Abstract
Background
DTNBP1 is associated with schizophrenia in many studies, but the associated alleles and haplotypes vary between samples.
Method
We assessed nine single nucleotide polymorphisms (SNPs) in this gene for association with schizophrenia in a new sample of 1021 cases and 626 controls from Ireland.
Results
Four SNPs give evidence of association (0.000018<p<0.045), most strongly with the common allele at rs760761. A haplotype of the common alleles of five markers (including rs760761) and the minor allele of rs2619538 overlapping the 5′ end of the DTNBP1 gene also gives evidence for association (p=0.0002). Secondary analyses showed no difference in the association signal based on sex or family history. These results are in agreement with the most consistently observed association with common alleles and common-allele haplotypes, reported in a previous study of Irish cases and controls but not in an Irish high-density family sample. Our results do not support the prior report from a Swedish sample of increased association in cases with a family history of psychotic illness. Comparison of human, chimpanzee and rhesus sequence suggest that rs760761 is a particularly variable position in the primate lineage.
Conclusion
This study provides further evidence from a large case/control sample for association of common DTNBP1 alleles and haplotypes with schizophrenia.
Keywords: schizophrenia, genetic, association, DTNBP1, dysbindin, case control
1. INTRODUCTION
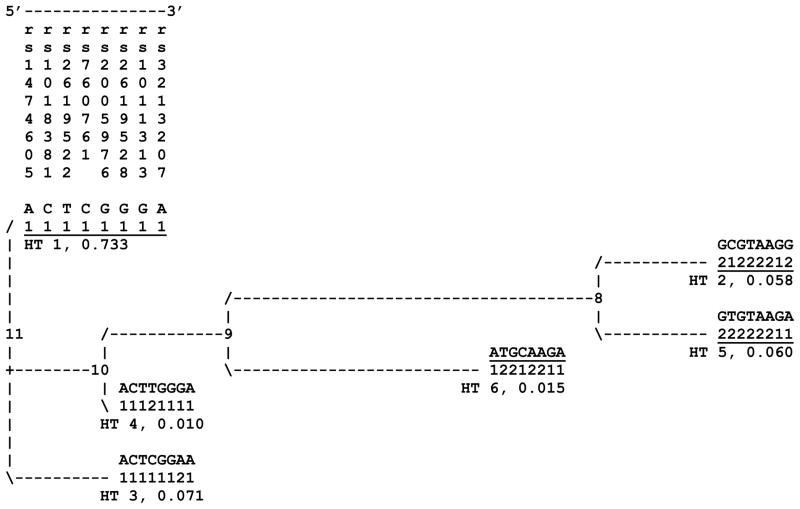
Schizophrenia has a complex etiology and lifetime prevalence of ~1% worldwide. Family, twin and adoption studies consistently demonstrate a strong genetic risk component (Kendler and Diehl, 1993; Maki et al., 2005). Schizophrenia is linked to 6p24-p22 (Straub et al., 1995) and associated with DTNBP1 (Straub et al., 2002; van den Oord et al., 2003). Reported associations vary considerably, but cluster in 8 commonly typed single nucleotide polymorphisms (SNPs). Data from these 8 SNPs yield a stable cladistic solution of 6 common haplotypes numbered throughout as in Figure 1.
Figure 1.
Original evolutionary tree produced from data for 8 DTNBP1 SNPs typed in the Irish Study of High Density Schizophrenia Families (ISHDSF) sample (van den Oord et al., 2003) and most commonly included in subsequent studies of this gene. Haplotype IDs and estimated population frequencies in this sample are shown below each haplotype for each in parentheses. The SNPs are arranged in genome sequence order and 3 ′-5 ′ in DTNBP1. The three underlined haplotypes (1, 2 and 5) have all been associated with schizophrenia in multiple independent samples.
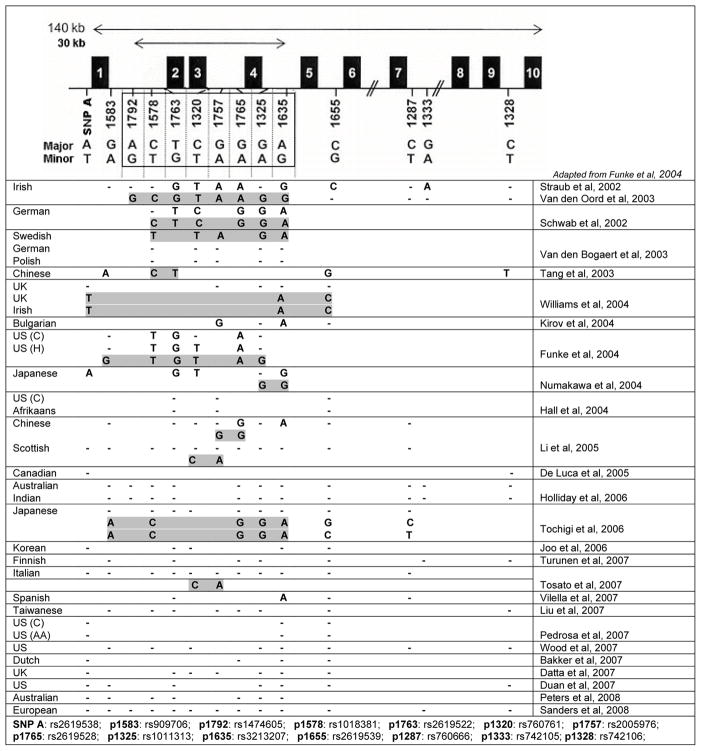
SNPs in DTNBP1 were associated with schizophrenia in 14 studies of 16 independent samples (Figure 2). Haplotype 2 was associated with schizophrenia in Irish high-density families (Straub et al., 2002; van den Oord et al., 2003) and Japanese case/controls (Numakawa et al., 2004). Swedish (Van Den Bogaert et al., 2003) and US (Funke et al., 2004) case/controls showed association with the closely related haplotype 5. Common alleles and haplotype 1 show association in 7 independent samples of families from Germany, Israel and Hungary (Schwab et al., 2003), China (Tang et al., 2003; Li et al., 2005), and Bulgaria (Kirov et al., 2004), and case/controls from the UK and Ireland (Williams et al., 2004) and Japan (Tochigi et al., 2006). Though initially negative, the Irish sample (Morris et al., 2003) was positive when additional SNPs were typed (Williams et al., 2004). In a small number of studies, other associations were observed: with the low frequency haplotype 6 in Scottish (Li et al., 2005) and Italian (Tosato et al., 2007) case/controls, and with rs3213207 and markers more 3′ in the gene in Spanish case/controls (Vilella et al., 2007) and mixed ancestry families (Duan et al., 2007).
