Abstract
DNA methylation is an epigenetic mark essential for mammalian development, genomic stability, and imprinting. DNA methylation patterns are established and maintained by three DNA methyltransferases: DNMT1, DNMT3A, and DNMT3B. Interestingly, all three DNMTs make use of alternative splicing. DNMT3B has nearly 40 known splice variants expressed in a tissue- and disease-specific manner, but very little is known about the role of these splice variants in modulating DNMT3B function. We describe here the identification and characterization of a novel alternatively spliced form of DNMT3B lacking exon 5 within the N-terminal regulatory domain. This variant, which we term DNMT3B3Δ5 because it is closely related in structure to the ubiquitously expressed DNMT3B3 isoform, is highly expressed in pluripotent cells and brain tissue, is downregulated during differentiation, and is conserved in the mouse. Creation of pluripotent iPS cells from fibroblasts results in marked induction of DNMT3B3Δ5. DNMT3B3Δ5 expression is also altered in human disease, with tumor cell lines displaying elevated or reduced expression depending on their tissue of origin. We then compared the DNA binding and subcellular localization of DNMT3B3Δ5 versus DNMT3B3, revealing that DNMT3B3Δ5 possessed significantly enhanced DNA binding affinity and displayed an altered nuclear distribution. Finally, ectopic overexpression of DNMT3B3Δ5 resulted in repetitive element hypomethylation and enhanced cell growth in a colony formation assay. Taken together, these results demonstrate that DNMT3B3Δ5 may play an important role in stem cell maintenance or differentiation and suggest that sequences encoded by exon 5 influence the functional properties of DNMT3B.
INTRODUCTION
DNA methylation, occurring at the C-5 position of cytosine within the CpG dinucleotide, represents a heritable mark of transcriptional repression important for regulating chromatin structure, genome stability, and genomic imprinting in mammals (1). Genomic methylation patterns are catalyzed by a family of three DNA methyltransferase (DNMTs), DNMT1, DNMT3A, and DNMT3B. In addition, a co-regulatory methyltransferase-like protein, DNMT3L, modulates activity and targeting of DNMT3A and DNMT3B (2). In general, DNMT1 acts predominantly as a maintenance methyltransferase, copying DNA methylation patterns during DNA replication from the parental to the newly synthesized daughter strand (3). DNMT3A and DNMT3B are critical for de novo DNA methylation during embryogenesis and germ cell development (4). The DNMT3s also play a role in maintenance DNA methylation, suggesting both overlapping and unique functions for each DNMT family member (5, 6). Dnmt3L resides in a complex with Dnmt3a and Dnmt3b in murine embryonic stem (ES) cells and the N-terminal domain of Dnmt3L interacts with the histone H3 tail only when unmethylated at the lysine 4 (K4) position (7).
Dnmt1 and Dnmt3b are essential for murine embryonic development, while Dnmt3a-deficient mice survive for several weeks after birth (4). Humans with hypomorphic mutations in DNMT3B display immunodeficiency, centromere instability, facial anomalies (ICF) syndrome, characterized by hypomethylation of the long satellite repeat tracks (satellite 2 and 3 repeats) within pericentromeric heterochromatin primarily affecting chromosomes 1, 9, and 16. ICF patients also display immune deficiencies, facial abnormalities, and a variable degree of mental retardation and developmental delay (4, 8, 9). While critical for mammalian development, properly regulated DNA methylation patterns are also required for normal cell growth control. For example, alterations in genome-wide patterns of DNA methylation are a hallmark of all transformed cells. Tumor cells display a global loss of DNA methylation from the repetitive fraction of the genome and less frequently from single copy genes. This contributes to aberrant gene expression and reduced genomic stability. In addition, CpG islands associated with tumor suppressor genes, which are normally unmethylated, acquire aberrant DNA hypermethylation resulting in potent long-term gene repression and loss of cell growth control (1). The exact mechanism by which cells establish normal DNA methylation patterns, and how these patterns become disrupted in cancer, remain largely unknown.
DNMT3B is of interest not only because of its important role in development, but also because of its strong links to cancer. Structurally, the N-terminal region of DNMT3B contains a PWWP domain (encoded by exons 6–7) and a plant homeodomain (PHD, encoded by exons 12–13). The PWWP domain is involved in recruiting DNMT3B to pericentromeric heterochromatin and non-specific DNA binding, while the PHD domain mediates protein-protein interactions and the ability of DNMT3B to repress transcription in a histone-deacetylase (HDAC)-dependent manner (10–12). DNMT3B is aberrantly over-expressed in many tumor types (13–15) and antisense-mediated depletion of DNMT3B results in apoptosis and demethylation of aberrantly methylated tumor suppressor genes in cancer cells (16). By overexpressing Dnmt3b in the ApcMin/+ murine colon cancer mouse model, Linhart et al. recently demonstrated that Dnmt3b contributes directly to aberrant methylation in cancer. Dnmt3b1 expression resulted in elevated numbers of tumors and an increase in the average size of colonic microadenomas (17).
Alternative splicing is a complex and highly regulated process, which, like genomic DNA methylation patterns, is frequently disrupted in cancer. At least 15% of all disease-causing single nucleotide base changes affect splicing. Alternative splicing is thought to occur in at least 70% of all genes and may be regulated differentially during development and in a tissue-specific manner. The four main types of alternative splicing events include exon skipping, alternative splice donor and acceptor sites, mutually exclusive exon usage, and intron retention. Exon skipping is most common in normal tissues, whereas intron retention, which may lead to reading frame shifts and premature termination, is more common in cancer cells (18). Interestingly, all three catalytically active DNA methyltransferases are subject to tissue-or developmental-stage specific alternative splicing. DNMT3B, however, has emerged as the one having the most alternatively spliced isoforms by far. Nearly 40 different DNMT3B isoforms generated by alternative splicing and/or alternative promoter usage have been reported (19–21). In normal tissues, for example, human testis expresses two additional isoforms, termed DNMT3B4 and DNMT3B5, which, due to alternative splicing, result in truncated and presumably inactive proteins. Interestingly, a DNMT3B splice variant arising from skipping of exons 21 and 22 within the C-terminal catalytic domain (DNMT3B3), also resulting in an inactive enzyme, is one of the most widely expressed forms of DNMT3B in somatic cells (14).
In the present study we sought to better understand a novel DNMT3B3-like splice variant lacking exon 5 immediately adjacent to the PWWP domain, which we identified and have termed DNMT3B3Δ5. This variant was initially isolated from a pluripotent embryonic carcinoma (EC) cell line and represented a significant fraction of the total DNMT3B transcripts. Consistent with its association with pluripotency, we detected DNMT3B3Δ5 expression in ES cells, neural stem cells, and induced pluripotent stem (iPS) cells. DNMT3B3Δ5 was also highly expressed in fetal and adult brain but present at low levels in most other somatic tissues. Expression of DNMT3B transcripts lacking exon 5 was elevated in tumor cell lines, particularly those derived from the liver and skin, but decreased in those derived from glia (glioma), breast, and colon. Functionally, purified recombinant DNMT3B3Δ5 displayed increased DNA binding affinity and when ectopically expressed in vivo, altered subcellular localization compared to DNMT3B3. Ectopic expression of DNMT3B3Δ5 in colon cancer cells resulted in hypomethylation of centromeric and pericentromeric repeats and elevated colony formation efficiency. These results therefore suggest that the exon 5 region of DNMT3B plays an important role in stem cell function and differentiation and that alternative splicing of DNMT3B generates proteins with distinct functions in vivo.
