Summary
Changes in chromatin composition accompany cellular differentiation in eukaryotes. While bulk chromatin is duplicated during DNA replication, Replication-Independent (RI) nucleosome replacement occurs in transcriptionally active chromatin and during specific developmental transitions where the genome is repackaged. In most animals, replacement uses the conserved H3.3 histone variant, but the functions of this variant have not been defined. Using mutations for the two H3.3 genes in Drosophila, we identify wide-spread transcriptional defects in H3.3-deficient animals. We show that mutant animals compensate for the lack of H3.3 in two ways: they up-regulate the expression of the major histone H3 genes, and maintain chromatin structure by using H3 protein for RI nucleosome replacement at active genes. Rescue experiments show that increased expression of H3 is sufficient to relieve transcriptional defects. In contrast, H3.3 is essential for male fertility, and germline cells specifically require the histone variant. Defects without H3.3 first occur around meiosis, resulting in a failure to condense, segregate, and reorganize chromatin. Rescue experiments with mutated transgenes demonstrate that H3.3-specific residues involved in RI nucleosome assembly – but not major histone modification sites – are required for male fertility. Our results imply that the H3.3 variant plays an essential role in chromatin transitions in the male germline.
Results and Discussion
H3.3 mutations are semi-lethal
The H3.3 histone variant comprises the bulk of nucleosomes in transcribed regions of the genome, while the major H3 histone packages bulk chromatin. This segregation is accomplished by the exclusive use of H3.3 by Replication-Independent (RI) nucleosome assembly systems that act at active genes [1]. The localization of a distinct histone variant to transcribed regions has raised the possibilities that the variant confers unique functional properties to chromatin or may maintain chromatin packaging after nucleosome disruption by elongating polymerases. To test these ideas, we generated null alleles of each of the two H3.3-encoding genes (His3.3A and His3.3B) in the Drosophila genome using nearby P elements (Supplementary Figure 1A). Individual homozygous mutations had little detrimental effect, but double-null mutants had reduced viability and were invariably sterile in both males and females. These phenotypes result specifically from the H3.3 mutations, because they were rescued in double-null animals carrying a His3.3A transgene (Supplementary Figure 1A, Table 1). Double-null animals exhaust maternal H3.3 supplies by the first instar stage (Supplementary Figure 1B), however most H3.3-deficient animals survive to adulthood and appear morphologically normal. This demonstrates that the H3.3 histone is not essential for viability, at least after the embryonic stage.
H3.3 is not required for nucleosome packaging
Biochemical fractionation has shown that H3.3 comprises ~25% of the total H3-type histones in Drosophila [2]. It has been suggested that H3.3 is needed to package chromatin regions where histones have been displaced by transcriptional activity [1]. However, we found no defects in bulk or highly transcribed chromatin (Supplementary Figure 2A), indicating that the histone variant is not required to retain nucleosomes in transcribed regions.
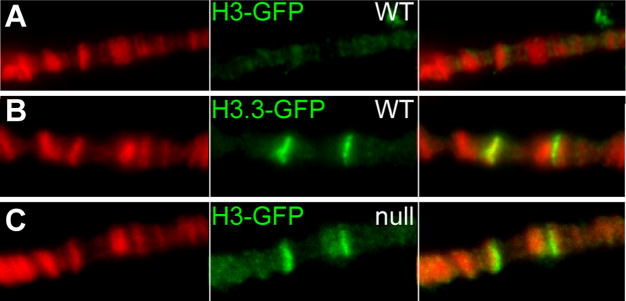
Nucleosomes in transcribed regions must be displaced to allow polymerase progression. RI assembly normally uses H3.3 to restore nucleosomes to transcribed regions [3]. We reasoned that H3.3-deficient animals may retain nucleosomes in transcribed regions if the major H3 histone is now used for RI assembly. To determine if this was the case, we used the inducible Hsp70 genes. These genes are rapidly induced by heat-shock, and high-level transcription displaces nucleosomes [4]. Transcription ceases when animals return to normal temperatures, and nucleosomes are restored to the gene. We used this system to track nucleosome displacement and replacement during gene induction. To test if H3.3-deficient animals allow H3 to be used for RI nucleosome assembly, we produced a pulse of GFP-tagged H3 or H3.3 histones during heat shock induction. The Hsp70 genes are induced in both wildtype and H3.3-deficient larvae (Supplementary Figure 2B). A pulse of H3.3-GFP intensely labels the HSP genes in polytene chromosomes from wild-type animals, while H3-GFP does not ([3]; Figure 1A,B). In contrast, H3-GFP does deposit at the Hsp70 loci in H3.3-deficient animals (Figure 1C). We conclude that RI assembly using the H3 histone restores nucleosomes to Hsp70 genes after transcription, and that the major H3 histone can contribute to maintaining nucleosome structure in H3.3-deficient cells.