Figure 2.
Summary of published association studies of DTNBP1 and schizophrenia. The 8 SNPs defining haplotypes in Figure 1 are shown boxed in the cartoon at the top. If association was observed for a marker, the associated allele is shown. Dashes (−) indicate markers tested but with no associated allele. Blank cells indicate marker not tested. Allele sets defining associated haplotypes are shown shaded, if any were reported. C: Caucasian; H: Hispanic; AA: African American. Note that the Duan et al. (2007) report observed association but not with markers common to any other study.
No association was reported in 13 studies of 17 independent samples: in German or Polish (Van Den Bogaert et al., 2003), Australian (Holliday et al., 2006), Korean (Joo et al., 2006), European- or African-American (Pedrosa et al., 2007), European-American (Wood et al., 2007), Dutch (Bakker et al., 2007), UK (Datta et al., 2007), Australian (Peters et al., 2008) and mixed European-ancestry (Sanders et al., 2008) case/controls, or in US or Afrikaans (Hall et al., 2004), Canadian (DeLuca et al., 2005), Australian or Indian (Holliday et al., 2006), Taiwanese (Liu et al., 2007) or Finnish (Turunen et al., 2007) families.
We tested DTNBP1 for association with schizophrenia in a new Irish case/control sample. We also assessed whether association varied by family history of illness or by sex as previously reported in some studies (Van Den Bogaert et al., 2003; Chen et al., 2007). We further assessed human and primate sequence data to determine the ancestral alleles of the SNPs studied here.
2. MATERIALS, METHODS AND SAMPLES
2.1 The Irish Case/Control Study of Schizophrenia (ICCSS) Sample
Cases were ascertained from in-patient and out-patient psychiatric facilities in Ireland and Northern Ireland, using the Structured Clinical Interview for DSM III-R, Patient version with an expanded psychosis section (Spitzer et al., 1987). DSM-III-R criteria were used for consistency with our family collection (Kendler et al., 1996). Detailed personal interviews and hospital record rating forms were completed for each proband. Subjects with a field diagnosis of schizophrenia or poor-outcome schizoaffective disorder were eligible if all four grandparents were born in Ireland or the UK. Schizoaffective disorder was defined as poor outcome if the subject was found on interview to have significant psychosocial deterioration and negative symptoms, and a course of illness consistent with chronic schizophrenia. The ICCSS sample includes 1021 cases.
Controls (N=626) were recruited from donors at the Northern Ireland Blood Transfusion Service (N=554) and from the Irish national police (N=38) and army reserve (N=34). Controls were asked about past history of psychotic illness and were eligible if they reported no history of psychotic illness and all four grandparents born in Ireland or the UK. The mode of sample collection did not allow for more detailed assessment. All participants gave appropriate informed consent. Recorded sex was verified by X/Y genotypes; cases were 68% male and 32% female, controls were 55% male, 45% female. This sample has ≥90% power to detect effects with minor allele frequency (MAF) ≥20% and a genotype relative risk (GRR) ≥1.3 at an alpha of 0.001.
Ireland has had a stable, homogeneous population (Cavalli-Sforza et al., 1994; Relethford, 1983; Sunderland et al., 1973) for ~15,000 years (Hill et al., 2000; Semino et al., 2000; McEvoy et al., 2004), with a genetic structure minimally influenced by human migrations over the last three millennia (McEvoy et al., 2004). In the experience of the blood bank staff, non-Irish donors are very rare, in agreement with the history of minimal in-migration to Ireland, and would have been excluded on the basis of questions about their grandparents.
2.2 Family History
Family history research diagnostic criteria (FH-RDC) (Andreasen et al., 1977) were assessed by the FH-RDC interview; cases were asked to report on psychotic illness in relatives (Endicott et al., 1978). The FH-RDC (kappa 0.49±0.02) compares favorably with best-estimate diagnosis (kappa 0.43±0.02) from interview of family members. Validated against best-estimate diagnosis, sensitivity was 0.37 and specificity was 0.996 (Roy et al., 1996). The FH-RDC criteria and instrument thus miss true illness in relatives but almost never produce a false positive.
We defined positive family history (FH+) by report of 1 or more first-degree relatives with schizophrenia or unspecified functional psychosis. We restrict the definition to first-degree relatives because we expect greater reliability of reporting for immediate family members. Unspecified functional psychosis required positive report of one or more specific symptoms (delusions, hallucinations, incoherence, or bizarre behavior) not due to a mood disorder. FH information is available for 739 cases (72.4%). Under our definition, there are 196 FH+ cases. Because family history information was not collected for controls, we cannot use a logistic regression approach to assess SNP×FH interaction. Rather, we compare these 196 FH+ cases to the most conservatively defined 478 FH − cases reporting no psychotic illness in either first or second degree relatives. This approach has ≥65% power to detect differences between the FH positive and negative subsamples assuming minor allele frequency (MAF) ≥20%, allelic odds ratio (OR) ≥1.3 and an alpha of 0.05.
2.3 Marker Selection and Genotyping
We genotyped 9 DTNBP1 SNPs (Table 1) allowing reconstruction of previously observed haplotypes (van den Oord et al., 2003). We included rs2619538 and rs2619539 because of their importance in defining association in a previous study of DTNBP1 in Irish case/controls (Williams et al., 2004). Markers were genotyped using fluorescence polarization detection of template-directed dye-terminator incorporation (FP-TDI) with appropriate AcycloPrime SNP detection kits for specific polymorphisms (PerkinElmer, Boston) and an automated allele scoring platform (Van den Oord et al., 2003). Primer sequences are available on request. We genotype 41 samples (11 cases and 30 controls) in duplicate, and use discordant genotypes in these duplicates to estimate genotyping error rates. DTNBP1 is transcribed in opposite orientation to the human genome sequence. All SNP data are presented 5′-3′ on the genomic (−) strand, in coding orientation for DTNBP1, with the exception of Figure S1 in genomic (+) orientation.
Table 1. Single marker analysis results.
