Abstract
Background
The epitope specificities and antiviral activities of class I HLA-restricted CD8+ T cells, especially those induced during human immunodeficiency virus type 1 (HIV-1) primary infection, are important considerations in designing HIV-1 vaccines. Conserved epitopes may be more commonly and persistently recognized than variable epitopes, as they may be more likely to be present in infecting viruses. However, some studies have shown preferential or similar targeting of variable versus conserved epitopes during primary infection.
Methods
We analyzed cytotoxic T-lymphocyte (CTL) responses toward predefined conserved and variable epitopes in 45 subjects during primary (n = 34) and/or chronic infection (n = 16).
Results
Conserved and variable CTL epitopes were recognized with similar probabilities, whereas conserved epitopes generally elicited subdominant responses during both primary and chronic infection. During primary infection, CTL responses against Gag versus responses against Env and variable epitopes tended to be associated with lower and higher viral loads, respectively. During chronic infection, Env-specific responses tended to be associated with lower CD4+ counts.
Conclusions
Subdominant CTL recognition of conserved HIV-1 epitopes commonly occurs from primary through chronic HIV-1 infection. These findings underscore the challenge in designing T cell based vaccines that can induce immunodominant CTL to conserved HIV-1 regions.
Keywords: HIV-1, CTL responses, conserved epitopes
Introduction
Extensive viral genetic variability found between and within individuals is a major obstacle to developing effective vaccines against HIV-1. While CTL responses [1–3] are critically involved in immune defense against HIV-1 infection, they are unable to eradicate the virus. Selective escape from CTL responses has been shown to be a major driving force of HIV-1 evolution [4–6]. However, escape mutations may be associated with replication fitness costs, that may therefore limit evolution in conserved CTL epitopes [7–13]. Indeed, HIV-1 control appears to be associated with CTL escape mutations in highly conserved regions of the virus in some instances [14]. This has led to HIV vaccine development efforts aimed at eliciting CTL responses exclusively towards highly conserved regions [15, 16].
Since epitopes in conserved regions of the HIV-1 proteome are more likely to be present in infecting viruses and may be less likely to undergo escape mutations, one might expect these epitopes to be more commonly and persistently recognized by CTL responses. Whole-proteome mapping of CD8+ CTL responses demonstrated that conserved 15- to 20-mer peptides were targeted more frequently than variable peptides [17]. Frequent targeting of conserved regions of Gag and Nef was also found in subtype-C infected Indian subjects [18]. These observations are consistent with a study of CTL responses to viral protease and integrase [19] and an in silico analysis of experimentally defined HIV CTL epitopes listed in the Los Alamos HIV Sequence and Immunology Database [20], both of which found CTL epitopes concentrated in relatively conserved regions. In contrast, studies in early/primary HIV-1 infection reported that Nef [21] and possibly Env [22] is preferentially recognized by CTL despite a high degree of genetic variation, and that peptides targeted by CTL responses were more variable [23] or that frequently and rarely targeted epitopes had similar variability [24]. Most studies, including some of those mentioned above [17, 19–21, 23], examined HIV-1 specific CTL responses using peptides derived from consensus sequences or reference strains of HIV-1. In a comprehensive study of viral evolution and CTL responses in one subject [6, 11], we found that 7/25 of the recognized epitopes were identified only by using autologous sequences, most of which were located in highly variable regions. Therefore, the lack of detection of CTL responses targeting epitopes in highly variable regions, especially Env, may be due to an inadequate array of testing peptides.
To compare the prevalence and immunodominance of CTL responses to conserved and variable epitopes, we analyzed CTL responses in 45 HIV-1 subtype B infected male subjects during primary and/or chronic infection. We found that conserved epitopes were recognized by CTL with similar probability as variable epitopes and generally elicited subdominant responses during both primary and chronic infection.
Materials and Methods
Study subjects
We measured CTL responses in 50 HIV-1 infected male subjects living in the State of Washington, US. These subjects were enrolled into a longitudinal study of HIV infection in the University of Washington Primary Infection Clinic. At the time of enrollment all subjects were either HIV antibody negative or HIV antibody positive with a negative or indeterminate Western blot, negative “detuned” antibody test, or negative HIV test within 365 days prior to screening [25]. Clinical information about these subjects is summarized in Table 1. The cutoff for assignment of primary infection in this study was set at 91 days post onset of acute symptoms of HIV-1 infection [such symptoms occurred, on average, within 2 weeks following HIV-1 exposure in this cohort (J.I.M., unpublished)], or about 105 days post exposure (corresponding to Fiebig stage V or VI of primary infection [26]), and samples obtained before or at this time were classified as being within primary infection. Samples from 39 subjects were obtained during primary infection. Samples from 16 subjects were obtained during chronic infection. In 5 subjects, samples from both primary and chronic infection were obtained. In 9 subjects, samples from two time points in chronic infection were obtained. All subjects provided written consent and the University of Washington Human Subjects Committees approved this study.
