Abstract
Metal uptake by the antioxidant defense metalloenzyme manganese superoxide dismutase (MnSOD) is an essential step in the functional maturation of the protein that is just beginning to be investigated in detail. We have extended earlier in vitro studies on metal binding by the dimeric Escherichia coli apo-MnSOD to investigate the mechanism of metal uptake by tetrameric human and Thermus thermophilus apo-MnSODs. Like the E. coli apo-MnSOD, these proteins also bind metal ions in vitro in a thermally-activated, pH-sensitive process. However, metal uptake by the tetrameric apo-MnSODs exhibits a number of important differences. In particular, there is no indication of conformational gating requirement for metal binding for these proteins, and the reaction is first-order in metal ion. The high concentration of metal ion that is required to achieve physiologically relevant metallation rates for tetrameric human apo-MnSOD in vitro suggests the possibility that co-translational metal binding or chaperone interactions may be required in vivo.
1. Introduction
Manganese superoxide dismutase (MnSOD) [1,2] is a crucial component of the cellular defense against oxidative stress [3–6] with an absolute requirement for a manganese metal cofactor for catalytic activity [7–9]. MnSOD is a member of the highly conserved and ubiquitous (Fe,Mn)-SOD enzyme superfamily, present in virtually every aerobic organism and differing mainly in the identity of the essential metal cofactor [9–11]. Proteins in this family typically assemble into symmetric multimers (dimers or tetramers) of identical subunits, and crystal structures are available for both dimeric and tetrameric proteins [12–20]. Each subunit adopts a conserved fold of two-domain architecture (including an N-terminal α-hairpin domain and a C-terminal mixed α/β-domain), with the metal binding site buried on the domain interface in the interior of the protein. In all cases, the metal ion is ligated by four amino acid side chains (three His and one Asp) arising from both N- and C-terminal regions, effectively cross-linking the two domains. Tetrameric MnSODs (including mesophilic human and thermophilic Thermus thermophilus MnSODs) are organized as a pair of dimers [14,15], whose individual structures are nearly identical to that found for the simpler dimeric SODs. Metal binding by MnSOD is fairly unselective and both Fe and Mn complexes form in vivo [20,21], but the enzyme exhibits a strict catalytic specificity and only the Mn complex is active [7–9]. This specificity appears to be the result of subtle structural differences between the Mn and Fe metal complexes that are reflected in their binding affinities and redox behavior [22–24]. Metal coordination by dimeric Escherichia coli MnSOD has been shown to exhibit moderately high binding affinity, but is kinetically irreversible due to a large protein activation barrier [25]. The binding of distinct metal ions (e.g., Mn2+ and Co2+) is mutually exclusive and kinetically competitive [7], indicating that there is a single high affinity binding site in each subunit.
We have previously investigated the mechanism of in vitro metal uptake by the dimeric apo-MnSOD from E. coli, using a continuous real-time fluorimetric metal uptake assay to monitor the progress of the metal binding reaction [25]. This method is based on quenching of the intrinsic protein fluorescence using Co2+ as a probe in place of Mn2+. The catalytic properties of protein reconstituted in vitro with Mn2+ under the same conditions have also been evaluated for comparison with the native metallated protein as a critical test of function. The biphasic metal uptake kinetics observed for E. coli apo-MnSOD have been interpreted in terms of a conformational gating mechanism for metal binding where the rate of metal uptake is determined by a structural transitions in the protein [25,26]. In this picture, the two kinetic phases reflect the presence of two distinct molecular species in the sample, interconverting in a temperature-dependent equilibrium: an “open” form that is able to bind metal ions rapidly and is associated with the fast phase, and a “closed” form that is unable to bind metal ion. The slow kinetic phase is thought to reflect to the rate-limiting conversion of “closed” to “open” form of the apoprotein. In the present work, we have extended the earlier studies to measure the kinetics of metal uptake by stable, folded, tetrameric human apo-MnSOD, together with Thermus thermophilus apo-MnSOD, investigating the effect of quaternary structure in the metal binding process.
2. Materials and Methods
Biochemical Materials
All reagents were from commercial sources and used without purification.
Culture media
Terrific Broth (TB) (12 g/L tryptone, 24 g/L yeast extract, 2.31 g/L potassium phosphate monobasic, 12.54 g/L potassium phosphate dibasic), Luria-Bertani medium (LB) (5 g/L NaCl, 5 g/L yeast extract, 10 g/L tryptone), supplemented with antibiotics as required for selection (carbenicillin, 100 mg/L; chloramphenicol, 25 mg/L). Super Optimal Broth (SOB) (0.5 g/L NaCl, 2.5 mL 1 M g/L KCl, 1.2 g/L MgSO4, 5 g/L yeast extract, 20 g/L tryptone, ; SOC (SOB supplemented with 3 g/L glucose).
Biological materials
Ultracompetent Escherichia coli XL2-Blue cells were from Stratagene (La Jolla, CA). Ultracompetent Escherichia coli BL21-AI cells (F− ompT hsdSB (rB−mB−) gal dcm araB::T7RNAP-tetA) [27] were from Invitrogen (Carlsbad, CA).