SNP IDs, published aliases, chromosome 6 genomic positions, major/minor alleles, Hardy-Weinberg equilibrium (HWE) p-values, case and control major allele frequencies, chi-square values, asymptotic p-values, permutation P-values and estimated odds ratios (OR) for the major allele of each SNP are shown for each of the 9 SNPs studied. Associated SNPs are shown in bold.
| SNP | Alias | Position | Alleles | HWE p | Frequency | Chi-sq | p-value | Perm. P | OR | |
|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | |||||||||
| rs2619538 | SNP A | 15773189 | A/T | 0.188 | 0.526 | 0.563 | 4.004 | 0.045 | 0.400 | 0.86 |
| rs1474605 | P1792 | 15766191 | A/G | 0.358 | 0.815 | 0.783 | 4.575 | 0.032 | 0.304 | 1.22 |
| rs1018381 | P1578 | 15765049 | C/T | 0.842 | 0.906 | 0.905 | 0.006 | 0.941 | 1.000 | 1.01 |
| rs2619522 | P1763 | 15761628 | T/G | 0.653 | 0.826 | 0.799 | 3.589 | 0.058 | 0.488 | 1.19 |
| rs760761 | P1320 | 15759111 | C/T | 0.177 | 0.809 | 0.742 | 18.404 | 1.8×10 −5 | 1.8×10 −4 | 1.47 |
| rs2005976 | P1757 | 15758781 | G/A | 0.008 | 0.834 | 0.806 | 3.644 | 0.056 | 0.475 | 1.21 |
| rs1011313 | P1325 | 15741411 | G/A | 1 | 0.891 | 0.899 | 0.518 | 0.472 | 1.000 | 0.92 |
| rs3213207 | P1635 | 15736081 | A/G | 0.540 | 0.903 | 0.881 | 4.192 | 0.041 | 0.364 | 1.26 |
| rs2619539 | P1655 | 15728834 | C/G | 0.669 | 0.520 | 0.519 | 0.001 | 0.978 | 1.000 | 1.00 |
2.4 Statistical Analysis
Single Marker and Haplotype Analyses
SNPs and haplotypes were analyzed in Haploview (Barrett et al., 2005). We used Haploview default settings and excluded any individual missing >50% of genotypes. We tested for significant (p<0.001) departures from Hardy-Weinberg Equilibrium (HWE) and compared linkage disequilibrium (LD) against HapMap and previous Irish family data. Primary haplotype analyses were performed within blocks defined by the default confidence-interval method (Gabriel et al., 2002). We assessed empirical significance with 100,000 permutations of case and control status because of the significance of the asymptotic p-value for rs760761/p1320.
Markers rs2619538 and rs2619539 have only modest LD with other markers assessed (Figure S1), but were important in defining the specific 3-marker haplotype (rs2619538/SNP A, rs3213207/p1635, rs2619539/p1655) associated in UK and Irish case/controls (Williams et al., 2004). We therefore directly tested haplotypes composed of these three markers; we assessed empirical significance with 5000 permutations of case and control status.
We also tested whether composite LD patterns differ between cases and controls using DPRIME (Zaykin et al., 2006) to compare differences between case and control LD matrices across a region. This test can detect differences in the extent of LD between cases and controls, which might be observed if interacting alleles are present in a gene and has higher power than haplotypic tests under certain disease models. We analyzed both r2 and D’ measures; because r2 is sensitive to low MAFs, results from r2 comparisons alone can be difficult to interpret. The significance of the LD difference is assessed by randomly permuting case/control status to create a series of null difference statistics against which the actual result is compared. We compared regional LD patterns between cases and controls across all 9 markers (rs2619538-rs2619539) and in a subset of 6 markers (rs2619538-rs2005976) in moderately high LD.
Secondary Analyses
To assess differences in association by sex, we coded each SNP in an additive framework and performed logistic regression analyses, including gender and a SNP×Sex interaction term. Given our sample size and assuming a MAF of .2, we have >80% power to detect interaction effects with a relative risk of >1.55 in the presence of no main effects beyond those induced by the interaction at a liberal alpha of .05. This is consistent with our actual results where our largest interaction effect was roughly 1.45.
To test for differences in evidence for association between sets of cases defined by FH status, we tested for allelic (1df) association by assessing Χ2 between FH+ and FH− cases. This analysis was performed using SAS (SAS Institute, 2002).
2.5 Human and Primate SNP Sequence Comparison
In order to orient phylogenetic trees, cladistic analyses and other studies of haplotypes generally make the simplifying assumption that common and ancestral alleles are the same. In order to define these patterns without assumptions, we sought to determine the derived and ancestral alleles at the 8 SNP positions studied here by comparison with chimpanzee and rhesus genome sequence data. Human DTNBP1 sequence covering the region studied here and shown in Figure 1 was extracted from genomic contig AL022343, which includes the 5′ 92 kb of DTNBP1 and all SNP positions in this study. To infer the ancestral alleles of these key DTNBP1 SNPs in the primate lineage, human sequence was aligned with syntenic extracts of chimpanzee (NW 107908) and rhesus (NW 001116478) genomic contigs using the MegAlign package within Lasergene7 (DNASTAR, Madison, WI).
2.6 Bioinformatic analysis
We used three bioinformatic approaches to assess evidence of functional significance for associated SNPs. First, we analyzed sequence conservation using VISTA (http://genome.lbl.gov/vista/index.shtml). Second, we searched for possible transcription factor binding sites (TFBSs) in 1000bp of sequence around a SNP of interest in the UCSC Genome Browser (http://genome.ucsc.edu/). Third, we examined overlap of expressed sequence tags (ESTs) with SNPs of interest.
3. RESULTS
3.1 Genotyping: missingness, error rates and LD
In total, 14/1647 samples (0.85%, 4 cases, 10 controls) were excluded due to missing data. Results are based on 1017 cases and 616 controls (N=1633), 1090 samples (66.18%) missing zero, 387 (23.50%) missing one, 111 (6.74%) missing two, 38 (2.31%) missing three and 7 (0.43%) missing four genotypes). By marker, average genotyping completion was 94.7% (89.3–98.7%).
We genotype 9 SNPs × 41 samples in duplicate (N=369 genotype pairs) and both genotypes were available for 344 (93.2%); 3 were discordant, estimating our genotyping error rate at 0.9%. All markers satisfied HWE criteria. LD patterns (Figure S1) are in close agreement with those observed previously in other samples. As previously observed, both rs2619538 (SNP A, 2 kb 5′ of start codon) and rs2619539 (p1655, intron 5) show reduced LD with all other markers studied.
3.2 Single Marker Analysis
The results of single marker analyses are shown in Table 1. We observed evidence of association between schizophrenia and 4 DTNBP1 SNPs at nominal significance levels of p<0.05. Common alleles were associated with schizophrenia for all but one marker. The most strongly associated SNP was rs760761 (p1320), with allele frequency 0.809 in cases and 0.742 in controls (Χ2=18.404, p<1.8×10−5, allelic OR 1.47). Smaller signals were observed for the minor allele of rs2619538 and the major alleles of rs1474605 and rs3213207 with case/control allele frequency differences between 2.2 and 4.1%. Only the result from rs760761/p1320 remained significant (P=1.8×10−4) after 100,000 permutations.