Table 1.
Study subjects and CTL responses to conserved and variable epitopes
| Primary infectiona | Chronic infectiona | |||
|---|---|---|---|---|
| Subjects | ||||
| Number | 34 | 16 | ||
| Median (range) days post onset of acute symptoms b | 27 (3 – 91) | 357 (109 – 1182) | ||
| Median (range) Log10 viral load (copies/ml) | 4.98 (2.88 –6.92) | 4.36 (<1.7 – 5.57) | ||
| Median (range) CD4+ T cell count (cells/μl) | 684 (241 – 1090) | 574 (130 – 1325) | ||
| Epitopes c | Conserved | Variable | Conserved | Variable |
| Fraction of subjects with CTL responses | 0.5 | 1 | 0.5 | 1 |
| Median (range) number of epitopes recognized | 1 (0–3) | 2 (1–8) | 1 (0–2) | 3 (1–9) |
| Probability of recognition | 0.145 | 0.173 | 0.143 | 0.239 |
Primary and chronic infection were defined in this study as occurring before or after 91 days of onset of acute symptoms of viral infection, respectively. These symptoms usually arise within about 2 weeks post exposure to HIV in this cohort (J.I. Mullins, data not shown).
PBMC specimens from these time points were derived and examined for CTL responses.
Conserved epitopes were defined as belonging to the upper quartile of the tested epitopes in terms of database frequency (conserved in greater than 78.68% of the sequences of HIV-1 subtype B in Las Alamos HIV Sequence Database of year 2005 [29]), while epitopes in the lower three quartiles were considered to be variable epitopes.
CTL responses were measured by IFN-γ ELISpot assays using predefined optimal epitopes [27, 28] (Table S1). All subjects’ HLA class I alleles were determined by sequence-specific primer molecular typing [30]. IFN-γ ELISpot assays were performed as previously described [27]. CTL responses in 21 subjects were examined during primary infection in a previous study by Cao et al [27], with a set of class I HLA-restricted HIV-1 epitopic peptides that were used in the study of Goulder et al [28] and matched the subjects’ HLA alleles. In addition, 460 overlapping 15-mer or 20-mer peptides spanning the entire HIV-1 proteome, derived from HXB2, MN or subtype B consensus sequences, were examined for CTL responses in these 21 subjects, and 15 previously unreported epitopes were defined and HLA restriction determined [27]. CTL responses to these 15 newly defined epitopes were also assessed in all 21 subjects. CTL responses were measured here in an additional 29 subjects, with defined epitopic peptides used by Goulder [28] and Cao et al [27] that matched the subjects’ HLA alleles. CTL responses were measured before combination antiretroviral therapy (ART) with two exceptions - responses in one subject were measured during failed ART (71 and 148 days after the start of ART) with viral loads consistently over 50,000 copies/ml, and in another subject 149 days after the termination of 11-months of ART.
Determination of epitope conservation and probability of recognition
For each epitope, the frequency of occurrence in HIV-1 subtype B sequences in the Los Alamos HIV Sequence Database of year 2005 [29] was calculated. The database frequency of an epitope represents the conservation of the entire epitope in the known HIV-1 subtype B population. Conserved epitopes were arbitrarily defined as belonging to the upper quartile of tested epitopes in terms of database frequency (conserved in ≥78.68% of the subtype B sequences), while epitopes in the lower three quartiles were considered variable epitopes. The probability of recognition of an epitope was taken as the number of subjects that recognized the epitope (xr) divided by the number of subjects in which the epitope was tested (xt). Therefore, the probability of recognition of conserved or variable epitopes is , n being the number of conserved or variable epitopes tested.
