Abstract
The sequential steps of neurogenesis are characterized by highly choreographed changes in transcription factor activity. In contrast to the well-studied mechanisms of transcription factor activation during neurogenesis, much less is understood regarding how such activity is terminated. We previously showed that MTGR1, a member of the MTG family of transcriptional repressors, is strongly induced by a proneural basic helix-loop-helix transcription factor, NEUROG2 in developing nervous system. In this study, we describe a novel feedback regulation of NEUROG2 activity by MTGR1. We show that MTGR1 physically interacts with NEUROG2 and represses transcriptional activity of NEUROG2. MTGR1 also prevents DNA binding of the NEUROG2/E47 complex. In addition, we provide evidence that proper termination of NEUROG2 activity by MTGR1 is necessary for normal progression of neurogenesis in the developing spinal cord. These results highlight the importance of feedback regulation of proneural gene activity in neurodevelopment.
Keywords: Development, Neurogenesis, Embryo, transcription, bHLH, spinal cord, MTGR1, NEUROG2, MTG/ETO proteins, nervy, transcriptional repressor
Introduction
Successive up-regulation and down-regulation of transcription factor expression is a characteristic feature of neurogenesis (Roztocil et al., 1997; Torii et al., 1999). For example, cells at different stages of neurogenesis in the developing spinal cord can be identified based on their mediolateral positions, and the stage of development of the cells correlates with the transcription factors that are active (summarized in Fig. 7). Postmitotic cells migrate through at least three distinct layers of the spinal cord as they progress through neurogenesis: the ventricular zone (VZ), the intermediate zone (IZ), and the mantle zone (MZ). Early proneural transcription factors such as NEUROG1 (previously NGN1, ATH4C), NEUROG2 (previously NGN2, ATH4A) and ASCL1 (previously cASH1) are first expressed in the VZ (Lo et al., 1991; Sommer et al., 1996; Ma et al., 1997; Murciano et al., 2002). Postmitotic cells migrate from the VZ into the IZ as the next group of transcription factors including PROX1, MTGR1 and NEUROD4 (previously ATH3) reach peak expression, and expression of the early factors begin to fall (Roztocil et al., 1997; Torii et al., 1999; Koyano-Nakagawa and Kintner, 2005). Finally, as cells reach the MZ, late transcription factors such as NEUROD1, MTG8, MTG16, PAX2 and ISL1 are expressed, and the earlier factors are lost. Each wave of transcription factors promotes expression of the next group. In order for expression of the intermediate group of transcription factors to fall as the cells enter the MZ, it is necessary that activity of the early transcription factors be terminated.
Figure 7. Model: NEUROG2 activity is limited to a narrower area compared to its expression domain by the function of MTGR1.
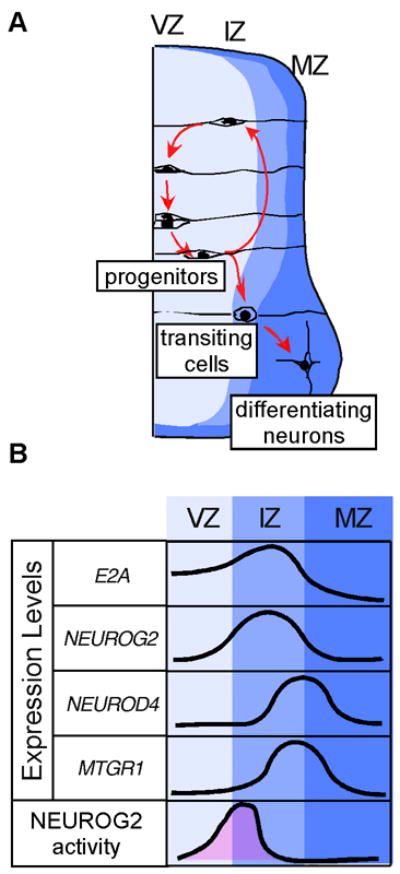
A: Diagram of cell movement during neurogenesis. Progenitors are located in the VZ. After the last mitosis, committed neuronal progenitors migrate to the IZ, and delaminate to the MZ. Note that similar gene expression pattern is observed in development of the mouse central nervous system (Alishahi et al., 2009). B: Relative expression levels of genes studied in this paper. The area shaded in pink represents conceptual transcriptional activity of NEUROG2 based on our findings. Note that the activity of NEUROG2 is limited to narrower window of time compared to the period of its gene expression.
The importance of terminating the activity of proneural transcription factors once they have fulfilled their roles was underscored by studies in which expression of such proteins was artificially extended. For example, in the developing brain, prolonged expression of proneural basic helix-loop-helix (bHLH) transcription factors in postmitotic neurons resulted in increased neuronal cell death and malformation of the cytoarchietecture (Isaka et al., 1999; Cai et al., 2000). Because it is estimated that newly born neurons migrate through the VZ and the IZ in a few hours (Koyano-Nakagawa and Kintner, 2005; Gui et al., 2007), cells should successively up- and down-regulate the expression of transcription factors rapidly within this short window of time.
Mechanisms that initiate transcription factor activity have been well investigated, whereas the mechanisms that terminate activity are poorly understood. Termination of transcription factor activity could be accomplished by at least three mechanisms that are not mutually exclusive: 1) ending further production of transcription factors at mRNA or protein level, 2) removal of already produced transcription factors by proteolysis, and 3) inhibiting the activity of transcription factors. While there is evidence indicating the employment of the first two mechanisms (Sriuranpong et al., 2002; Vosper et al., 2007), the third, inhibition of bHLH transcriptional activity, is less well established. Members of the MTG family of transcriptional repressors are strongly induced during neurogenesis, and it has been suggested that they inhibit transcription factor activity (Koyano-Nakagawa and Kintner, 2005).
MTG8 (RUNXT1, ETO), MTG16 (CBFA2T3) and MTGR1 (CBFA2T2) are members of the MTG/ETO/CBFA2T protein family, a small group of transcriptional repressors (for reviews, see Davis et al., 2003; Hug and Lazar, 2004). They are reported to act as a “protein scaffold”, bridging different transcription factors. Several groups including ours have previously reported that MTGR1 is strongly induced in Xenopus laevis by various proneural bHLH proteins including X-NGNR-1, Xash3, Xath3 (XNeuroD4), Xath5 (XAtoh7) and XNeuroD (Cao et al., 2002; Koyano-Nakagawa and Kintner, 2005; Logan et al., 2005; Seo et al., 2007). The strong induction by proneural bHLH proteins as well as its highly conserved structure across species suggests that MTGR1 may play an important role in neuronal differentiation. In this report, we show that MTGR1 physically interacts with NEUROG2 and inhibits its transcriptional activity. We provide evidence that this inhibition takes place in vivo in the developing spinal cord, and is necessary for normal progression of the neurogenic program. Our results indicate that termination of NEUROG2 activity is ensured by transcriptional repression by MTGR1, in addition to down-regulation of NEUROG2 expression and degradation of NEUROG2 protein. These three independent mechanisms are used to warrant proper progression of neurogenesis.
Materials and Methods
Cells
Cell lines were maintained according to the protocol by American Type Culture Collection. P19 embryonal carcinoma cells were cultured in alpha-MEM (Cellgro, Herndon, VA) supplemented with 2.5% fetal calf serum (FCS) and 7.5% newborn calf serum with 100 units/ml penicillin, 100μg/ml streptomycin, and 0.24μg/ml of amphotericin B as Funigizone ® (Gibco, Grand Island, NY). HEK293T cells were cultured in Dulbecco’s Modification of Eagle’s Medium (Cellgro, Herndon, VA) supplemented with 10% FCS with 1g/L glucose, 100 units/ml penicillin, 100μg/ml streptomycin, and 0.24μg/ml of amphotericin B as Funigizone ® (Gibco, Grand Island, NY).
Plasmids
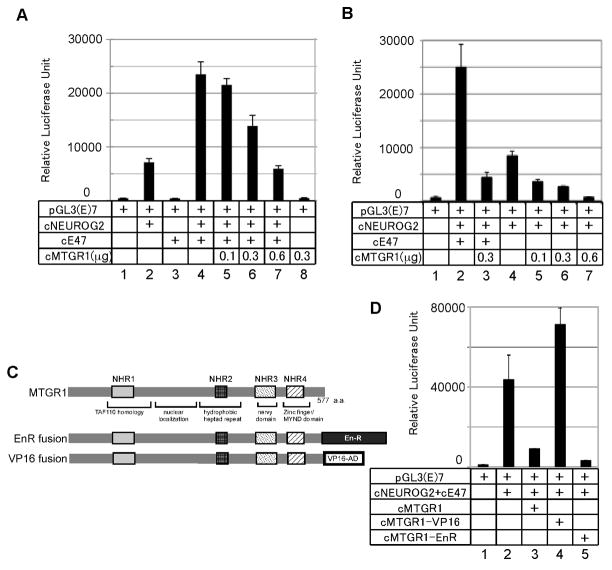
Chick and Xenopus MTGR1 clones were previously described (Koyano-Nakagawa and Kintner, 2005). All MTG constructs were cloned into the pCS2(+) vectors with or without the myc epitope tag. pCS2(+) cMTGR1-VP16 and pCS2(+) cMTGR1-EnR were constructed by fusing the activation domain of VP16 protein and the repressor domain of engrailed, respectively, to the 3′ side of the entire open reading frame of cMTGR1 in-frame. Expression of the fusion proteins was confirmed by analyzing expression of myc epitope-tagged molecules in a parallel reaction (Fig. S3C). Expression vectors for GST fusion molecules, pGEX-cNEUROG2, pGEX-cE47 were constructed by shuttling the entire coding frames of the factors in-frame into the pGEX vectors (GE Healthcare Technologies, Waukesha, WI). The following plasmids were kind gifts from the laboratories listed below. pcDNA3 HA-mNeurog2, pcDNA3 HA-cNEUROG2, pcDNA3 HA-hE47 and in situ probe for chicken NEUROD4: Soo-Kyung Lee and Sam Pfaff (Lee et al., 2005); DLL4: Domingos Henrique (Henrique et al., 1995); EMSV-mE12: Atsushi Asakura; pcDNA3 cE47: Klemens Meyer (Conlon and Meyer, 2004). pGL3P(E)7Luc reporter gene was made by shuttling the E-box fragment of pGL2P(E)7Luc (given by Masato Nakafuku) into the pGL3P backbone. The E-box fragment contains seven tandem repeats of sequence, AGGCAGGTGGC (E-box is underlined). pRL-CMV is from Promega (Madison, WI). siRNA vector for MTGR1 (psiRNA-cMTGR1) was constructed by inserting 66 mer oligonucleotide GTA CCT CGC ACT ACA CCC TGG AAG ACA TTC AAG AGA TGT CTT CCA GGG TGT AGT GCT TTT TGG AAA (only the top strand is shown) into the HindIII-Acc65I fragment of psiRNA-h7SKGFPzeo (InvivoGen, San Diego, CA). This makes a bi-cistronic vector expressing siRNA and GFP. An siRNA vector targeted to a different part of cMTGR1 was constructed using 66 mer, GTA CCT CGC CCG TGG AAG TGA AGA TAC ATC AAG AGT GTA TCT TCA CTT CCA CGG GCT TTT TGG AAA, which yielded essentially the same results. In this paper results with the first siRNA construct are shown. psiRNA-h7SKgz-Scr, a vector expressing siRNA of “scramble” sequence that is not complementary to any genomic sequence in the database was used as a control (InvivoGen, San Diego, CA).
