Abstract
Aromatic amines such as 4-aminobiphenyl (ABP) require biotransformation to exert their carcinogenic effects. Genetic polymorphisms in biotransformation enzymes such as N-acetyltransferase 2 (NAT2) may modify cancer risk following exposure. Nucleotide excision repair-deficient Chinese hamster ovary (CHO) cells stably transfected with human cytochrome P4501A1 (CYP1A1) and a single copy of either NAT2*4 (rapid acetylator), NAT2*5B (common Caucasian slow acetylator), or NAT2*7B (common Asian slow acetylator) alleles (haplotypes) were treated with ABP to test the effect of NAT2 polymorphisms on DNA adduct formation and mutagenesis. ABP N-acetyltransferase catalytic activities were detectable only in cell lines transfected with NAT2 and were highest in cells transfected with NAT2*4, lower in cells transfected with NAT2*7B, and lowest in cells transfected with NAT2*5B. Following ABP treatment, N-(deoxyguanosin-8-yl)-4-aminobiphenyl (dG-C8-ABP) was the primary adduct formed. Cells transfected with both CYP1A1 and NAT2*4 showed the highest concentration-dependent cytotoxicity, hypoxanthine phosphoribosyl transferase (hprt) mutants, and dG-C8-ABP adducts. Cells transfected with CYP1A1 and NAT2*7B showed lower levels of cytotoxicity, hprt mutagenesis, and dG-C8-ABP adducts. Cells transfected with CYP1A1 only or cells transfected with both CYP1A1 and NAT2*5B did not induce cytotoxicity, hprt mutagenesis or dG-C8-ABP adducts. ABP DNA adduct levels correlated very highly (r > 0.96) with ABP-induced hprt mutant levels following each treatment. The results of the present study suggest that investigations of NAT2 genotype or phenotype associations with disease or toxicity could be more precise and reproducible if heterogeneity within the “slow” NAT2 acetylator phenotype is considered and incorporated into the study design.
Keywords: N-acetyltransferase 2, Acetylator genotype, 4-aminobiphenyl, DNA adducts
1. Introduction
4-Aminobiphenyl (ABP) is present in both mainstream and side stream cigarette smoke [1] and recently was reaffirmed as a human urinary bladder carcinogen [2]. ABP-induced DNA adduct formation and mutagenesis require metabolic activation. P450s 1A1 and 1A2 catalyze the N-hydroxylation of arylamines in human tissues [3]. Although CYP1A2 is considered the primary isozyme for hepatic activation, CYP1A1 may be particularly important in urinary bladder, the target organ for ABP carcinogenesis in humans [4]. Cyp1a2 knockout mice do not differ in ABP DNA adduct levels in either urinary bladder or liver compared to wildtype mice [5,6]. Genetic polymorphisms in CYP1A1 have been shown to modify DNA adduct levels in normal breast tissues of women with breast cancer, especially in smokers [7]. As depicted in Figure 1, N-acetylation of ABP catalyzed by arylamine N-acetyltransferase 2 (NAT2) is generally considered a detoxification step since it competes with P450-catalyzed N-hydroxylation. Following N-hydroxylation, O-acetylation of N-hydroxy-ABP catalyzed by arylamine N-acetyltransferase 2 (NAT2) generates acetoxy-derivatives that are highly unstable, leading to the formation of an arylnitrenium ion that binds to DNA and ultimately leads to mutagenesis and carcinogenesis [3]. In humans, the major ABP-induced DNA adduct is N-(deoxyguanosin-8-yl)-4-aminobiphenyl (dG-C8-ABP) [8,9].
Fig. 1.

Metabolic pathways for 4-aminobiphenyl.
Epidemiological data on the role of NAT2 genetic polymorphism in susceptibility to various cancers suggest that the role of NAT2 genetic polymorphism varies both with carcinogen and organ site [10]. PubMed search (conducted April 2009) of “NAT2” and “cancer” yields over 500 literature citations. Because DNA adduct levels are a function of environmental exposure and polymorphism in genes involved in carcinogen metabolism, DNA adducts are an informative biomarker for investigations of genetic variation in carcinogen metabolism. There is a critical need to characterize genotype and associated phenotypic catalytic activities of NAT2 enzymes at physiological concentrations of co-factors and environmental exposure levels of carcinogen.
NAT2 genetic polymorphisms resulting in rapid and slow acetylator phenotypes are frequent in human populations [10]. The NAT2*4 allele (haplotype) is associated with rapid acetylator phenotype, whereas the NAT2*5B and NAT2*7B alleles are associated with slow acetylator phenotype [10]. Relative to the NAT2*4 reference allele, NAT2*5B possesses three single nucleotide polymorphisms (SNPs) in the NAT2 open reading frame: T341C (I114T), C481T (synonymous) and A803G (K268R) whereas NAT2*7B possesses two polymorphisms in the NAT2 open reading frame: C282T (synonymous) and G857A (G286E) [10,11]. Epidemiologic studies suggest a role for NAT2 genetic polymorphism in susceptibility to various cancers related to arylamine exposures [10], but inconsistent results in these and other molecular epidemiology studies [11,12] suggest the need for laboratory-based experiments to support the biological plausibility and the conclusions inferred from these epidemiologic studies [13]. For example Ambrosone et al [14] reported that slow acetylator NAT2 genotype increased risk for breast cancer in postmenopausal smokers but studies to replicate these findings have been inconsistent. Whereas some studies are supportive of this initial report [15–18], other studies [19–25] reported no appreciable difference between rapid and slow NAT2 acetylator smokers for breast cancer risk, while three others reported that rapid acetylator NAT2 smokers were possibly at higher risk [26–28].
We expected ABP-induced DNA adducts and mutants would be higher in slow than in rapid NAT2 acetylators. To test this hypothesis, we measured ABP-induced DNA adducts and mutants in nucleotide excision repair-deficient Chinese hamster ovary (CHO) cell lines transfected with human CYP1A1 and rapid or slow acetylator NAT2 alleles.
2. Materials and methods
2.1 Cell Culture
The UV5-CHO cell line, a nucleotide excision repair-deficient derivative of the AA8 line [29], was obtained from the ATCC (Catalog number: CRL-1865). Since UV5-CHO lacks nucleotide excision repair due to a mutation in the XPD (ERCC2) gene [30], it is hypersensitive to bulky adduct mutagens and belongs to the excision repair cross complementation group 2. All cells were grown in alpha-modified minimal essential medium (Cambrex) without L-glutamine, ribosides, and deoxyribosides supplemented with 10% fetal bovine serum (Hyclone), 100 units/ml penicillin, 100 micrograms/ml streptomycin (Cambrex), and 2 mM L-glutamine (Cambrex) at 37 °C in 5% CO2. Media were supplemented with appropriate selective agents to maintain stable transfectants.
