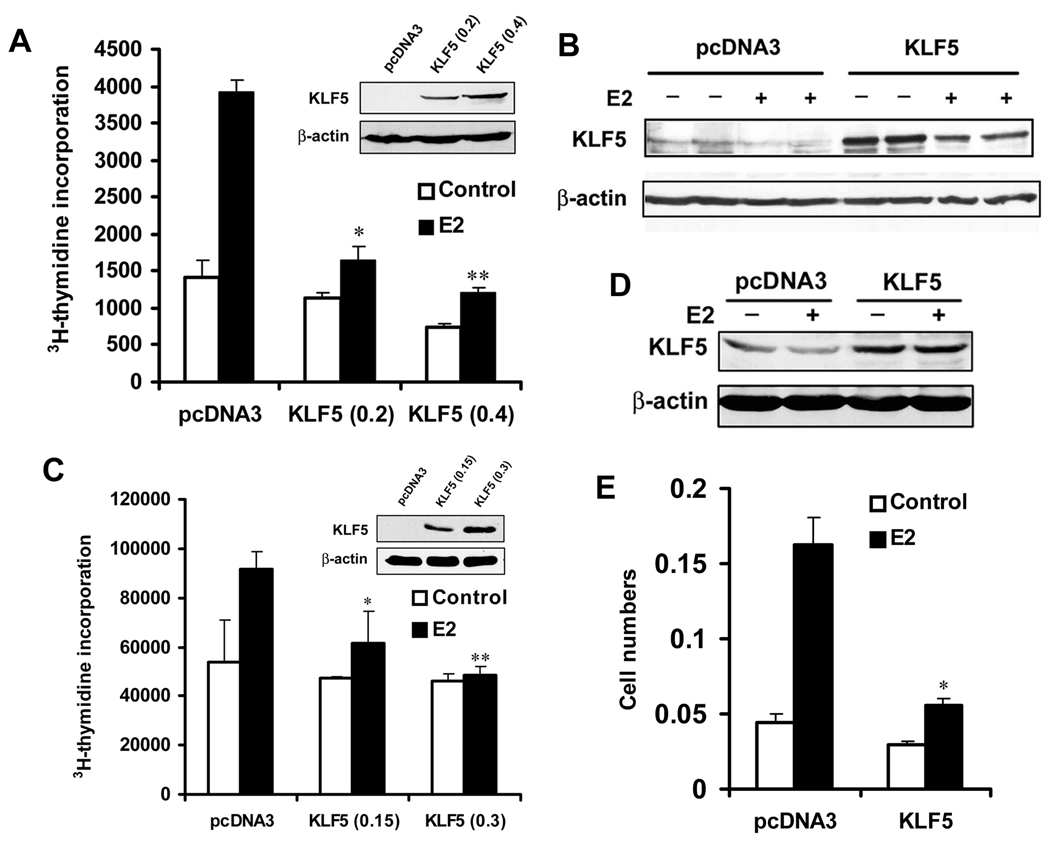
We first measured rates of DNA synthesis, as indicated by the incorporation of tritiated thymidine (3H-TdR) into DNA, for cell proliferation. In the ER-positive MCF-7 breast cancer cell line, KLF5 expression was increased by transfecting different amounts of KLF5 expression plasmid, and estrogen treatment (E2 at 1 nM) was applied for 20 hours. As shown in Figure 1A, estrogen-induced cell proliferation was inhibited by up to 69% (P < 0.01) in KLF5-expressing MCF-7 cells. In cells without E2 treatment, cell proliferation was only slightly inhibited (33%, P = 0.06). Protein expression of KLF5 in transfected MCF-7 cells was verified by western blotting (Fig. 1B). Using the same method, we also analyzed T47D cells, another ER-positive breast cancer cell line. Again, estrogen-induced cell proliferation was inhibited by KLF5 expression (Fig. 1C, 1D). We then performed a colony formation assay in MCF-7 cells, and found that estrogen-promoted cell growth was obviously suppressed by KLF5 in cells transfected with KLF5 plasmid and selected in the presence of neomycin for 16 days and E2 treatment during the last 12 days (Fig. 1E). These results indicate that KLF5 inhibits estrogen-induced cell proliferation in ER-positive breast cancer cells.
Figure 1. KLF5 inhibits estrogen-induced cell proliferation in ER-positive breast cancer cell lines.