EMSA and in vitro degradation assay
The following probe was synthesized for EMSA. E-box motifs are underlined; (Ebox)3: GATCCAGGCAGGTGGCAGGCAGGTGGCAGGCAGGTGGCAGATC. Unlabeled transcription factors were synthesized using TNT® coupled Reticulocyte Lysate Systems (Promega, Madison, WI). Parallel reactions including 35S-methionine were set up to monitor protein synthesis. Fifteen μl of binding reaction containing 30% glycerol, 40 mM HEPES (pH 7.9), 10 mM MgCl2, 0.2 mM DTT, 0.02% TritonX-100, 4 μl in vitro translated protein, 200 ng sheared salmon sperm DNA and 1 ng of labeled probe was set up. Competition experiments included 20 ng of unlabeled oligonucleotide. Reactions were incubated at 23°C and separated on a 1.5 mm thick 4% acrylamide gel (acrylamide:bisacrylamide=80:1, 6.75mM Tris pH7.9, 3.3mM NaOAc, 1mM EDTA, 2.5% glycerol). The gel was run at 4°C with buffer circulation. For in vitro degradation assay, EMSA reactions were set up with unlabeled oligonucleotide. Ten μl of reaction was removed periodically and boiled immediately in 1X sample buffer. All gels were dried after electrophoresis and imaged on Storm Phosphorimager (GE Healthcare Technologies, Waukesha, WI).
GST Pull-down and immunoprecipitation assays
GST fusion proteins induced in BL21 cells were purified using glutathione sepharose beads (GE Healthcare Technologies, Waukesha, WI). Proteins bound to beads were analyzed by SDS-PAGE and similar amounts of protein based on Coomasie staining were used for binding reactions. In a typical reaction, 25–50 μl of 35S-labeled transcription factors were diluted in 800 μl of binding buffer (Zhang et al., 2004) and mixed with GST fusion proteins preadsorbed to glutathione beads. After three washes in binding buffer, proteins remaining on the beads were separated by SDS-PAGE and autoradiographed. For immunoprecipitation, HEK293T cells were transfected with expression vectors of transcription factors using Fugene 6 (Roche) and cultured for 24 hrs. Proteosomal inhibitors MG132 (23 μM) and lactacystine (4.7 μM) were added to the culture and further incubated for 6 hrs before harvest. The treatment with proteosomal inhibitors was necessary because NEUROG2 protein has a short half-life and it was not possible to detect the protein without the treatment. The samples were then immunoprecipitated with either 2 μg of anti-HA antibody (Santa Cruz SC 805) or 0.2 μg of anti-myc tag antibody (Upstate 4A6).
In ovo electroporation, immunohistochemistry and in situ hybridization
In ovo electroporation and immunohistochemistry were performed as described previously (Koyano-Nakagawa and Kintner, 2005). Embryos were electroporated at Hamburger-Hamilton (HH) stage 13 (day 2 of incubation) and harvested at stage 16 (24 hours after electroporation) or stage 22 (48 hours after electroporation). Antibodies and dilutions were as follows: rabbit anti-GFP (Molecular Probes, 1:400), mouse anti-myc epitope (Upstate 4A6, 1:5000), rat anti-HA (Roche 3F10, 1:2000), rabbit anti-Pax2 (Zymed, 1:200), mouse anti-Isl1 (Developmental Studies Hybridoma Bank, 1:10), mouse anti-HuC/D (Molecular Probes, 1:100), TuJ1 (Covance, 1:500), rat anti-BrdU (Harlan, 1:500), rat anti-NEUROD4 (a gift from Thomas Jessell, 1:2000), rabbit anti-cNEUROG2 (a gift from David Anderson, 1:500) and rabbit anti-hMTGR1 (a gift from Issay Kitabayashi, 1:5000). In situ hybridization on 20μm thick cryosections were done following standard protocol (Tuttle et al., 1999). For double fluorescent in situ hybridization, digoxigenin- and fluorescein-labeled RNA probes were used for hybridization and the hybridized probes were detected as previously described (Fior and Henrique, 2005).
Quantification of in situ hybridization signals
Areas positive for NEUROD4 and DLL1 were measured using Image J (NIH) following the instruction to the software. Briefly, TIFF images were converted to 8-bit and smoothed for median, with a radius of 2 pixels. Multithresholder plugin was used to set a threshold with maximum entropy. Pixels of the positive areas were counted and converted to square microns. The boundaries of the VZ, the IZ, and the MZ were morphologically identified by the arrangement of nuclei as visualized by DAPI nuclear staining.
Luciferase assay
P19 cells were plated at 7×104 per well (24-well cluster) and transfected in triplicates with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) with pRL-CMV as an internal control. In all experiments, 0.2 μg of the reporter genes, 0.3 μg of bHLH expression vectors, 0.1 μg of E12 expression vector was used. Total amount of DNA transfected per well was adjusted with an empty vector, so that the DNA:Lipofectamine ratio is constant for all samples. Twenty-four hours later, cells were harvested and luciferase activity was measured using Dual Luciferase Assay Reagent (Promega, Madison, WI). A relative luciferase unit (RLU) was calculated by dividing the luciferase activity by Renilla luciferase activity and averaged. Data were normalized to the mean control value to calculate fold change.
Western blot
Ten microliter each of the lysate used for luciferase assay was separated on a SDS polyacrylamide gel. Western blotting and development was done as described (Ausubel, 1994). Mouse anti-myc epitope antibody (Upstate 4A6) was used at 1:20000 dilution.
Results
MTGR1 is transiently expressed in differentiating neurons
MTG proteins are known to interact with sequence specific transcription factors and repress transcription. We aimed to identify the target transcription factor whose activity is repressed by MTGR1 in the developing nervous system. Since MTGR1 is induced during neurogenesis, MTGR1 is likely to interact with transcription factors that are expressed in differentiating neurons. A report showing the interaction of MTG8 with HEB, a bHLH protein related to E2A (Zhang et al., 2004), prompted us to investigate whether MTGR1 similarly targets a bHLH protein, NEUROG2. We first compared the temporal pattern of MTGR1 gene expression with that of bHLH transcription factors, NEUROG2 and NEUROD4, in chick embryos (Fig. 1). Two color fluorescent in situ hybridization combined with immunohistochemistry showed that NEUROG2 is induced the earliest among these three genes in the VZ and continues to be expressed in the medial half of the IZ (Fig. 1A, C). MTGR1 follows NEUROG2 expression and peaks in the IZ (Fig. 1A). Its expression level decreases when cells reach the MZ and begin to express TuJ1. NEUROD4 shows similar pattern of overlap with NEUROG2, induced after NEUROG2 and peaking in the IZ (Fig. 1C). Double-labeling experiments show that both mRNA and proteins of MTGR1 are co-localized with those of NEUROD4 in the VZ, the IZ and the MZ, particularly in the ventral spinal cord (Fig. 1B, D). These results indicate the temporal sequence of gene expression in the differentiating neurons; NEUROG2 is expressed first, followed by MTGR1 and NEUROD4, and later by a neuronal marker TuJ1. The same sequence of gene induction was observed throughout the neurogenic stages from embryonic day (E)2 to E7 (Fig. S1). Thus, we conclude that MTGR1 is expressed in differentiating neurons, closely following NEUROG2 expression. This temporal sequence of gene expression was consistent with that observed in development of the mouse central nervous system (Alishahi et al., 2009). Because over-expression of MTGR1 in the chick spinal cord at E2 by in vivo electroporation did not suppress NEUROG2 expression (Fig. S2), we hypothesized that MTGR1 acts at the protein level to negatively regulate the NEUROG2 activity during neurogenesis.
Figure 1. Expression of NEUROG2, MTGR1, and NEUROD4 in the chick embryo spinal cord.