Figure 1. The major H3 histone can be used for RI nucleosome assembly in H3.3-deficient animals.
(A–C) Deposition of newly-produced histone-GFP protein at induced Hsp70 genes in polytene chromosomes. Larvae carrying inducible histone-GFP genes were heat shocked for 1 hour at 37°, allowed to recover for 2 hours, and then stained for GFP (green) and DNA (red). The genotypes of larvae are indicated. (A) Wild-type larvae carrying an inducible H3-GFP construct show no RI deposition, while (B) wild-type larvae carrying an inducible H3.3-GFP construct show efficient deposition of GFP at the induced Hsp70 genes. (E) Double-null larvae carrying an inducible H3-GFP construct show deposition of the tagged histone.
Transcription requires ongoing nucleosome replacement
While the H3 histone can maintain nucleosomal packaging of transcribed genes, the reduced viability of H3.3-deficient animals indicates that this is not sufficient. As the H3.3 histone is normally enriched in proportion to transcription [5], we examined the expression of the highly-expressed Rp49, RpS18, and Cp15 genes by Northern analysis. The abundance of mRNA from these three genes was reduced 25–90% in H3.3-deficient adults (data not shown). To extend these results, we measured genome-wide changes in gene expression using Affymetrix Drosophila 2.0 gene microarrays. To control for background differences between strains, we compared profiles from double-null, H3.3-deficient adult males to those from double-null males that also carried the His3.3A rescue transgene. We identified 99 transcripts were significantly down-regulated and 288 that were up-regulated (Supplementary Figure 3 and Supplementary Table 1). Down-regulated genes were predominantly normally highly expressed genes, consistent with the idea that the H3.3 histone is required for proper transcription of very active genes.
The lack of H3.3 histones may have two molecular consequences for transcribed genes. First, transcribed chromatin is now packaged exclusively with the major H3 histone. Second, the rate of nucleosome replacement at transcribed genes may be reduced, as H3.3 is normally the substrate for replacement. A lack of H3.3 might reduce gene expression if either the variant or if replacement promotes DNA access and transcription. The first explanation implies that the histone subtypes are non-redundant, while the second implies that continual expression of any H3-type histone would be sufficient for proper transcriptional regulation. We distinguished between these two possibilities by constitutively expressing H3 or H3.3 histones in H3.3 double-null animals. We used the His3.3A rescue transgene, and constructed a second line carrying the same genomic fragment recoded at 4 codons to produce the H3 protein (His3.3AH3). Both the His3.3A or His3.3AH3 transgenes rescued viability of double-null animals (Table 1), indicating that the semi-lethality of H3.3-deficient animals is due to reduced nucleosome replacement. To determine the effects of constitutive H3 production on expression changes, we profiled gene expression from double-null adult males carrying the His3.3AH3 transgene. Expression values from the three matched strains (H3.3-deficient, His3.3A-rescued, and His3.3AH3-rescued) are compared in a scatter-plot in Figure 2A. Expression from the two transgenes was identical, thus any difference in transcriptional regulation can be attributed to the type of histone produced. We found that almost all genes mis-regulated in H3.3-deficient males fall on the diagonal of the scatter-plot, indicating that expression of these genes are rescued by either transgene. Thus, we conclude that expression changes in H3.3-deficient animals are due to reduced nucleosome replacement, and not the lack of the histone variant per se. The attenuated expression of these genes when histone supplies are limiting suggests that maintaining full nucleosome packaging is necessary for high-level transcription.