(A–D) DNA synthesis, as indicated by the incorporation of 3H-thymidine (3H-TdR), was used to indicate cell proliferation in both MCF-7 (A) and T-47D (C) breast cancer cell lines. KLF5 expression was restored by transfecting 0.4 µg plasmid DNA including different amounts of pcDNA3-KLF5 and vector control into cells. Expression of KLF5 in transfected cells was confirmed by western blot analysis in both MCF-7 (B) and T-47D (D) cells. E2 treatment was at 1 nM for 20 hours. (E) Cell proliferation was analyzed by colony formation assay in MCF-7 cells. Cells were treated with 1 nM E2 for the last 12 days. Asterisks indicate statistically significant differences in cell growth between cells transfected with pcDNA3 and cells transfected with pcDNA3-KLF5 in the presence of estrogen (p< 0.05).
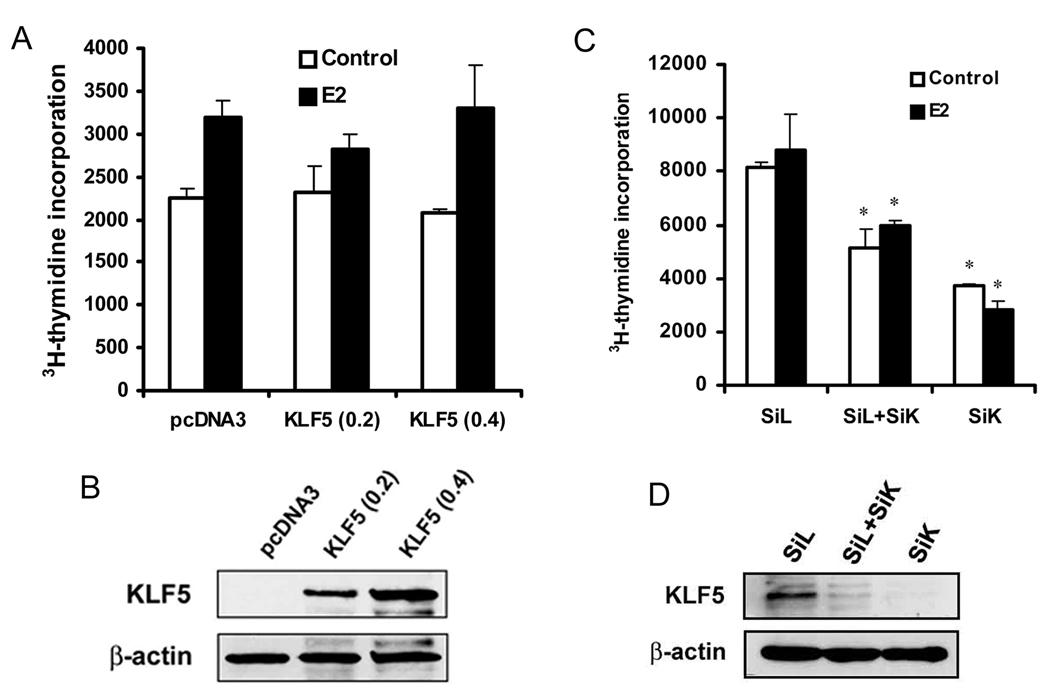
We also evaluated the effect of KLF5 on cell proliferation in ER-negative breast cancer cells. Different amounts of KLF5 plasmid were transfected into SK-BR-3 cells, which were then treated with 1 nM E2 or DMSO control for 20 hours. The purpose of E2 treatment in SK-BR-3 cells was to better compare the effect of KLF5 on E2 effect in different breast cancer cells. While E2 had little (P > 0.05) effect on cell proliferation, KLF5 had no effect regardless of E2 treatment (Fig. 2A). Protein expression of KLF5 was markedly increased by transfection, as demonstrated by western blotting (Fig. 2B). In the MCF 10A mammary epithelial cell line, which is immortalized but not transformed, KLF5 is expressed at a higher level 26 but ER is not expressed 27. To further examine the function of KLF5 in the proliferation of mammary epithelial cells, KLF5 was knocked down by siRNA in MCF 10A cells, and DNA synthesis was measured. The proliferation of MCF 10A cells was significantly suppressed by the knockdown of KLF5, which is consistent with previous findings that show a stimulatory role of KLF5 in the proliferation of epithelial cells 12, 28, 29. MCF 10A cells do not respond to estrogen treatment (Fig. 2C, D). These results indicate that KLF5 has a different effect on the proliferation of ER-negative mammary epithelial cells including cancer cells and non-tumorigenic cells.
Figure 2. KLF5 has no detectable effect on the function of estrogen in cell proliferation in ER-negative breast epithelial cells.