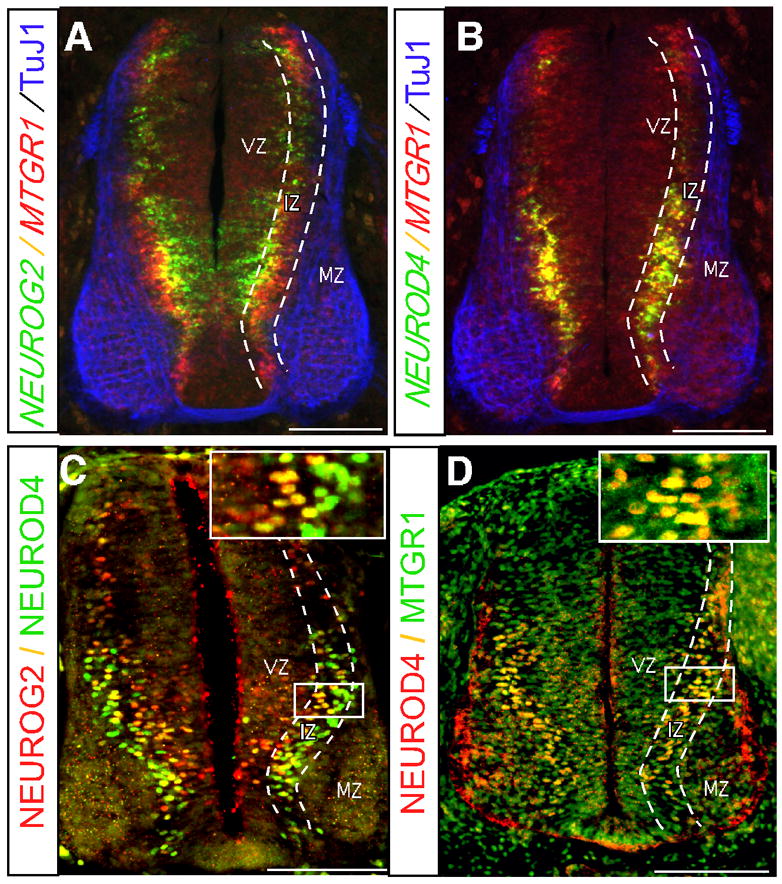
Cross sections of embryos at HH stage 24 at the brachial level are shown. A, B: Fluorescent in situ hybridization of NEUROG2, NEUROD4 and MTGR1 combined with immunohistochemistry of TuJ1. C, D: Double immunohistochemistry of NEUROG2, NEUROD4 and MTGR1. Colors of the probes are indicated next to figures. Boundaries of the VZ, the IZ, and the MZ are indicated by dotted lines. Boxed areas are enlarged in the insets of C and D. Bars: 100 μm. Note that NEUROG2 is expressed most medially, closest to the ventricle, followed by MTGR1 and NEUROD4, appearing at the same medio-lateral position. Both genes are down-regulated in the MZ.
MTGR1 inhibits transcriptional activity of NEUROG2
We first tested the hypothesis that MTGR1 inhibits the function of NEUROG2 by using a transient transfection assay in P19 cells (Fig. 2A, B). As a reporter, we used the plasmid, pGL3(E)7-Luc, which contains seven repeats of an E-box sequence, a recognition sequence of bHLH proteins, linked to the luciferase reporter gene (Lee et al., 2005). NEUROG2 activates this reporter by heterodimerizing with its binding partner E12 or E47, which are splice variants produced from the E2A gene. This reporter gene alone had very low activity that is indistinguishable from the background luciferase activity from mock transfection control (Fig 2A, B; lane 1). NEUROG2 alone activated the pGL3(E)7-Luc reporter significantly, presumably by acting together with the endogenous E proteins (Fig. 2A, lane 2; 2B, lane 4). Adding E47 augmented NEUROG2 activity by three fold (Fig. 2A, lane 4; 2B, lane 2), whereas E47 alone did not activate the reporter (Fig. 2A, lane 3). Transcriptional activity by the combination of NEUROG2 and E47 was inhibited by MTGR1 in a dose-dependent manner (Fig. 2A lanes 4–7). Transcriptional activity by NEUROG2 alone was also inhibited dose-dependently (Fig. 2B, lanes 4–7). MTGR1 alone without NEUROG2 and E47 did not change transcriptional activity (Fig. 2A, lane 8). Similarly, MTGR1 did not affect transcription activity of a control reporter without E-box motifs either with or without co-transfection of vectors encoding NEUROG2 and E47 (data not shown). These results suggest that MTGR1 inhibits E-box dependent NEUROG2-E47 activity by interacting with this protein complex. Similar inhibitory activity of MTGR1 was observed using combinations of mouse Neurog2 (mNeurog2), mouse E12 (mE12) and Xenopus MTGR1 (XMTGR1) (Fig. S3A), or mNeurog2, human E47 (hE47) and chicken MTGR1 (Fig. S3B). Thus inhibition of NEUROG2-E47 activity by MTGR1 occurs across species, and it is likely the protein-protein interaction interfaces are conserved.
Figure 2. MTGR1 represses transcriptional activity of the NEUROG2-E47 complex.
A, B, D: Transient transfection assay. C: Diagram of the fusion molecules of chick MTGR1. A, B: P19 cells were transfected with the indicated vectors and increasing amounts of the MTGR1 expression vector (0.1, 0.3 and 0.6 μg). MTGR1 repressed transcription activity of NEUROG2-E47 (A) or NEUROG2 alone (B) dose dependently. Total amount of DNA transfected per sample was adjusted with an empty vector. D: Transcription activity of MTGR1 fusion proteins. P19 cells were transfected with the indicated vectors and 0.3μg of vectors encoding MTGR1 fusion molecules. Bars show SD for all paels. Essentially the same results were obtained from P19 and HEK293T cells for all assays, and the results from P19 cells are shown.
To examine if MTGR1 represses transcription of the NEUROG2-E47 complex bound to DNA or by other mechanisms such as competitively inhibiting binding to DNA or to co-factors, we made fusion proteins of MTGR1 with the repressor domain of engrailed (EnR) or the activation domain of VP16 (Fig. 2C). If MTGR1 interacts with the NEUROG2-E47 complex bound to DNA, a forced fusion with EnR should repress, and fusion with VP16 should augment the transcriptional activity of NEUROG2-E47. As shown in Figure 2D, lanes 4 and 5, VP16 fusion enhanced and EnR fusion of MTGR1 inhibited the activity of NEUROG2-E47, respectively. Either construct did not affect transcriptional activity in the absence of NEUROG2, E47, or the target E-box sequence (data not shown). Thus, these results indicate that MTGR1 inhibits transcriptional activity at least in part by interacting with the NEUROG2-E47 complex bound to the E-box sequence. These results, however, do not eliminate the possibility that MTGR1 also inhibits the NEUROG2-E47 complex by other mechanisms such as inhibition of DNA binding.
MTGR1 physically interacts with NEUROG2 and E47
The reporter gene assays described above suggested that MTGR1 might act by interacting with the NEUROG2-E47 complex. Thus, we used a GST pull-down assay to test if MTGR1 directly interacts with NEUROG2 and/or E47. We found that MTGR1, MTGR1-EnR, and MTGR1-VP16 co-purified with GST-NEUROG2 but not with the control GST protein (Fig. 3A, lanes 1–4). The efficiency of co-purification was comparable to E47 protein purified with GST-NEUROG2 (lane 5). MTGR1 and MTGR1-EnR also co-purified with GST-E47, but the recovery was not as efficient as with GST-NEUROG2, and the interaction between MTGR1-VP16 and E47 was very weak compared to other interactions (lanes 6–8). These results indicated that MTGR1 physically interacts with NEUROG2, and less efficiently with E47 in vitro.
Figure 3. MTGR1 directly interacts with NEUROG2.
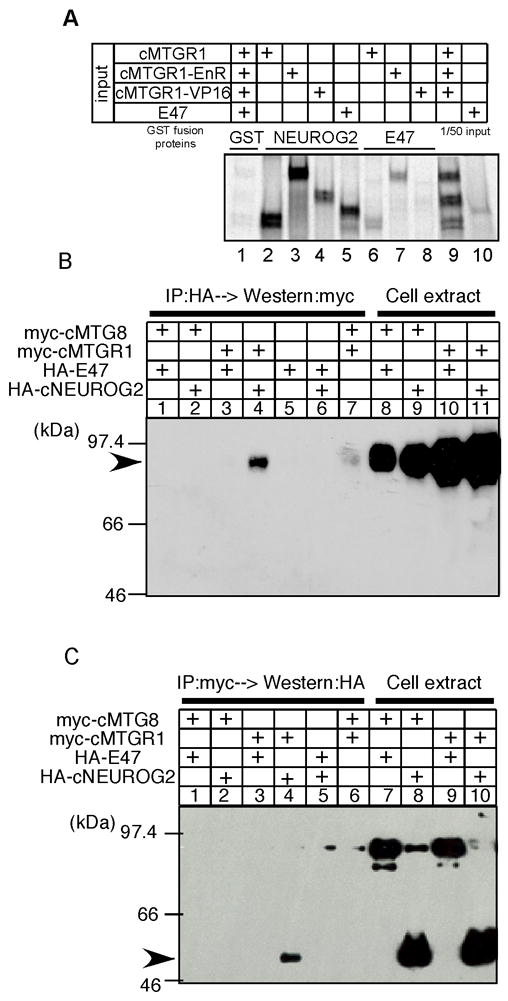
A: GST pull-down assay. Radiolabeled MTG proteins or E47 were pulled down with glutathione sepharose beads coated with GST molecule alone (lane 1), GST-NEUROG2 fusion (lanes 2–5) or GST-E47 fusion molecules (lanes 6–8). Lanes 9–10 contain 1/50 of input proteins. B, C: Immunoprecipitation assays. HEK293T cells were transfected with expression vectors of the indicated proteins. Cell lysates were immunoprecipitated with antibodies against the HA (B) or the myc epitopes (C), and recovered proteins were analyzed by Western blot using anti-myc (B) or anti-HA (C) antibodies. Whole cell extracts were analyzed in lanes 8–11 of B and lanes 7–10 of C to confirm protein expression.
Next, we tested if this interaction takes place in cultured cells by immunoprecipitation assays. Expression vectors of MTGR1 tagged with the myc epitope and NEUROG2 or E47 tagged with the haemagglutinin epitope (HA) were transfected into HEK293T cells. When anti-HA antibody was used for immunoprecipitation, MTGR1 co-precipitated with NEUROG2, but not with E47 (Fig. 3B lanes 3, 4). In contrast, myc epitope-tagged MTG8, another MTG family member, did not co-purify with either NEUROG2 or E47 under the same condition (lanes 1, 2). A reverse experiment was done by immunoprecipitating the cell extracts with anti-myc epitope antibody (Fig. 3C). As expected from the results in Fig. 3B, NEUROG2 but not E47 co-purified with MTGR1 (lanes 3, 4), and neither of the proteins co-purified with MTG8 (lanes 1, 2). Thus, our results showed that the interaction between NEUROG2 and MTGR1 is specific and takes place in a cellular environment. Interaction between E47 and MTGR1 was not detected under this condition. We attempted to detect interaction of endogenous proteins using nuclear extracts of embryonic day 14.5 mouse cortex, in which both NEUROG2 and MTGR1 are abundantly expressed (Alishahi et al., 2009), but were not able to carry out the experiment due to the low sensitivity of anti NEUROG2 and anti MTGR1 antibodies.