Table 1.
Viability of males carrying mutations in His3.3 genes.
| genotype |
Viabilitya |
Fertilityb |
|---|---|---|
| His3.3B+/Y ; His3.3A+ | 1.10 ± 0.1 | fertile |
| His3.3B0/Y ; His3.3A2X1 | 0.42 ± 0.4 | sterile |
| His3.3B0/Y ; His3.3A2X1 ; P[His3.3A] | 0.98 ± 0.1 | fertile |
| His3.3B0/Y ; His3.3A2X1 ; P[His3.3AH3] | 0.70 ± 0.2 | sterile |
| His3.3B0/Y ; His3.3A2X1 ; P[His3.3AK4R] | 0.20 ± 0.1 | fertile |
| His3.3B0/Y ; His3.3A2X1 ; P[His3.3AK9R] | 0.50 ± 0.3 | fertile |
| His3.3B0/Y ; His3.3A2X1 ; P[His3.3AS31A] | 0.78 ± 0.2 | fertile |
Control males were generated by crossing w1118 mothers to w1118/Y ; +/S2CyO males. Mutant males were generated from crosses of w/Y ; His3.3A2X1/CyO fathers with or without transgenes on the 3rd chromosome to y w His3.3B0 ; His3.3A2X1/S2CyO mothers.
Viability was calculated as the fraction of wild-type to curly-winged males, and at least 100 control males were counted for each cross.
Fertility was assayed by crossing males to w1118 females. Fertile genotypes gave >100 progeny, and sterile genotypes gave none.
Figure 2. Rescue of transcriptional mis-regulation by H3.3 or H3 histones.
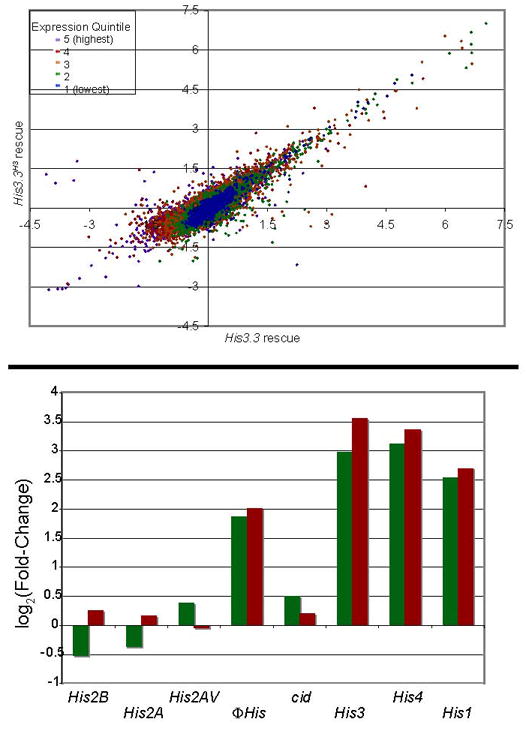
(A) Comparison of transcriptional changes in adult males carrying rescue transgenes. The x-axis displays fold-changes in gene expression between H3.3-deficient males and males carrying the His3.3A rescue transgene, and the y-axis displays fold-changes between H3.3-deficient males and males carrying the His3.3AH3 transgene. Genes are color-coded in quantiles of transcript abundance. Genes with equal expression in His3.3A and His3.3AH3 –rescued males are expected to lie on the diagonal. Expression of most genes is rescued by either transgene. Genes not rescued by H3 expression are listed in Supplementary Table 2. (B) Expression of histone genes in H3.3-deficient and rescued adult males. The relative change in expression between wild-type-rescued animals and null animals (green), and between wild-type-rescued animals and H3-rescued animals (red) is shown). The mutated His3.3A and His3.3B genes are not shown.