3.3 Haplotype Analysis
Results of haplotype analyses are shown in Table 2. Haplotypes were analyzed within the blocks defined by Haploview (Table 2A), and in the specific combination (rs2619538/SNP A, rs3213207/p1635, rs2619539/p1655, Table 2B) associated in UK and Irish case/controls (Williams et al., 2004). We observed association on the haplotype defined by the major alleles of rs1474605-rs2005976 (markers 2–6, Χ2=17.919, p=2.31×10−5, Perm P=0.0003, haplotypic OR 1.44). The observed case/control difference in frequency of this haplotype (6.3%) is almost identical to the allele frequency difference observed for rs760761/p1320 in single marker analyses (6.7%). We observed no evidence for association in the ICCSS sample with any haplotype composed of alleles of rs2619538/SNP A, rs3213207/p1635, rs2619539/p1655 (Table 2B).
Table 2. Haplotype analysis results.
SNPs included, specific haplotypes, haplotype frequencies in cases and controls, chi-sq values, asymptotic p-values, empirical P-values from 100000 (A.) or 5000 (B.) permutations and odds ratios (OR) are shown for each haplotype. A. Analysis of haplotypes in HAPLOVIEW defined LD blocks. The haplotype showing evidence of association with schizophrenia in the ICCSS sample is shown in bold. Block numbers are as shown in Figure S1. Numbers in parentheses indicate equivalent structures in Figure 1. B. Analysis of haplotypes defined by rs2619538/SNP A, rs3213207/p1635 and rs2619539/p1655. The haplotype associated in Williams et al, 2004 is noted by an asterisk but shows no evidence of association in this sample.
| Block | SNPs | HT | Case Freq. | Con. Freq. | Chi-sq | p-value | Perm. P | OR |
|---|---|---|---|---|---|---|---|---|
| A. | ||||||||
| 2 | rs1474605-rs2005976 | ACTCG (1,3) | 0.805 | 0.742 | 17.919 | 2.31×10 −5 | 0.0003 | 1.44 |
| (p1792-p1757) | GCGTA (2) | 0.083 | 0.104 | 4.078 | 0.044 | 0.407 | 0.78 | |
| 1005 cases, 605 cons | GTGTA (5) | 0.086 | 0.092 | 0.311 | 0.577 | 1 | 0.93 | |
| GCTTG | 0.011 | 0.017 | 1.662 | 0.197 | 0.929 | 0.67 | ||
| ACTTG (4) | 0.002 | 0.030 | 45.178 | 1.80×10 −11 | 0.000 | 0.08 | ||
| 1 | rs3213207-rs2619539 | AC | 0.520 | 0.521 | 0.004 | 0.947 | 1 | 1.00 |
| (p1635-p1655) | AG | 0.383 | 0.359 | 1.942 | 0.163 | 0.882 | 1.11 | |
| 1017 cases, 614 cons | GG | 0.097 | 0.120 | 4.364 | 0.037 | 0.358 | 0.79 | |
| B. | ||||||||
| N/A | rs2619538-rs3213207-rs2619539 | AAC | 0.334 | 0.345 | 0.476 | 0.490 | 0.960 | 0.95 |
| (SNP A-p1635-p1655) | TAG | 0.277 | 0.248 | 3.225 | 0.073 | 0.295 | 1.16 | |
| 1017 cases, 616 cons | TAC* | 0.186 | 0.176 | 0.482 | 0.488 | 0.959 | 1.07 | |
| AAG | 0.107 | 0.109 | 0.057 | 0.811 | 0.999 | 0.97 | ||
| AGG | 0.087 | 0.106 | 3.412 | 0.065 | 0.266 | 0.80 | ||
| TGG | 0.01 | 0.015 | 1.274 | 0.259 | 0.754 | 0.70 | ||
3.4 Case/Control LD Comparison
We compared LD patterns between cases and controls across all 9 markers (rs2619538-rs2619539) and 6 markers in moderate-high LD (rs2619538-rs2005976), using both r2 and D′ measures. We used 1K–100K permutations to generate an appropriate null distribution. In the 9 marker comparison, we found a significant difference between cases and controls for the r2 (Z2=0.0011; empirical P=0.01) but not the D′ based (Z2=0.0016; empirical P=0.22) composite LD comparisons. In the 9 marker comparison, the observed difference may result from the sensitivity of r2 to MAF noted above. However, in the 6 marker window, we found significant differences comparing both D′ (Z2=0.0033; empirical P=5×10−4) and r2 (Z2=0.0026; empirical P=1×10−5). These results indicate that there is a difference in the composite LD between cases and controls in DTNBP1, particularly in the 6 SNPs rs2619538-rs2005976 (where both tests were significant). Because both cases and controls are Irish, this difference seems unlikely to be due to population stratification, and instead to reflect a real difference in the composite LD structure in this region between cases and controls.
3.5 SNP × Sex Interaction Analysis
In logistic regression tests of gender differences in association (Table S1), 4 markers gave nominally significant evidence of association, 3 of which were also associated in primary analyses. However, the SNP × Sex interaction term was not significant for any marker tested, providing no evidence for differential association depending on the sex of the case.
3.6 Family History Analysis
Based on the single marker results above, we limited FH analyses to the most significantly associated marker, rs760761 (Table S2). Using the categorical definitions described in Methods, there were 196 FH+ and 478 conservatively defined FH− cases (no illness reported in first or second degree family members) in the ICCSS sample. A genotype at rs760761 was missing for 47 individuals with family history (28 FH− and 19 FH+), leaving a total analyzed sample N=627 (1254 alleles), 450 FH− and 177 FH+. There was no evidence of any difference in association of rs760761 between FH+ and FH− cases (Table S2).
3.7 Human and Primate SNP Sequence Analysis
We sought to infer the ancestral allele at key DTNBP1 SNPs. We determined the alleles present in chimpanzee and rhesus reference sequences at the positions corresponding to the 8 haplotype-defining SNPs in Figure 1 (rs1474605-rs3213207, Table 3a). The data are consistent with the human major allele in European populations being ancestral in 3 cases (rs1474605/p1792, rs1018381/p1578, rs1011313/p1325) and derived in 2 cases (rs2619522/p1763, rs2619528/p1765). In 2 cases (rs2005976/p1757, rs3213207/p1635) the human major allele in European populations is present in the chimpanzee and minor allele present in the rhesus reference sequences. Neither human allele at rs760761 is present in chimpanzee and rhesus sequence.
Table 3. Comparison of human, chimpanzee and rhesus sequence in DTNBP1.