It is common that protein association changes the affinity of those proteins to a third molecule. Thus, we tested whether heterodimerization of NEUROG2 and E47 changes their ability to interact with MTGR1 by pulling down co-synthesized NEUROG2 and E47 with MTGR1-GST fusion proteins. The recovery rate was the same as separately synthesized proteins, indicating that heterodimerization of NEUROG2 and E47 did not change the affinity to MTGR1 (data not shown).
Taken together, our immunoprecipitation assay showed specific interaction of MTGR1 and NEUROG2 both in vitro and in cultured cells. MTGR1 also interacted with E47 as a synthetic protein in vitro, but this interaction was much weaker than the interaction between MTGR1 and NEUROG2. This interaction was not detectable in HEK293T cells, suggesting that the interaction is weak, if any happens in vivo.
MTGR1 inhibits DNA binding of the NEUROG2-E47 complex, but does not dissociate the preformed complex from DNA
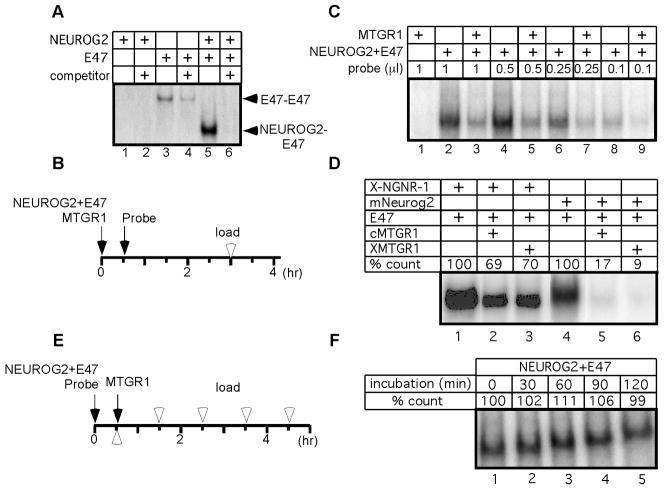
Next, we examined if transcriptional inhibition by MTGR1 involves a change in DNA binding activity of the NEUROG2-E47 complex. To examine the DNA binding activity, electrophoretic mobility shift assay (EMSA) was performed with proteins synthesized in vitro. In our experimental condition, E47 but not NEUROG2 bound to the E-box sequence as a homodimer (Fig 4A, lanes 1, 3). When NEUROG2 and E47 were co-translated, they preferentially formed a heterodimer, and a homodimer of E47 was not detected (Fig 4A, lane 5). Since the combination of NEUROG2 and E47 activates transcription strongly (Fig. 2), we used co-translated proteins of NEUROG2 and E47 in the following assays. First, we asked if MTGR1 prevents DNA association of the NEUROG2-E47 complex. When we pre-incubated NEUROG2, E47 and MTGR1 before adding the probe, the DNA binding activity of the NEUROG-E47 complex was reduced (Fig. 4B, C lanes 2, 3). To examine if MTGR1 forms a higher-order complex with NEUROG2 and E47 that binds to DNA, we titrated down the amount of the radiolabeled DNA probe and made it the limiting reagent. Even after a long exposure to the film, we did not observe formation of additional bands that show different mobility from the one formed by NEUROG2-E47 alone (Fig. 4C and data not shown). Therefore, when MTGR1 complexes with NEUROG2-E47 before DNA binding, it primarily inhibits the association of the NEUROG2-E47 complex with DNA. It should be noted, however, that EMSA requires the protein-DNA complex to stay associated throughout electrophoresis, and does not detect a complex that has short half-life. Thus, it remains possible that MTGR forms a minor population of unstable DNA binding complexes with NEUROG2-E47. The inhibition of DNA binding activity was also observed using combinations of X-NGNR-1 and chicken or Xenopus MTGR1 (Fig. 4D lanes 1–3), and mouse Neurog2 and chicken or Xenopus MTGR1 (lanes 4–6), indicating that the protein interaction can take place across species.
Figure 4. MTGR1 blocks DNA binding activity of the NEUROG2-E47 complex.
A: EMSA of NEUROG2 and E47, either synthesized individually (lanes 1–4) or in the same reaction (lanes 5–6). Twenty-fold excess of cold competitor was added to test the specificity (lanes 2, 4, 6). Arrowheads point to bands containing E47 homodimer (E47-E47) and NEUROG2-E47 heterodimer (NEUROG2-E47). B, E: Experimental schedule testing the effect of MTGR1 on association (B) and dissociation (E) of NEUROG2-E47 complex to/from DNA, respectively. Black arrows and white arrowheads indicate the times when proteins and probes were mixed and loaded on a gel, respectively. C: EMSA was carried out using schedule shown in B with varying amounts of probes. D: Various MTG molecules were examined following the schedule shown in B. Intensity of each band was quantified on a phosphorimager and shown as percentage of the band without MTGR1. F: EMSA was done as diagrammed in E. Relative radioactivity compared to lane 1 is shown. Since samples were applied every 30 min on a continuously running gel, migration distances appear different from each other.
It is possible that association of the NEUROG2-E47 complex with MTGR1 induced degradation of NEUROG2 or E47 and that resulted in reduction of the DNA binding activity. To examine this possibility, we radiolabeled the three proteins and monitored the protein levels in a reaction identical to EMSA. The levels of NEUROG2 and E47 proteins remained constant throughout the experiment, either in the presence or absence of MTGR1, indicating that inhibition of DNA binding was not due to degradation of NEUROG2 or E47 (Fig. S4A).
To determine whether MTGR1 disrupted the heterodimer of NEUROG2 and E47 and inhibited its DNA binding ability, we carried out an immunoprecipitation assay (Fig. S4B). NEUROG2 and E47 were co-synthesized and immunoprecipitated with the antibody against NEUROG2. E47 co-purified with NEUROG2 regardless of the presence or absence of MTGR1, indicating that binding to MTGR1 did not inhibit heterodimerization of NEUROG2 and E47.
Finally, we asked if MTGR1 promotes dissociation of the NEUROG2-E47 complex already bound to DNA (Fig. 4E, F). In this experiment, the NEUROG2-E47 complex was mixed with the probe first and then MTGR1 was added. The same volumes were removed from the reaction every 30 minutes and immediately applied to a running non-denaturing gel. In contrast to our experiments above, we did not observe a decrease in DNA binding activity even after 120 min of incubation. No additional DNA binding complex was detected in this assay (data not shown). Therefore, if NEUROG2-E47 was already bound to DNA, MTGR1 does not dissociate the complex from DNA.
Collectively, our results show that MTGR1 can inhibit DNA association of the NEUROG2-E47 complex, but it cannot induce dissociation of an already formed complex on DNA.
NEUROG2 activity is inhibited by MTGR1 in vivo
We next asked if suppression of NEUROG2-E47 activity by MTGR1 takes place in vivo. To address this question, we chose to use NEUROD4 and DLL1 as a readout of NEUROG2 activity. Xath-3, the Xenopus laevis counterpart of NEUROD4 is demonstrated as a direct target of neurogenin (Logan et al., 2005; Seo et al., 2007). In the mouse telencephalon, Neurod4 gene expression is completely lost in Neurog2 null animals (Mattar et al., 2008). In the chick and mouse nervous system, NEUROD4 closely follows NEUROG2 expression throughout the developmental period (Fig. 1, S1 and Alishahi et al., 2009), and NEUROD4 expression is induced by ectopic expression of NEUROG2 (Fig. S5C–d). Furthermore, we noticed that the 5′ upstream region of the NEUROD4 gene contains 16 putative E-box sequences within the 4 kb region upstream of the transcription initiation site (data not shown). In particular, E-box sequences were 12 fold overrepresented compared to random distribution of nucleotides between −3.3 and −2.8 kb relative to the transcription start site. EMSA analysis showed that the NEUROG2-E47 complex binds efficiently to the E-box sequence found in this promoter region (data not shown). Thus, evidences suggest that, similar to other species, chicken NEUROD4 is induced by NEUROG2 and can be used as readout of NEUROG2 activity. Similarly, DLL1 is a known direct target of Neurog2 in mouse and frog (Castro et al., 2006; Seo et al., 2007), and the conserved expression patterns of these genes in chick embryo show that it is highly likely that chicken DLL1 is also directly regulated by NEUROG2. Thus, NEUROD4 and DLL1 expression was used as an indicator of NEUROG2 activity.
We previously reported that over-expression of MTGR1 alone at day 2 of incubation does not significantly affect neuronal differentiation of chick spinal cord, likely due to regulatory response by endogenous mechanisms or compensation by the growth of the untransfected cells (Koyano-Nakagawa and Kintner, 2005). Thus, to further examine the interaction of NEUROG2 and MTGR1 in vivo, we used a gene knockdown approach. When an expression vector for short interference RNA against cMTGR1 (siRNA-cMTGR1) was electroporated into E2 chick spinal cord, NEUROD4 was up-regulated after 24 h hours in the electroporated area (Fig. S5 A, a). This up-regulation of NEUROD4 following knock down of MTGR1, an indication of increased NEUROG2 activity, supports our hypothesis that MTGR1 inhibits NEUROG2 activity in vivo. At 48 h, MTGR1 expression was reduced to 53% of the control side (Fig. S5, B, b), but NEUROD4 expression did not differ significantly from the control side (Fig. 5, B1, B2, D, E). We reasoned that because endogenous NEUROG2 is expressed transiently in vivo, reducing expression level of MTGR1 alone might not dramatically affect the expression pattern of NEUROD4. Thus, in the next series of experiments, we reduced the expression level of MTGR1 while over-expressing NEUROG2.