Statistical analysis
Statistical analyses were conducted using GraphPad Prism software (GraphPad Software, San Diego, CA). Categorical data were compared using Fisher’s exact test. Paired continuous variables were compared using Wilcoxon signed rank test. Correlations were tested using Spearman’s rank coefficient. For all tests, p values of <0.05 were considered significant. Chronic CTL responses were measured at a single time point in 7 subjects and at two time points in the other 9 subjects. Therefore, during chronic infection, two parallel analyses were conducted unless specifically noted: Analysis 1 used data from the former seven subjects and the data of the first time points from the latter nine subjects; while Analysis 2 used data from the former seven subjects and the data of the second time points from the latter nine subjects. If results from both analyses were similar, only the result from Analysis 1 is reported. Otherwise, results from both analyses are reported.
Results
Half of the epitopes tested were recognized by CTL
ELISpot assays were performed using 120 predefined optimal epitopes with samples from primary infection, and 84 of those peptides were also tested in samples from chronic infection (Figure 1). The 120 peptides corresponded to 90 (75%) variable epitopes restricted by 31 different Class I HLA alleles and 30 (25%) conserved epitopes restricted by 20 different Class I HLA alleles (Figure 1,Table S1). Of the 120 epitopes, 86 corresponded to the subtype B consensus sequence reported in year 2005 [29].
Figure 1.
Numbers of HIV-1 epitopes tested and recognized. Black bars represent epitopes tested and gray bars represent epitopes recognized. A and B) all epitopes, C and D) conserved epitopes, E and F) variable epitopes during primary and chronic infection, respectively.
Three of the 21 subjects tested in the Cao et al study [27] and two of the 29 subjects tested here did not show responses and were excluded from our analyses. CTL responses were only measured during primary infection in these five subjects. For the remaining 45 subjects, CTL responses were detected at all time points examined. Therefore, CTL responses were analyzed in 34 subjects during primary infection and 16 subjects during chronic infection. All subjects analyzed had at least one HLA allele restricting at least one conserved epitope and one HLA allele restricting at least one variable epitope. Epitopes recognized during primary and chronic infections are summarized in Figure 1. About half of the epitopes tested were recognized, 66 or 55% in primary and 40 or 48% in chronic infection, respectively. During primary infection, the largest numbers of epitopes recognized were in Env. During chronic infection, however, more Nef and Gag epitopes were recognized than Env epitopes.
Similar probability of recognition for conserved and variable epitopes
Although all subjects analyzed had at least one HLA allele restricting conserved epitopes, only half (not including the two with ART) recognized at least one conserved epitope, while all recognized at least one variable epitope during both primary and chronic infection (Table 1). Nonetheless, the proportions of the tested epitopes that were recognized were not significantly different between conserved and variable epitopes during both primary and chronic infection (Figure 1). During primary infection, 43% of the conserved and 59% of the variable epitopes tested were recognized (p = 0.15, Fisher’s exact test); and during chronic infection, 33% of the conserved and 52% of the variable epitopes tested were recognized (p = 0.21; for the nine subjects with two time point measurements, epitopes recognized at one or both time points were counted). Because epitopes were tested in different number of subjects, we then measured the probabilities of recognition and found that they were similar for conserved and variable epitopes during both primary (p = 0.46, Fisher’s exact test; Table 1) and chronic infection (p = 0.12).
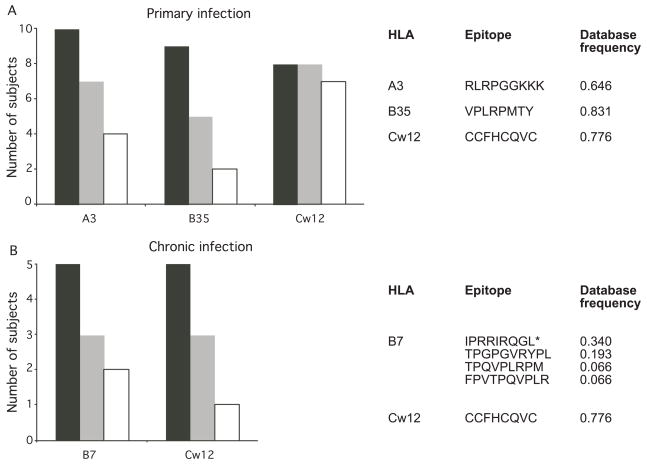
A subset of CTL epitopes are consistently reported to be targeted during early HIV-1 infection [24, 31]. We next examined whether a similar subset of epitopes were recognized in our 45 subjects, and the level of conservation of these commonly recognized epitopes. Epitopes were considered to be commonly recognized if they were tested in more than five subjects and were recognized in more than half of the tested subjects. Eight epitopes fit this criteria (Figure 2,Table S1), with only the B*3501-restricted VPLRPMTY being conserved. VPLRPMTY and the A3-restricted RLRPGGKKK epitope were also commonly targeted during early infection in previous studies [24, 31]. Another commonly reported early epitope, the B7-restriced IPRRIRQGL, was commonly detected only in chronic infection in our study.