2.2. Construction and characterization of UV5/CHO cell lines
Construction of UV5/CHO cells expressing human CYP1A1 and either NAT2*4 or NAT2*5B was recently reported and characterized [31]. An identical method was used for construction of UV5/CHO cells expressing CYP1A1 and NAT2*7B. Briefly, the NAT2*7B open reading frame was amplified by polymerase chain reaction (PCR), digested with NheI and XhoI (New England Biolabs), and inserted into the similarly prepared pcDNA5/FRT vector (Invitrogen). The pcDNA5/FRT plasmid containing human NAT2*7B was co-transfected with pOG44, a Flp recombinase expression plasmid, into UV5FRT/CYP1A1 cells constructed and characterized previously [31]. Integration of the pcDNA5/FRT construct into the FRT site was confirmed by PCR. Geneticin-and hygromycin B-resistant clones containing the NAT2*7B/CYP1A1 were expanded and assayed for CYP1A1 activity by measuring 7-ethoxyresorufin O-deethylase (EROD) activity as described previously [32]. NAT2*7B/CYP1A1 clones with similar levels of EROD activity to the NAT2*4/CYP1A1 and NAT2*5B/CYP1A1 cell lines were selected for survival, mutagenesis, and DNA adduct studies.
Cell lysates from the UV5 and each of the transfected CHO cell lines were tested for sulfamethazine (SMZ; an NAT2-specific substrate) and ABP N-acetyltransferase activities as previously described [33,34].
2.3. Cell survival and mutagenesis
Assays for cytotoxicity and mutagenesis were carried out as described previously [32]. Briefly, cells were grown for 12 doublings, with selective agents in complete hypoxanthine-aminopterin-thymidine medium (30 µM hypoxanthine, 0.1 µM aminopterin, and 30 µM thymidine). Cells were plated at a density of 5 × 105 cells/T-25 flask and incubated for 24 h, after which media were changed and the cells were treated separately for 48 h under cover with - 1 to 4 µM ABP or vehicle control (0.5% dimethyl sulfoxide). Survival was determined by colony-forming assay and expressed as percent of vehicle control. The remaining cells were replated and subcultured. After 7 days of growth, cultures were plated for cloning efficiency in complete media and for hypoxanthine phosphoribosyl transferase (hprt) mutations in complete media containing 40 µM 6-thioguanine (Sigma). Dishes were seeded with 1 × 105 cells/100 mm dish (10 replicates) and incubated for 7 days; cloning efficiency dishes were seeded with 100 cells/well/six-well plate in triplicate and incubated for 6 days.
2.4. Identification and quantitation of ABP-DNA adducts
dG-C8-ABP and dG-C8-ABP-D5 adduct standard (>95% purity) were obtained from Toronto Research Chemicals. Cells grown in 15-cm plates were treated separately with ABP as described above for the cell survival and mutagenesis assays. Cells were harvested after 48 h of ABP treatment and DNA was extracted and quantified as previously described [32]. One-tenth volumes each of proteinase K solution (20 mg/mL) and 10% SDS were added to the cell lysate, and the mixture was incubated at 37°C for 60 min. One volume of phenol, equilibrated with 10 mM Tris HCl (pH 8.0), was added to the mixture, which was then vortexed and centrifuged at 3,600 × g for 15 min. The aqueous layer was removed and added to 1 volume of phenol/chloroform/isoamyl alcohol (25:24:1) saturated with 10 mM Tris HCl (pH 8.0), which was vortexed and centrifuged. The aqueous layer was removed and added to 1 volume of cold (−20°C) isopropanol, and the mixture was vortexed and centrifuged. The DNA pellet was washed with 70% ethanol and redissolved in 5 mM Tris HCl (pH 7.4) containing 1 mM CaCl2, 1 mM ZnCl2, and 10 mM MgCl2. DNA was quantified by UV spectroscopy using A260 nm. DNA quality was monitored by UV spectroscopy using A260/280 nm and this ratio was consistently above 1.9. DNA samples (200 µg) added to 1 ng (3.3 adducts per 106 DNA bases) deuterated internal standard (dG-C8-ABP-D5) were digested at 37ºC with 10 units DNAse I (US Biological) for 1 h followed by 5 units micrococcal nuclease (Sigma), 5 units nuclease P1 (US Biological), 0.01 units spleen phosphodiesterase (Sigma), and 0.01 units snake venom phosphodiesterase (Sigma) for 6 h followed by 5 units alkaline phosphatase (Sigma) overnight. Two volumes of acetonitrile were added to the digest, which was then filtered and concentrated to 100 µl in a speed vacuum.
Samples were subjected to binary gradient high performance liquid chromatography (HPLC) and introduced into a Micromass Quattro LC triple quadrupole mass spectrometer using a custom-built nanospray as described previously [32,35]. Samples were loaded onto a Inertsil C18 precolumn (5 mm x 300 m i.d., 5 µm; LC Packings) using Perkin Elmer ABI 140D syringe pumps and a Hewlett Packard 1100 Series autosampler. Multiple reaction monitoring (dwell time, 0.5 s; span, 0.4 Da) was used to measure the [M+H]+ to [(M-116) + H]+ (loss of deoxyribose) mass transition. Multiple reaction monitoring in the electrospray ionization-positive ion mode was carried out using argon as the collision gas. Capillary and cone voltages and collision energies were optimized for cleavage of the glycosidic bond. The dG-C8-ABP adduct was monitored using the transition from m/z 435 to m/z 319 and the deuterated internal standard (dG-C8-ABP-D5) was monitored using the transition from m/z 440 to m/z 324.
Results
The CYP1A1-, CYP1A1/NAT2*5B-, CYP1A1/NAT2*7B- and CYP1A1/NAT2*4- transfected cell lines catalyzed EROD activity at 100–120 relative fluorescence units (RFU)/ million cells that did not differ significantly (p>0.05) from each other. CYP1A1 EROD catalytic activity was not detected (<5 RFU/million cells) in the untransfected UV5 cells.