Less than 30 mis-regulated transcripts are not rescued by the His3.3AH3 transgene (Figure 2A and Supplementary Table 2). These include the genes encoding the major H3, H4, and H1 histones (His3, His4, and His1), but not the major H2A and H2B genes or the variant genes for H2AV or Cid (Figure 2B). All five major histone genes are located in an array at one locus, and are normally coordinately regulated and highly expressed only in S phase of the cell cycle [6]. However, our results demonstrate that in H3.3-deficient adult males some histone genes can be de-regulated while others are not. Major histone transcripts normally lack a poly-A tail and instead carry a 3′ stem-loop structure that is used for S phase regulation of the transcript [6]. In contrast, His3 transcripts were detected in H3.3-deficient animals by qPCR from both oligo-dT-primed and random-primed cDNA pools, indicating that the up-regulated transcripts are poly-adenylated (not shown, and Figure 2B). Poly-adenylation of histone transcripts improves their stability and translation outside of S phase [7]. This change in mRNA processing supports the idea that the endogenous genes now produce H3 histones throughout the cell cycle. We infer that the endogenous H3 genes are up-regulated and used for nucleosome replacement as two parts of a compensatory response in H3.3-deficient cells to maintain chromatin structure. Transcriptional activation of the H3 genes in H3.3-deficient adults can account for induction of the H4 genes, because the H3 and H4 genes are divergent from a shared promoter region [6]. There is no obvious explanation for the induction of the H1 linker histone or the H1 pseudogene ΦHis. However, the specific induction of H1 genes in H3.3-deficient animals suggests that H1 may contribute to chromatin packaging and to the survival of mutant animals.
The major histone genes are normally only expressed during S phase of the cell cycle to provide histones for DNA replication [6]. However, the specific up-regulation of some histones – but not others – implies that cells monitor levels of H3-type histones. The levels of soluble histone-chaperone complexes regulate histone gene expression during S phase of the cell cycle, when vast quantities of histones are required for chromatin duplication [8]. A similar mechanism may detect levels of histones in gap phases, and modulate transcription of histone genes, poly-adenylation of transcripts, or both.
H3.3 is required for male meiosis
Both the His3.3A and the His3.3AH3 transgenes rescue viability and most transcriptional defects of double-null animals, but two differences between the transgenic genotypes imply a unique role for the H3.3 histone. First, only the His3.3A transgene suppresses over-expression of the endogenous H3 genes (Figure 2B). H3.3-deficient animals must continue to sense the lack of the histone variant even when H3 is abundant. Second, only the His3.3A transgene restores fertility to double-null males; those carrying the His3.3AH3 transgene are sterile (Table 1). To determine specific roles of the H3.3 histone, we examined development of the male germline in detail.