Figure 2.
Epitopes commonly recognized during A) primary and B) chronic infection. Only those epitopes that were tested in more than five subjects and were recognized in more than half of the tested subjects were evaluated. Black bars represent the number of subjects tested and gray bars represent the number of subjects that recognized the epitope. White bars represent the number of subjects with an immunodominant response to the epitope. The right panel lists the commonly recognized epitopes and their database frequencies. For HLA B7, the four epitopes listed were recognized by same number of subjects, and the epitope found to elicit immunodominant CTL responses is labeled with *. Thus, only single gray and white bars are presented for B7.
Duration of CTL recognition of conserved and variable epitopes
Of the five subjects with CTL responses measured during both primary and chronic infection, three recognized a total of five epitopes (two variable and three conserved) only during primary infection, whereas in the remaining two subjects, all epitopes were recognized during both primary and chronic infection. Again of these five subjects, three recognized a total of six epitopes (five variable and one conserved) only during chronic infection. Of the nine subjects with CTL responses measured at two time points during chronic infection, five recognized a total of nine epitopes (eight variable and one conserved) only at the first time points. In the remaining four subjects, all epitopes were recognized at both time points. One variable epitope was recognized only at the second time point in one subject. In total, responses to 4/20 (20%) conserved epitopes and 10/42 (24%) variable epitopes waned over time (p = 1.0, Fisher’s exact test). New responses were detected at second time points in a total of four subjects and seven epitopes (11% of all epitopes recognized in these 14 subjects).
Subdominant CTL responses to conserved epitopes
We next examined the relationship between epitope conservation and within-subject dominant CTL responses. We defined the CTL response of the highest magnitude in a subject as the dominant response, and all others as subdominant. During primary infection, dominant epitopes were located in Gag, Vpr, Tat, Env and Nef in 10, 3, 7, 7, 7 subjects, respectively. Fifty percent (17/34) of subjects recognized conserved epitopes, yet these corresponded to the dominant responses in only 12% (n = 4, the B27-restricted KRWIILGLNK in two subjects and the B*3501-restricted VPLRPMTY in the other two). During chronic infection, dominant epitopes were located in Gag, Pol, Tat, Env and Nef in 4, 3, 1, 4, 4 subjects, respectively (using the data of the first time points if subjects had two CTL measurements during chronic infection). Fifty percent (8/16) of subjects recognized conserved epitopes, however, they were dominant in only 25% (n = 4, the B27-restricted KRWIILGLNK and the B51-restricted EKEGKISKI in one each, and the A26-restricted EVIPMFSAL in two others). Therefore, in both primary and chronic infection, dominant responses within a subject were mostly elicited by variable epitopes. Notably, only two subjects examined during primary infection and another subject examined during chronic infection had the protective HLA-B27 allele, and all three dominantly recognized the conserved B27-restricted epitope KRWIILGLNK. In addition, the three epitopes commonly recognized during primary infection in this study were also the dominant epitopes in 40% to 88% of the subjects that recognized them (Figure 2).
We next compared the magnitudes of CTL responses to conserved and variable epitopes. During primary infection, the within-subject maximum (Figure 3A) and median (Figure 3C) magnitudes of CTL responses were both significantly lower to conserved epitopes compared to variable epitopes (p < 0.0001, Wilcoxon signed rank test). During chronic infection, the within-subject maximum magnitudes of CTL responses were significantly lower to conserved epitopes than to variable epitopes (p = 0.015; Figure 3B), while median magnitudes were not significantly different (p = 0.15; Figure 3D). For subjects whose CTL recognized both conserved and variable epitopes, the maximum magnitudes of responses were also significantly lower to conserved epitopes than to variable epitopes during primary infection (p = 0.01).
Figure 3.
Maximum and median magnitudes of CTL responses to conserved and variable HIV-1 epitopes within subjects. A and B) comparison of the maximum magnitudes, C and D) comparison of the median magnitudes of CTL responses to conserved and variable epitopes during primary and chronic infection. Each line represents results from an individual subject. The symbols were highlighted red when the within-subject dominant CTL responses were elicited by conserved epitopes. The horizontal bars represent the mean for each data set. Only data from the first time point are presented if chronic CTL responses were measured at two time points. In one subject, the dominant CTL response elicited by a conserved epitope (the A26-restricted EVIPMFSAL) was observed at the second time point measurement, and is thus not presented in the figure.