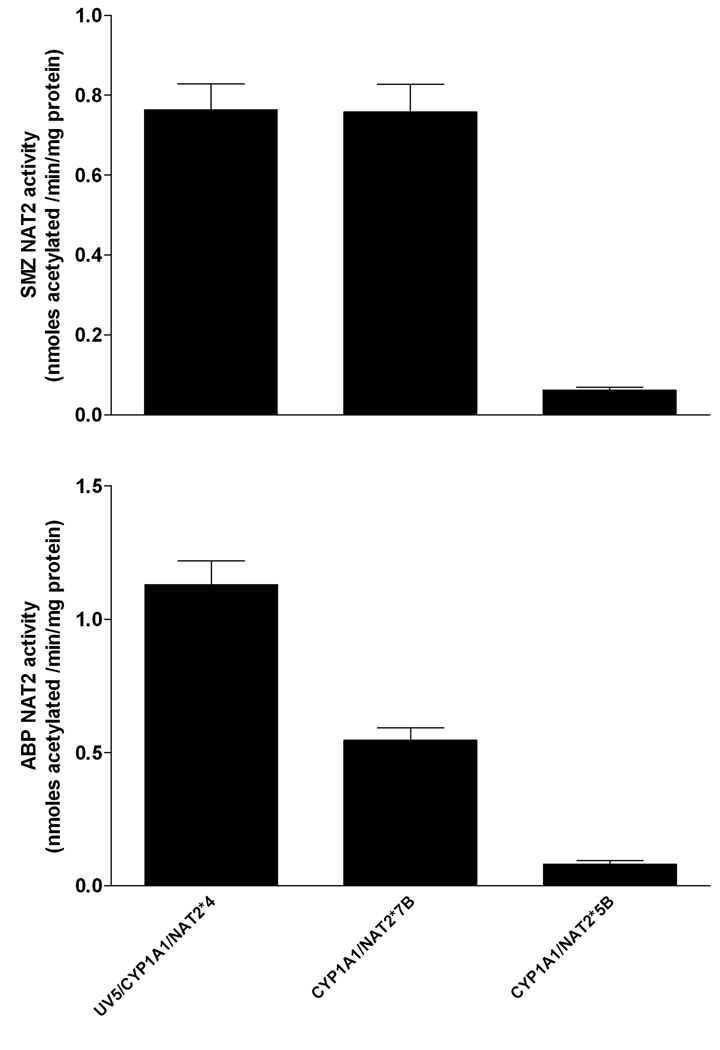
Transfection of NAT2*7B was confirmed by measurement of human NAT2-specific SMZ N-acetyltransferase catalytic activity. SMZ NAT2 activity in the UV5/CYP1A1/NAT2*7B cell line was similar to the UV5/CYP1A1/NAT2*4 cell line and over 12-fold greater (p<0.001) than the UV5/CYP1A1/NAT2*5B cell line (Fig. 2). SMZ N-acetyltransferase catalytic activity was not detected in the UV5 or UV5/CYP1A1 cell lines (<20 pmole/min/mg). Cell lysates from UV5 and each of the transfected CHO cell lines were tested for ABP NAT2 catalytic activity. ABP NAT2 catalytic activity in UV5/CYP1A1/NAT2*4 cells was about 2-fold higher (p<0.01) than in the UV5/CYP1A1/NAT2*7B cell line, which was about 7-fold higher (p<0.01) than the UV5/CYP1A1/NAT2*5B cell line (Fig. 2). ABP NAT2 activity was not detected in the UV5 and UV5/CYP1A1 cell lines (<10 pmol/min/mg protein).
Fig. 2.
NAT2 catalytic activities in cell lysates of CYP1A1/NAT2*4, CYP1A1/NAT2*7B- and CYP1A1/NAT2*5B-transfected CHO cells. Each bar represents Mean ± S.E.M. for three determinations. NAT2 activities in the UV5 and UV5/CYP1A1 cell lines were not detected (<20 pmole/min/mg). Sulfamethazine (SMZ) NAT2 catalytic activities (top) were significantly lower (p<0.001) in the CYP1A1/NAT2*5B- transfected cell line. ABP NAT2 catalytic activities (bottom) differed significantly (p<0.01) between all NAT2-transfected cell lines. ABP NAT2 activity was not detected in the UV5 and UV5/CYP1A1 cell lines (<10 pmol/min/mg protein).
As shown in Figure 3A, ABP treatment resulted in dose-dependent reductions in survival of the CYP1A1/NAT2*4- and CYP1A1/NAT2*7B–transfected CHO cell lines. Cell survival following 2–4 µM ABP treatment differed significantly (p<0.001) in the order UV5/CYP1A1/NAT*4 < UV5/CYP1A1/NAT2*7B < UV5, UV5/CYP1A1, and UV5/CYP1A1/NAT2*5B. As shown in Figure 3B, ABP treatment resulted in dose-dependent production of hprt mutants in the CYP1A1/NAT2*4- and CYP1A1/NAT2*7B–transfected CHO cell lines. ABP -induced hprt mutants differed significantly in the order UV5/CYP1A1/NAT2*4 > UV5/CYP1A1/NAT2*7B > UV5, UV5/CYP1A1, and UV5/CYP1A1/NAT2*5B following 1 µM (p<0.05) and 2–4 µM (p<0.001) ABP treatment.
Fig. 3.
A: Cell survival in UV5 CHO cell lines following treatment with ABP. Percent survival on the ordinate is plotted versus ABP treatment concentration on the abscissa. Each data point represents Mean ± S.E.M. for three to six experiments in UV5 (open circles), UV5/CYP1A1 (closed circles), UV5/CYP1A1/NAT2*5B (open squares), UV5/CYP1A1/NAT2*7B (closed squares), and UV5/CYP1A1/NAT2*4 (open triangles) cell lines. Cell survival differed significantly (p<0.001) in the order UV5/CYP1A1/NAT*4 < UV5/CYP1A1/NAT2*7B < UV5, UV5/CYP1A1, and UV5/CYP1A1/NAT2*5B following 2–4 µM ABP treatment.
B: ABP -induced hprt mutants in UV5 CHO cell lines. ABP-induced hprt mutants are plotted on the ordinate versus treatment concentration on the abscissa. Each data point represents Mean ± S.E.M. for two to five experiments in UV5 (open circles), UV5/CYP1A1 (closed circles), UV5/CYP1A1/NAT2*5B (open squares), UV5/CYP1A1/NAT2*7B (closed squares), and UV5/CYP1A1/NAT2*4 (open triangles) cell lines. ABP -induced hprt mutants differed significantly in the order UV5/CYP1A1/NAT*4 > UV5/CYP1A1/NAT2*7B > UV5, UV5/CYP1A1, and UV5/CYP1A1/NAT2*5B following 1 µM (p<0.05) and 2–4 µM (p<0.001) ABP treatment.
dG-C8-ABP standard was characterized by HPLC-tandem mass spectrometry (LC-MS/MS) and used to verify the identity of DNA adducts formed in cell culture. dG-C8-ABP corresponding to m/z 435, with a major aglycone ion of m/z 319 was identified in CHO cells treated with ABP (Fig 4). dG-C8-ABP was quantitated by comparison to dG-C8-ABP-D5 used as an internal standard. Identical patterns of fragmentation were observed for dG-C8-ABP and dG-C8-ABP-D5, except that the latter were shifted by 5 Da. As shown in Figure 5, dG-C8-ABP DNA adduct levels were dose-dependent only in the CYP1A1/NAT2*4- and CYP1A1/NAT2*7B–transfected cell lines. ABP-DNA adduct levels were not detected in UV5 CHO cell lines, nor in lines transfected with CYP1A1 or CYP1A1/NAT2*5B. ABP-DNA adduct levels were significantly (p<0.001) higher in UV5/CYP1A1/NAT2*4 than UV5/CYP1A1/NAT2*7B cell lines after each ABP treatment. ABP DNA adduct levels correlated very highly (r > 0.96) with ABP-induced hprt mutant levels following each treatment. Transfection with NAT2 was required for significant detection of both adducts and mutants in the order NAT2*4 ≫ NAT2*7B ≫NAT2* 5B.