RESULTS
Identification and cloning of a novel DNMT3B splice variant in human cells
While cloning full-length DNMT3B and its known splice variants from human embryonic carcinoma (EC) cells and fetal brain tissue by PCR (which were used because both are known to express high levels of DNMT3B), we noted two slightly smaller forms consistently amplifying (Figs. 1A–1B). We cloned and sequenced these and, upon alignment to the sequence of full-length DNMT3B, noted that these two isoforms were related to DNMT3B3 (a commonly expressed splice variant lacking exons 21 and 22 (14)), but lacked exon 5 or both exons 4 and 5 (Figs. 1A–1B (upper panel), Online Supplemental Fig. S1). We have termed these two variants DNMT3B3Δ5 and DNMT3B3Δ(4+5), respectively. We also use ‘DNMT3BΔ5’ or ‘DNMT3BΔ(4+5)’ for situations where the splicing in other regions of DNMT3B is not determined since we cannot exclude the possibility that exon 4-exon 5 alternative splicing also occurs in the context of other DNMT3B isoforms. The fusion of exon 4 to exon 6 in DNMT3BΔ5 preserves the normal DNMT3B open reading frame. Ectopic expression of DNMT3B3Δ5 upon transient transfection of HCT116 cells confirmed that alternative splicing yields an in-frame product of the expected size that appeared as stable as other DNMT3B isoforms (Fig. 1B, lower panel). Exons 4 and 5 are immediately N-terminal to the PWWP domain, which is encoded by sequence in exons 6 and 7 (Fig. 1A and Fig S1). This region also has relatively little homology with DNMT3A (not shown). Since DNMT3BΔ(4+5) was detectable only in human fetal brain (Fig. 1B, and data not shown), we focused our remaining studies on DNMT3BΔ5.
Fig. 1.
Identification of a novel alternative splice variant in the human DNMT3B locus whose expression correlates with pluripotency. (A) Schematic showing the splicing structure of the human DNMT3B gene with 23 exons. Translation begins in exon 2 (open boxes are untranslated exons). The alternatively spliced regions examined in the paper are denoted below this with bold arrows. A blowup of the exon 2–7 region of DNMT3B shows the location of the PWWP domain, the primers used in semi-quantitative RT-PCR (bold arrows below exons) and quantitative RT-PCR (bold arrows above exons). ‘*’ putative sumoylation site. (B) Representative ethidium bromide stained agarose gel photo showing expression of the newly identified splice variants DNMT3BΔ5 and DNMT3BΔ(4+5). Ectopic expression of DNMT3B3Δ5 in HCT116 cells yields a stable protein product of the expected molecular weight. DNMT3B1, DNMT3B3, and DNMT3B3Δ5 were expressed in HCT116 cells following transient transfection and detected with the Express antibody epitope tag (right). ‘*’ non-specific amplification product confirmed by DNA sequencing. (C) DNMT3BΔ5 expression is regulated during differentiation. Expression of DNMT3B isoforms during differentiation of NCCIT EC cells over twenty days with retinoic acid, relative to GAPDH expression. (D) Quantitative RT-PCR analysis of DNMT3B1-6 and DNMT3BΔ5 expression, relative to GAPDH, in human pluripotent cell lines (left panel), two neural stem cell lines (SCP’s) and one glioma tumor initiating cell line (H1228, middle panel), and the relative expression for each of the cell lines (right panel). ‘hES’ human ES cells.
DNMT3BΔ5 expression is dynamically regulated during stem cell differentiation
Given the association of DNMT3BΔ5 expression with pluripotent EC cells, where it was first identified, we sought to investigate its expression during differentiation in several human model systems. We therefore designed both semi-quantitative (located in exon 3 and exon 6) and quantitative real-time RT-PCR primers (which specifically recognize the DNMT3BΔ5 isoform by having one primer span the unique exon 4-exon 6 junction) to examine expression of DNMT3BΔ5. Moderate to high-level expression of DNMT3BΔ5 was detected not only in human EC cells prior to differentiation, but also a human embryonic stem cell line and two human neural stem cell (Figs. 1C–1D). To study this further, we differentiated NCCIT EC cells with retinoic acid for 20 days and monitored expression of DNMT3B. DNMT3BΔ5 expression increased up to day 10, relative to exon 5-containing transcripts, then declined markedly (Fig. 1C). Upon differentiation of neural stem cells with retinoic acid there was a marked decrease in expression of DNMT3BΔ5 relative to exon 5-containing DNMT3B transcripts (DNMT3B1–6), consistent with the NCCIT data. In contrast, the glioma tumor initiating cell line (tumor stem cell) H1228 displayed the opposite trend, where DNMT3BΔ5 increased upon differentiation with retinoic acid (Fig. 1D, middle and right panels). Given the strong association of DNMT3BΔ5 expression with pluripotency, we confirmed that another DNMT3B isoform associated with pluripotency, DNMT3B1 (22), was co-expressed in cells expressing DNMT3BΔ5 (Fig. S2). Taken together, these results demonstrate the existence of a novel DNMT3B splice variant whose expression is strongly associated with pluripotency.
Conservation of alternative splicing in the homologous region of murine Dnmt3b (exon 6) and its association with pluripotency
The murine and human DNMT3B genes are highly conserved (88% identity at the amino acid level), with murine Dnmt3b possessing one extra non-coding exon at its 5’-end (Fig. 2A). The region equivalent to exon 5 in human DNMT3B is therefore exon 6 in murine Dnmt3b. These regions are approximately 83% identical at the amino acid level. To examine the possible conservation of a DNMT3BΔ5-like transcript in murine cells (except lacking exon 6 instead of exon 5), we designed semi-quantitative (located in exon 5 and exon 7) and quantitative real-time RT-PCR primers (spanning the unique exon 5-exon 7 junction, Fig. 2A). We were indeed able to detect Dnmt3bΔ6 (homologous to DNMT3BΔ5 in human) expression in the murine P19 EC cell line (Figs. 2A–2B).
Fig. 2.
Conservation of alternative splicing at the murine Dnmt3b locus and its expression during differentiation. (A) Schematic structure of the murine DNMT3b gene. Labeling is in Fig. 1A. A blowup of exons 3–8 is also shown. (B) Representative ethidium bromide stained agarose gel showing expression of Dnmt3bΔ6, the murine homolog of DNMT3BΔ5, in the P19 EC cell line using semi-quantitative RT-PCR. (C) Expression of murine Dnmt3b1–6 (all transcripts including exon 6) and Dnmt3bΔ6 during in vitro differentiation of ES cells by LIF withdrawal and embryoid body formation, relative to Gapdh (left panel). Ratio of expression of Dnmt3bΔ6 to Dnmt3b1–6 (black bars) and Oct4 expression (gray bars) during ES cell differentiation. Values are set relative to those at day zero (set at 100%, middle panel). Ratio of Dnmt3bΔ6 to Dnmt3b1–6 expression in ES cells, MEFs, and iPS cells derived from MEFs.
We then extended these studies by using murine R1 ES cells to further examine Dnmt3bΔ6 expression during differentiation. Dnmt3bΔ6 expression was initially high but declined up to day four of differentiation, after which it began to increase again out to 18 days (Fig. 2C). This expression pattern is consistent with a number of other differentiation-associated genes such as osteocalcin (23). Oct4, a gene required for maintenance of pluripotency, declined over the entire differentiation period as expected (Fig. 2C center panel, gray bars). Finally, it has recently been demonstrated that pluripotent ES-cell like cells can be derived from differentiated somatic cells by the ectopic expression of four pluripotency-associated transcription factors, Oct4, Sox2, Klf4, and c-Myc (24). Since Dnmt3bΔ6 is most highly expressed in ES cells and downregulated during differentiation, we expected that an iPS line would show elevated Dnmt3bΔ6 expression compared to the parental culture. We therefore generated iPS cells from mouse embryo fibroblasts (MEFs) and confirmed that they were bona fide iPS cells by criteria such as expression of Nanog, alkaline phosphatase positivity, and morphology (Fig. S3). MEFs expressed very little Dnmt3bΔ6 compared to Dnmt3b transcripts containing exon 6, while the iPS line expressed significantly elevated levels of Dnmt3bΔ6 that exceeded even the established R1 ES cells (Fig. 2C, right panel). These results therefore demonstrate that expression of DNMT3BΔ5 and the homologous Dnmt3bΔ6 splice variant is strongly associated with ‘stem-ness’ and is dynamically regulated during differentiation. These variants may therefore have important functions in maintaining the stem cell phenotype or regulating differentiation.