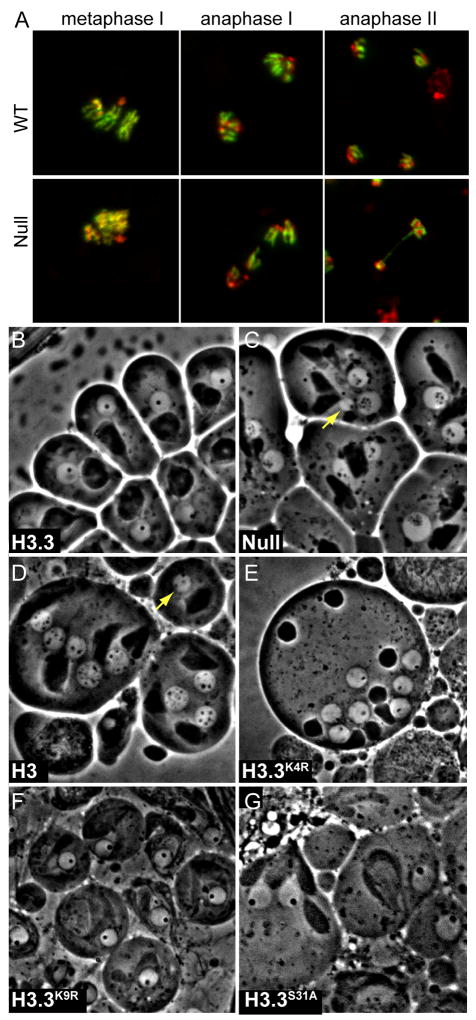
The adult testis contains a cluster of germline stem cells at its apical tip. Cell division produces a spermatogonial cell, which then undergoes 4 mitotic divisions, an extensive primary spermatocyte growth phase, meiosis, and finally differentiation into individualized haploid sperm [9]. We first compared testes from wildtype and double-null males by phase-contrast imaging and DNA-staining. Round developing spermatids were present in testes from H3.3-deficient animals, but varied in size, and late elongating nuclei were never observed (Supplementary Figure 3A,B). This suggested that chromosome segregation was defective in H3.3-deficient germlines. We therefore examined chromosome spreads to determine how meiosis proceeds in the absence of H3.3. As cells enter meiosis and Metaphase I, chromosomes condense into distinct rods, where sister-chromatids and homologs are tightly associated (Figure 3A). Clear defects in H3.3-deficient cells are apparent by this stage, with diffuse chromosomes that are only partially condensed. This is the earliest identified defect in H3.3-deficient testes.
Figure 3. Condensation and segregation defects in H3.3-deficient meiotic cells.
(A) Meiotic chromosome spreads in wild-type (top) and H3.3-deficient (bottom) testes. Testes were stained with anti-H3K4trime antibodies (green) to distinguish euchromatic and heterochromatic regions and orient chromosomes. DAPI-stained DNA is in red. In Metaphase I, chromosomes from wild-type testes are compacted and distinct, while those from H3.3-deficient testes are poorly resolved. In Anaphase I in wild-type testes, similar amounts of chromatin segregate to each pole. In H3.3-deficient testes, unequal numbers of chromosomes segregate between the two poles of division, and chromosomes are trapped between the poles. In Anaphase II in wildtype testes, sister chromatids segregate equally between the poles of division (four products from a single meiosis are shown). In Anaphase II in H3.3-deficient testes, chromatin bridges span the two poles of division. (B–G) Modification sites of H3.3 are not required for male germline function. Phase-contrast microscopy of post-meiotic round spermatid nuclei. Genotypes and mutant rescue constructs are indicated. (B) In wild-type testes, each nucleus contains similar amounts of chromatin, and a single nuclear body (phase-dark spot). (C) In H3.3-deficient testes, some round nuclei are larger or smaller than wild-type (yellow arrow indicates a micronucleus), resulting from aberrant segregation of chromosomes in meiosis. The nuclear body in many nuclei is fragmented into numerous freckles. (D) Double-null mutants carrying a His3.3AH3 rescue transgene show large and small nuclei (yellow arrow indicates a micronucleus), and fragmented nuclear bodies. (E) Double-null mutants carrying a His3.3AK4R rescue transgene show equal-sized nuclei and a single nuclear body in each nucleus. (F) Double-null mutants carrying a His3.3AK9R rescue transgene show equal-sized nuclei and a single nuclear body in each nucleus. (G) Double-null mutants carrying a His3.3AS31A rescue transgene show equal-sized nuclei and a single nuclear body in each nucleus.
We found that Metaphase I with incompletely condensed chromosomes is followed by an aberrant Anaphase I in H3.3-deficient cells. Anaphase I in wildtype cells segregates homologs to opposite poles of division (Figure 3A), but without H3.3 chromosomes frequently lag between the two poles. All chromosomes can be affected, because we observed meiotic figures with variable numbers of chromosomes trapped between the two poles of division. Lagging chromosomes may occur because chromosomes incompletely condense at Metaphase I, resulting in the entanglement of homologs as segregation begins.