Fig. 4.

Electrospray ionization spectra of dG-C8-ABP and dG-C8-ABP-D5. Collision induced dissociation fragmentation of dG-C8-ABP and dG-C8-ABP-D5 at −50 V collision energy (middle panels) shows fragmentation of the ABP and guanosine moieties dominated by the major fragment produced from loss of deoxyribose: the aglycone ion of m/z 319 or m/z 324 for dG-C8-ABP and dG-C8-ABP-D5, respectively. The collision energy was −25 V for multiple reaction monitoring transitions used during quantitation (lower panels).
Fig. 5.
ABP -induced dG-C8-ABP adduct levels in UV5 CHO cell lines. Adduct levels are plotted on the ordinate versus ABP treatment concentration on the abscissa. Each data point represents Mean ± S.E.M. for three experiments (the S.E.M. sometimes falls within the symbol) in UV5/CYP1A1/NAT2*5B (open squares), UV5/CYP1A1/NAT2*7B (closed squares), and UV5/CYP1A1/NAT2*4 (open triangles) cell lines. dG-C8-ABP DNA adduct levels were not detected in UV5 CHO cell lines, nor in CYP1A1- or CYP1A1/NAT2*5B- transfected cell lines (data not shown). dG-C8-ABP DNA adduct levels were significantly (p<0.001) higher in UV5/CYP1A1/NAT2*4 than UV5/CYP1A1/NAT2*7B cell lines at each ABP treatment concentration.
Discussion
Our studies clearly show that metabolic activation of ABP can be catalyzed by CYP1A1 and that mutagenesis and DNA adduct formation require further metabolic activation by NAT2. Metabolic O-acetylation by NAT2 substantially increased levels of DNA adducts and mutants and rapid acetylator NAT2 catalyzed this activation to a substantially greater extent than slow acetylator NAT2. The primary DNA adduct formed was dG-C8-ABP, which is the primary adduct that has been identified in human urinary bladder [8,36,37], breast [38–40], lung [41] and pancreas [9] and is consistent with N-hydroxylation followed by O-acetylation pathway as depicted in Figure 1. The ABP concentration-dependent increases in dG-C8-ABP adducts and hprt mutants were similar and dG-C8-ABP adduct levels correlated very highly (r > 0.96) with hprt mutant frequency providing further evidence for this pathway. Taken together, these results strongly suggest that ABP-induced mutagenesis follows formation of the dG-C8-ABP adduct which in turn is dependent upon O-acetylation catalyzed by NAT2 as depicted in Figure 1.
Previous studies have shown that the effect of NAT2 polymorphism on the levels of DNA adducts and mutations varies with carcinogen. Following treatment of human CYP1A1- or CYP1A2-transfected CHO cells with 0–2 µM 2-amino-1-methyl-6-phenylimidazo [4,5-b] pyridine, additional transfection with rapid and slow acetylator NAT2 had little further effect [32,42]. In contrast, following treatment of human CYP1A1 transfected CHO cells with 0–3 µM 2-amino-3,8-dimethylimidazo-[4,5-f]quinoxaline [31], and of human CYP1A2-transfected CHO cells with 0–6 µM 2-amino-9H–pyrido[2,3-b]indole [43], additional transfection with rapid and slow acetylator NAT2 substantially increased levels of both DNA adducts and mutants. However, these arylamines are heterocyclic amine carcinogens which undergo N-acetylation very poorly [44–46]. Thus, it is not unexpected that O-acetylation of the N-hydroxy metabolites leads to higher levels of DNA adducts and mutants in rapid NAT2 acetylators. In contrast, ABP is an aromatic amine carcinogen that readily undergoes both N-acetylation and following N-hydroxylation, O-acetylation both catalyzed by NAT2 [47,48]. Thus, NAT2 could decrease or increase DNA adducts and mutants as depicted in Figure 1. Consistent with current literature and established theory, we expected ABP-induced DNA adducts and mutants would be higher in slow than in rapid NAT2 acetylators. However, our results clearly showed that they were both actually higher following transfection with NAT2*4, the rapid acetylator allele. Our results are consistent with earlier studies reporting higher levels of DNA damage in hepatocytes from rapid NAT2 acetylator rabbits [49], and with urinary bladder DNA adduct levels in rapid NAT2 acetylator congenic Syrian hamsters [50] following exposure to another aromatic amine (2-aminofluorene). In addition, the results are consistent with higher levels of breast ABP-DNA adducts [40], and higher levels of bulky DNA adducts in urothelial [51] and white blood cells [52] of human rapid NAT2 acetylators.
Our most novel finding was the significant differences observed between CYP1A1/NAT2*7B-and CYP1A1/NAT2*5B–transfected CHO cells for both DNA adducts and mutants following ABP treatment. The differences were significant at all ABP concentrations (1–4 µM) tested but increased as the ABP concentration increased. This may reflect treatment concentrations below the affinity of ABP or N-hydroxy-ABP for human NAT2. An apparent Km of 26 µM for ABP [53] and 108 µM for N-hydroxy-ABP [54] have been reported for human NAT2. Although both NAT2*4 and NAT2*7B are associated with “slow” NAT2 acetylator phenotype, previous studies have shown that recombinant expression of the NAT2*5B allele results in strikingly lower 2-aminofluorene N-acetyltransferase [55] and N-hydroxy-ABP O-acetyltransferase [56] catalytic activities than does recombinant expression of the NAT2*7B allele.