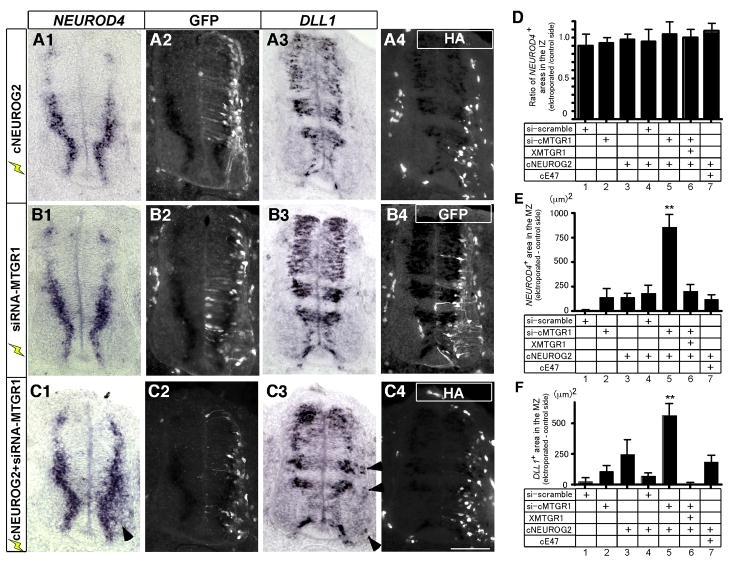
Figure 5. NEUROG2 activity is inhibited by MTGR1 in vivo.
A–C: Chick spinal cord was electroporated with expression vectors of HA-tagged cNEUROG2 (A1–4), siRNA-MTGR1 (B1–4) or combination of both (C1–4) at E2 and harvested at E4. The siRNA-MTGR1 vector has a GFP gene that allows identification of the electroporated cells. For A1–4, an expression vector for GFP was co-electroporated as a tracer. Sections at the brachial level were analyzed for NEUROD4 expression by in situ hybridization (A1, B1, C1), anti-GFP immunohistochemistry (A2, B2, B4, C2), DLL1 in situ hybridization (A3, B3, C3), and immunohistochemistry against the HA tag (A4, C4). The left two and the right two pictures of each line are from the same section, and the two pairs of pictures are from the neighboring sections. In situ hybridization signal is visible in the immunofluorescent channels as dark areas due to absorption of light by color pigments. The electroporated sides are oriented to the right in all figures. Arrowheads in C1 and C3 point to ectopically expressed NEUROD4 and DLL1 in the MZ. Scale bar: 100 μm. D–F: Quantification of the areas expressing NEUROD4 in the IZ (D), NEUROD in the MZ (E) and DLL1 in the MZ (F). D. Pixels of NEUROD4 positive areas within the IZ were counted and the ratios of electroporated and un-electroporated sides were calculated. E, F. Pixels of positive areas for each marker in the MZ was quantified and the difference between electroporated and control sides is shown. Error bars: SEM.
First, we examined the induction profile of NEUROD4 in response to exogenous NEUROG2. When NEUROG2 alone was introduced into the chick spinal cord, NEUROD4 expression was up-regulated at 24h (Fig. S5C–d). At this stage, the MZ has not formed been yet and expression of NEUROD4 that corresponds to the IZ is found at the lateral edge of the spinal cord. In contrast, after 48 h of incubation, when most electroporated cells have already migrated to the MZ, NEUROD4 was down-regulated in these MZ cells even though they still expressed high amounts of NEUROG2 (Fig. 5A1, A4, E). These cells were post-mitotic and expressed neuronal markers (Lamar et al., 2001 and data not shown). To test if the failure of NEUROG2 to activate NEUROD4 in the MZ is due to the lack of its binding partner E47, which is normally down-regulated in the MZ (Fig. S1), we co-expressed cE47 with NEUROG2. Similar to the experiments with NEUROG2 alone, cells migrated to the MZ after 48 hrs but still did not express NEUROD4 (Fig. 5E, column7). These observations indicated two things: 1) that neurogenic gene cascade is activated and cells begin to differentiate in response to exogenous NEUROG2 (as shown by early induction of NEUROD4 at 24 h); and 2) that transcriptional activity of NEUROG2 is suppressed by the time cells reach the MZ (at 48 h post-electroporation) by an endogenous mechanism induced in this gene cascade.
Thus, using this paradigm, we asked if MTGR1 is involved in suppressing the transcriptional activity of NEUROG2. We co-expressed NEUROG2 along with siRNA-MTGR1 in a developing chick spinal cord. (Fig. 5C1–4, D, E). This creates an environment where exogenous NEUROG2 is supplied and MTGR1 expression is reduced. In contrast to individual electroporation of NEUROG2 or siRNA-MTGR1 alone, NEUROD4 was ectopically induced in the MZ (Fig. 5C1, arrowhead, and E, column 5). This ectopic induction of NEUROD4 was not observed in control experiments expressing NEUROG2 and scrambled siRNA (siRNA-scramble) (Fig. 5E, column 4), and was partly prevented by co-expression of Xenopus MTGR1, which is not affected by siRNA-MTGR1 (Fig. 5E, column 6).
Analysis of neighboring sections by in situ hybridization with the DLL1probe, another readout of NEUROG2 activity, showed essentially the same results (Fig. 5, A3, B3, C3, F). DLL1 has restricted expression along the dorso-ventral axis of the spinal cord, but in the levels that have ventricular/intermediate DLL1 expression, ectopic expression in the MZ was observed when NEUROG2 and siRNA-MTGR1 was co-expressed (Fig. 5C3, arrowheads and F).
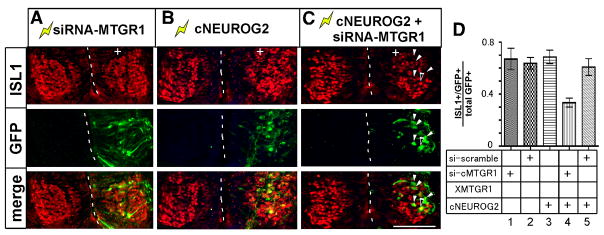
These cells expressing ectopic NEUROD4 or DLL1 in the MZ have prolonged and/or enhanced NEUROG2 activity, because NEUROG2 is ectopically expressed but MTGR1 level is decreased. To examine the outcome of prolonged NEUROG2 activity, adjacent sections were examined for expression of ISL1, a marker for postmitotic motorneurons (Fig. 6). Electroporation of siRNA-MTGR1 or cNEUROG2 vector alone did not cause change in the overall number of ISL1 positive cells at 48 h post electroporation, however co-electroporation of siRNA-MTGR1 vector with cNEUROG2 vector reduced overall number of ISL1 positive cells (Fig. 6A–C). We quantified the proportion of GFP positive cells that co-labeled with ISL1 (Fig. 6D). In embryos electroporated with siRNA-MTGR1, control siRNA or cNEUROG2 vectors, 60–70% of GFP positive cells co-labeled with ISL1. In contrast, in embryos co-electroporated with siRNA-MTGR1 and cNEUROG2 vectors, only 34% of cells co-labeled with ISL1 (Fig. 6C, arrowheads and Fig. 6D, lane 4). Similar results were obtained when scored for co-expression of GFP with PAX2 or HuC/D (data not shown).
Figure 6. Cells with elevated NEUROG2 activity in the MZ show impaired neurogenesis.
A–C: Electroporated embryos were analyzed for expression of ISL1 (red) and GFP (green). Electroporated sides are oriented to the right and indicated by (+). The midlines of the spinal cord are demarcated by dotted lines. White arrowheads in C point to GFP positive, ISL1 negative cells. Black arrowhead points to a double positive cell. Bar: 100 μm. D: Quantification of the ratio of cells co-labeled with ISL1 and GFP. GFP positive cells within the postmitotic motorneuron domain were counted. Each bar represents average of 10–15 sections. Error bars: SEM.
In summary, our data suggest that MTGR1 functions in vivo to terminate NEUROG2 activity as neurogenesis progresses. When MTGR1 function was attenuated by siRNA in the presence of ectopic NEUROG2, cells initiated the differentiation program, but continued to express target genes of NEUROG2, NEUROD4 and DLL1, even in the MZ and failed to fully induce postmitotic markers. Migration of the cells to the MZ, in contrast, appeared to happen normally and did not seem to require down-regulation of NEUROG2 activity by MTGR1.
Discussion
A negative feedback mechanism to terminate NEUROG2 activity is required for progression of neurogenesis
The neurogenic gene cascade is characterized by transient expression of transcription factors in a stereotypical order (Fig. 7). In addition to those examined in this paper, many transcription factors are known to be transiently expressed during neurogenesis. Examples include Neurog1, Prox1, Hes6, Ascl1, NeuroD, PHD1, and MyT1/NZF-2b (Saito et al., 1996; Sommer et al., 1996; Ma et al., 1997; Torii et al., 1999; Koyano-Nakagawa et al., 2000; Matsushita et al., 2002; Misra et al., 2008). After the final mitosis, neuronal progenitors move from the ventricular surface through the IZ to the MZ within 6–8 hrs (Koyano-Nakagawa and Kintner, 2005). Thus, it is estimated that these cells switch the array of transcription factors expressed every 2–3 hours. Although the significance of the changing array of transcription factor expression is not completely understood, it is proposed that the proper termination of gene activity used in early steps of differentiation is necessary for normal progression of neurogenesis (Isaka et al., 1999; Cai et al., 2000).
We show in the study presented here that MTGR1, a transcriptional repressor induced by proneural bHLH proteins, feeds back to terminate transcriptional activity of the NEUROG2-E47 complex. It is already known that NEUROG2 mRNA is expressed in a narrow window of time during development (Figs. 1, S1), and NEUROG2 protein is degraded quickly by a ubiquitin-mediated mechanism (Vosper et al., 2007). Our current study has identified a third mechanism, termination of NEUROG2 activity by MTGR1, which itself is a transcriptional target of NEUROG2. This third mechanism is likely to further limit the duration of NEUROG2 activity even if the NEUROG2 protein is still present (Fig. 7B). We propose that such a feedback system ensures rapid progress through the sequential steps required for neurogenesis.