DNMT3BΔ5 expression in normal tissues and transformed cell lines
DNMT3B transcripts lacking exon 5, as assessed by qRT-PCR, represented a significant fraction of the total DNMT3B transcripts in EC and ES cells. To examine DNMT3BΔ5 expression more broadly, we assessed its levels in a panel of normal human tissues, which revealed that DNMT3BΔ5 was moderately expressed in several tissues, particularly human brain (Figs. 3A–3C). Examination of the ratio of transcripts containing exon 5 to those lacking exon 5 confirmed that DNMT3BΔ5 represented a major fraction of DNMT3B transcripts in adult neural tissues. Fetal brain expressed the highest level of DNMT3BΔ5 of all tissues we examined. Several other tissues, including colon, heart, and breast also expressed significant levels of DNMT3BΔ5 relative to transcripts that include exon 5 (Fig. 3C).
Fig. 3.
Expression of DNMT3BΔ5 in normal human and murine tissues. (A) Representative semi-quantitative RT-PCR amplification of DNMT3B transcripts containing exon 5 (DNMT3B1-6, top-most band) and those lacking exon 5 (DNMT3BΔ5, bottom-most band) for select human tissues. ‘*’ non-specific amplification product confirmed by DNA sequencing. (B) Expression of DNMT3B transcripts with (black bars) and without (gray bars) exon 5 by quantitative real-time RT-PCR for each of the human tissues shown. Values are presented relative to amplification of GAPDH as a loading control. (C) Ratio of expression of DNMT3B transcripts with exon 5 to DNMT3BΔ5 to emphasize the amount of DNMT3B transcripts lacking exon 5 relative to the total DNMT3B mRNA level. (D) Expression of Dnmt3bΔ6 in murine tissues and cell lines relative to Gapdh (top panel) and the ratio of Dnmt3b transcripts with exon 6 relative to those lacking exon 6 (bottom panel). Values are the average of at least three independent PCR amplifications and the bar represents the standard deviation from the mean. ‘DPC’ days post-coitum.
Given that splicing is often altered in transformed cells (18) and that altered splicing of DNMT3B in tumor cells has been well documented (20, 21), we examined expression of DNMT3B transcripts with and without exon 5 by qRT-PCR in a panel of 58 tumor cell lines (Figs. 4A–4B). There were several notable changes in expression in tumor cells compared to the respective normal tissue from which they were derived (Fig. 3). For example, colorectal carcinoma cell lines generally expressed moderate levels of DNMT3B transcripts with exon 5 but very low levels of DNMT3BΔ5 compared to normal colon (Fig. 4C). Similar results were observed for lines derived from breast and brain tumors. In contrast, expression of DNMT3BΔ5 was elevated, relative to transcripts containing exon 5, in tumor cell lines derived from liver, skin (melanoma), and lung (Fig. 4C). Cell lines like MOLT-4, HEK293, and G-401 expressed high levels of both forms of DNMT3B (Fig. 4B). Taken together, this analysis demonstrates that the ratio of exon 5-containing to exon 5-lacking DNMT3B transcripts is markedly altered in some tumor cells, which could contribute to the DNA methylation abnormalities in tumor cells if the lack of exon 5 alters the functional properties of DNMT3B.
Fig. 4.
Expression of DNMT3BΔ5 in human tumor cell lines. (A) Representative semi-quantitative RT-PCR amplification of DNMT3B transcripts with (upper arrow) and without exon 5 (bottom arrow) for select cell lines. (B) Expression of DNMT3B transcripts containing exon 5 (black bars) compared to transcripts lacking exon 5 (gray bars) by quantitative RT-PCR for each of the 58 human cell lines shown (upper portion of each panel). Values are presented relative to amplification of GAPDH as a loading control. Ratio of expression of DNMT3B transcripts with exon 5 to DNMT3BΔ5 (lower portion of each panel, thick gray bars). Values are the average of at least three independent PCR amplifications and the bar represents the standard deviation from the mean. U2-OS is derived from an osteosarcoma, HEC59 from endometrial cancer, MOLT-4 from acute lymphoblastic leukemia, and HEK293 and G-401 from kidney cancer. (C) The ratio of expression of DNMT3BΔ5 relative to DNMT3B1-6 (‘+Ex 5’) transcripts in tumor cell lines derived from each tissue (having at least four cell lines) from Fig. 4 was averaged (gray bars) and graphed relative to the same ratio from corresponding normal tissues (two independent tissue preparations, black bars, Fig. 3).
We also investigated expression of Dnmt3bΔ6, the murine homolog of DNMT3BΔ5, in a panel of murine cell lines and tissues. Consistent with human EC cells, the murine P19 EC cell line expressed moderate levels of Dnmt3bΔ6 as well as Dnmt3b transcripts that include exon 6, while Dnmt3bΔ6 expression was nearly undetectable in the immortalized fibroblast line NIH3T3 (Fig. 3D). In fetal tissues, overall expression of Dnmt3b transcripts was generally higher and Dnmt3bΔ6 represented a small but significant fraction of the total Dnmt3b mRNA level in fetal brain, liver, whole embryo, and adult testis (Fig. 3D).
Functional differences between DNMT3B3 and DNMT3B3Δ5 in vitro and in vivo
The exon 5 region of DNMT3B does not contain any known functional motifs, but is immediately adjacent to the PWWP domain. Exclusion of exon 5 could alter an as yet unknown function of the extreme N-terminal region of DNMT3B or change the amino acid context of the PWWP domain and therefore alter its properties. To assess the role of exon 5 in DNA binding, we expressed and purified full-length recombinant human 6XHis-tagged DNMT3B3 and DNMT3B3Δ5 by infection of Sf9 insect cells with recombinant baculovirus followed by lysis and metal chelating affinity purification. This method yielded highly pure preparations of both proteins (Fig. 5). We then compared their DNA binding affinities using an electrophoretic mobility shift assay (EMSA) with a fragment of human satellite 2 (Sat2) derived from chromosome 1 as the DNA probe. Sat2 is a known DNMT3B target sequence that is consistently hypomethylated in ICF syndrome patients (8, 25). A range of protein concentrations was used with a fixed amount of Sat2 DNA probe. EMSA reactions were resolved on agarose gels and bandshifts visualized by SYBR Green staining. Each DNMT3B isoform exhibited a concentration-dependent increase in the amount of shifted probe (Fig. 5A). EMSA bandshifts were quantitated and the percent shift, relative to the concentration of DNMT3B, was plotted and the KD calculated using the Hill equation. Interestingly, DNMT3B3Δ5 (KD=119 nM) possessed significantly higher affinity for the Sat2 DNA probe compared to DNMT3B3 (KD=246 nM, Fig. 5B). The sigmoidal shape of both binding curves also suggested cooperative binding of DNMT3B. These results therefore indicate that sequences encoded by exon 5 play a role in DNA binding or influence the three dimensional structure of the adjacent PWWP domain.
Fig. 5.