A. Human major and minor alleles with reference chimpanzee and rhesus sequence for 8 key haplotype defining SNPs in DTNBP1. Alleles at rs760761 are shown in bold. B. Local sequence around the position of rs760761 in humans and orthologous regions in primates (rs760761 at position 21). The ambiguous 3 bp deletion region in the rhesus sequence is shaded.
| A. | |||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Alias | Human | Chimp | Rhesus | |||||||||||||||||||||||||||||||||||||
| major | minor | ||||||||||||||||||||||||||||||||||||||||
| rs1474605 | p1792 | A | G | A | A | ||||||||||||||||||||||||||||||||||||
| rs1018381 | p1578 | C | T | C | C | ||||||||||||||||||||||||||||||||||||
| rs2619522 | p1763 | T | G | G | G | ||||||||||||||||||||||||||||||||||||
| rs760761 | p1320 | C | T | A | G/– | ||||||||||||||||||||||||||||||||||||
| rs2005976 | p1757 | G | A | G | A | ||||||||||||||||||||||||||||||||||||
| rs2619528 | p1765 | G | A | A | A | ||||||||||||||||||||||||||||||||||||
| rs1011313 | p1325 | G | A | G | G | ||||||||||||||||||||||||||||||||||||
| rs3213207 | p1635 | A | G | A | G | ||||||||||||||||||||||||||||||||||||
| B. | 10 | 20 | 30 | 40 | |||||||||||||||||||||||||||||||||||||
| Human | T | A | T | A | A | C | A | T | C | A | A | A | A | G | C | A | C | A | A | A | Y | A | A | C | A | A | G | A | T | A | A | A | A | A | T | T | A | G | A | C | A |
| Chimp | T | A | T | A | A | C | A | T | C | A | A | A | A | G | C | A | C | A | A | A | A | A | A | C | A | A | G | A | T | A | A | A | A | A | T | T | A | G | A | C | A |
| Rhesus | T | A | T | A | A | C | A | T | C | A | A | A | A | G | C | A | C | A | A | A | G | A | A | — | — | — | G | A | T | A | A | A | A | A | T | T | A | G | A | C | A or |
| T | A | T | A | A | C | A | T | C | A | A | A | A | G | C | A | C | A | A | A | — | — | — | G | A | A | G | A | T | A | A | A | A | A | T | T | A | G | A | C | A | |
Sequence around rs760761/p1320 in human, chimpanzee and rhesus is shown in Table 3b; the SNP is shown at position 21. Comparisons of the three primate sequences show that neither human allele (C/T) is present in the reference chimpanzee sequence (A). The sequence around this position is ambiguous (G or −) in rhesus due to a 3 bp deletion relative to the human and chimpanzee sequences. The sequence context and alignment are consistent with the deletion occurring at either positions 21–23 (with the position of rs760761 deleted and a rhesus-specific variant (G) at position 24) or positions 24–26 (as shown in Table 3b, with G at the site of rs760761). In either case, diversity is higher than average at the position of rs760761 based on the limited number of SNPs we have assessed so far, but rs760761 is intronic and there is as yet no evidence that this SNP has any functional significance.
3.8 Bioinformatic analyses
The VISTA analysis shows that rs760761 is not conserved between human and mouse, and is located in a LINE repeat element, both arguing against any functional significance. No TFBS was identified anywhere in the 1000 bp of sequence surrounding the rs760761, and no known EST overlaps this position. These results are in general agreement with prior bioinformatic analyses of the gene and of specific key SNPs (see Discussion).
4. DISCUSSION
We observed evidence for association of schizophrenia with SNPs in DTNBP1, particularly with the common allele of rs760761/p1320 (the only result to remain significant after permutation testing) and a haplotype composed of the common alleles of five markers (including rs760761) and the minor allele of rs2619538. Frequency differences observed for the haplotype (6.3%) and the best SNP rs760761 (6.7%) are similar. The pattern of LD and specific haplotypes were very similar to those observed previously. This large sample provides further support for the involvement of DTNBP1 in schizophrenia.
We observed no evidence for differential association of DTNBP1 by family history. Even with the reduced sample size in our case-only analysis, we examined a substantially larger sample than the 142 cases and 272 controls from Sweden previously reported (Van Den Bogaert et al., 2003). All definitions of positive family history in current use have fairly low sensitivity, as described above, so our non-replication of this finding is unlikely to be due to substantial differences in the sensitivity of the family history definition between studies. We also observed no difference in association by sex, as reported for a locus on chromosome 5 in the ICCSS sample (Chen et al., 2007).
Our results agree with those reported previously in German families and Chinese and Japanese case/control samples. Few studies included rs2619538, so data for comparison are limited; there is no evidence for association of this marker in our data. LD between rs2619538 and other markers is consistently lower than between markers in the defined block. Otherwise, our results also agree with those reported in the study of 2 independent samples of UK and Irish case/controls.
The results of our bioinformatic analyses of rs760761 are in general agreement with the few published reports of such studies of DTNBP1. So far there has been no evidence for function ascribed to any of the intronic SNPs in the gene (including those studied here). A ChIP-chip study of fetal brain tissue identified only one promoter region immediately 5′ of the gene containing the specific histone modifications for which antibodies were included; the closest SNP studied, rs2619538, is ~1.5 kb outside of this putative promoter region (Pedrosa et al., 2009). A study of expression differences by SNP genotype and haplotype detected no evidence of any effect of rs760761 (Bray et al., 2005). Therefore, in common with most reported intronic or intergenic associations, LD between the associated SNPs and other functionally significant variation remains the most likely explanation for our results. Our results are also in keeping with a recent survey of 180 genomewide association studies (http://www.genome.gov/27529020) showed that association of complex traits with SNPs in non-coding regions of the genome is commonly observed: among 782 unique SNPs with P < 10−6, only 67 (8.6%) were in coding or regulatory regions, while 340 (43.5%) were in intronic regions and 354 (45.3%) were in intergenic regions.
In comparative assessments of human and primate sequence, the pattern we observe is complex and does not support any simple explanation of the ages and origins of haplotypes observed in the region. There are 5 positions where chimpanzee and rhesus sequence are informative about the likely ancestral alleles with respect to human ones. Considering only these five positions, haplotypes 2 and 6 (Fig. 1) display four ancestral alleles, haplotypes 1, 4 and 5 contain three ancestral alleles, and haplotype 3 contains two ancestral alleles. Neither human allele at rs760761 is present in chimpanzee and rhesus sequence. This SNP was the most significantly associated marker in the present study, and it also gave significant evidence of heterogeneity in allelic association across samples in a recent meta-analysis (Maher et al., 2009) using new methodology suitable for detecting and quantifying between sample differences (the “flip-flop” phenomenon (Lin et al., 2007) in the scale and/or direction of allelic association.