Correlations between CTL responses and viral load and CD4+ T cell counts
Finally, we examined the relationship of each of the following factors to viral load and CD4+ T cell counts: The number of recognized epitopes across the viral proteome; the number of recognized conserved and variable epitopes; the fraction of epitopes recognized (total, conserved and variable), taking the number of tested epitopes into account; and the total and mean magnitude of CTL responses against all tested epitopes, as well as conserved and variable epitopes.
As shown in Table 2, during primary infection, increased numbers of Gag epitopes recognized were associated with lower viral loads (p = 0.0499, Spearman’s rank coefficient), while increased number of Env epitopes recognized and increased total magnitudes of responses to Env and variable epitopes, as well as mean magnitude of responses to variable epitopes were associated with higher viral loads (p = 0.0273, p = 0.0096, p = 0.0188 and p = 0.0483, respectively). During chronic infection, increased numbers of Env epitopes recognized were associated with lower CD4+ T cell counts (p = 0.0309). We also found increased total magnitudes of responses to Gag as well as increased numbers of recognized epitopes from auxiliary proteins associated with lower viral load, when data of the second time points in the nine subjects with two measurements were used (p = 0.0421 and p = 0.0498, respectively). No other significant correlations of CTL responses with viral loads or CD4+ T cell counts were found (Table 2S). Given the large number of tests performed, it should be noted that after conservative Bonferroni correction for multiple testing, the correlations identified in this section were rendered non-significant.
Table 2.
Significant correlations of CTL responses with viral loads and CD4+ T cell counts a.
| CTL responses | Spearman r | p | ||
|---|---|---|---|---|
| Primary infection | Number of epitopes recognized | |||
| log10VL b | Gag | −0.3390 | 0.0499 | |
| log10 VL | Env | 0.3786 | 0.03 | |
| Total magnitude | ||||
| log10 VL | Env | 0.4380 | 0.01 | |
| log10 VL | variable | 0.4010 | 0.02 | |
| Mean magnitude | ||||
| log10 VL | variable | 0.3412 | 0.048 | |
| Chronic infection | Number of epitopes recognized | |||
| CD4+ T cell counts | Env | −0.5398 | 0.03 | |
| log10 VL | Aux c | −0.4976 | 0.05 | |
| Total magnitude | ||||
| log10 VL | Gag c | −0.5130 | 0.04 | |
The correlations were determined by Spearman’s rank coefficient. Chronic CTL responses were measured in 16 subjects: at a single time point in 7 subjects and at two time points in 9 subjects. Two parallel analyses were conducted. Analysis 1 used data from the former seven subjects and the data of the first time points from the latter nine subjects, and Analysis 2 used data from the former seven subjects and the data of the second time points from the latter nine subjects. Results from both analyses were generally similar. Hence, only the results from Analysis 1 are reported, unless specifically noted.
VL, viral load.
Result from Analysis 2. Data from the second time points were used for subjects with chronic CTL responses measured twice.
Discussion
We analyzed CTL responses in 45 subjects during primary and/or chronic HIV-1 infection using peptides corresponding to known epitopes for each subject as a function of their restricting Class I HLA. We found that conserved epitopes were recognized with similar probability as variable epitopes, however, the epitopes that were commonly recognized in subjects that shared the restricting HLA alleles were predominantly variable. Within a subject, CTL responses to conserved epitopes were generally subdominant, especially during primary infection. In addition, we found that increased numbers of recognized Gag epitopes, decreased numbers of recognized Env epitopes and decreased magnitudes of CTL responses to Env and variable epitopes tended to be correlated with lower viral loads during primary infection. Conversely, increased number of recognized Env epitopes tended to be correlated with lower CD4+ T cell counts during chronic infection.
One limitation of our study is that we examined CTL responses using only predefined optimal epitopes, most of which are derived from consensus sequences. Our assays will underestimate HIV-1-specific CTL responses to non-consensus, or variable epitopes, as we do not know whether the viruses infecting subjects contained the tested epitopes or not. Ideally, the testing of CTL responses to highly variable regions should include specific autologous peptides. Without testing autologous peptides, the rate of missed variable epitopes can be as high as 1/3 of the total epitopes recognized as we previously reported [6, 11]. However, we would expect to have observed a greater bias toward recognition of variable epitopes, or more profound dominant responses to variable epitopes if a more comprehensive epitope panel were assessed.