Comparison of the DNA binding affinities of DNMT3B3 and DNMT3B3Δ5 for a human Sat2 DNA probe. (A) Representative electrophoretic mobility shift assays (EMSAs) using purified recombinant DNMT3B3 (left panel) and DNMT3B3Δ5 (right panel). An increasing amount of each protein is used (indicated by the wedge, concentrations from left to right are 50, 100, 150, 200, 250, 300, 350, 400, 450, and 500 nM) with a fixed amount of Sat2 DNA probe (3 ng). Mobility of the free and DNMT3B-bound probes is indicated at the right. (B) Calculation of DNA binding affinity (KD). EMSAs for each protein and concentration (50–700 nM range) were run at least in triplicate and the percent shift at each protein concentration calculated. Values were fitted and the KD was calculated using the Hill equation. Error bars are the standard deviation from the mean. (C) Coomassie blue stained 10% SDS-PAGE gel showing the purified 6X-His-tagged DNMT3B3 and DNMT3B3Δ5 used in the EMSA reactions.
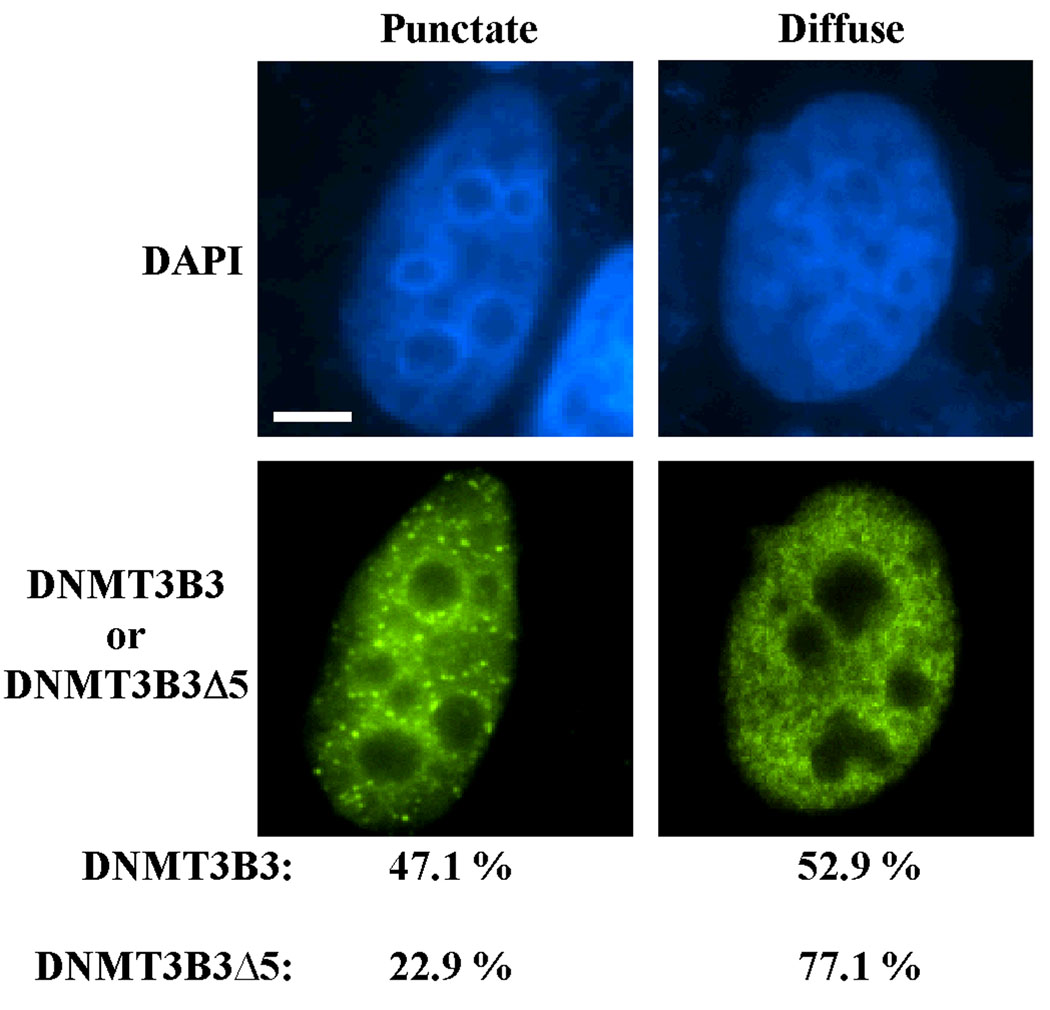
We next examined the subcellular localization of DNMT3B3Δ5 compared to DNMT3B3 by transiently transfecting tagged versions of each into HeLa cells followed by immunofluorescence microscopy. In HeLa cells, DNMT3B3 displayed two exclusively nuclear localization patterns: (1) punctate with enrichment in heterochromatic regions, and (2) diffuse nuclear accumulation (Fig. 6). DNMT3B3Δ5 also showed these two staining patterns, however the frequency of each pattern differed markedly. DNMT3B3 displayed the punctate distribution much more frequently than DNMT3B3Δ5 (Fig. 6). In addition, DNMT3B3Δ5 accumulated less at heterochromatic regions in the diffuse staining pattern. Taken together, the EMSA and immunofluorescence studies reveal that DNMT3B3Δ5 has properties distinct from DNMT3B3 and may therefore regulate genomic methylation patterns differently from other DNMT3B isoforms.
Fig. 6.
Immunofluorescence localization of DNMT3B3 and DNMT3B3Δ5 in human cells. GFP-DNMT3B3 and FLAG-DNMT3B3Δ5 expression plasmids were transfected into HeLa cells. Forty-eight hours later cells were fixed, stained with anti-FLAG antibody, and photographed with a fluorescence microscope. All images were deconvolved and a representative of each is shown. Quantitation of the percent of transfected cells expressing either the diffuse or the punctate localization patterns of DNMT3B3 and DNMT3B3Δ5 is indicated below the images. Blue - DNA, green - DNMT3B3 or DNMT3B3Δ5. Bar = 10 microns.