(A) ER-negative breast cancer cell line SK-BR-3 was subjected to 3H-TdR incorporation assay after transfection, following the same procedures as in Figure 1A. (B) Confirmation of KLF5 expression in SK-BR-3 cells after transfection, as detected by western blot analysis. (C) 3H-TdR incorporation in the MCF 10A human mammary epithelial cell line, which is immortal but non-tumorigenic, after knockdown of KLF5 by transfection of siRNA against KLF5 (SiK) or luciferase control (SiL) into cells. Total siRNA concentration was 200 nM, with 100 nM each for SiL and SiK in the group of SiL+SiK. (D) Confirmation of KLF5 knockdown in MCF 10A cells by western blot analysis. Asterisks indicate statistically significant differences in cell growth between cells transfected with negative control siRNA and cells transfected with KLF5 siRNA (P < 0.05).
KLF5 inhibits the function of ERα in gene regulation in ER-positive breast cancer cells
Based on the fact that KLF5 only inhibited estrogen-induced cell proliferation in ER positive breast cancer cells, we hypothesized that the inhibition is caused by KLF5 inhibiting the function of estrogen-ER signaling. To assess the effect of KLF5 on estrogen response, we measured the ability of KLF5 to modulate the transcriptional activity of ERα in ER-positive MCF-7 cells transiently transfected with KLF5 expression plasmid. Using an estradiol (E2)-stimulated estrogen-responsive promoter luciferase reporter, ERE-TK-Luc 21, we found that KLF5 inhibited E2-mediated reporter activity in a dose-dependent manner, whereas empty pcDNA3 plasmid had no effect (Fig. 3A). This result indicates that KLF5 suppresses the transcriptional activity of ER induced by estrogen.
Estrogen receptor regulates gene transcription either by direct binding to the promoter of a target gene such as CTSD, which encodes for cathepsin D 30, or by indirect binding through an interaction with other transcription factors including Sp1 and AP1 31. Genes regulated by indirect binding of ER include c-Myc 32, whose promoter does not contain a typical ER responsive element (ERE). We examined the effect of KLF5 on estrogen-induced expression of CSTD and c-Myc in MCF-7 and T47D cells. In MCF-7 cells, when pcDNA3 empty vector was transfected, E2 induced c-Myc mRNA expression rapidly in 1 to 2 hours, as detected by real time PCR assay (Fig. 3B). When KLF5 was transfected, however, E2-induced c-Myc transcription was significantly inhibited (Fig. 3B). E2-induced c-Myc mRNA expression was also suppressed by KLF5 in T-47D cells (Fig. 3C).
For the CSTD gene, E2 also induced mRNA expression rapidly in both MCF-7 and T-47D cells, and E2-induced CSTD expression was also significantly inhibited by re-expression of KLF5 (Fig. 3E, 3F). The results in Figure 3 demonstrate that KLF5 inhibits the function of estrogen-ER signaling in gene transcription.
KLF5 reduces the binding of ERα to promoters of target genes
To explore how KLF5 inhibits E2-induced c-Myc expression, chromatin immunoprecipitation (ChIP) assay was performed in MCF-7 cells transfected with KLF5 expression plasmid (pcDNA3-KLF5) or vector control, treated with 25 nM E2 for 1 hour. An antibody against ERα was used to precipitate the ER-DNA complex. We found that E2 induced strong recruitment of ERα to c-myc promoter, and the recruitment was decreased by KLF5 (Fig. 3D). Similarly, E2 induced strong binding of ERα to CSTD promoter, and the binding was also decreased by KLF5 (Fig. 3G).