It has not been readily recognized that termination of bHLH activity is necessary for normal progression of neurogenesis, partly because over-expression of NEUROG2 in the developing nervous system promoted neurogenesis and did not appear to cause aberrant differentiation (Lamar et al., 2001; Novitch et al., 2001; Garcia-Dominguez et al., 2003). We demonstrate that, in these experiments, the normal course of the neurogenic gene cascade was activated, and the activity of NEUROG2 was inhibited by endogenous MTGR1. Our results clearly show that if activity of NEUROG2 is not properly terminated, cells continue to express markers of intermediate steps of neurogenesis and do not fully differentiate to express postmitotic markers (Fig. 5, 6). This finding has implications for efforts to induce various types of stem cells to differentiate as neurons for therapeutic use. One approach being investigated is misexpression of transcription factors that induce neurogenesis. Our finding indicates that it will be important to ensure the activity of neurogenic factors is terminated in a timely manner.
In light of sequential induction of transcription factors, it is interesting to consider the seemingly discrepant observations regarding the function of MTGR1. Cao et al. reported that over-expression of MTGR1 in Xenopus embryos expanded the neural plate but inhibited neurogenesis (Cao et al., 2002), whereas we reported in chick embryos that MTGR1 is a positive regulator of neurogenesis (Koyano-Nakagawa and Kintner, 2005). In fly, precocious expression of Nvy, a MTG homolog, is inhibitory to sensory organ precursor differentiation but isochronic over-expression is not (Wildonger and Mann, 2005). We think that these discrepant observations can be explained by the relative timing of gene expression: since NEUROG2 function is required to initiate neurogenesis, if MTGR1 is over-expressed before the onset of neurogenesis, then it blocks NEUROG2 from initiating the differentiation program resulting in expansion of the progenitor pool. Over-expression of MTGR1 after the onset of neurogenesis does not block neurogenesis because the NEUROG2-induced differentiation program has already turned on the necessary set of genes. However, as we observed in this study, if NEUROG2 activity is sustained after the onset of neurogenesis by blocking MTGR1, cells fail to terminate NEUROG2 activity and cells do not differentiate properly.
Molecules that control progression of neurogenesis
Two main processes, cell fate specification and differentiation, operate during neurogenesis. The transcription factor code that determines cell fate has been well studied in many systems (Guillemot, 2007; Dessaud et al., 2008; Ohsawa and Kageyama, 2008). In contrast, only a handful of transcription factors are known to be involved in the control of progression of neurogenesis, especially the transition from proliferating progenitors to differentiated neurons. One emerging concept from these studies, nevertheless, is that exit from the cell cycle, lateral migration and expression of postmitotic genes are regulated by different sets of transcription factors. For example, Ebf1 is required for lateral migration and differentiation of spinal neurons, but not for cell cycle exit (Garcia-Dominguez et al., 2003). Sox4 and Sox11 promote expression of neuronal markers, but do not induce cell cycle withdrawal (Bergsland et al., 2006). In contrast, Prox1 and p57 are required for cell cycle exit, but are not sufficient to induce the expression of a full array of neuronal markers (Gui et al., 2007; Misra et al., 2008). There are still many transcription factors downstream of proneural bHLHs whose target pathways are not fully understood (Roztocil et al., 1997; Sandberg et al., 2005; Schmid et al., 2007; Soustelle and Giangrande, 2007). Attenuation of MTGR1 function in the presence of NEUROG2 inhibited expression of postmitotic markers, caused prolonged expression of progenitor characteristics, but did not significantly affect cell migration. It would be of interest to investigate the molecular interaction of MTGR1 with the transcription factors involved in regulating those aspects of neurogenesis.
Mechanism of inhibiting NEUROG2 activity by MTGR1
MTG family proteins are known to repress transcription either by bridging sequence-specific transcription factors and the HDAC complex (acting as an “active” inhibitor) or by competing for interaction with CBP/p300 (acting as an “passive” or competitive inhibitor) (Hug and Lazar, 2004; Rossetti et al., 2004). In either case, they do not directly bind to DNA but function by interacting with other proteins. We show that MTGR1 inhibits NEUROG2 activity by at least two mechanisms, which are not mutually exclusive.
We propose that one of the mechanisms of inhibition is through recruiting inhibitory co-factors to the NEUROG2-E47 complex already formed on DNA. Our transcription assay showing that repression requires the presence of the NEUROG2-E47 complex and the E-box target sequence supports this model. In addition, MTGR1-EnR fusion protein inhibited, and MTGR1-VP16 augmented the NEUROG2-E47 activity through the E box. This result is consistent with the model that this inhibitory activity takes place by binding to the NEUROG2-E47 complex formed on DNA. Based on the report that mouse MTGR1 interacts with mSin3A, N-CoR and HDAC3 (Amann et al., 2005), we speculate that this repression activity involves recruitment of histone deacetylases. The fact that we did not detect the formation of a higher order complex containing NEUROG2, E47 and MTGR1 in EMSA seemingly contradicts the results of the transcription assay. We speculate this is due to the short half-life of the complex or to the heterogeneity of the higher order complex (Rossetti et al., 2004). Future experiments using crosslinking techniques and new antibodies that can be used for immunoprecipitation are needed to clarify this issue.
In contrast, the EMSA experiment suggests a second mode of inhibiting NEUROG2-E47 activity by MTGR1. This experiment clearly showed inhibition of DNA binding activity of the NEUROG2-E47 complex when MTGR1 is associated with the complex before DNA binding. It should be noted, however, that MTGR1 did not dissociate the NEUROG2-E47 complex that was already bound to DNA. Thus, in this mode of inhibition, MTGR1 represses the activity of only the NEUROG2-E47 species that are not bound to DNA. It is possible, in our transfection assays with VP16 or EnR fusion molecules of MTGR1, that some of the fusion molecules have inhibited the DNA binding of unbound NEUROG2-E47 complexes. However, VP16 fusion molecule of MTGR1 strongly activated the transcriptional activity of NEUROG2-E47 further beyond the normal activity of the NERUOG2-E47 complex. Thus, we think that the activity we observed in this experiment reflects the action of VP16 and EnR fusion proteins on the NEUROG2-E47 complex that was already bound to DNA before the fusion proteins associated with them.
Because MTGR1 is induced by NEUROG2 and follows the expression of NEUROG2 with a delay (Figs. 1, S1 and Alishahi et al., 2009), we speculate that these two modes of inhibition may take place sequentially in vivo. In the early phase of neuronal differentiation, NEUROG2 and E47 are expressed first, and the expression level of MTGR1 is low. When MTGR1 levels becomes high, it may bind to the NEUROG2-E47 complex that is already bound to DNA and inhibit transcription using its repressor activity. In the later phase, when NEUROG2, E47 and MTGR1 are present, MTGR1 may bind to new NEUROG2 and E47 proteins being synthesized and prevent them from binding to DNA. We think that these two mechanisms of inhibition will efficiently terminate activity of the NEUROG2-E47 complex (Fig. 7B).
Up to now, E proteins have been the only bHLH transcription factors reported to interact with MTG proteins (Plevin et al., 2006; Wei et al., 2007). We provide evidence that activity of NEUROG2-E47 or NEUROG2 alone is inhibited by MTGR1, showing that NEUROG2 is a novel target of MTGR1-mediated repression. This does not eliminate the possibility that E proteins are also targeted by MTGR1, because endogenous E proteins expressed in P19 cells could potentially contribute to the transcriptional activity we observed. We think, however, the main target of MTGR1 is NEUROG2, because immunoprecipitation assays of over-expressed proteins in HEK293T cells showed binding of MTGR1 to NEUROG2 but not to E47, and interaction of MTGR1-E47 was significantly weaker than that of MTGR1-NEUROG2 in the GST pull-down assay.
The interaction between MTG8 and HEB takes place through the N-terminal activation domain (AD1) of HEB (Zhang et al., 2004). Since there is no sequence in the NEUROG2 protein that resembles AD1, it is likely that there is a distinct MTGR1 interaction surface within the NEUROG2 protein. This is not surprising, since MTG proteins are known to function as a protein “scaffold”, interacting with various transcription factors (Hug and Lazar, 2004; Rossetti et al., 2004). It would be of interest to map the interaction domains of MTGR1 and NEUROG2. Since MTG family proteins are highly conserved, there may be similar but specific interactions between other MTG family members and other proneural bHLH proteins. Indeed, our data showed that NEUROG2 binds MTGR1 but not MTG8. Whether such specific interactions contribute to functional difference of bHLH proteins is under investigation.
Modulators of bHLH protein activity during development
Transcriptional activity of proneural bHLH proteins is modulated by many mechanisms. For example, X-NGNR-1 interacts with CBP/p300, members of histone acetyltransferases (Koyano-Nakagawa et al., 1999). Brg1, a component of the SWI/SNF chromatin remodeling complex, has also been reported as a necessary co-factor of X-NGNR-1 and NeuroD activity in Xenopus (Seo et al., 2005). Id and Hes family proteins are known inhibitors of proneural bHLH transcription factors (Kageyama et al., 1997; Yokota, 2001). They heterodimerize with bHLH proteins and inhibit transcription by forming non-DNA binding complexes. We report for the first time that MTGR1 associates with NEUROG2 and inhibits its function. This adds a new group of repressor proteins that could potentially interact with proneural bHLH proteins. We speculate that proneural bHLH proteins interact with different sets of transcriptional co-factors at different stages of development to rapidly switch on or off their transcriptional activity. This also predicts that specific timing of co-factor expression is important to regulate proneural protein activity.
Recent studies have shown that Neurogenins have late functions after the initial phase of neurogenesis. Such activities do not involve their transactivation capacities and include control of cell fate, cell migration, axon projections and dendrite morphologies (Sun et al., 2001; Seibt et al., 2003; Schuurmans et al., 2004; Hand et al., 2005; Hand and Polleux, 2006). It will be of interest to know if inhibition of transcriptional activity by MTG proteins is required for neurogenins to exert such late functions. In addition to the spinal cord, MTGR1 is induced by bHLH proteins in other systems such as retina and small intestine (Amann et al., 2005; Logan et al., 2005). Thus, it is likely that feedback regulation of bHLH protein by MTGR1 is a widely used mechanism to promote cellular differentiation.