Ectopic expression of DNMT3BΔ5 alters repetitive element DNA methylation and enhances clonogenic growth
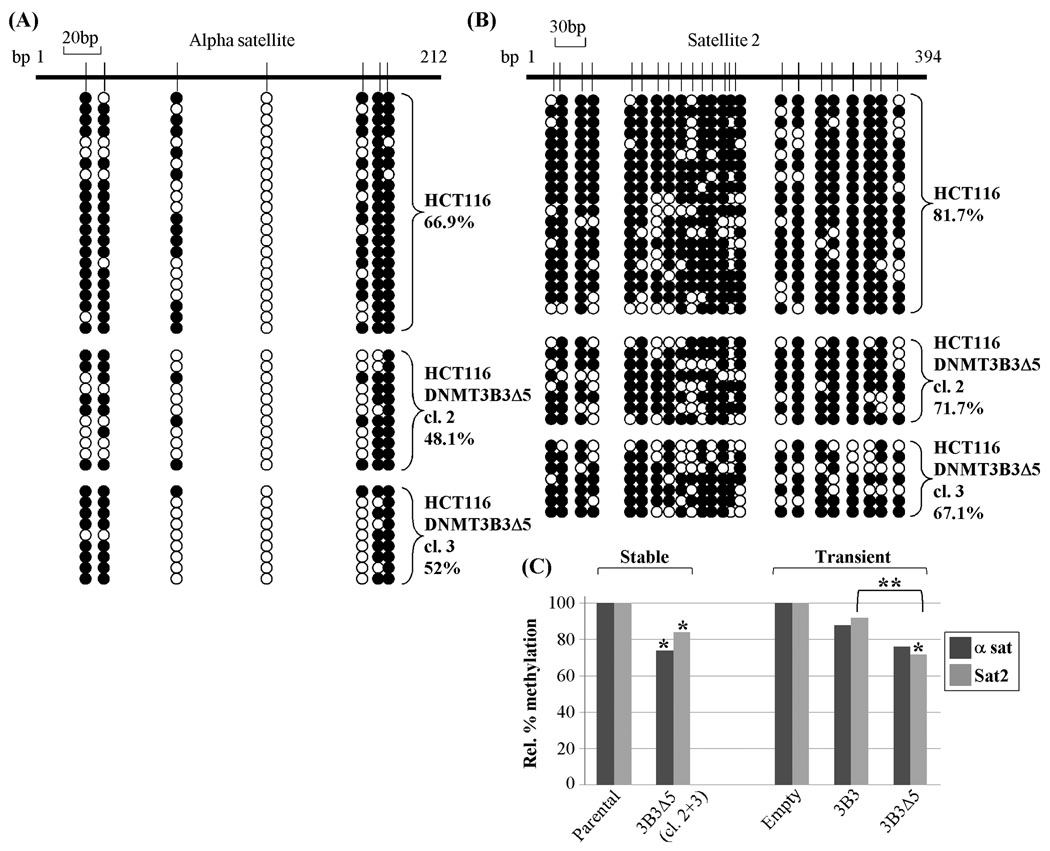
To determine how DNMT3B3Δ5 expression may affect genomic DNA methylation patterns in vivo, we created HCT116 colon cancer cell lines stably expressing DNMT3B3Δ5. Parental HCT116 cells expressed DNMT3B3Δ5 from the endogenous locus at extremely low levels (Fig. 4). We confirmed two HCT116 clones with significantly elevated expression of DNMT3B3Δ5 by quantitative RT-PCR and western blotting (Fig. S4). Interestingly, the level of endogenous exon 5-containing DNMT3B transcripts was suppressed in the stable DNMT3B3Δ5 clones compared to the parental HCT116 cells, suggestive of a regulatory interplay between the endogenous and ectopically expressed forms of DNMT3B. For reasons that are unclear we were unable to establish HCT116 cell lines stably expressing DNMT3B3. We then examined two repetitive regions of the genome known to be DNMT3B targets, the pericentromeric Sat2 repeat (also analyzed by EMSA above) and the centromeric alpha satellite (αSat) repeat (25). Sat2 is invariantly hypomethylated in ICF syndrome patients, while a subset of ICF patients also lose methylation from the αSat repeat (26, 27). The equivalent regions (major and minor satellite) in murine Dnmt3b knockouts and an ICF mouse model also become hypomethylated (28). Using bisulfite genomic sequencing (BGS), we determined that both αSat and Sat2 repeats were densely methylated in parental HCT116 cells (~67% and 82%, respectively, Figs. 7A–7B). The DNMT3B3Δ5-expressing clones, however, demonstrated ~17% and ~12% reductions in DNA methylation at the αSat and Sat2 repeats, respectively (Fig. 7). These decreases were significant for both regions (p=0.0014 for αSat and p<0.001 for Sat2) as assessed by the Chi-square test. The relative changes in total DNA methylation at each region examined is also summarized graphically in Fig. 7C. These results therefore suggest that aberrant expression of DNMT3BΔ5 during tumorigenesis could contribute to the genomic DNA methylation defects that are a hallmark of transformed cells.
Fig. 7.
DNMT3B3Δ5 expression alters DNA methylation in vivo. Bisulfite genomic sequencing (BGS) analysis of (A) centromeric alpha satellite, and (B) pericentromeric satellite 2 (Sat2) repeats in parental HCT116 cells and two HCT116 clones stably overexpressing DNMT3B3Δ5 (clones 2 and 3). Each row of circles represents the DNA methylation pattern (open circle – unmethylated, black circle – methylated CpG) of an individual cloned and sequenced DNA molecule. The total percent methylation for all analyzed clones is shown at the right. Two independent DNMT3B3Δ5 clones were examined with comparable levels of transgene expression (Fig. S4). (C) Summary of BGS data with statistical significance shown for the stably transfected DNMT3B3Δ5 lines (left portion) and HCT116 3BKO cells transiently transfected with DNMT3B3 or DNMT3B3Δ5 (right portion). Data is presented relative to the parental line or empty vector transfected cells (set at 100%). * significance relative to parental or empty vector, ** significance for DNMT3B3Δ5 relative to DNMT3B3 using the Chi-square test. For the transient transfection experiments, 12–16 clones were sequenced for each construct/transfection (not shown) and used in the calculations.
To further examine DNMT3B3Δ5’s capacity to modulate centromeric DNA methylation levels and compare this to the closely related and also catalytically inactive DNMT3B3 splice variant, we employed a series of transfections into HCT116 cells in which the DNMT3B gene has been genetically inactivated by homologous recombination (3BKO, (29)). These cells retain most of their DNA methylation and there is no endogenous DNMT3B to confound our interpretations. The transient transfection method also allows us to directly compare the effect of ectopic expression of DNMT3B3Δ5 with DNMT3B3, which we were unable to do with the stable cell lines. We therefore transiently transfected expression plasmids for DNMT3B3, DNMT3B3Δ5, or empty parental expression vector into HCT116 3BKO cells. After 48 hours of incubation, we analyzed the DNA methylation status of the αSat and Sat2 regions by BGS. Results from this experiment, which are summarized as the percent methylation relative to the empty vector control, reveal that ectopic expression of DNMT3B3Δ5 reduced centromeric and pericentromeric DNA methylation levels by 24 and 28%, respectively. Ectopic expression of DNMT3B3, which is also catalytically inactive, resulted in 12% and 8% decreases in DNA methylation at the αSat and Sat2 regions, respectively, nearly half the reduction we observed in the DNMT3B3Δ5 transfections (Fig. 7C). RT-PCR analysis indicated that DNMT3B3 and DNMT3B3Δ5 were expressed at comparable levels in the transiently transfected populations (data not shown). Although the DNA hypomethylation at the αSat region did not reach statistical significance, the reduction in methylation at Sat2 did reach significance for the DNMT3B3Δ5 transfections (p=0.001, Chi-square). In addition, the degree of DNA hypomethylation at Sat2 was significantly greater in the DNMT3B3Δ5 transfection than in the DNMT3B3 transfection (p=0.001). Taken together, these results suggest that expression of catalytically inactive DNMT3B splice variants is indeed capable of modulating genomic DNA methylation levels. In addition, our data demonstrate that not all splice variants behave similarly just because they lack catalytic activity. In the case of DNMT3B3Δ5, this may be due to its enhanced DNA binding affinity, potentially blocking access of catalytically active forms of DNMT3B or DNMT3A (which interacts with DNMT3B) to their target sites in the genome.
We observed tumor cell lines with increased and decreased DNMT3BΔ5 expression relative to corresponding normal tissues (Fig. 4), suggesting that this variant may possess growth modulatory properties dependent on the cell or tissue type. To examine this further, we performed colony formation assays using the same expression plasmids used for the transient transfection/ BGS analysis described above. In this case we used both HCT116 parental and 3BKO cells. Plasmids were transfected and expressing cells were selected with G418 for 14 days. Following this period, surviving colonies were stained and counted. Ectopic expression of DNMT3B3Δ5 resulted in a small but significant increase in colony number in parental HCT116 cells (p=0.005, Student’s t-test). Although the change in colony number in DNMT3B3 transfected HCT116 cells was not significantly different than the empty vector control, it was significantly different from the DNMT3B3Δ5 transfected HCT116 cells (p=0.02, Fig. 8 top panel). Results in 3BKO HCT116 cells showed a similar trend although the magnitude of the increases was greater, especially for the DNMT3B3Δ5 transfected cells (Fig. 8). DNMT3B3Δ5 expression resulted in nearly double the number of colonies compared to empty vector transfected 3BKO cells (Fig. 8 bottom panel, all differences were significant). Taken together, these results indicate that expression of catalytically inactive DNMT3B splice variants influence genomic DNA methylation patterns and cell growth suggesting that improper levels of these variants may contribute to both the global DNA methylation defects as well and the deregulated growth phenotypes that are hallmarks of tumor cells.