Protein interaction between KLF5 and ERα occurs and is inducible by E2
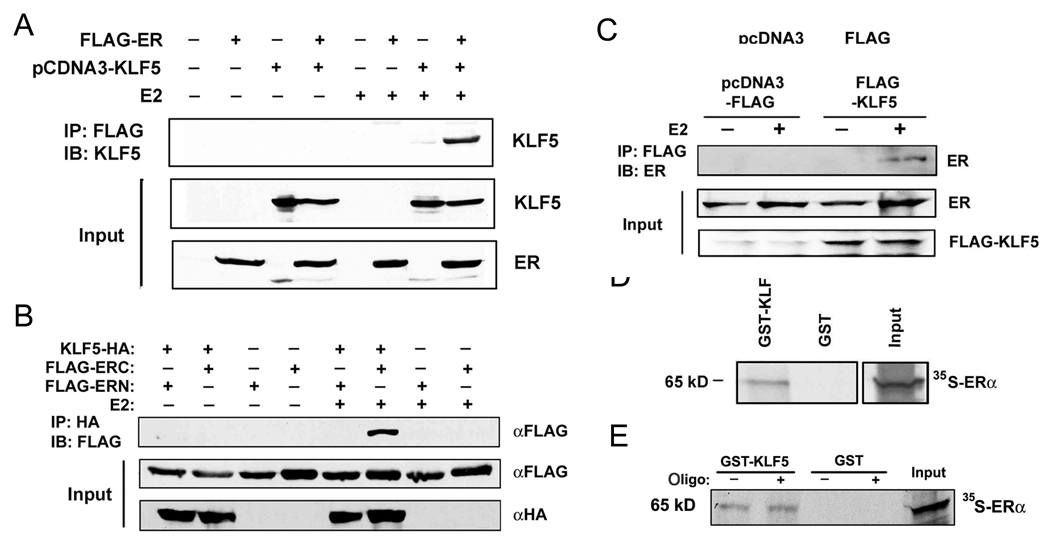
As a transcription factor, KLF5 may suppress the function of ERα through protein interaction in the nucleus. To test this hypothesis, HEK-293 cells were transfected with FLAG-tagged ERα and KLF5 expression construct, and were treated with E2 at 1 nM for 20 hours. Cell lysates were subjected to immunoprecipitation with anti-FLAG antibody and immunoblotting with antibody against KLF5. As shown in Figure 4A, KLF5 protein was only detected in cells co-transfected with FLAG-ER and KLF5 and treated with E2. This result suggests that E2 induced the interaction between ERα and KLF5. To confirm this interaction, HEK-293 cells were transfected with HA-tagged KLF5 and the FLAG-tagged N-terminal part of ER (ERN, residues 1 to 320) or C-terminal ER (ERC, residues 252 to 595), and treated with E2 at 1 nM for 20 hours. Cell lysates were subjected to immunoprecipitation with anti-HA antibody and immunoblotting with anti-FLAG antibody. We found that E2-induced protein interaction between KLF5 and ER occurred in ERC but not in ERN (Fig. 4B), which could be due to lack of estrogen binding domain in ERN. Alternatively, it is possible that ERN cannot enter the nucleus of a cell. To test whether the interaction occurs with endogenous ER, MCF-7 cells transfected with pcDNA3 empty vector or FLAG-tagged KLF5 were treated with E2 (5 nM for 1 hour) and subjected to immunoprecipitation with anti-FLAG antibody and immunoblotting with antibody against ERα. As shown in Figure 4C, interaction between KLF5 and endogenous ERα was also induced by estrogen. Furthermore, using in-vitro-translated 35S-labeled full-length ERα protein, a GST pull-down assay showed that ERα was pulled down by GST-fused KLF5 (Fig. 4D) but not GST alone, indicating that ERα can interact with KLF5 directly. To investigate whether the binding of ERα to DNA interferes with the interaction between ERα and KLF5 proteins, double strand oligonucleotides containing a typical ERE was also added to the GST pull-down reaction. The ERE DNA did not affect the interaction between ERα and KLF5 (Fig. 4E). Collectively, these results demonstrate that KLF5 directly interacts with ERα protein, and KLF5 binds to ERα through the C-terminal part of ERα.
Figure 4. Detection of protein interaction between KLF5 and ERα.
(A) E2 induces protein interaction between KLF5 and ERα in HEK-293 cells transfected with FLAG-tagged ERα and pcDNA3-KLF5 and treated with E2 (1 nM for 20 hour), as detected by immunoprecipitation (IP) with anti-FLAG antibody followed by immunoblotting (IB) with antibody against KLF5. (B) E2-induced interaction between KLF5 and ERα involves the C-terminal part of ERα. HEK-293 cells transfected with HA-tagged KLF5 and FLAG-tagged ER fragments (ERN or ERC) were treated with E2 (1 nM for 20 hour), and cell lysates were subjected to IP with anti-HA antibody and IB with anti-FLAG antibody. (C) Interaction between KLF5 and ERα also occurs with endogenous ERα. MCF-7 cells transfected with pcDNA3 empty vector or FLAG-tagged KLF5 and treated with E2 (5 nM for 1 hour) and 10 µM MG132 were subjected to IP with anti-FLAG antibody and IB with antibody against ERα. (D, E) Pull-down of in vitro translated ERα (35S-ERα) by GST-fused KLF5. GST alone serves as a negative control. Annealed double strand oligonucleotides (oligo) containing an ERE were added to the mixture of GST-KLF5 and 35S- ERα (E).