All commonly recognized epitopes we found during primary infection were also the immunodominant epitopes in multiple subjects. These results are in agreement with previous studies showing that the magnitudes of CTL responses positively correlated with their frequencies of recognition in the population [34], and the CTL responses against the most frequently targeted epitopes tended to be of the highest magnitude [24]. Similarly, these prior studies also examined only predefined epitopes, and thus underestimated overall CTL responses.
It is not clear why variable epitopes tend to be dominant. Our results suggest that the conservation level of an epitope plays a minor role in determining the frequency of recognition in the HIV-1 infected population and the immunodominance within a subject. Factors, including the kinetics of viral protein expression [35], antigen processing and presentation [36–38], and interactions between T cell receptors (TCR) and epitope–HLA complexes [39], might have contributed to the dominant recognition of variable epitopes. Of the subjects examined during primary infection, 41% showed immunodominant CTL responses towards Tat or Nef. As previously hypothesized [40], early expression of Tat and Nef during viral replication may have accelerated presentation of Tat and Nef epitopes to CD8+ T cells, and thus might be associated with immunodominant CTL responses preferentially towards these variable proteins during primary infection. The highly conserved Pol protein is expressed at a relatively low level in HIV-1 infection [41], which might also contribute to the less frequent immunodominant targeting of conserved epitopes. In our testing epitope panel, seven of the eight epitopes that can be presented by more than one HLA alleles are variable. Perhaps variable epitopes are more likely to be presented by multiple HLA alleles and thus in sum be more likely to be dominant. It is also possible that variable epitopes might have higher affinity to HLA alleles or higher avidity to CTL. Or perhaps responses to conserved epitopes are partially suppressed by regulatory factors reacting to chronic CTL activity. All these hypothesis need to be confirmed by further detailed studies.
Although early dominant CTL responses tend to exact strong selective pressure [6], our results lead us to hypothesize that the relatively low conservation of epitopes dominantly recognized early during infection may allow the rapid accumulation of escape mutants, and thereby CTL responses to these early epitopes might contribute little to the durable control of HIV-1 infection after initial curtailment of viremia. In addition, our observation of conserved epitopes generally eliciting subdominant CTL responses may provide some explanation to previous findings showing subdominant CD8+ T cell responses to be involved in control of SIV/HIV virus replication in both monkey [42] and humans [33] studies during chronic infection.
Previous studies of chronic HIV-1 infection, including in a subtype C infected heterosexual population from South Africa [43] and in subtype B infected populations from Peru and US [44], have shown that CTL responses to Gag are associated with viral control, while responses to Env are associated with higher levels of viremia. Our study of HIV-1 subtype B infected homosexual men from US, further suggests beneficial effect of CTL responses to Gag and deleterious effect of responses to Env and variable epitopes during primary infection.
Although it has recently been proposed that it could be desirable for an HIV vaccine to have immunodominant CTL responses towards conserved regions of HIV-1 [15, 16]; most vaccine strategies have so far sought to replicate and build on patterns of responses seen in natural infection. However, our results suggest that the immunogenicity of an effective HIV vaccine should not mimic most natural infection, unless it mimics responses induced in subjects who exhibit exquisite control of viral replication over many years of infection. Overcoming natural immunodominance and direct immunodominant CTL responses towards conserved regions of HIV-1 will be an essential challenge to overcome in HIV vaccine design [15].
Supplementary Material
Acknowledgments
We thank Dr. Nicole Frahm and Stephen Maley for editing and helpful comments. This work was supported by grants from the US Public Health Service to JIM (P01 AI57005) and the Computational Biology Core of the University of Washington Center for AIDS Research (P30 AI27757), and by grant M01-RR-00037 for leukapheresis.
Financial support: Grants from the US Public Health Service to JIM (P01 AI57005) and the University of Washington Center for AIDS Research (P30 AI27757), and grant M01-RR-00037 for leukapheresis.
Footnotes
Potential conflicts of interest: none reported.
Meeting(s) where the information has previously been presented: none.