As recently reviewed [11,57], different mechanisms have been reported for the reductions in NAT2 activity resulting from SNPs within the NAT2*5B and NAT2*7B alleles. The T341C (I114T) polymorphism in NAT2*5B yields slow acetylator phenotype by enhanced protein degradation [58], whereas the G857A (G286E) polymorphism in NAT2*7B yields slow acetylator phenotype through altered affinity for some but not all substrates and acetyl coenzyme A cofactor [48]. Interestingly, G857A (G286E) has been shown to decrease apparent Km towards N-hydroxy-ABP from 108 to 50 µM [54]. Since single nucleotide polymorphisms effect changes via variant mechanisms, it follows that there may be heterogeneity within the “slow” acetylator phenotype [59] and such observations have appeared in the scientific literature. For example, isoniazid -induced hepatotoxicity is highest in “super slow” acetylators [60,61]. Individuals with slow acetylator NAT2 phenotype experienced more severe hepatotoxicity than rapid acetylators and less risk was observed in individuals possessing NAT2*7 haplotypes [62]. English chemical dye workers with documented exposure to aromatic amine carcinogens showed a striking association between urinary bladder cancer and slow acetylator phenotypes [63]. The NAT2 phenotype data was not separated into two phenotypes (rapid and slow), but rather into eight phenotypes of metabolic ratios. Five of these ratios correspond to rapid acetylators, and the other three correspond to various degrees of slow acetylator phenotype. As reviewed previously [64], urinary bladder cancer risk increased as NAT2 metabolic ratio (phenotype) decreased and the risk was markedly increased in the “slowest” NAT2 phenotype. Four studies found that urinary bladder cancer risk was highest in individuals possessing NAT2*5 haplotypes [65–68]. NAT2*5 alleles also were associated with increased risk for breast cancer in women smokers [69–70]. These results suggest that NAT2 slow acetylator phenotype is not homogeneous, but rather that multiple slow acetylator phenotypes exist resulting from different mechanisms inferred by various SNPs and haplotypes.
An increased understanding of NAT2 genotype/phenotype relationships should lead to improved individualized risk assessments to disease and/or toxicities associated with carcinogen exposures. Lack of replication regarding the role of NAT2 genotype or phenotype on cancer risk following arylamine exposures may lead many to conclude that NAT2 genetic polymorphisms are not sufficiently biologically relevant to merit attention. The results of the present study suggest that the investigations of NAT2 genotype or phenotype associations with disease or toxicity could be more precise and reproducible if heterogeneity within the “slow” NAT2 acetylator phenotype is considered and incorporated into the study design.
Acknowledgements
This research was supported in part by United States Public Health Service grants R01-CA034627, T32-ES011564 and P30-ES014443.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stabbert R, Schafer KH, Biefel C, Rustemeier K. Analysis of aromatic amines in cigarette smoke. Rapid Commun Mass Spectrom. 2003;17:2125–2132. doi: 10.1002/rcm.1161. [DOI] [PubMed] [Google Scholar]
- 2.Baan R, Straif K, Grosse Y, Secretan B, Ghissassi FEl, Bouvard V, Benbrahim-Tallaa L, Cogliano V. Carcinogenicity of some aromatic amines, dyes, organic dyes, and related exposures. Lancet Oncol. 2008;9:322–323. doi: 10.1016/S1470-2045(08)70089-5. [DOI] [PubMed] [Google Scholar]
- 3.Kim D, Guengerich FP. Cytochrome P450 activation of arylamines and heterocyclic amines. Annu Rev Pharmacol Toxicol. 2005;45:27–49. doi: 10.1146/annurev.pharmtox.45.120403.100010. [DOI] [PubMed] [Google Scholar]
- 4.Dorrenhaus, Muller T, Roos PH. Increased CYP1A1 expression in human exfoliated urothelial cells of cigarette smokers compared to non-smokers. Arch Toxicol. 2007;81:19–25. doi: 10.1007/s00204-006-0134-9. [DOI] [PubMed] [Google Scholar]
- 5.Tsuneoka Y, Dalton TP, Miller ML, Clay CD, Shertzer HG, Talaska G, Medvedovic M, Nebert DW. 4-aminobiphenyl-induced liver and urinary bladder DNA adduct formation in Cyp1a2(−/−) and Cyp1a2(+/+) mice. J Natl Cancer Inst. 2003;95:1227–1237. doi: 10.1093/jnci/djg025. [DOI] [PubMed] [Google Scholar]
- 6.Talaska G, Ginsburg D, LaDow K, Puga A, Dalton T, Warshawsky D. Impact of Cyp1a2 or Ahr gene knockout in mice: implications for biomonitoring studies. Toxicol Lett. 2006;162:246–249. doi: 10.1016/j.toxlet.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 7.Firozi PF, Bondy ML, Sahin AA, Chang P, Lukmanji F, Singletary ES, Hassan MM, Li D. Aromatic DNA adducts polymorphisms of CYP1A1, NAT2, and GSTM1 in breast cancer. Carcinogenesis. 2002;23:301–306. doi: 10.1093/carcin/23.2.301. [DOI] [PubMed] [Google Scholar]
- 8.Talaska G, al-Juburi AZ, Kadlubar FF. Smoking related carcinogen-DNA adducts in biopsy samples of human urinary bladder: identification of N-(deoxyguanosin-8-yl)-4-aminobiphenyl as a major adduct. Proc Natl Acad Sci U S A. 1991;88:5350–5354. doi: 10.1073/pnas.88.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ricicki EM, Soglia JR, Teitel C, Kane R, Kadlubar F, Vouros P. Detection and quantification of N-(deoxyguanosin-8-yl)-4-aminobiphenyl adducts in human pancreas tissue using capillary liquid chromatography-microelectrospray mass spectrometry. Chem Res Toxicol. 2005;18:692–699. doi: 10.1021/tx049692l. [DOI] [PubMed] [Google Scholar]
- 10.Hein DW, Doll MA, Fretland AJ, Leff MA, Webb SJ, Xiao GH, Devanaboyina US, Nangju NA, Feng Y. Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol Biomarkers Prev. 2000;9:29–42. [PubMed] [Google Scholar]
- 11.Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet. 2002;3:391–397. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]