DISCUSSION
Transcriptional regulation of target genes by nuclear receptors (NRs) is a complex, multistep and tightly controlled process. One of the major breakthroughs in understanding NRs was the discovery of the interacting coregualtor proteins that can either positively (coactivitor) or negatively (corepressors) modulate NR activity 16. NR corepressors have been defined as factors that ‘interact with nuclear receptors and lower the transcriptional rate at their target genes’ 33. More than twenty molecules have been identified as ERα corepressors, including NCoR, MSRT, the repressor of ERα activity (REA), and BRCA1 34. In the present study, we found that KLF5 inhibited estrogen-induced cell proliferation and expression of ER target genes, and that KLF5 decreased the binding of ER to its target gene promoters in ER positive breast cancer cells (Fig. 1–3). Furthermore, KLF5 interacted with ER directly, and the interaction was not affected by ER-binding promoter DNA (Fig. 4). These results indicate that KLF5 is a modulator of ERα that could be recruited to the ER transcriptional complex induced by E2 to inhibit the binding of ERα to its target gene promoters, thus inhibiting the function of ERα.
As a modulator of ERα, KLF5 not only inhibits the E2-ER-ERE signaling but also suppresses estrogen-induced cell proliferation in MCF-7 and T47D cells (Fig. 1). The E2-ERα signaling induces the expression of c-Myc 32, which has an important function in promoting cell-cycle progression 35. As shown in Figure 3, KLF5 repressed c-Myc transcription induced by E2 in MCF-7 and T47D cells. In addition, KLF5 inhibited the recruitment of ERα to c-myc promoter in MCF-7 cells. These results suggest that KLF5 inhibits estrogen-induced cell proliferation through the repression of estrogen-induced transcription of ER target genes.
Estrogen and ERα have been implicated in the development of breast cancer, based on data from both clinical and animal studies. Cumulative exposure of the breast epithelium to estrogen is a risk factor associated with breast cancer 36. Transcriptional coregulators are involved in ER-dependent actions of estrogen in breast cancer 37. For example, the ERα coactivitor SRC3, which is overexpressed in some breast tumors, mediates hormone-dependent cellular proliferation in cultured cells and tumor initiation and growth in mouse models of mammary tumorigenesis 38. On the other hand, ERα corepressor BRCA1 not only inhibits ERα activity 21 but is also responsible for DNA repair 39. Therefore, loss of BRCA1 could facilitate breast tumorigenesis. Previously it was reported that KLF5 mRNA was expressed at high levels in non-neoplastic breast epithelial cells and in normal human mammary tissues but at lower levels in various breast cancer cell lines 4. Combined with our finding that KLF5 suppresses ER function in ER positive breast cancer cells, we hypothesize that loss or down-regulation of KLF5 facilitates the initiation and development of breast cancer. We are testing this hypothesis using KLF5 knockout mice.
Tong et al. reported that a higher level of KLF5 mRNA is significantly associated with poorer disease-free survival and overall survival in sporadic breast cancer 13. Moreover, KLF5 expression was significantly correlated with expression of HER2 and MK167 but not ER status. These findings may be related to the pro-proliferative and ER-independent function of KLF5 in epithelial cells. Our findings in this report may be specific to ER-positive breast cancer cells. On the other hand, KLF5 is a target of E3 ubiquitin ligases including WWP1 22, which can be amplified and overexpressed in breast cancer 40. Therefore, protein level of KLF5 maybe not correlated with mRNA level in some cancers. It should be interesting to investigate whether KLF5 protein level correlates with overall survival in breast cancer patients.
Physiologically, estrogen-ER signaling plays an essential role in the development and homeostasis of the breast by causing permanent changes in the architecture and biological characteristics of the gland 41. KLF5, on the other hand, has been implicated in the renewal and maintenance of stem/progenitor cells in recent studies 29, 42. KLF5 has also been shown to function in the regulation of epithelial proliferation and differentiation 43–45. Taken together with our results showing estrogen-caused protein degradation of KLF5 (Zhao et al., manuscript submitted), we speculate that the interaction between KLF5 and ER also plays a role in normal development and epithelial homeostasis of the breast.
In conclusion, our results suggest that KLF5 inhibits the function of ERα in gene regulation and cell proliferation through protein interaction that interrupts the binding of ERα to the promoter of its target genes and subsequent target gene induction. Consistent with frequent down-regulation and/or deletion of KLF5 in breast cancer, our findings further provide evidence that KLF5 has a tumor suppressor role in ER-positive breast cancers.
Abbreviations
- ChIP
chromatin immunoprecipitation
- CTSD
cathepsin D
- ER
estrogen receptor
- KLF5
Kruppel like factor 5
- NR
nuclear receptors
- PCR
polymerase chain reaction
- RAR
retinoid acid receptor
- SiRNA
small interfering RNA
Footnotes
In the present study, for the first time KLF5 was identified as a modulator of ERα in estrogen-induced transcription. Moreover, the findings provide evidence that KLF5 could have a tumor suppressor role in ER-positive breast cancer.