References
- 1.Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koup RA, Safrit JT, Cao Y, et al. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–5. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musey L, Hughes J, Schacker T, Shea T, Corey L, McElrath MJ. Cytotoxic-T-cell responses, viral load, and disease progression in early human immunodeficiency virus type 1 infection. N Engl J Med. 1997;337:1267–74. doi: 10.1056/NEJM199710303371803. [DOI] [PubMed] [Google Scholar]
- 4.Jones NA, Wei X, Flower DR, et al. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J Exp Med. 2004;200:1243–56. doi: 10.1084/jem.20040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen TM, Altfeld M, Geer SC, et al. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J Virol. 2005;79:13239–49. doi: 10.1128/JVI.79.21.13239-13249.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, McNevin J, Cao J, et al. Selection on the human immunodeficiency virus type 1 proteome following primary infection. J Virol. 2006;80:9519–29. doi: 10.1128/JVI.00575-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedrich TC, Dodds EJ, Yant LJ, et al. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat Med. 2004;10:275–81. doi: 10.1038/nm998. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez CS, Stratov I, De Rose R, et al. Rapid viral escape at an immunodominant simian-human immunodeficiency virus cytotoxic T-lymphocyte epitope exacts a dramatic fitness cost. J Virol. 2005;79:5721–31. doi: 10.1128/JVI.79.9.5721-5731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leslie AJ, Pfafferott KJ, Chetty P, et al. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med. 2004;10:282–9. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Picado J, Prado JG, Fry EE, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, McNevin J, Zhao H, et al. Evolution of human immunodeficiency virus type 1 cytotoxic T-lymphocyte epitopes: fitness-balanced escape. J Virol. 2007;81:12179–88. doi: 10.1128/JVI.01277-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneidewind A, Brockman MA, Yang R, et al. Escape from the Dominant HLA-B27 Restricted CTL Response in Gag is Associated with a Dramatic Reduction in HIV-1 Replication. J Virol. 2007 doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneidewind A, Brockman MA, Sidney J, et al. Structural and functional constraints limit options for cytotoxic T-lymphocyte escape in the immunodominant HLA-B27-restricted epitope in human immunodeficiency virus type 1 capsid. J Virol. 2008;82:5594–605. doi: 10.1128/JVI.02356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang YE, Li B, Carlson JM, et al. Protective HLA class I alleles that restrict acute-phase CD8+ T-cell responses are associated with viral escape mutations located in highly conserved regions of human immunodeficiency virus type 1. J Virol. 2009;83:1845–55. doi: 10.1128/JVI.01061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rolland M, Nickle DC, Mullins JI. HIV-1 Group M Conserved Elements Vaccine. PLoS Pathogens. 2007;3:e157. doi: 10.1371/journal.ppat.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altfeld M, Allen TM. Hitting HIV where it hurts: an alternative approach to HIV vaccine design. Trends Immunol. 2006;27:504–10. doi: 10.1016/j.it.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Frahm N, Korber BT, Adams CM, et al. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J Virol. 2004;78:2187–200. doi: 10.1128/JVI.78.5.2187-2200.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thakar MR, Bhonge LS, Lakhashe SK, et al. Cytolytic T lymphocytes (CTLs) from HIV-1 subtype C-infected Indian patients recognize CTL epitopes from a conserved immunodominant region of HIV-1 Gag and Nef. J Infect Dis. 2005;192:749–59. doi: 10.1086/432547. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez WR, Addo MM, Rathod A, et al. CD8+ T lymphocyte responses target functionally important regions of Protease and Integrase in HIV-1 infected subjects. J Transl Med. 2004;2:15. doi: 10.1186/1479-5876-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yusim K, Kesmir C, Gaschen B, et al. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J Virol. 2002;76:8757–8768. doi: 10.1128/JVI.76.17.8757-8768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lichterfeld M, Yu XG, Cohen D, et al. HIV-1 Nef is preferentially recognized by CD8 T cells in primary HIV-1 infection despite a relatively high degree of genetic diversity. Aids. 2004;18:1383–92. doi: 10.1097/01.aids.0000131329.51633.a3. [DOI] [PubMed] [Google Scholar]