- 12.Colhoun HM, McKeigue PM, Davey G. Smith Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361:865–872. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- 13.Hein DW. N-acetyltransferase single nucleotide polymorphisms: Emerging concepts serve as a paradigm for understanding complexities of personalized medicine. Exp Opin Drug Metab Toxicology. 2009;5:353–366. doi: 10.1517/17425250902877698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambrosone CB, Freudenheim JL, Graham S, Marshall JR, Vena JE, Brasure JR, Michalek AM, Laughlin R, Nemoto T, Gillenwater KA, Shields PG. Cigarette smoking, N-acetyltransferase 2 genetic polymorphisms, and breast cancer risk. JAMA. 1996;276:1494–1501. [PubMed] [Google Scholar]
- 15.Huang CS, Chern HD, Shen CY, Hsu SM, Chang KJ. Association between N-acetyltransferase 2 (NAT2) genetic polymorphism and development of breast cancer in post-menopausal Chinese women in Taiwan, an area of great increase in breast cancer incidence. Int J Cancer. 1999;82:175–179. doi: 10.1002/(sici)1097-0215(19990719)82:2<175::aid-ijc4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 16.Alberg AJ, Daudt A, Huang HY, Hoffman SC, Comstock GW, Helzlsouer KJ, Strickland PT, Bell DA. N-acetyltransferase 2 (NAT2) genotypes, smoking, cigarette and the risk of breast cancer. Cancer Detect Prev. 2004;28:187–193. doi: 10.1016/j.cdp.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Sillanpaa P, Hirvonen A, Kataja V, Eskelinen M, Kosma VM, Uusitupa M, Vainio H, Mitrunen K. NAT2 slow acetylator genotype as an important modifier of breast cancer risk. Int J Cancer. 2005;114:579–584. doi: 10.1002/ijc.20677. [DOI] [PubMed] [Google Scholar]
- 18.Ambrosone CB, Kropp S, Yang J, Yao S, Shields PG, Chang-Claude J. Cigarette smoking, N-acetyltransferase 2 genotypes, and breast cancer risk: pooled analysis and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2008;17:15–26. doi: 10.1158/1055-9965.EPI-07-0598. [DOI] [PubMed] [Google Scholar]
- 19.Hunter DJ, Hankinson SE, Hough H, Gertig DM, Garcia-Closas M, Spiegelman D, Manson JE, Colditz GA, Willett WC, Speizer FE, Kelsey K. A prospective study of NAT2 acetylation genotype, cigarette smoking, and risk of breast cancer. Carcinogenesis. 1997;18:2127–2132. doi: 10.1093/carcin/18.11.2127. [DOI] [PubMed] [Google Scholar]
- 20.Delfino RJ, Smith C, West JG, Lin HJ, White E, Liao SY, Gim JS, Ma HL, Butler J, Anton-Culver H. Breast cancer, passive and active cigarette smoking and N-acetyltransferase 2 genotype. Pharmacogenetics. 2000;10:461–469. doi: 10.1097/00008571-200007000-00009. [DOI] [PubMed] [Google Scholar]
- 21.Krajinovic M, Ghadirian P, Richer C, Sinnett H, Gandini S, Perret C, Lacroix A, Labuda D, Sinnett D. Genetic susceptibility to breast cancer in French-Canadians: role of carcinogen-metabolizing enzymes and gene-environment interactions. Int J Cancer. 2001;92:220–225. doi: 10.1002/1097-0215(200102)9999:9999<::aid-ijc1184>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 22.Egan KM, Newcomb PA, Titus-Ernstoff L, Trentham-Dietz A, Mignone LI, Farin F, Hunter DJ. Association of NAT2 and smoking in relation to breast cancer incidence in a population-based case-control study (United States) Cancer Causes Control. 2003;14:43–51. doi: 10.1023/a:1022517506689. [DOI] [PubMed] [Google Scholar]
- 23.Lash TL, Bradbury BD, Wilk JB, Aschengrau A. A case-only analysis of the interaction between N-acetyltransferase 2 haplotypes and tobacco smoke in breast cancer etiology. Breast Cancer Research. 2005;7:R385–R393. doi: 10.1186/bcr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lissowska J, Brinton LA, Zatonski W, Blair A, Bardin-Mikolajczak A, Peplonska B, Sherman ME, Szeszenia-Dabrowska N, Chanock S, Garcia-Closas M. Tobacco smoking, NAT2 acetylation genotype and breast cancer risk. Int J Cancer. 2006;119:1961–1969. doi: 10.1002/ijc.22044. [DOI] [PubMed] [Google Scholar]
- 25.Ochs-Balcom HM, Wiesner G, Elston RC. A meta-analysis of the association of N-acetyltransferase 2 gene (NAT2) variants with breast cancer. Am J Epidemiol. 2007;166:246–254. doi: 10.1093/aje/kwm066. [DOI] [PubMed] [Google Scholar]
- 26.Millikan RC, Pittman GS, Newman B, Tse CK, Selmin O, Rockhill B, Savitz D, Moorman PG, Bell DA. Cigarette smoking, N-acetyltransferases 1 and 2, and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 1998;7:371–378. [PubMed] [Google Scholar]
- 27.Morabia A, Bernstein MS, Bouchardy I, Kurtz J, Morris MA. Breast cancer and active and passive smoking: the role of the N-acetyltransferase 2 genotype. Am J Epidemiol. 2000;152:226–232. doi: 10.1093/aje/152.3.226. [DOI] [PubMed] [Google Scholar]
- 28.Chang-Claude J, Kropp S, Jager B, Bartsch H, Risch A. Differential effect of NAT2 on the association between active and passive smoke exposure and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:698–704. [PubMed] [Google Scholar]
- 29.Thompson LH, Rubin JS, Cleaver JE, Whitmore GF, Brookman K. A screening method for isolating DNA repair-deficient mutants of CHO cells. Somatic Cell Genet. 1980;6:391–405. doi: 10.1007/BF01542791. [DOI] [PubMed] [Google Scholar]
- 30.Weber CA, Salazar EP, Stewart SA, Thompson LH. Molecular cloning and biological characterization of a human gene, ERCC2, that corrects the nucleotide excision repair defect in CHO UV5 cells. Mol Cell Biol. 1988;8:1137–1146. doi: 10.1128/mcb.8.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bendaly J, Zhao S, Neale JR, Metry KJ, Doll MA, States JC, Pierce WM, Jr, Hein DW. 2-Amino-3,8-dimethylimidazo-[4,5-f]quinoxaline-induced DNA adduct formation and mutagenesis in DNA repair-deficient Chinese hamster ovary cells expressing human cytochrome P4501A1 and rapid or slow acetylator N-acetyltransferase 2. Cancer Epidemiol Biomarkers Prev. 2007;16:1503–1509. doi: 10.1158/1055-9965.EPI-07-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metry KJ, Zhao S, Neale JR, Doll MA, States JC, McGregor WG, Pierce WM, Jr, Hein DW. 2-amino-1-methyl-6-phenylimidazo [4,5-b] pyridine-induced DNA adducts and genotoxicity in chinese hamster ovary (CHO) cells expressing human CYP1A2 and rapid or slow acetylator N-acetyltransferase 2. Mol Carcinog. 2007;46:553–563. doi: 10.1002/mc.20302. [DOI] [PubMed] [Google Scholar]