REFERENCES
- 1.Dang DT, Bachman KE, Mahatan CS, Dang LH, Giardiello FM, Yang VW. Decreased expression of the gut-enriched Kruppel-like factor gene in intestinal adenomas of multiple intestinal neoplasia mice and in colonic adenomas of familial adenomatous polyposis patients. FEBS letters. 2000;476:203–207. doi: 10.1016/s0014-5793(00)01727-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. Journal of cellular physiology. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 3.Sun R, Chen X, Yang VW. Intestinal-enriched Kruppel-like factor (Kruppel-like factor 5) is a positive regulator of cellular proliferation. J Biol Chem. 2001;276:6897–6900. doi: 10.1074/jbc.C000870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C, Bhalala HV, Qiao H, Dong JT. A possible tumor suppressor role of the KLF5 transcription factor in human breast cancer. Oncogene. 2002;21:6567–6572. doi: 10.1038/sj.onc.1205817. [DOI] [PubMed] [Google Scholar]
- 5.Chen C, Bhalala HV, Vessella RL, Dong JT. KLF5 is frequently deleted and down-regulated but rarely mutated in prostate cancer. The Prostate. 2003;55:81–88. doi: 10.1002/pros.10205. [DOI] [PubMed] [Google Scholar]
- 6.Shindo T, Manabe I, Fukushima Y, Tobe K, Aizawa K, Miyamoto S, Kawai-Kowase K, Moriyama N, Imai Y, Kawakami H, Nishimatsu H, Ishikawa T, et al. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nature medicine. 2002;8:856–863. doi: 10.1038/nm738. [DOI] [PubMed] [Google Scholar]
- 7.Bateman NW, Tan D, Pestell RG, Black JD, Black AR. Intestinal tumor progression is associated with altered function of KLF5. J Biol Chem. 2004;279:12093–12101. doi: 10.1074/jbc.M311532200. [DOI] [PubMed] [Google Scholar]
- 8.Nandan MO, Yoon HS, Zhao W, Ouko LA, Chanchevalap S, Yang VW. Kruppel-like factor 5 mediates the transforming activity of oncogenic H-Ras. Oncogene. 2004;23:3404–3413. doi: 10.1038/sj.onc.1207397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bieker JJ. Krüppel-like factors: fhree fingers in many pies. J. Biol. Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 10.Miyamoto S, Suzuki T, Muto S, Aizawa K, Kimura A, Mizuno Y, Nagino T, Imai Y, Adachi N, Horikoshi M, Nagai R. Positive and negative regulation of the cardiovascular transcription factor KLF5 by p300 and the oncogenic regulator SET through interaction and acetylation on the DNA-binding domain. Mol. Cell. Biol. 2003;23:8528–8541. doi: 10.1128/MCB.23.23.8528-8541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aizawa K, Suzuki T, Kada N, Ishihara A, Kawai-Kowase K, Matsumura T, Sasaki K, Munemasa Y, Manabe I, Kurabayashi M, Collins T, Nagai R. Regulation of platelet-derived growth factor-A chain by Kruppel-like factor 5: new pathway of cooperative activation with nuclear factor-kappaB. J. Biol. Chem. 2004;279:70–76. doi: 10.1074/jbc.M306621200. [DOI] [PubMed] [Google Scholar]
- 12.Chanchevalap S, Nandan MO, Merlin D, Yang VW. All-trans retinoic acid inhibits proliferation of intestinal epithelial cells by inhibiting expression of the gene encoding Kruppel-like factor 5. FEBS Lett. 2004;578:99–105. doi: 10.1016/j.febslet.2004.10.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong D, Czerwenka K, Heinze G, Ryffel M, Schuster E, Witt A, Leodolter S, Zeillinger R. Expression of KLF5 is a prognostic factor for disease-free survival and overall survival in patients with breast cancer. Clin Cancer Res. 2006;12:2442–2448. doi: 10.1158/1078-0432.CCR-05-0964. [DOI] [PubMed] [Google Scholar]
- 14.Johnston SR, Dowsett M. Aromatase inhibitors for breast cancer: lessons from the laboratory. Nat Rev Cancer. 2003;3:821–831. doi: 10.1038/nrc1211. [DOI] [PubMed] [Google Scholar]