Further meiotic aberrations occur in Anaphase II. In wild-type cells sister-chromatids segregate equally to the two poles of division, producing 4 haploid nuclei (Figure 3A). In H3.3-deficient testes, chromosome bridges were observed at the Anaphase II stage. These must result from attachments between sister-chromatids.
H3.3 is essential for remodeling of germline chromatin
Lagging chromosomes and bridges during meiosis produce meiotic products containing unequal amounts of chromatin, and this is apparent by phase-contrast microscopy. In wildtype testes, post-meiotic nuclei are very similar in size (Figure 4B). H3.3-deficient testes contain post-meiotic nuclei that are larger and smaller than wildtype (Figure 4C).
Phase-contrast imaging of post-meiotic nuclei revealed a second nuclear defect in H3.3-deficient germlines. In wild-type testes, single phase-dark nuclear body transiently appears in early haploid nuclei, and then disappears as spermatid differentiation proceeds (Figure 3B). This structure is thought to be an aggregate of basic proteins, including histones [9]. Strikingly, in H3.3-deficient testes the post-meiotic nuclear body is often fragmented into 2–15 smaller bodies (Figure 3C). Fragmentation of this structure is not observed in other meiotic segregation mutants [10]. The disruption of this structure implies that there are defects in chromatin arrangement within the post-meiotic nucleus.
To determine if H3.3 is specifically required during meiosis, we examine testes in rescued males. The His3.3A rescue construct restored all aspects of normal germlines (meiotic segregation, the single nuclear body in post-meiotic nuclei, production of motile sperm, and male fertility) to double-null males. In contrast, meiotic defects and fragmentation of the post-meiotic nuclear body persisted in double-null males carrying the recoded His3.3AH3 transgene (Figure 3D). Thus, while continuous production of H3 is sufficient to rescue transcriptional defects in these animals, it is not sufficient for germline meiotic functions.
We then mapped what amino acid residues are required for H3.3 germline functions. H3.3 differs from H3 at the exposed residue-31 in the N-terminal tail, and at a cluster of three residues within the histone-fold domain (HFD; [11]). The tail difference between the two histones confers a unique modification site to the H3.3 histone (H3.3S31P) that has been suggested to be a binding site for trans-acting factors [12]. Mutational analysis has identified that the cluster of differences within the HFD are required for RI assembly of H3.3 [13]. We found that a His3.3A rescue transgene carrying the H3 identity at position 31 (His3.3AS31A) rescues the viability and fertility of double-null males (Table 1). Testes from these males contained post-meiotic nuclei of equal sizes, demonstrating that meiotic segregation was restored in this genotype (Figure 3G). We conclude that the H3.3S31 residue is not required for mitotic or meiotic functions in Drosophila, and germline functions must depend on the HFD differences that specify RI nucleosome replacement.
Recent work from another group has also identified that H3.3 is required for male fertility in Drosophila [14], but did not conclude that histone replacement in the germline was required. Instead, this group argued that the germline function of H3.3 involves epigenetic methylation of lysine-4, because they found that an alanine substitution in a rescue construct (H3.3K4A) did not rescue fertility. We examined the germlines of double-null males carrying an arginine substitution at this position (H3.3K4R). Surprisingly, we found that the H3.3K4R transgene rescues fertility, and germline cells show no defects (Figure 3E). H3.3K4R also eliminates K4 methylation on this histone, and we conclude methylation of H3.3 is not required for male fertility. The differing rescue with K4R and K4A substitutions suggests that a positive charge at this position is critical for fertility. Such effects have been described with budding yeast histone mutants [15]. This requirement is specific for the K4 position, because both changes of K9 to alanine or to arginine in H3.3 transgenes rescue male fertility ([14]; Figure 3F). These experiments rule out any essential germline role for modification of H3.3 at either K4 or K9 residues. Notably, the H3.3K4R construct does not rescue the semi-lethality of H3.3 null mutations (Table 1), consistent with a role of methylation at this site in transcriptional regulation in somatic cells.