Supplementary Material
Cross sections of embryos from 3rd to 7th day of incubation at the brachial level are shown. Boundaries of the VZ, the IZ, and the MZ are indicated by dotted lines. Probes used are NEUROG2 (A1–5), NEUROD4 (B1–5), MTGR1 (C1–5) and E47 (D1–5). Note that relative order of gene expression is conserved throughout development. Bar: 100 μm.
Chick embryos were electroporated into the right side of the spinal cords with vectors expressing cMTGR1 and GFP. The left sides serve as un-electroporated controls. A, B: Immunohistochemistry using antibodies against NEUROG2 (A) and GFP (B). Note that NEUROG2 expression is unchanged after ectopic expression of MTGR1. C: Merge of A and B. Boundaries of VZ, IZ and MZ are indicated by dotted lines. Bar: 100 μm.
A, B: P19 cells were transfected with vectors encoding factors from the indicated species. Mouse Neurog2, mouse E12 and Xenopus MTGR1 were used in A, and mouse Neurog2, human E47, chick MTGR1 were used in B. E12 is a splice variant of E47. The amount of MTGR1 plasmids transfected is shown in micrograms. In all combinations tested, MTGR1 repressed transcriptional activity of Neurog2, suggesting that protein interaction surface is conserved across species. C: Western blot of myc epitope-tagged fusion MTGR1 molecules. Vectors encoding full-length cMTGR1 (lane 1), cMTGR1-VP16 (lane 2), cMTGR1-EnR (lane 3) were transfected into P19 cells in the same condition as the luciferase assay, and 10 μl of cell extracts were analyzed using anti-myc epitope antibody.
A: MTGR1 does not affect the stability of NEUROG2 or E47 in the EMSA reaction. Radiolabeled NEUROG2 and E47 were incubated with (bottom panel) or without (top panel) radiolabeled MTGR1. Samples were processed the same way as the EMSA reaction except for using an unlabeled DNA. Equal amounts of reaction were removed every hour and analyzed on an SDS-PAGE. B: MTGR1 does not dissociate the NEUROG2-E47 heterodimer. Radiolabeled NEUROG2 and E47 synthesized by co-translation were incubated with non-radioactive MTGR1 synthesized in a separate reaction. The binding condition was the same as the GST pull down assay. After incubation, the samples were immunoprecipitated with anti-NEUROG2 antibody and examined by autoradiography. Note that E47 co-immunoprecipitates only in the presence of NEUROG2, and this binding is not disrupted by preincubation with MTGR1 (compare lanes 4 and 6). Arrowhead indicates the E47 band.
siRNA-MTGR1 vector was electroporated into E2 chick spinal cord and analyzed for NEUROD4 expression after 24 h (A, a) or 48 h for MTGR1 expression (B, b). A: At 24h, NEUROD4 is transiently induced on the electroporated area (arrowheads), but the expression level goes back to normal after 48 h (compare with Fig 5, B1, D, E). B, b: At 48h, endogenous MTGR1 expression level was reduced to 53% (arrowheads). Expression vector of HA-tagged NEUROG2 protein was electroporated into E2 chick spinal cord and analyzed 24 hours later (C–d). Sections were doubly labeled with in situ hybridization with NEUROD4 probe (C, D) and immunohistochemistry using anti-HA antibody (c, d). Electroporated side is oriented to the right. Two examples of ectopic induction are shown in C and D. Arrowheads and the bracket indicate ectopically induced NEUROD4. Bars: 100 μm.
Acknowledgments
We thank the following people for the kind gift of plasmids; Soo-Kyung Lee (Baylor College of Medicine) Sam Pfaff (Salk Institute), Masato Nakafuku (Cincinnati Children’s Hospital), Atsushi Asakura (Univ. Minnesota), Klemens Meyer (Univ. Cambridge). NEUROD4 antibody is a kind gift from Thomas Jessell (Columbia Univ.) and Bennet Novitch (UCLA), anti-cNEUROG2 antibody is from David Anderson (Caltech), and anti hMTGR1 antibody is from Issay Kitabayashi (National Cancer Center Research Institute, Japan). We thank Zibing Jiang and David Hernandez for technical assistance, and Atsushi Asakura and Jonathan Slack for helpful discussions and critically reading the manuscript. DNA sequence analyses were done using resources of the Supercomputing Institute at the Univ. of Minnesota. This work was supported by Grants to NKN from Minnesota Medical Foundation, Academic Health Center, Univ. of Minnesota, Hough Parkinson’s Awards, and NIH (MH078998).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alishahi A, Koyano-Nakagawa N, Nakagawa Y. Regional expresion of MTG genes in the developing mouse central nervous system. Dev Biol. 2009 doi: 10.1002/dvdy.22021. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann JM, Chyla BJI, Ellis TC, Martinez A, Moore AC, Franklin JL, McGhee L, Meyers S, Ohm JE, Luce KS, Ouelette AJ, Washington MK, Thompson MA, King D, Gautam S, Coffey RJ, Whitehead RH, Hiebert SW. Mtgr1 is a transcriptional corepressor that is required for maintenance of the secretory cell lineage in the small intestine. Molecular and Cellular Biology. 2005;25:9576–9585. doi: 10.1128/MCB.25.21.9576-9585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, editor. Current protocols in molecular biology. John Wiley & Sons, Inc; 1994. [Google Scholar]
- Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Gene Dev. 2006;20:3475–3486. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Morrow EM, Cepko CL. Misexpression of basic helix-loop-helix genes in the murine cerebral cortex affects cell fate choices and neuronal survival. Development. 2000;127:3021–3030. doi: 10.1242/dev.127.14.3021. [DOI] [PubMed] [Google Scholar]
- Cao Y, Zhao H, Grunz H. XETOR regulates the size of the proneural domain during primary neurogenesis in Xenopus laevis. Mech Develop. 2002;119:35–44. doi: 10.1016/s0925-4773(02)00285-x. [DOI] [PubMed] [Google Scholar]
- Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Gottgens B, Matter JM, Guillemot F. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Developmental Cell. 2006;11:831–844. doi: 10.1016/j.devcel.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Conlon TM, Meyer KB. Cloning and functional characterisation of avian transcription factor E2A. BMC Immunol. 2004;14:11. doi: 10.1186/1471-2172-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JN, McGhee L, Meyers S. The ETO (MTG8) gene family. Gene. 2003;303:1–10. doi: 10.1016/s0378-1119(02)01172-1. [DOI] [PubMed] [Google Scholar]
- Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- Fior R, Henrique D. A novel hes5/hes6 circuitry of negative regulation controls Notch activity during neurogenesis. Developmental Biology. 2005;281:318–333. doi: 10.1016/j.ydbio.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Garcia-Dominguez M, Poquet C, Garel S, Charnay P. Ebf gene function is required for coupling neuronal differentiation and cell cycle exit. Development. 2003;130:6013–6025. doi: 10.1242/dev.00840. [DOI] [PubMed] [Google Scholar]
- Gui HX, Li SK, Matise MP. A cell-autonomous requirement for Cip/Kip cyclin-kinase inhibitors in regulating neuronal cell cycle exit but not differentiation in the developing spinal cord. Developmental Biology. 2007;301:14–26. doi: 10.1016/j.ydbio.2006.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F. Cell fate specification in the mammalian telencephalon. Prog Neurobiol. 2007;83:37–52. doi: 10.1016/j.pneurobio.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Hand R, Polleux F. The transcriptional regulation of glutamatergic pyramidal neurons by Neurogenin2. International Journal of Developmental Neuroscience. 2006;24:578–578. [Google Scholar]
- Hand R, Bortone D, Mattar P, Nguyen L, Heng JIT, Guerrier S, Boutt E, Peters E, Barnes AP, Parras C, Schuurmans C, Guillemot F, Polleux F. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- Hug BA, Lazar MA. ETO interacting proteins. Oncogene. 2004;23:4270–4274. doi: 10.1038/sj.onc.1207674. [DOI] [PubMed] [Google Scholar]
- Isaka F, Ishibashi M, Taki W, Hashimoto N, Nakanishi S, Kageyama R. Ectopic expression of the bHLH gene Math1 disturbs neural development. European Journal of Neuroscience. 1999;11:2582–2588. doi: 10.1046/j.1460-9568.1999.00699.x. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Ishibashi M, Takebayashi K, Tomita K. bHLH transcription factors and mammalian neuronal differentiation. Int J Biochem Cell Biol. 1997;29:1389–1399. doi: 10.1016/s1357-2725(97)89968-2. [DOI] [PubMed] [Google Scholar]
- Koyano-Nakagawa N, Kintner C. The expression and function of MTG/ETO family proteins during neurogenesis. Developmental Biology. 2005;278:22–34. doi: 10.1016/j.ydbio.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Koyano-Nakagawa N, Wettstein D, Kintner C. Activation of Xenopus Genes Required for Lateral Inhibition and Neuronal Differentiation during Primary Neurogenesis. Mol Cell Neurosci. 1999;14:327–339. doi: 10.1006/mcne.1999.0783. [DOI] [PubMed] [Google Scholar]
- Koyano-Nakagawa N, Kim J, Anderson D, Kintner C. Hes6 acts in a positive feedback loop with the neurogenins to promote neuronal differentiation. Development. 2000;127:4203–4216. doi: 10.1242/dev.127.19.4203. [DOI] [PubMed] [Google Scholar]
- Lamar E, Kintner C, Goulding M. Identification of NKL, a novel Gli-Kruppel zinc-finger protein that promotes neuronal differentiation. Development. 2001;128:1335–1346. doi: 10.1242/dev.128.8.1335. [DOI] [PubMed] [Google Scholar]
- Lee SK, Lee B, Ruiz EC, Pfaff SL. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes & Development. 2005;19:282–294. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo LC, Johnson JE, Wuenschell CW, Saito T, Anderson DJ. Mammalian achaete-scute homolog 1 is transiently expressed by spatially restricted subsets of early neuroepithelial and neural crest cells. Genes Dev. 1991;5:1524–1537. doi: 10.1101/gad.5.9.1524. [DOI] [PubMed] [Google Scholar]