- 15.Thiantanawat A, Long BJ, Brodie AM. Signaling pathways of apoptosis activated by aromatase inhibitors and antiestrogens. Cancer research. 2003;63:8037–8050. [PubMed] [Google Scholar]
- 16.McDonnell DP, Norris JD. Connections and regulation of the human estrogen receptor. Science. 2002;296:1642–1644. doi: 10.1126/science.1071884. [DOI] [PubMed] [Google Scholar]
- 17.Velarde MC, Zeng Z, McQuown JR, Simmen FA, Simmen RC. Molecular endocrinology (Baltimore, Md. Vol. 21. 2007. Kruppel-like factor 9 is a negative regulator of ligand-dependent estrogen receptor alpha signaling in Ishikawa endometrial adenocarcinoma cells; pp. 2988–3001. [DOI] [PubMed] [Google Scholar]
- 18.Kada N, Suzuki T, Aizawa K, Munemasa Y, Matsumura T, Sawaki D, Nagai R. Acyclic retinoid inhibits functional interaction of transcription factors Kruppel-like factor 5 and retinoic acid receptor-alpha. FEBS letters. 2008;582:1755–1760. doi: 10.1016/j.febslet.2008.04.040. [DOI] [PubMed] [Google Scholar]
- 19.Fujiu K, Manabe I, Ishihara A, Oishi Y, Iwata H, Nishimura G, Shindo T, Maemura K, Kagechika H, Shudo K, Nagai R. Synthetic retinoid Am80 suppresses smooth muscle phenotypic modulation and in-stent neointima formation by inhibiting KLF5. Circulation research. 2005;97:1132–1141. doi: 10.1161/01.RES.0000190613.22565.13. [DOI] [PubMed] [Google Scholar]
- 20.Sun X, Frierson HF, Chen C, Li C, Ran Q, Otto KB, Cantarel BL, Vessella RL, Gao AC, Petros J, Miura Y, Simons JW, et al. Frequent somatic mutations of the transcription factor ATBF1 in human prostate cancer. Nature genetics. 2005;37:407–412. doi: 10.1038/ng1528. [DOI] [PubMed] [Google Scholar]
- 21.Fan S, Wang J, Yuan R, Ma Y, Meng Q, Erdos MR, Pestell RG, Yuan F, Auborn KJ, Goldberg ID, Rosen EM. BRCA1 inhibition of estrogen receptor signaling in transfected cells. Science. 1999;284:1354–1356. doi: 10.1126/science.284.5418.1354. [DOI] [PubMed] [Google Scholar]
- 22.Chen C, Sun X, Guo P, Dong XY, Sethi P, Cheng X, Zhou J, Ling J, Simons JW, Lingrel JB, Dong JT. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J. Biol. Chem. 2005;280:41553–41561. doi: 10.1074/jbc.M506183200. [DOI] [PubMed] [Google Scholar]
- 23.Dong XY, Chen C, Sun X, Guo P, Vessella RL, Wang RX, Chung LW, Zhou W, Dong JT. FOXO1A is a candidate for the 13q14 tumor suppressor gene inhibiting AR signaling in prostate cancer. Cancer research. 2006;66:6998–7006. doi: 10.1158/0008-5472.CAN-06-0411. [DOI] [PubMed] [Google Scholar]
- 24.Shang Y, Brown M. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295:2465–2468. doi: 10.1126/science.1068537. [DOI] [PubMed] [Google Scholar]
- 25.Chen C, Sun X, Guo P, Dong XY, Sethi P, Cheng X, Zhou J, Ling J, Simons JW, Lingrel JB, Dong JT. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J Biol Chem. 2005;280:41553–41561. doi: 10.1074/jbc.M506183200. [DOI] [PubMed] [Google Scholar]
- 26.Chen C, Sun X, Ran Q, Wilkinson KD, Murphy TJ, Simons JW, Dong JT. Ubiquitin-proteasome degradation of KLF5 transcription factor in cancer and untransformed epithelial cells. Oncogene. 2005;24:3319–3327. doi: 10.1038/sj.onc.1208497. [DOI] [PubMed] [Google Scholar]
- 27.Lane MA, Romagnoli L, Cruise B, Cohn GM. Spontaneous conversion to estrogen receptor expression by the human breast epithelial cell line, MCF-10A. Oncology reports. 1999;6:507–511. doi: 10.3892/or.6.3.507. [DOI] [PubMed] [Google Scholar]
- 28.Bateman NW, Tan D, Pestell RG, Black JD, Black AR. Intestinal tumor progression is associated with altered function of KLF5. J. Biol. Chem. 2004;279:12093–12101. doi: 10.1074/jbc.M311532200. [DOI] [PubMed] [Google Scholar]