Neither of the two reported Irish case/control samples support the results from the Irish high-density family sample, the outlier among samples from Ireland, perhaps more consistent with underlying differences in the risk variants between the two sample types. Multiplex schizophrenia families are relatively rare, and affected individuals in such families may carry different and perhaps more highly penetrant risk variants. Conversely, supportive data from a Japanese case/control sample argue against an allele specific to a set of Irish high density families with a common founder. The results of these two studies along with those in the Swedish and US samples occur on related haplotypes sharing substantial variation. This close evolutionary relationship between haplotypes 2 and 5 raises the possibility of a variant common to both. If this was true, however, both haplotypes should be associated in the samples where one or other is; this has generally not been observed.
Rare variants and particularly structural variation in the human genome have been a focus of some interest in schizophrenia genetic studies recently. Three copy-number variants have been identified in the genomic interval around DTNBP1 (chromosome 6 variations 2620, 37543 and 7536 from the Database of Genomic Variation, http://projects.tcag.ca/variation/?source=hg18). Variants in this region were observed on only 1/540 chromosomes in the largest of the three studies reporting CNVs in this area (Redon et al., 2006), and none overlap the DTNBP1 gene. It thus seems unlikely that structural variation is a major contributor to or confounder of the association signals observed in this region.
The present study has some limitations. First, even though our sample is among the largest in which DTNBP1 has been studied, our power is high for MAF ≥20% and/or OR ≥1.3, but these values represent the largest effect sizes currently expected in complex traits. Effects below these thresholds would have lower probability of being detected, declining as either or both of the MAF and OR decrease. Second, we genotyped SNPs that had been included in prior studies because these should define the same haplotypes and haplotype blocks (barring any difference in the underlying LD in our sample) as previously reported. This makes our study data directly comparable with prior reports and provided reasonable power but our genotyped markers do not include SNPs selected on functional grounds. Three nonsynonymous SNPs in DTNBP1 are reported in dbSNP, but all have low MAFs, in the range 1–6% where our sample power is lower. Finally, as we note above, in common with most association studies, our results are likely to reflect LD between the markers typed (for which no functional role has been suggested) and currently unknown variation in the locus.
Fluctuation in association results is common in studies of complex traits, but may not necessarily indicate false positive findings (Gruber et al., 2007; Lin et al., 2007). Certainly, such results are not consistent with an underlying hypothesis of a single risk variant in a gene. The collective data from studies outlined above are most consistent with multiple liability variants in DTNBP. The evolutionary distance between haplotype 1 and haplotypes 2 and 5 (Figure 1) is consistent with this idea. In conclusion, the results from the present study of DTNBP1 and schizophrenia in a large, ethnically homogeneous sample add further support for the association of common alleles and common haplotypes with schizophrenia.
Supplementary Material
Footnotes
Supplementary information is available at the Schizophrenia Research website.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Andreasen NC, Endicott J, Spitzer RL, Winokur G. The family history method using diagnostic criteria. Reliability and validity. Arch Gen Psychiatry. 1977;34:1229–1235. doi: 10.1001/archpsyc.1977.01770220111013. [DOI] [PubMed] [Google Scholar]
- Bakker SC, Hoogendoorn ML, Hendriks J, Verzijlbergen K, Caron S, Verduijn W, Selten JP, Pearson PL, Kahn RS, Sinke RJ. The PIP5K2A and RGS4 genes are differentially associated with deficit and non-deficit schizophrenia. Genes Brain Behav. 2007;6:113–119. doi: 10.1111/j.1601-183x.2006.00234.x. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bray NJ, Preece A, Williams NM, Moskvina V, Buckland PR, Owen MJ, O’Donovan MC. Haplotypes at the dystrobrevin binding protein 1 (DTNBP1) gene locus mediate risk for schizophrenia through reduced DTNBP1 expression. Hum Mol Genet. 2005;14:1947–1954. doi: 10.1093/hmg/ddi199. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton University Press; Princeton, NJ: 1994. [Google Scholar]
- Chen X, Wang X, Hossain S, O’Neill FA, Walsh D, van den OE, Fanous A, Kendler KS. Interleukin 3 and schizophrenia: the impact of sex and family history. Mol Psychiatry. 2007;12:273–282. doi: 10.1038/sj.mp.4001932. [DOI] [PubMed] [Google Scholar]
- Datta SR, McQuillin A, Puri V, Choudhury K, Thirumalai S, Lawrence J, Pimm J, Bass N, Lamb G, Moorey H, Morgan J, Punukollu B, Kandasami G, Kirwin S, Sule A, Quested D, Curtis D, Gurling HM. Failure to confirm allelic and haplotypic association between markers at the chromosome 6p22.3 dystrobrevin-binding protein 1 (DTNBP1) locus and schizophrenia. Behav Brain Funct. 2007;3:50. doi: 10.1186/1744-9081-3-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca V, Voineskos D, Shinkai T, Wong G, Kennedy JL. Untranslated region haplotype in dysbindin gene: analysis in schizophrenia. J Neural Transm. 2005;112:1263–1267. doi: 10.1007/s00702-005-0338-9. [DOI] [PubMed] [Google Scholar]
- Duan J, Martinez M, Sanders AR, Hou C, Burrell GJ, Krasner AJ, Schwartz DB, Gejman PV. DTNBP1 (Dystrobrevin binding protein 1) and schizophrenia: association evidence in the 3′ end of the gene. Hum Hered. 2007;64:97–106. doi: 10.1159/000101961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endicott J, Andreasen N, Spitzer RL. Family History Research Diagnostic Criteria Biometrics Research Department. New York State Psychiatric Institute; New York: 1978. [Google Scholar]