Fig. 8.
Ectopic expression of DNMT3B splice variants modulates clonogenic growth of HCT116 parental and 3BKO cells. Cell lines were transfected with the indicated expression plasmids and expressing cells were selected with G418. Surviving colonies were stained and counted. All transfections were repeated at least three times and values are the average and the bar is the standard deviation from the mean. ‘Empty’ – unmodified parental expression vector, * significance relative to parental or empty vector, ** significance for DNMT3B3Δ5 relative to DNMT3B3 using the Student’s t-test.
DISCUSSION
In the present study we have identified and characterized a novel conserved splice variant of DNMT3B, DNMT3B3Δ5, that is associated with ‘stem-ness’ and with adult and fetal neural tissue. This transcript is similar to the widely expressed DNMT3B3 variant but also lacks exon 5 in human DNMT3B or exon 6 in murine Dnmt3b. Although not yet tested, both DNMT3B3Δ5 and Dnmt3b3Δ6 would be expected to be catalytically inactive since DNMT3B3 is also inactive (30) but exon 5 skipping may occur in the context of other DNMT3B isoforms which are active. Aside from brain and testis, DNMT3BΔ5 expression is limited in adult tissues and is altered in tumor cell lines. Generally, cell lines derived from low-expressing tissues display elevated DNMT3BΔ5 expression while those derived from moderate/high-expressing tissues display reduced DNMT3BΔ5 expression. DNMT3BΔ5 and Dnmt3bΔ6 are dynamically regulated during differentiation of ES, EC, and neural stem cells and conversion of differentiated, low Dnmt3bΔ6-expressing fibroblasts to iPS cells results in marked upregulation of Dnmt3bΔ6. Finally, we demonstrate that DNMT3B3Δ5 possesses distinct biochemical properties compared to its ‘parent’ isoform, DNMT3B3, and that elevated expression of DNMT3B3Δ5 causes centromeric hypomethylation and enhances clonogenic growth, suggesting that it may differentially regulate DNA methylation or chromatin structure in stem cells and contribute to methylation defects in cancer if inappropriately expressed.
Interestingly, DNMT3B is extensively alternatively spliced with nearly 40 isoforms identified thus far (14, 20, 21). A recent study also reported the identification of a transcript with a similar structure to Dnmt3bΔ6 in murine ES cells, supporting our findings (31). The role of these splice variants in establishing normal tissue DNA methylation patterns and possibly contributing to aberrant DNA methylation characteristic of tumor cells is largely unknown. DNMT3B4, a variant highly expressed in human testis, is over-expressed in hepatocellular cancers. Elevated DNMT3B4 expression results in pericentromeric satellite DNA hypomethylation, which is associated with genomic instability, and increased cell proliferation (32). In keeping with this result, we have shown that ectopic expression of DNMT3B3Δ5 in cells that normally express it at a very low level, results in hypomethylation of sequences at and adjacent to the centromere. In another study, seven novel DNMT3B transcripts originating from within intron 4 and retaining a portion of exon 5 were detected in non-small cell lung cancer (NSCLC) cells. Some of these transcripts (termed the ΔDNMT3B family) extended through the end of the gene and thus may be catalytically active, while others displayed variable inclusion of exons 7–9 and premature termination at exon 11 or 17 due to frameshifts (20). Expression of ΔDNMT3B transcripts was associated with poorer clinical outcome and with hypermethylation of the RASSF1A tumor suppressor gene in NSCLC patients (33). In the most extensive study to date, Ostler et al. identified over 20 new DNMT3B isoforms in cancer cells. Most of the alternative splicing they reported resulted in premature translation stop signals that truncate DNMT3B prior to the catalytic domain. Other transcripts involved alternative usage of exons 4–6, indicating that this area of DNMT3B, like the exon 21–22 region, is a hotbed of alternative splicing activity. The most abundant splice variant identified by Ostler et al., termed DNMT3B7, was ectopically over-expressed in HEK293 cells and resulted in altered gene expression and promoter CpG island hypermethylation (21, 34). Although the reductions in DNA methylation that we observed at the centromeric and pericentromeric satellite regions upon ectopic expression of DNMT3B3Δ5 would appear relatively minor, hypomethylation of this magnitude was recently reported in a bisulfite genomic sequencing-based analysis of the Sat2 region in glioma (35) and the alpha satellite region is also a well-established target of DNA hypomethylation in many cancers (36). Results from these studies, along with our colony formation assay data, suggest that hypomethylation mediated by inappropriate expression of certain DNMTB splice variants may lead to oncogenic changes that could contribute to transformation. Elevated expression of truncated catalytically inactive splice variants of DNMT3B was also highly predictive of ovarian cancer compared to normal ovarian epithelium (37). These studies therefore indicate that expression of certain DNMT3B splice variants in tumor cells results in functional consequences to cell growth and both gene-specific and repetitive region DNA methylation patterns.
What are the functions of alternatively spliced forms of DNMT3B such as DNMT3B3Δ5 or DNMT3B4? One possibility is that they result in proteins with altered functions, either gain or loss. Many of the DNMT3B variants are expected to be catalytically inactive because they lack exons 20 and 21 in the catalytic domain or lack the catalytic domain entirely (21). One advantage of expressing an inactive variant in differentiated somatic cells is that it may help keep unauthorized de novo methylation, and the consequent aberrant gene silencing, in check. Since many of these transcripts, including DNMT3B3Δ5, are differentially expressed during development and differentiation, it would appear more plausible that they have altered functions, due to exclusion of particular exons that are important for modulating methylation patterns during these processes. Such functions may include altered DNA binding, as we have shown here, or altered DNA target site preference. A catalytically inactive isoform of DNMT3B, such as DNMT3B3Δ5, with enhanced DNA binding affinity but retaining a similar genomic site preference as the catalytically active DNMT3B isoforms (which we can only hypothesize is the case at this time), may be able to block access of the active isoforms to the DNA and consequently lead to hypomethylation of DNMT3B target sites. Since DNMT3B interacts with both DNMT3A and DNMT3L, and DNMT3L stimulates DNMT3 activity (38–41), another intriguing possibility is that certain splice variants differ in their ability to make these interactions. Mutations that eliminate Dnmt3a-Dnmt3a or Dnmt3a-Dnmt3L interactions eliminate Dnmt3a catalytic activity (41). Catalytically inactive DNMT3B variants that still retain their ability to interact with DNMT3A might reduce DNMT3A’s catalytic activity or alter its target site preference. Such a mechanism may account for the hypomethylation we observe upon DNMT3B3Δ5 overexpression. Alternatively, if transcripts like DNMT3B3Δ5 still interact with DNMT3L, they may titrate DNMT3L away from active isoforms of DNMT3A and DNMT3B, thus indirectly modulating genomic DNA methylation levels. Comparison of the activities and interaction partners of DNMT3B splice variants will be an important area of future research that should provide important insights into the regulation and maintenance of genomic DNA methylation patterns in mammalian cells.
MATERIALS AND METHODS
Tissue culture and cell lines
All tumor cell lines were purchased from the American Type Culture Collection and grown in McCoy’s 5-A media supplemented with 10% fetal bovine serum and 2 mM L-glutamine (Invitrogen). Creation of induced pluripotent stem (iPS) cells and the growth and differentiation conditions of neural stem cell lines (SCP23 and SCP27), murine ES cells, and NCCIT EC cells is described in the online supplemental material. Creation and characterization of stable DNMT3B3Δ5-expressing HCT116 cells is also described in the supplemental methods.