- 33.Leff MA, Fretland AJ, Doll MA, Hein DW. Novel human N-acetyltransferase 2 alleles that differ in mechanism for slow acetylator phenotype. J Biol Chem. 1999;274:34519–34522. doi: 10.1074/jbc.274.49.34519. [DOI] [PubMed] [Google Scholar]
- 34.Fretland AJ, Doll MA, Zhu Y, Smith L, Leff MA, Hein DW. Effect of nucleotide substitutions in N-acetyltransferase-1 on N-acetylation (deactivation) and O-acetylation (activation) of arylamine carcinogens: implications for cancer predisposition. Cancer Detect Prev. 2002;26:10–14. doi: 10.1016/s0361-090x(02)00005-3. [DOI] [PubMed] [Google Scholar]
- 35.Neale JR, Smith NB, Pierce WM, Hein DW. Methods for aromatic and heterocyclic amine carcinogen-DNA adduct analysis by liquid chromatography-tandem mass spectrometry. Polycycl Aromat Compd. 2008;28:402–417. doi: 10.1080/10406630802377773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartsch H, Malaveille C, Friesen M, Kadlubar FF, Vineis P. Black (air-cured) and blond (flue-cured) tobacco cancer risk. IV: Molecular dosimetry studies implicate aromatic amines as bladder carcinogens. Eur J Cancer. 1993;29A:1199–1207. doi: 10.1016/s0959-8049(05)80315-6. [DOI] [PubMed] [Google Scholar]
- 37.Zayas B, Stillwell SW, Wishnok JS, Trudel LJ, Skipper P, Yu MC, Tannenbaum SR, Wogan GN. Detection and quantification of 4-ABP adducts in DNA from bladder cancer patients. Carcinogenesis. 2007;28:342–349. doi: 10.1093/carcin/bgl142. [DOI] [PubMed] [Google Scholar]
- 38.Gorlewska-Roberts K, Green B, Fares M, Ambrosone CB, Kadlubar FF. Carcinogen-DNA adducts in human breast epithelial cells. Environ Mol Mutagen. 2002;39:184–192. doi: 10.1002/em.10060. [DOI] [PubMed] [Google Scholar]
- 39.Faraglia B, Chen SY, Gammon MD, Zhang Y, Teitelbaum SL, Neugut AI, Ahsan H, Garbowski GC, Hibshoosh H, Lin D, Kadlubar FF, Santella RM. Evaluation of 4-aminobiphenyl-DNA adducts in human breast cancer: the influence of tobacco smoke. Carcinogenesis. 2003;24:719–725. doi: 10.1093/carcin/bgg013. [DOI] [PubMed] [Google Scholar]
- 40.Ambrosone CB, Abrams SM, Gorlewska-Roberts K, Kadlubar FF. Hair dye use, meat intake, and tobacco exposure and presence of carcinogen-DNA adducts in exfoliated breast ductal epithelial cells. Arch Biochem Biophys. 2007;464:169–175. doi: 10.1016/j.abb.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 41.Culp SJ, Roberts DW, Talaska G, Lang NP, Fu PP, Lay JO, Jr, Teitel CH, Snawder JE, Von Tungeln LS, Kadlubar FF. Immunochemical, 32P–postlabeling,, GC/MS detection of 4-aminobiphenyl-DNA adducts in human peripheral lung in relation to metabolic activation pathways involving pulmonary N-oxidation conjugation, and peroxidation. Mutat Res. 1997;378:97–112. doi: 10.1016/s0027-5107(97)00101-2. [DOI] [PubMed] [Google Scholar]
- 42.Bendaly J, Metry KJ, Doll MA, Jiang G, States JC, Smith NB, Neale JR, Holloman JL, Pierce WM, Jr, Hein DW. Role of human CYP1A1 and NAT2 in 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine-induced mutagenicity and DNA adducts. Xenobiotica. 2009;39:399–406. doi: 10.1080/00498250902748953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turesky RJ, Bendaly J, Yasa I, Doll MA, Hein DW. The impact of NAT2 acetylator genotype on mutagenesis and DNA adducts from 2-amino-9H–pyrido[2,3-b]indole. Chem Res Toxicol. 2009;22:726–733. doi: 10.1021/tx800473w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Minchin RF, Reeves PT, Teitel CH, McManus ME, Mojarrabi B, Ilett KF, Kadlubar FF. N-and O-acetylation of aromatic and heterocyclic amine carcinogens by human monomorphic and polymorphic acetyltransferases expressed in COS-1 cells. Biochem Biophys Res Commun. 1992;185:839–844. doi: 10.1016/0006-291x(92)91703-s. [DOI] [PubMed] [Google Scholar]
- 45.Hein DW, Rustan TD, Ferguson RJ, Doll MA, Gray K. Metabolic activation of aromatic and heterocyclic N-hydroxyarylamines by wild-type and mutant recombinant human NAT1 and NAT2 acetyltransferases. Arch Toxicol. 1994;68:129–133. doi: 10.1007/s002040050045. [DOI] [PubMed] [Google Scholar]
- 46.Lau EY, Felton JS, Lightstone FC. Insights into the o-acetylation reaction of hydroxylated heterocyclic amines by human arylamine N-acetyltransferases: a computational study. Chem Res Toxicol. 2006;19:1182–1190. doi: 10.1021/tx0600999. [DOI] [PubMed] [Google Scholar]
- 47.Hein DW, Doll MA, Rustan TD, Gray K, Feng Y, Ferguson RJ, Grant DM. Metabolic activation and deactivation of arylamine carcinogens by recombinant human NAT1 and polymorphic NAT2 acetyltransferases. Carcinogenesis. 1993;14:1633–1638. doi: 10.1093/carcin/14.8.1633. [DOI] [PubMed] [Google Scholar]
- 48.Zang Y, Doll MA, Zhao S, States JC, Hein DW. Functional characterization of single-nucleotide polymorphisms and haplotypes of human N-acetyltransferase 2. Carcinogenesis. 2007;28:1665–1671. doi: 10.1093/carcin/bgm085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McQueen CA, Maslansky CJ, Glowinski IB, Crescenzi SB, Weber WW, Williams GM. Relationship between the genetically determined acetylator phenotype and DNA damage induced by hydralazine and 2-aminofluorene in cultured rabbit hepatocytes. Proc Natl Acad Sci USA. 1982;79:1269–1272. doi: 10.1073/pnas.79.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng Y, Jiang W, Hein DW. 2-Aminofluorene-DNA adduct levels in tumor-target and nontarget organs of rapid and slow acetylator Syrian hamsters congenic at the NAT2 locus. Toxicol Appl Pharmacol. 1996;141:248–255. doi: 10.1006/taap.1996.0281. [DOI] [PubMed] [Google Scholar]