- Funke B, Finn CT, Plocik AM, Lake S, DeRosse P, Kane JM, Kucherlapati R, Malhotra AK. Association of the DTNBP1 Locus with Schizophrenia in a U.S. Population. Am J Hum Genet. 2004;75:891–898. doi: 10.1086/425279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumensteil B, Higgins J, DeFelice M, Lochner A, Faggart M, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altschuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gruber JD, Genissel A, Macdonald SJ, Long AD. How repeatable are associations between polymorphisms in achaete-scute and bristle number variation in Drosophila? Genetics. 2007;175:1987–1997. doi: 10.1534/genetics.106.067108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D, Gogos JA, Karayiorgou M. The contribution of three strong candidate schizophrenia susceptibility genes in demographically distinct populations. Genes Brain Behav. 2004;3:240–248. doi: 10.1111/j.1601-183X.2004.00078.x. [DOI] [PubMed] [Google Scholar]
- Hill EW, Jobling MA, Bradley DG. Y-chromosome variation and Irish origins. Nature. 2000;404:351–352. doi: 10.1038/35006158. [DOI] [PubMed] [Google Scholar]
- Holliday EG, Handoko HY, James MR, McGrath JJ, Nertney DA, Tirupati S, Thara R, Levinson DF, Hayward NK, Mowry BJ, Nyholt DR. Association study of the dystrobrevin-binding gene with schizophrenia in Australian and Indian samples. Twin Res Hum Genet. 2006;9:531–539. doi: 10.1375/183242706778025035. [DOI] [PubMed] [Google Scholar]
- Joo EJ, Lee KY, Jeong SH, Ahn YM, Koo YJ, Kim YS. The dysbindin gene (DTNBP1) and schizophrenia: no support for an association in the Korean population. Neurosci Lett. 2006;407:101–106. doi: 10.1016/j.neulet.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Diehl SR. The genetics of schizophrenia: A current, genetic-epidemiologic perspective. Schizophrenia Bulletin. 1993;19:261–285. doi: 10.1093/schbul/19.2.261. [DOI] [PubMed] [Google Scholar]
- Kendler KS, O’Neill FA, Burke J, Murphy B, Duke F, Straub RE, Shinkwin R, Ni NM, MacLean CJ, Walsh D. Irish study on high-density schizophrenia families: field methods and power to detect linkage. Am J Med Genet. 1996;67:179–190. doi: 10.1002/(SICI)1096-8628(19960409)67:2<179::AID-AJMG8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Kirov G, Ivanov D, Williams NM, Preece A, Nikolov I, Milev R, Koleva S, Dimitrova A, Toncheva D, O’Donovan MC, Owen MJ. Strong evidence for association between the dystrobrevin binding protein 1 gene (DTNBP1) and schizophrenia in 488 parent-offspring trios from Bulgaria. Biol Psychiatry. 2004;55:971–975. doi: 10.1016/j.biopsych.2004.01.025. [DOI] [PubMed] [Google Scholar]
- Li T, Zhang F, Liu X, Sun X, Sham PC, Crombie C, Ma X, Wang Q, Meng H, Deng W, Yates P, Hu X, Walker N, Murray RM, St Clair D, Collier DA. Identifying potential risk haplotypes for schizophrenia at the DTNBP1 locus in Han Chinese and Scottish populations. Mol Psychiatry. 2005;10:1037–1044. doi: 10.1038/sj.mp.4001718. [DOI] [PubMed] [Google Scholar]
- Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip-flop phenomenon. Am J Hum Genet. 2007;80:531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, Liu YL, Fann CS, Yang WC, Wu JY, Hung SI, Chen WJ, Chueh CM, Liu WM, Liu CC, Hsieh MH, Hwang TJ, Faraone SV, Tsuang MT, Hwu HG. No association evidence between schizophrenia and dystrobrevin-binding protein 1 (DTNBP1) in Taiwanese families. Schizophr Res. 2007 doi: 10.1016/j.schres.2007.02.003.. [DOI] [PubMed] [Google Scholar]
- Maher BS, Reimers MA, Riley BP, Kendler KS. Allelic Heterogeneity in Genetic Association Meta-Analysis: An application to DTNBP1 and Schizophrenia. Human Heredity. 2009 doi: 10.1159/000264445. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki P, Veijola J, Jones PB, Murray GK, Koponen H, Tienari P, Miettunen J, Tanskanen P, Wahlberg KE, Koskinen J, Lauronen E, Isohanni M. Predictors of schizophrenia--a review. Br Med Bull. 2005;73–74:1–15. doi: 10.1093/bmb/ldh046. [DOI] [PubMed] [Google Scholar]
- McEvoy B, Richards M, Forster P, Bradley DG. The Longue Duree of genetic ancestry: multiple genetic marker systems and Celtic origins on the Atlantic facade of Europe. Am J Hum Genet. 2004;75:693–702. doi: 10.1086/424697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DW, McGhee KA, Schwaiger S, Scully P, Quinn J, Meagher D, Waddington JL, Gill M, Corvin AP. No evidence for association of the dysbindin gene [DTNBP1] with schizophrenia in an Irish population-based study. Schizophr Res. 2003;60:167–172. doi: 10.1016/s0920-9964(02)00527-3. [DOI] [PubMed] [Google Scholar]
- Numakawa T, Yagasaki Y, Ishimoto T, Okada T, Suzuki T, Iwata N, Ozaki N, Taguchi T, Tatsumi M, Kamijima K, Straub RE, Weinberger DR, Kunugi H, Hashimoto R. Evidence of novel neuronal functions of dysbindin, a susceptibility gene for schizophrenia. Hum Mol Genet. 2004;13:2699–2708. doi: 10.1093/hmg/ddh280. [DOI] [PubMed] [Google Scholar]
- Pedrosa E, Locker J, Lachman HM. Survey of schizophrenia and bipolar disorder candidate genes using chromatin immunoprecipitation and tiled microarrays (ChIP-chip) J Neurogenet. 2009;23:341–352. doi: 10.1080/01677060802669766. [DOI] [PubMed] [Google Scholar]
- Pedrosa E, Ye K, Nolan KA, Morrell L, Okun JM, Persky AD, Saito T, Lachman HM. Positive association of schizophrenia to JARID2 gene. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:45–51. doi: 10.1002/ajmg.b.30386. [DOI] [PubMed] [Google Scholar]
- Peters K, Wiltshire S, Henders AK, Dragovic M, Badcock JC, Chandler D, Howell S, Ellis C, Bouwer S, Montgomery GW, Palmer LJ, Kalaydjieva L, Jablensky A. Comprehensive analysis of tagging sequence variants in DTNBP1 shows no association with schizophrenia or with its composite neurocognitive endophenotypes. Am J Med Genet B Neuropsychiatr Genet. 2008 doi: 10.1002/ajmg.b.30741. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relethford JH. Genetic structure and population history of Ireland: a comparison of blood group and anthropometric analyses. Ann Hum Biol. 1983;10:321–333. doi: 10.1080/03014468300006481. [DOI] [PubMed] [Google Scholar]