- 22.Troyer RM, McNevin J, Liu Y, et al. Variable Fitness Impact of HIV-1 Escape Mutations to Cytotoxic T Lymphocyte (CTL) Response. PLoS Pathog. 2009;5:e1000365. doi: 10.1371/journal.ppat.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bansal A, Gough E, Sabbaj S, et al. CD8 T-cell responses in early HIV-1 infection are skewed towards high entropy peptides. Aids. 2005;19:241–50. [PubMed] [Google Scholar]
- 24.Altfeld M, Kalife ET, Qi Y, et al. HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8(+) T Cell Response against HIV-1. PLoS Med. 2006;3:e403. doi: 10.1371/journal.pmed.0030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stekler JD, Sycks BJ, Holte S, et al. HIV Dynamics in Seminal Plasma during Primary HIV Infection. AIDS Res Hum Retro. 2008;24:1269. doi: 10.1089/aid.2008.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiebig EW, Wright DJ, Rawal BD, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. Aids. 2003;17:1871–9. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 27.Cao J, McNevin J, Holte S, Fink L, Corey L, McElrath MJ. Comprehensive analysis of human immunodeficiency virus type 1 (HIV-1)-specific gamma interferon-secreting CD8+ T cells in primary HIV-1 infection. J Virol. 2003;77:6867–78. doi: 10.1128/JVI.77.12.6867-6878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goulder PJ, Altfeld MA, Rosenberg ES, et al. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J Exp Med. 2001;193:181–94. doi: 10.1084/jem.193.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leitner T, Foley B, Hahn B, et al. HIV Sequence Compendium 2005. Los Alamos, N. M.: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, NM; 2006. p. 648+viii. [Google Scholar]
- 30.Bunce M, Fanning GC, Welsh KI. Comprehensive, serologically equivalent DNA typing for HLA-B by PCR using sequence-specific primers (PCR-SSP) Tissue Antigens. 1995;45:81–90. doi: 10.1111/j.1399-0039.1995.tb02422.x. [DOI] [PubMed] [Google Scholar]
- 31.Yu XG, Addo MM, Rosenberg ES, et al. Consistent patterns in the development and immunodominance of human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T-cell responses following acute HIV-1 infection. J Virol. 2002;76:8690–701. doi: 10.1128/JVI.76.17.8690-8701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reche PA, Keskin DB, Hussey RE, Ancuta P, Gabuzda D, Reinherz EL. Elicitation from virus-naive individuals of cytotoxic T lymphocytes directed against conserved HIV-1 epitopes. Med Immunol. 2006;5:1. doi: 10.1186/1476-9433-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frahm N, Kiepiela P, Adams S, et al. Control of human immunodeficiency virus replication by cytotoxic T lymphocytes targeting subdominant epitopes. Nat Immunol. 2006;7:173–8. doi: 10.1038/ni1281. [DOI] [PubMed] [Google Scholar]
- 34.Bihl F, Frahm N, Di Giammarino L, et al. Impact of HLA-B alleles, epitope binding affinity, functional avidity, and viral coinfection on the immunodominance of virus-specific CTL responses. J Immunol. 2006;176:4094–101. doi: 10.4049/jimmunol.176.7.4094. [DOI] [PubMed] [Google Scholar]
- 35.Probst HC, Tschannen K, Gallimore A, et al. Immunodominance of an antiviral cytotoxic T cell response is shaped by the kinetics of viral protein expression. J Immunol. 2003;171:5415–22. doi: 10.4049/jimmunol.171.10.5415. [DOI] [PubMed] [Google Scholar]
- 36.Chen W, Norbury CC, Cho Y, Yewdell JW, Bennink JR. Immunoproteasomes shape immunodominance hierarchies of antiviral CD8(+) T cells at the levels of T cell repertoire and presentation of viral antigens. J Exp Med. 2001;193:1319–26. doi: 10.1084/jem.193.11.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Gall S, Stamegna P, Walker BD. Portable flanking sequences modulate CTL epitope processing. J Clin Investigation. 2007;117:3563–3575. doi: 10.1172/JCI32047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sette A, Vitiello A, Reherman B, et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J Immunol. 1994;153:5586–92. [PubMed] [Google Scholar]
- 39.Yewdell JW, Bennink JR. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
- 40.Addo MM, Yu XG, Rathod A, et al. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol. 2003;77:2081–92. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hill M, Tachedjian G, Mak J. The packaging and maturation of the HIV-1 Pol proteins. Curr HIV Res. 2005;3:73–85. doi: 10.2174/1570162052772942. [DOI] [PubMed] [Google Scholar]
- 42.Friedrich TC, Valentine LE, Yant LJ, et al. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J Virol. 2007;81:3465–76. doi: 10.1128/JVI.02392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiepiela P, Ngumbela K, Thobakgale C, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 44.Zuniga R, Lucchetti A, Galvan P, et al. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J Virol. 2006;80:3122–5. doi: 10.1128/JVI.80.6.3122-3125.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.