- 51.Vermeulen R, Talaska G, Schumann B, Bos RP, Rothman N, Kromhout H. Urothelial cell DNA adducts in rubber workers. Environ Mol Mutagen. 2002;39:306–313. doi: 10.1002/em.10078. [DOI] [PubMed] [Google Scholar]
- 52.Agudo A, Peluso M, Sala N, Capella G, Munnia A, Piro S, Marin F, Ibanez R, Amiano P, Tormo MJ, Ardanaz E, Barricarte A, Chirlaque MD, Dorronsoro M, Larranaga N, Martinez C, Navarro C, Quiros JR, Sanchez MJ, Gonzalez CA. Aromatic DNA adducts and polymorphisms in metabolic genes in healthy adults: findings from the EPIC-Spain cohort. Carcinogenesis. 2009;30:968–976. doi: 10.1093/carcin/bgp062. [DOI] [PubMed] [Google Scholar]
- 53.Hein DW, Doll MA, Rustan TD, Gray K, Feng Y, Ferguson RJ, Grant DM. Metabolic activation and deactivation of arylamine carcinogens by recombinant human NAT1 and polymorphic NAT2 acetyltransferases. Carcinogenesis. 1993;14:1633–1638. doi: 10.1093/carcin/14.8.1633. [DOI] [PubMed] [Google Scholar]
- 54.Zang Y, Doll MA, States JC, Hein DW. The effect of the G857A(K268R) polymorphism on N-acetyltransferase 2-mediated carcinogen metabolism is substrate-dependent. Proc Amer Assoc Cancer Res. 2006;47:2033. (abstract). [Google Scholar]
- 55.Hein DW, Ferguson RJ, Doll MA, Rustan TD, Gray K. Molecular genetics of human polymorphic N-acetyltransferase: enzymatic analysis of 15 recombinant wild-type, mutant, and chimeric NAT2 allozymes. Hum Mol Genet. 1994;3:729–734. doi: 10.1093/hmg/3.5.729. [DOI] [PubMed] [Google Scholar]
- 56.Hein DW, Doll MA, Rustan TD, Ferguson RJ. Metabolic activation of N-hydroxyarylamines and N-hydroxyarylamides by 16 recombinant human NAT2 allozymes: effects of 7 specific NAT2 nucleic acid substitutions. Cancer Res. 1995;55:3531–3536. [PubMed] [Google Scholar]
- 57.Walraven JM, Zang Y, Trent JO, Hein DW. Structure/function evaluations of single nucleotide polymorphisms in human N-acetyltransferase 2. Curr Drug Metab. 2008;9:471–486. doi: 10.2174/138920008784892065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zang Y, Zhao S, Doll MA, States JC, Hein DW. The T341C (Ile114Thr) polymorphism of N-acetyltransferase 2 yields slow acetylator phenotype by enhanced protein degradation. Pharmacogenetics. 2004;14:717–723. doi: 10.1097/00008571-200411000-00002. [DOI] [PubMed] [Google Scholar]
- 59.Hein DW. N-acetyltransferase 2 genetic polymorphism: effects of carcinogen and haplotype on urinary bladder cancer risk. Oncogene. 2006;25:1649–1658. doi: 10.1038/sj.onc.1209374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang YS, Chern HD, Su WJ, Wu JC, Lai SL, Yang SY, Chang FY, Lee SD. Polymorphism of the N-acetyltransferase 2 gene as a susceptibility risk factor for antituberculosis drug-induced hepatitis. Hepatology. 2002;35:883–889. doi: 10.1053/jhep.2002.32102. [DOI] [PubMed] [Google Scholar]
- 61.Makarova SI. Human N-acetyltransferases and drug-induced hepatotoxicity. Curr Drug Metab. 2008;9:538–545. doi: 10.2174/138920008784892047. [DOI] [PubMed] [Google Scholar]
- 62.Cho HJ, Koh WJ, Ryu YJ, Ki CS, Nam MH, Kim JW, Lee SY. Genetic polymorphisms of NAT2 and CYP2E1 associated with antituberculosis drug-induced hepatotoxicity in Korean patients with pulmonary tuberculosis. Tuberculosis (Edinb) 2007;87:551–556. doi: 10.1016/j.tube.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 63.Cartwright RA, Glashan RW, Rogers HJ, Ahmad RA, Barham-Hall D, Higgins E, Kahn MA. Role of N-acetyltransferase phenotypes in bladder carcinogenesis: a pharmacogenetic epidemiological approach to bladder cancer. Lancet. 1982;2:842–845. doi: 10.1016/s0140-6736(82)90810-8. [DOI] [PubMed] [Google Scholar]
- 64.Hein DW. Molecular genetics and function of NAT1 and NAT2: role in aromatic amine metabolism and carcinogenesis. Mutat Res. 2002;506–507:65–77. doi: 10.1016/s0027-5107(02)00153-7. [DOI] [PubMed] [Google Scholar]
- 65.Brockmoller J, Cascorbi I, Kerb R, Roots I. Combined analysis of inherited polymorphisms in arylamine N-acetyltransferase 2, glutathione S-transferases M1, and T1, microsomal epoxide hydrolase, and cytochrome P450 enzymes as modulators of bladder cancer risk. Cancer Res. 1996;56:3915–3925. [PubMed] [Google Scholar]
- 66.Okkels H, Sigsgaard T, Wolf H, Autrup H. Arylamine N-acetyltransferase 1 (NAT1) and 2 (NAT2) polymorphisms in susceptibility to bladder cancer: the influence of smoking. Cancer Epidemiol Biomarkers Prev. 1997;6:225–231. [PubMed] [Google Scholar]
- 67.Filiadis IF, Georgiou I, Alamanos Y, Kranas V, Giannakopoulos X, Lolis D. Genotypes of N-acetyltransferase-2 and risk of bladder cancer: a case-control study. J Urol. 1999;161:1672–1675. [PubMed] [Google Scholar]
- 68.Desoky ESEl, AbdelSalam YM, Salama RH, Akkad MAEl, Atanasova S, Ahsen Nvon, Armstrong VW, Oellerich M. NAT2*5/*5 genotype (341T>C) is a potential risk factor for schistosomiasis-associated bladder cancer in Egyptians. Ther Drug Monit. 2005;27:297–304. doi: 10.1097/01.ftd.0000164197.95494.aa. [DOI] [PubMed] [Google Scholar]
- 69.van der Hel OL, Peeters PHM, Hein DW, Doll MA, Grobbee DE, Kromhout D, de Mesquita BB. NAT2 slow acetylation and GSTM1 null genotypes may increase postmenopausal breast cancer risk in long-term smoking women. Pharmacogenetics. 2003;13:399–407. doi: 10.1097/00008571-200307000-00005. [DOI] [PubMed] [Google Scholar]
- 70.Baumgartner KB, Schlierf TJ, Yang D, Doll MA, Hein DW. N-acetyltransferase 2 genotype modification of active cigarette smoking on breast cancer risk among Hispanic and non-Hispanic white women. Toxicological Sciences. 2009 doi: 10.1093/toxsci/kfp199. doi:10.1093/toxsci/kfp199 (Epub August 19, 2009) [DOI] [PMC free article] [PubMed] [Google Scholar]