DNMT3B isoform cloning and sequencing
Total RNA was prepared from cell lines by Trizol extraction (Invitrogen). Multiscribe reverse transcriptase (ABI) was used to prepare cDNA from 2 µg of total RNA according to the manufacturer’s protocol. RT-PCR primers used to amplify full-length human DNMT3B are 5’-GGC TGG ATT CAT GAA GGG AGA CAC CAG GCA T-3’ (F) and 5’- GCC TGT CGA CCT ATT CAC ATG CAA AGT AGT CCT TCA GAG G- 3’(R). Use of these primers led to the identification of DNMT3B3Δ5. All PCR products were cloned using the TOPO TA cloning kit (Invitrogen) and sequenced using the M13 forward and reverse primers and gene-specific internal primers (available upon request). Apart from the major DNMT3B splice variants DNMT3B1 and DNMT3B3, a new smaller PCR product was observed, which lacked exons 5, 20, and 21 (DNMT3B3Δ5). The exon 4–6 junction resulted in a transcript that was in-frame with the normal DNMT3B coding region. While analyzing the expression of DNMT3BΔ5 in normal tissues by semi-quantitative RT-PCR, we identified an additional novel PCR product in fetal brain, which was also cloned and sequenced. This product, which lacked both exons 4 and 5, was termed DNMT3BΔ(4+5). Sequences were aligned with existing NCBI Genbank entries for DNMT3B and DNMT3B splice variant sequences with accession numbers NM_006892, NM_175848, NM_175849, NM_175850.
Recombinant protein expression and purification
The DNMT3B3 and DNMT3B3Δ5 coding sequences were cloned into the pFastBacHT vector (Invitrogen) and used to produce recombinant hexahistidine (6X-His)-tagged DNMT3B3 and DNMT3B3Δ5 upon infection of Sf9 insect cells cultured in Sf900II medium (Invitrogen). Recombinant baculovirus production and titering using the Bac-to-Bac Baculovirus Expression System (Invitrogen) was performed according to the manufacturer’s instructions and as we have described previously (42). Recombinant 6X-His-tagged proteins were subsequently purified with Ni-NTA agarose beads (Novagen) as described (42).
Electrophoretic mobility shift assays (EMSA)
Creation of the satellite 2 probe for EMSA is described in the supplemental material. Recombinant DNMT3s were incubated with 3 ng human Sat2 probe in reaction buffer containing 10 mM HEPES (pH 7.5), 50 mM KCl, 1 mM EDTA, 0.1% Triton-X 100, 5% glycerol, 0.1 mM DTT and 5 µM S-adenosyl-L-methionine (SAM, New England Biolabs). Enzyme concentrations used in the binding reactions ranged from 0–700 nM. The binding reaction was performed on ice for 30 minutes after which the samples were resolved on a 0.7% agarose gel in 1X HEE running buffer (10 mM HEPES (pH 7.5), 1 mM EDTA, 0.5 mM EGTA) at 65V for 2.5 hours. Gels were stained for 30 minutes with SYBR Green I (Invitrogen) and visualized with a Bio-Rad gel documentation system. The percent shift was quantitated using Quantity One 4.6.1 (Bio-Rad). All EMSAs were repeated at least in triplicate. The relative quantity of shifted material was calculated in Quantity One using the sum of the intensity of the bands in each lane. The curve was fit and the KD calculated using the Hill equation in SigmaPlot 8.0.
Reverse Transcriptase (RT)-PCR
Two micrograms of total RNA per sample was reverse transcribed with Multiscribe reverse transcriptase (ABI) using the manufacturer’s protocol. One microliter of cDNA was used per reaction for both semi-quantitative and quantitative (Q) RT-PCR reactions. Semi-quantitative RT-PCR primers are: human DNMT3B 5’-CAG AGG CCG AAG ATC AAG CTC-3’ (F) and 5’- TCC ACT GTC TGC CTC CAC CTG- 3’ (R), and murine Dnmt3b 5’-CAA GCT CCC GGC TGT CTA AGA-3’ (F) and 5’-CAT ACC CTC CTG ATC TCC ATC-3’ (R). We also used other DNMT3B primers specific for alternative splicing within the catalytic region (exons 21–22) and the N-terminal region of DNMT3B (exons 10–11) that have been described previously (14, 22). As an internal loading control, human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was measured using primers 5′-TTG GTA TCG TGG AAG GAC TC-3′ (F) and 5′-ACA GTC TTC TGG GTG GCA GT-3’ (R). Murine Gapdh expression was measured using primers 5’-CAC TTG AAG GGT GGA GCC AAA AG-3’ (F) and 5’- GTG GAT GCA GGG ATG ATG TTC TG-3’(R). The same human and murine GAPDH primers were used for both the semi-quantitative and qRT-PCR reactions. QPCR was performed using human DNMT3B full-length specific primer 5-AGA AAG CCC AGA TGT CCG AAC-3’(F) or human DNMT3BΔ5-specific primer spanning the junction between exons 4 and 6 (unique to this variant) using primer 5’-GAA AGC CCA GCT TCC CTG AGA C-3’ (F) and the common reverse primer 5’-AGT TGT GTC CTC TGT GTC GTC TGT-3’ (R). Murine Dnmt3b full-length specific primer 5’-CGC CAC CAT GTG CAG GAG TAC-3’ (F) or a Dnmt3bΔ6-specific primer spanning the junction between exons 5 and 7 (specific for Dnmt3bΔ6, the murine equivalent of human DNMT3BΔ5) 5’-GAA AGC CCG GCT TCT CGG AGA C-3’ (F) and the common reverse primer 5’-GAC GCT CTT AGG TGT CAC TTC TTC C-3’ (R) were used. DNMT3B expression was normalized to GAPDH expression using the formula (2(Ct(GAPDH)-Ct(DNMT3B)). Fold expression of DNMT3BΔ5 was calculated by dividing the relative expression of the splice variant (lacking exon 5/6) by the relative expression of DNMT3B isoforms including these exons. Oct4 qRT-PCR primers are 5’-GGT GGA GGA AGC CGA CAA CAA T-3’ (F) and 5’-CAC CAG GGT CTC CGA TTT-3’ (R). Normal tissue RNAs were purchased from Clontech.
Transfections, western blotting, and immunofluorescence microscopy
The pcDNA4 HisMax-DNMT3B constructs (described in the supplemental methods) were transiently transfected into HCT116 cells using TransIT LT-1 (Mirus) according to the manufacturer’s instructions. Forty-eight hours after transfection, cells were harvested for whole cell extract using lysis buffer (10 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton-X 100). Whole cell extracts were resolved on a 10% SDS-PAGE gel and transferred to PVDF membrane overnight for western blotting. Protein expression was detected using the anti-Express antibody from Invitrogen (1:1000). For immunofluorescence, cells were grown on 22-mm2 glass coverslips in six-well plates and transfected with GFP-DNMT3B3 or FLAG-DNMT3B3Δ5 expression constructs (supplemental methods) using TransIT LT-1. Cells were fixed with 4% paraformaldehyde in 1X PBS (pH 7.0) forty-eight hours post-transfection, permeabilized with 0.5% Triton X-100, and incubated with anti-FLAG primary antibody (Sigma anti-FLAG M2) diluted 1:50 in 1X PBS with 0.1% Tween-20 for one hour at room temperature. Cells were then washed three times with PBST, subsequently incubated with anti-mouse-TRITC labeled secondary antibody, then counterstained with Hoechst 33342 to visualize DNA. Images were captured using a Nikon TE2000 inverted microscope and deconvolved using Nikon Elements software.