- Roy MA, Walsh D, Kendler KS. Accuracies and inaccuracies of the family history method: a multivariate approach. Acta Psychiatr Scand. 1996;93:224–234. doi: 10.1111/j.1600-0447.1996.tb10639.x. [DOI] [PubMed] [Google Scholar]
- Sanders AR, Duan J, Levinson DF, Shi J, He D, Hou C, Burrell GJ, Rice JP, Nertney DA, Olincy A, Rozic P, Vinogradov S, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Crowe RR, Cloninger CR, Martinez M, Gejman PV. No significant association of 14 candidate genes with schizophrenia in a large European ancestry sample: implications for psychiatric genetics. Am J Psychiatry. 2008;165:497–506. doi: 10.1176/appi.ajp.2007.07101573. [DOI] [PubMed] [Google Scholar]
- SAS Institute. SAS. [v9] Cary; NC: 2002. Ref Type: Computer Program. [Google Scholar]
- Schwab SG, Knapp M, Mondabon S, Hallmayer J, Borrmann-Hassenbach M, Albus M, Lerer B, Rietschel M, Trixler M, Maier W, Wildenauer DB. Support for association of schizophrenia with genetic variation in the 6p22.3 gene, dysbindin, in sib-pair families with linkage and in an additional sample of triad families. Am J Hum Genet. 2003;72:185–190. doi: 10.1086/345463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De BG, Francalacci P, Kouvatsi A, Limborska S, Marcikiae M, Mika A, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA. The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science. 2000;290:1155–1159. doi: 10.1126/science.290.5494.1155. [DOI] [PubMed] [Google Scholar]
- Spitzer RL, Williams JB, Gibbon J. Structured Clinical Interview for DSM-III-R - Patient Version (SCID-P, 4/1/87) New York State Psychiatric Institute; New York: 1987. [Google Scholar]
- Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, Cesare AT, Gibberman A, O’Neill FA, Walsh D, Kendler KS. Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of mouse dysbindin, is associated with schizophrenia. Am J Hum Genet. 2002;71:337–348. doi: 10.1086/341750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub RE, MacLean CJ, O’Neill FA, Burke J, Murphy B, Duke F, Shinkwin R, Webb BT, Zhang J, Walsh D, Kendler KS. A potential vulnerability locus for schizophrenia on chromosome 6p24–22: Evidence for genetic heterogeneity. Nature Genetics. 1995;11:287–293. doi: 10.1038/ng1195-287. [DOI] [PubMed] [Google Scholar]
- Sunderland E, Tills D, Bouloux C, Doyl J. Genetic studies in Ireland. In: Roberts DF, Sunderland E, editors. Genetic Variation in Britain. XII. Taylor & Francis Ltd; London: 1973. pp. 141–170. [Google Scholar]
- Tang JX, Zhou J, Fan JB, Li XW, Shi YY, Gu NF, Feng GY, Xing YL, Shi JG, He L. Family-based association study of DTNBP1 in 6p22.3 and schizophrenia. Mol Psychiatry. 2003;8:717–718. doi: 10.1038/sj.mp.4001287. [DOI] [PubMed] [Google Scholar]
- Tochigi M, Zhang X, Ohashi J, Hibino H, Otowa T, Rogers M, Kato T, Okazaki Y, Kato N, Tokunaga K, Sasaki T. Association study of the dysbindin (DTNBP1) gene in schizophrenia from the Japanese population. Neurosci Res. 2006;56:154–158. doi: 10.1016/j.neures.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Tosato S, Ruggeri M, Bonetto C, Bertani M, Marrella G, Lasalvia A, Cristofalo D, Aprili G, Tansella M, Dazzan P, Diforti M, Murray RM, Collier DA. Association study of dysbindin gene with clinical and outcome measures in a representative cohort of Italian schizophrenic patients. Am J Med Genet B Neuropsychiatr Genet. 2007 doi: 10.1002/ajmg.b.30484. [DOI] [PubMed] [Google Scholar]
- Turunen JA, Peltonen JO, Pietilainen OP, Hennah W, Loukola A, Paunio T, Silander K, Ekelund J, Varilo T, Partonen T, Lonnqvist J, Peltonen L. The role of DTNBP1, NRG1, and AKT1 in the genetics of schizophrenia in Finland. Schizophr Res. 2007;91:27–36. doi: 10.1016/j.schres.2006.11.028. [DOI] [PubMed] [Google Scholar]
- Van Den Bogaert A, Schumacher J, Schulze TG, Otte AC, Ohlraun S, Kovalenko S, Becker T, Freudenberg J, Jonsson EG, Mattila-Evenden M, Sedvall GC, Czerski PM, Kapelski P, Hauser J, Maier W, Rietschel M, Propping P, Nothen MM, Cichon S. The DTNBP1 (dysbindin) gene contributes to schizophrenia, depending on family history of the disease. Am J Hum Genet. 2003;73:1438–1443. doi: 10.1086/379928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Oord E, Sullivan PF, Chen X, Kendler KS, Riley B. Identification of a high risk haplotype for the dystrobrevin binding protein 1 (DTNBP1) gene in the Irish study of high density schizophrenia families. Mol Psychiatry. 2003;8:499–510. doi: 10.1038/sj.mp.4001263. [DOI] [PubMed] [Google Scholar]
- Van den Oord EJCG, Jiang Y, Riley BP, Kendler KS, Chen X. FP-TDI SNP genotype scoring by manual and statistical procedures: A study of error rates and types. Biotechniques. 2003;34:610–624. doi: 10.2144/03343dd04. [DOI] [PubMed] [Google Scholar]
- Vilella E, Costas J, Sanjuan J, Guitart M, De DY, Carracedo A, Martorell L, Valero J, Labad A, De FR, Najera C, Molto MD, Toirac I, Guillamat R, Brunet A, Valles V, Perez L, Leon M, de Fonseca FR, Phillips C, Torres M. Association of schizophrenia with DTNBP1 but not with DAO, DAOA, NRG1 and RGS4 nor their genetic interaction. J Psychiatr Res. 2007 doi: 10.1016/j.jpsychires.2007.02.005.. [DOI] [PubMed] [Google Scholar]
- Williams NM, Preece A, Morris DW, Spurlock G, Bray NJ, Stephens M, Norton N, Williams H, Clement M, Dwyer S, Curran C, Wilkinson J, Moskvina V, Waddington JL, Gill M, Corvin AP, Zammit S, Kirov G, Owen MJ, O’Donovan MC. Identification in 2 independent samples of a novel schizophrenia risk haplotype of the dystrobrevin binding protein gene (DTNBP1) Arch Gen Psychiatry. 2004;61:336–344. doi: 10.1001/archpsyc.61.4.336. [DOI] [PubMed] [Google Scholar]
- Wood LS, Pickering EH, Dechairo BM. Significant support for DAO as a schizophrenia susceptibility locus: examination of five genes putatively associated with schizophrenia. Biol Psychiatry. 2007;61:1195–1199. doi: 10.1016/j.biopsych.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Zaykin DV, Meng Z, Ehm MG. Contrasting linkage-disequilibrium patterns between cases and controls as a novel association-mapping method. Am J Hum Genet. 2006;78:737–746. doi: 10.1086/503710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.