Our work resolves the paradox of why the H3.3 variant conserved, when either H3-type histone suffices for nucleosome replacement at transcribed genes. The variant must be maintained for its essential germline function, and its contribution to maintaining chromatin of transcribed regions must be acquired. Indeed, a specialized role of H3.3 in gametogenesis appears conserved across eukaryotes. In Tetrahymena, the analogous hv2 histone variant is also dispensable in vegetative cells, and essential for sexual cycles [16]. In Drosophila, we have identified the essential role of H3.3 involves nucleosome replacement preceding meiosis, perhaps to dis-entangle condensing chromatids before segregation. This raises the possibility that H3.3-dependent nucleosome replacement has roles in chromatin architecture in specialized cell types that are distinct from its role in transcribed genes.
Supplementary Material
Acknowledgments
We thank Arielle Schlitt. A.S. was a recipient of a post-doctoral fellowship for research abroad from the Japan Society for the Promotion of Science.
Footnotes
Experimental Procedures
Detailed methods are provided in the Supplementary Information.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Henikoff S, Ahmad K. Assembly of variant histones into chromatin. Ann Rev Cell Dev Biol. 2005;21:133–153. doi: 10.1146/annurev.cellbio.21.012704.133518. [DOI] [PubMed] [Google Scholar]
- 2.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci U S A. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwartz BE, Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 2005;19:804–14. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu C, Wong YC, Elgin SC. The chromatin structure of specific genes: II. Disruption of chromatin structure during gene activity. Cell. 1979;16:807–814. doi: 10.1016/0092-8674(79)90096-5. [DOI] [PubMed] [Google Scholar]
- 5.Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- 6.Marzluff WF, Duronio RJ. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol. 2002;14:692–699. doi: 10.1016/s0955-0674(02)00387-3. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan E, Santiago C, Parker ED, Dominski Z, Yang X, Lanzotti DJ, Ingledue TC, Marzluff WF, Duronio RJ. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 2001;15:173–187. doi: 10.1101/gad.862801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutton A, Bucaria J, Osley MA, Sternglanz R. Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics. 2001;158:587–596. doi: 10.1093/genetics/158.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuller MT. Spermatogenesis. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1993. pp. 71–147. [Google Scholar]
- 10.Wakimoto BT, Lindsley DL, Herrera C. Toward a comprehensive genetic analysis of male fertility in Drosophila melanogaster. Genetics. 2004;167:207–16. doi: 10.1534/genetics.167.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akhmanova AS, Bindels PC, Xu J, Miedema K, Kremer H, Hennig W. Structure and expression of histone H3.3 genes in Drosophila melanogaster and Drosophila hydei. Genome. 1995;38:586–600. doi: 10.1139/g95-075. [DOI] [PubMed] [Google Scholar]
- 12.Hake SB, Garcia BA, Kauer M, Baker SP, Shabanowitz J, Hunt DF, Allis CD. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc Natl Acad Sci U S A. 2005;102:6344–6349. doi: 10.1073/pnas.0502413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 14.Hodl M, Basler K. Curr Biol. Vol. 19. 2009. Transcription in the absence of histone H3.3; pp. 1221–1226. [DOI] [PubMed] [Google Scholar]
- 15.Thompson JS, Ling X, Grunstein M. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature. 1994;369:245–247. doi: 10.1038/369245a0. [DOI] [PubMed] [Google Scholar]
- 16.Cui B, Liu Y, Gorovsky MA. Deposition and function of histone H3 variants in Tetrahymena thermophila. Mol Cell Biol. 2006;26:7719–7730. doi: 10.1128/MCB.01139-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.