Bisulfite genomic sequencing (BGS)
Genomic DNA extracted from stably or transiently transfected cells was bisulfite treated as described (43). Bisulfite treated DNA was used as template to amplify PCR products corresponding to satellite alpha with primers 5’-GGA TAT GTG GAT AGT TTT GAA G-3’ (F) and 5’-TTC CTT TTT CAC CAT AAA CCT C-3’(R) and satellite 2 (chr. 1) with primers 5’- GAA TTA TTG AAT AGA ATT GAA TGG-3’ (F) and 5’- TAA ATA ATA ACT CCT TTC ATT T-3’ (R). PCR products were gel purified using the Qiaex II gel extraction kit (Qiagen) and cloned using the TA Cloning Kit (Invitrogen). Cloned products were sequenced in a 96-well plate format using the M13 reverse primer at the University of Florida Interdisciplinary Center for Biotechnology Research and methylation levels were analyzed.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant R01CA114229 (KDR). We thank Dr. James Resnick for providing murine tissues, Dr. Melanie Ehrlich for providing the pUC1.77 plasmid, and Dr. Howard Fine for providing H1228 cells. We also thank Jason Orr Brant and Jianghui Zhu for their help with the statistical analysis.
REFERENCES
- 1.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 2.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 3.Leonhardt H, Page AW, Weier H, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 4.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 5.Liang G, Chan MF, Tomigahara Y, et al. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol. 2002;22:480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson M, Krassowska A, Gilbert N, et al. Severe global DNA hypomethylation blocks differentiation and induces hyperacetylation in embryonic stem cells. Mol Cell Biol. 2004;24:8862–8871. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ooi SKT, Qiu C, Bernstein E, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu G-L, Bestor TH, Bourc'his D, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 9.Hansen RS, Wijmenga C, Luo P, et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachman KE, Rountree MR, Baylin SB. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J Biol Chem. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- 11.Qiu C, Sawada K, Zhang X, Cheng X. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat Struct Biol. 2002;9:217–224. doi: 10.1038/nsb759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen T, Tsujimoto N, Li E. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol Cell Biol. 2004;24:9048–9058. doi: 10.1128/MCB.24.20.9048-9058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S. Expression of mRNA for DNA methyltransferases and methyl-CpG-binding proteins and DNA methylation status on CpG islands and pericentromeric satellite regions during human hepatocarcinogenesis. Hepatology. 2001;33:561–568. doi: 10.1053/jhep.2001.22507. [DOI] [PubMed] [Google Scholar]
- 14.Robertson KD, Uzvolgyi E, Liang G, et al. The human DNA methyltransferases (DNMTs) 1, 3a, and 3b: Coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanai Y, Ushijima S, Nakanishi Y, Hirohashi S. DNA methyltransferase expression and DNA methylation of CpG islands and peri-centromeric satellite regions in human colorectal and stomach cancers. Int J Cancer. 2001;91:205–212. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1040>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 16.Beaulieu N, Morin S, Chute IC, Robert M-F, Nguyen H, MacLeod AR. An essential role for DNA methyltransferase DNMT3B in cancer cell survival. J Biol Chem. 2002;277:28176–28181. doi: 10.1074/jbc.M204734200. [DOI] [PubMed] [Google Scholar]
- 17.Linhart HG, Lin H, Yamada Y, et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim E, Goren A, Ast G. Insights into the connection between cancer and alternative splicing. Trends Genet. 2008;24:7–10. doi: 10.1016/j.tig.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Robertson KD, Jones PA. Tissue-specific alternative splicing in the human INK4a/ARF cell cycle regulatory locus. Oncogene. 1999;18:3810–3820. doi: 10.1038/sj.onc.1202737. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Wang J, Sun S, et al. A novel DNMT3B subfamily, ΔDNMT3B, is the predominant form of DNMT3B in non-small cell lung cancer. Int J Oncol. 2006;29:201–207. [PubMed] [Google Scholar]
- 21.Ostler KR, Davis EM, Payne SL, et al. Cancer cells express aberrant DNMT3B transcripts encoding truncated proteins. Oncogene. 2007;26:5553–5563. doi: 10.1038/sj.onc.1210351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weisenberger DJ, Velicescu M, Cheng JC, Gonzales FA, Liang G, Jones PA. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol Canc Res. 2004;2:62–72. [PubMed] [Google Scholar]
- 23.Candeliere GA, Rao Y, Floh A, Sandler SD, Aubin JE. cDNA fingerprinting of osteoprogenitor cells to isolate differentiation stage-specific genes. Nucleic Acids Res. 1999;27:1079–1083. doi: 10.1093/nar/27.4.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–376. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Gopalakrishnan S, Sullivan BA, Trazzi S, Della Valle G, Robertson KD. DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp256. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miniou P, Jeanpierre M, Bourc'his D, Barbosa ACC, Blanquet V, Viegas-Pequignot E. α-satellite DNA methylation in normal individuals and in ICF patients: heterogeneous methylation of constitutive heterochromatin in adult and fetal tissues. Hum Genet. 1997;99:738–745. doi: 10.1007/s004390050441. [DOI] [PubMed] [Google Scholar]
- 27.Ehrlich M, Jackson K, Weemaes C. Immunodeficiency, centromeric region instability, facial anomalies syndrome (ICF) Orphanet J Rare Dis. 2006:1. doi: 10.1186/1750-1172-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ueda Y, Okano M, Williams C, Chen T, Georgopoulos K, Li E. Roles for Dnmt3b in mammalian development: a mouse model for the ICF syndrome. Development. 2006;133:1183–1192. doi: 10.1242/dev.02293. [DOI] [PubMed] [Google Scholar]
- 29.Rhee I, Bachman KE, Park BH, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 30.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nature Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 31.Gowher H, Stuhlmann H, Felsenfeld G. Vezf1 regulates genomic DNA methylation through its effects on expression of DNA methyltransferase Dnmt3b. Genes Dev. 2008;22:2075–2084. doi: 10.1101/gad.1658408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H, Hirohashi S. Overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, associated with DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis. Proc Natl Acad Sci USA. 2002;99:10060–10065. doi: 10.1073/pnas.152121799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Walsh G, Liu DD, Lee JJ, Mao L. Expression of ΔDNMT3B variants and its association with promoter methylation of p16 and RASSF1A in primary non-small cell lung cancer. Cancer Res. 2006;66:8361–8366. doi: 10.1158/0008-5472.CAN-06-2031. [DOI] [PubMed] [Google Scholar]
- 34.Fackenthal JD, Godley LA. Aberrant RNA splicing and its functional consequences in cancer cells. Dis Model Mech. 2008;1:37–42. doi: 10.1242/dmm.000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cadieux B, Ching T-T, Van den Berg SR, Costello JF. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66:8469–8476. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- 36.Narayan A, Ji W, Zhang X-Y, et al. Hypomethylation of pericentromeric DNA in breast adenocarcinomas. Int J Cancer. 1998;77:833–838. doi: 10.1002/(sici)1097-0215(19980911)77:6<833::aid-ijc6>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 37.Klinck R, Bramard A, Inkel L, et al. Multiple alternative splicing markers for ovarian cancer. Cancer Res. 2008;68:657–663. doi: 10.1158/0008-5472.CAN-07-2580. [DOI] [PubMed] [Google Scholar]
- 38.Chedin F, Lieber MR, Hsieh C-L. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci USA. 2002;99:16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Margot JB, Ehrenhofer-Murray AE, Leonhardt H. Interactions within the mammalian DNA methyltransferase family. BMC Mol Bio. 2003;4:7. doi: 10.1186/1471-2199-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through direct interaction. J Biol Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 41.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yokochi T, Robertson KD. Preferential methylation of unmethylated DNA by mammalian de novo DNA methyltransferase Dnmt3a. J Biol Chem. 2002;277:11735–11745. doi: 10.1074/jbc.M106590200. [DOI] [PubMed] [Google Scholar]
- 43.Jin B, Tao Q, Peng J, et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum Mol Genet. 2008;17:690–709. doi: 10.1093/hmg/ddm341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.