- Logan MA, Steele MR, Van Raay TJ, Vetter ML. Identification of shared transcriptional targets for the proneural bHLH factors Xath5 and XNeuroD. Developmental Biology. 2005;285:570–583. doi: 10.1016/j.ydbio.2005.06.033. [DOI] [PubMed] [Google Scholar]
- Ma Q, Sommer L, Cserjesi P, Anderson DJ. Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. J Neurosci. 1997;17:3644–3652. doi: 10.1523/JNEUROSCI.17-10-03644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita F, Kameyama T, Marunouchi T. NZF-2b is a novel predominant form of mouse NZF-2/MyT1, expressed in differentiated neurons especially at higher levels in newly generated ones. Mech Develop. 2002;118:209–213. doi: 10.1016/s0925-4773(02)00250-2. [DOI] [PubMed] [Google Scholar]
- Mattar P, Langevin LM, Markham K, Klenin N, Shivji S, Zinyk D, Schuurmans C. Basic helix-loop-helix transcription factors cooperate to specify a cortical projection neuron identity. Mol Cell Biol. 2008;28:1456–1469. doi: 10.1128/MCB.01510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra K, Gui HX, Matise MP. Prox1 regulates A transitory state for interneuron neurogenesis in the spinal cord (vol 237, pg 393, 2008) Dev Dynam. 2008;237:1214–1214. doi: 10.1002/dvdy.21422. [DOI] [PubMed] [Google Scholar]
- Murciano A, Zamora J, Lopez-Sanchez J, Frade JM. Interkinetic nuclear movement may provide spatial clues to the regulation of neurogenesis. Mol Cell Neurosci. 2002;21:285–300. doi: 10.1006/mcne.2002.1174. [DOI] [PubMed] [Google Scholar]
- Novitch BG, Chen AI, Jessell TM. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Ohsawa R, Kageyama R. Regulation of retinal cell fate specification by multiple transcription factors. Brain Research. 2008;1192:90–98. doi: 10.1016/j.brainres.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Plevin MJ, Zhang J, Guo C, Roeder RG, Ikura M. The acute myeloid leukemia fusion protein AML1-ETO targets E proteins via a paired amphipathic helix-like TBP-associated factor homology domain. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10242–10247. doi: 10.1073/pnas.0603463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetti S, Hoogeveen AT, Sacchi N. The MTG proteins: chromatin repression players with a passion for networking. Genomics. 2004;84:1–9. doi: 10.1016/j.ygeno.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Roztocil T, Matter-Sadzinski L, Alliod C, Ballivet M, Matter JM. NeuroM, a neural helix-loop-helix transcription factor, defines a new transition stage in neurogenesis. Development. 1997;124:3263–3272. doi: 10.1242/dev.124.17.3263. [DOI] [PubMed] [Google Scholar]
- Saito T, Lo L, Anderson DJ, Mikoshiba K. Identification of novel paired homeodomain protein related to C. elegans unc-4 as a potential downstream target of MASH1. Dev Biol. 1996;180:143–155. doi: 10.1006/dbio.1996.0291. [DOI] [PubMed] [Google Scholar]
- Sandberg M, Kallstrom M, Muhr J. Sox21 promotes the progression of vertebrate neurogenesis. Nat Neurosci. 2005;8:995–1001. doi: 10.1038/nn1493. [DOI] [PubMed] [Google Scholar]
- Schmid T, Kruger M, Braun T. NSCL-1 and-2 control the formation of precerebellar nuclei by orchestrating the migration of neuronal precursor cells. J Neurochem. 2007;102:2061–2072. doi: 10.1111/j.1471-4159.2007.04694.x. [DOI] [PubMed] [Google Scholar]
- Schuurmans C, Armant O, Nieto M, Stenman JM, Britz O, Klenin N, Brown C, Langevin LM, Seibt J, Tang H, Cunningham JM, Dyck R, Walsh C, Campbell K, Polleux F, Guillemot F. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. Embo Journal. 2004;23:2892–2902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibt J, Schuurmans C, Gradwhol G, Dehay C, Vanderhaeghen P, Guillemot F, Polleux F. Neurogenin2 specifies the connectivity of thalamic neurons by controlling axon responsiveness to intermediate target cues. Neuron. 2003;39:439–452. doi: 10.1016/s0896-6273(03)00435-5. [DOI] [PubMed] [Google Scholar]
- Seo S, Richardson GA, Kroll KL. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005;132:105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- Seo S, Lim JW, Yellajoshyula D, Chang LW, Kroll KL. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. Embo J. 2007;26:5093–5108. doi: 10.1038/sj.emboj.7601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer L, Ma Q, Anderson DJ. neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- Soustelle L, Giangrande A. Glial differentiation and the Gcm pathway. Neuron Glia Biology. 2007;3:5–16. doi: 10.1017/S1740925X07000464. [DOI] [PubMed] [Google Scholar]
- Sriuranpong V, Borges MW, Strock CL, Nakakura EK, Watkins DN, Blaumueller CM, Nelkin BD, Ball DW. Notch signaling induces rapid degradation of achaete-scute homolog 1. Mol Cell Biol. 2002;22:3129–3139. doi: 10.1128/MCB.22.9.3129-3139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, Greenberg ME. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Torii M, Matsuzaki F, Osumi N, Kaibuchi K, Nakamura S, Casarosa S, Guillemot F, Nakafuku M. Transcription factors Mash-1 and Prox-1 delineate early steps in differentiation of neural stem cells in the developing central nervous system. Development. 1999;126:443–456. doi: 10.1242/dev.126.3.443. [DOI] [PubMed] [Google Scholar]
- Tuttle R, Nakagawa Y, Johnson JE, O’Leary DD. Defects in thalamocortical axon pathfinding correlate with altered cell domains in Mash-1-deficient mice. Development. 1999;126:1903–1916. doi: 10.1242/dev.126.9.1903. [DOI] [PubMed] [Google Scholar]
- Vosper JMD, Fiore-Heriche CS, Horan I, Wilson K, Wise H, Philpott A. Regulation of neurogenin stability by ubiquitin-mediated proteolysis. Biochemical Journal. 2007;407:277–284. doi: 10.1042/BJ20070064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Liu S, Lausen J, Woodrell C, Cho S, Biris N, Kobayashi N, Wei Y, Yokoyama S, Werner MH. A TAF4-homology domain from the corepressor ETO is a docking platform for positive and negative regulators of transcription. Nat Struct Mol Biol. 2007;14:653–661. doi: 10.1038/nsmb1258. [DOI] [PubMed] [Google Scholar]
- Wildonger J, Mann RS. Evidence that nervy, the Drosophila homolog of ETO/MTG8 promotes mechanosensory organ development by enhancing Notch signaling. Dev Biol. 2005;286:507–520. doi: 10.1016/j.ydbio.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Yokota Y. Id and development. Oncogene. 2001;20:8290–8298. doi: 10.1038/sj.onc.1205090. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Yamamura S, Chait BT, Roeder RG. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science. 2004;305:1286–1289. doi: 10.1126/science.1097937. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cross sections of embryos from 3rd to 7th day of incubation at the brachial level are shown. Boundaries of the VZ, the IZ, and the MZ are indicated by dotted lines. Probes used are NEUROG2 (A1–5), NEUROD4 (B1–5), MTGR1 (C1–5) and E47 (D1–5). Note that relative order of gene expression is conserved throughout development. Bar: 100 μm.
Chick embryos were electroporated into the right side of the spinal cords with vectors expressing cMTGR1 and GFP. The left sides serve as un-electroporated controls. A, B: Immunohistochemistry using antibodies against NEUROG2 (A) and GFP (B). Note that NEUROG2 expression is unchanged after ectopic expression of MTGR1. C: Merge of A and B. Boundaries of VZ, IZ and MZ are indicated by dotted lines. Bar: 100 μm.
A, B: P19 cells were transfected with vectors encoding factors from the indicated species. Mouse Neurog2, mouse E12 and Xenopus MTGR1 were used in A, and mouse Neurog2, human E47, chick MTGR1 were used in B. E12 is a splice variant of E47. The amount of MTGR1 plasmids transfected is shown in micrograms. In all combinations tested, MTGR1 repressed transcriptional activity of Neurog2, suggesting that protein interaction surface is conserved across species. C: Western blot of myc epitope-tagged fusion MTGR1 molecules. Vectors encoding full-length cMTGR1 (lane 1), cMTGR1-VP16 (lane 2), cMTGR1-EnR (lane 3) were transfected into P19 cells in the same condition as the luciferase assay, and 10 μl of cell extracts were analyzed using anti-myc epitope antibody.
A: MTGR1 does not affect the stability of NEUROG2 or E47 in the EMSA reaction. Radiolabeled NEUROG2 and E47 were incubated with (bottom panel) or without (top panel) radiolabeled MTGR1. Samples were processed the same way as the EMSA reaction except for using an unlabeled DNA. Equal amounts of reaction were removed every hour and analyzed on an SDS-PAGE. B: MTGR1 does not dissociate the NEUROG2-E47 heterodimer. Radiolabeled NEUROG2 and E47 synthesized by co-translation were incubated with non-radioactive MTGR1 synthesized in a separate reaction. The binding condition was the same as the GST pull down assay. After incubation, the samples were immunoprecipitated with anti-NEUROG2 antibody and examined by autoradiography. Note that E47 co-immunoprecipitates only in the presence of NEUROG2, and this binding is not disrupted by preincubation with MTGR1 (compare lanes 4 and 6). Arrowhead indicates the E47 band.
siRNA-MTGR1 vector was electroporated into E2 chick spinal cord and analyzed for NEUROD4 expression after 24 h (A, a) or 48 h for MTGR1 expression (B, b). A: At 24h, NEUROD4 is transiently induced on the electroporated area (arrowheads), but the expression level goes back to normal after 48 h (compare with Fig 5, B1, D, E). B, b: At 48h, endogenous MTGR1 expression level was reduced to 53% (arrowheads). Expression vector of HA-tagged NEUROG2 protein was electroporated into E2 chick spinal cord and analyzed 24 hours later (C–d). Sections were doubly labeled with in situ hybridization with NEUROD4 probe (C, D) and immunohistochemistry using anti-HA antibody (c, d). Electroporated side is oriented to the right. Two examples of ectopic induction are shown in C and D. Arrowheads and the bracket indicate ectopically induced NEUROD4. Bars: 100 μm.