- 29.Guo P, Dong XY, Zhang X, Zhao KW, Sun X, Li Q, Dong JT. Pro-proliferative Factor KLF5 Becomes Anti-proliferative in Epithelial Homeostasis upon Signaling-mediated Modification. J Biol Chem. 2009;284:6071–6078. doi: 10.1074/jbc.M806270200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Augereau P, Miralles F, Cavailles V, Gaudelet C, Parker M, Rochefort H. Characterization of the proximal estrogen-responsive element of human cathepsin D gene. Molecular endocrinology(Baltimore, Md. 1994;8:693–703. doi: 10.1210/mend.8.6.7935485. [DOI] [PubMed] [Google Scholar]
- 31.Saville B, Wormke M, Wang F, Nguyen T, Enmark E, Kuiper G, Gustafsson JA, Safe S. Ligand-, cell-, and estrogen receptor subtype (alpha/beta)-dependent activation at GC-rich (Sp1) promoter elements. J Biol Chem. 2000;275:5379–5387. doi: 10.1074/jbc.275.8.5379. [DOI] [PubMed] [Google Scholar]
- 32.Dubik D, Shiu RP. Mechanism of estrogen activation of c-myc oncogene expression. Oncogene. 1992;7:1587–1594. [PubMed] [Google Scholar]
- 33.McKenna NJ, Lanz RB, O'Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocrine reviews. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 34.Dobrzycka KM, Townson SM, Jiang S, Oesterreich S. Estrogen receptor corepressors -- a role in human breast cancer? Endocrine-related cancer. 2003;10:517–536. doi: 10.1677/erc.0.0100517. [DOI] [PubMed] [Google Scholar]
- 35.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 36.Henderson BE, Feigelson HS. Hormonal carcinogenesis. Carcinogenesis. 2000;21:427–433. doi: 10.1093/carcin/21.3.427. [DOI] [PubMed] [Google Scholar]
- 37.Xu J, Li Q. Review of the in vivo functions of the p160 steroid receptor coactivator family. Molecular endocrinology(Baltimore, Md. 2003;17:1681–1692. doi: 10.1210/me.2003-0116. [DOI] [PubMed] [Google Scholar]
- 38.Liao L, Kuang SQ, Yuan Y, Gonzalez SM, O'Malley BW, Xu J. Molecular structure and biological function of the cancer-amplified nuclear receptor coactivator SRC-3/AIB1. The Journal of steroid biochemistry and molecular biology. 2002;83:3–14. doi: 10.1016/s0960-0760(02)00254-6. [DOI] [PubMed] [Google Scholar]
- 39.Welcsh PL, King MC. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum Mol Genet. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- 40.Chen C, Sun X, Guo P, Dong XY, Sethi P, Zhou W, Zhou Z, Petros J, Frierson HF, Jr, Vessella RL, Atfi A, Dong JT. Ubiquitin E3 ligase WWP1 as an oncogenic factor in human prostate cancer. Oncogene. 2007;26:2386–2394. doi: 10.1038/sj.onc.1210021. [DOI] [PubMed] [Google Scholar]
- 41.Russo J, Hasan Lareef M, Balogh G, Guo S, Russo IH. Estrogen and its metabolites are carcinogenic agents in human breast epithelial cells. The Journal of steroid biochemistry and molecular biology. 2003;87:1–25. doi: 10.1016/s0960-0760(03)00390-x. [DOI] [PubMed] [Google Scholar]
- 42.Ema M, Mori D, Niwa H, Hasegawa Y, Yamanaka Y, Hitoshi S, Mimura J, Kawabe Y, Hosoya T, Morita M, Shimosato D, Uchida K, et al. Kruppel-like factor 5 is essential for blastocyst development and the normal self-renewal of mouse ESCs. Cell stem cell. 2008;3:555–567. doi: 10.1016/j.stem.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Sur I, Rozell B, Jaks V, Bergstrom A, Toftgard R. Epidermal and craniofacial defects in mice overexpressing Klf5 in the basal layer of the epidermis. J. Cell Sci. 2006;119:3593–3601. doi: 10.1242/jcs.03070. [DOI] [PubMed] [Google Scholar]
- 44.Guo P, Dong XY, Zhang X, Zhao KW, Sun X, Li Q, Dong JT. Pro-proliferative factor KLF5 becomes anti-proliferative in epithelial homeostasis upon signaling-mediated modification. J. Biol. Chem. 2009 doi: 10.1074/jbc.M806270200. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wan H, Luo F, Wert SE, Zhang L, Xu Y, Ikegami M, Maeda Y, Bell SM, Whitsett JA. Kruppel-like factor 5 is required for perinatal lung morphogenesis and function. Development. 2008;135:2563–2572. doi: 10.1242/dev.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]