Expression cloning
DNA encoding the mature human mitochondrial superoxide dismutase polypeptide (lacking the mitochondrial targeting sequence) was amplified from a cDNA clone (pHMNSOD-4) [28] obtained from the American Type Culture Collection (Manassas, VA), using primers HSOD-1 (5′-GCACATATGAAGCACAGCACAGCCTCCCCGACCTGC-3′) and HSOD-2 (5′-GCAAAGCTTACTTTTTGCAAGCCATGTATCTTTCAGTTACATTCTCCC-3′) designed based on the cDNA sequence. The PCR product was digested with Nde I and Hind III restriction enzymes, and ligated into a similarly digested pET23a expression vector (Novagen, Madison, WI). The ligation mixture was transformed into XL-2 Blue ultracompetent E. coli cells, recovered in SOC medium, selected on LB/carbenicillin agar, and an insert-containing plasmid (pET23HSOD) isolated. The sequence of the insert was verified by nucleotide sequence analysis (Molecular Biology Core, OHSU). pET23HSOD was transformed into competent E. coli BL21-AI cells (Invitrogen, Carlsbad, CA), recovered in SOC medium, and transformants were selected on LB/carbenicillin agar.
Expression
High level expression of recombinant human MnSOD was achieved by growing the BL21-AI | pET23HSOD expression strain to high cell density in 3 L Terrific Broth containing 0.2% glucose and 100 mg/L carbenicillin for 24 h at 37°C with shaking. The cell mass of this uninduced culture was isolated by sterile centrifugation, and resuspended in fresh TB (without phosphate buffer) supplemented with 0.2% glycerol and 0.2% L-arabinose, with 100 mg/L carbenicillin and 5 mM MnCl2. The induced culture was incubated at 37°C with shaking for 24 h, and the cells recovered by centrifugation. High level expression of E. coli MnSOD and recombinant Thermus thermophilus HB8 MnSOD was performed as previously described [21,29].
Protein purification
Recombinant human MnSOD was purified from the E. coli expression host. Cell-free extract prepared by sonication of 40 g of cells was loaded onto a DE-52 ion exchange column (5×45 cm) equilibrated with 5 mM potassium phosphate buffer pH 7.8 containing 1 mM EDTA. The SOD-containing flow-through fractions were filtered and concentrated, then dialyzed against 10 mM sodium acetate buffer (pH 5.5). The dialysate was further fractionated on a CM-52 column (2.5×45 cm) eluted with a linear gradient of 10 – 200 mM sodium acetate (pH 5.5). The peak fractions containing human MnSOD, identified by SDS-PAGE, were pooled, yielding ~60 mg of purified protein. E. coli MnSOD and recombinant Thermus thermophilus HB8 MnSOD were purified as previously described [21,29].
Preparation of apoprotein
Human apomanganese superoxide dismutase (apo-MnSOD) was prepared by a modification of the procedure previously described for preparation of E. coli apo-MnSOD [23,30]. Human MnSOD (approximately 30 mL of 1 mg/mL in 3.5 M guanidinium HCl containing 10 mM each of EDTA and mercaptoethanol, pH 3.5) was dialyzed against 500 mL of the same solution for 24 h at 4°C. The sample was transferred to a second guanidinium HCl solution (2.5 M guanidinium HCl with EDTA and mercaptoethanol) for a further 24 h. The sample was transferred to 1 L of 20 mM Tris HCl (pH 7.6) containing 10 mM each of EDTA and mercaptoethanol, and stirred gently for 8 h. Finally, the sample was transferred to 1 L of 20 mM Tris HCl (pH 8) containing 1 mM EDTA (three changes total), and the precipitate was removed by centrifugation. The apoprotein product was concentrated by ultrafiltration over a 25 mm diameter YM10 membrane. EDTA was removed from the apoprotein by desalting over Bio-Gel P-6. The quaternary structure of the product was determined by size exclusion chromatography. E. coli apo-MnSOD was prepared as previously described [23,30], and Thermus thermophilus HB8 apo-MnSOD was isolated directly from the E. coli expression host [29].
In vitro reconstitution
Human apo-MnSOD (0.1 mM) in 40 mM HEPES buffer (pH 8.2) containing 5 mM MnCl2 or CoCl2 was incubated at 37°C for 2 h. After cooling on ice, potassium phosphate monobasic and EDTA were added to a final concentration of 20 mM each, and the protein was desalted by gel filtration (Bio Gel P-30) in 50 mM potassium phosphate, (pH7.0). Reconstitution was performed either aerobically, or under argon to prevent oxidation of the metal ion. Recombinant Thermus thermophilus HB8 apo-MnSOD was reconstituted in 40 mM MOPS buffer (pH 7.6) at 65°C.
Fluorimetric analysis
Fluorescence measurements were performed using a Cary Eclipse spectrofluorimeter (Varian, Inc., Walnut Creek, CA) equipped with a Cary temperature controller and a Peltier 4-position multicell holder. The pH-dependent emission intensity at 333 nm was obtained by excitation of 25 μg/mL of recombinant human apo-MnSOD in 20 mM buffers, 50 μg/mL of Escherichia coli apo-MnSOD, or 50 μg/mL of Thermus thermophilus HB8 apo-MnSOD at 280 nm (37°C), using a dynode voltage of 650 V. The pH of each sample was measured directly in the fluorescence cuvette. For metal uptake measurements, metal binding timecourses were initiated by adding an aliquot of CoCl2 stock solution to a thermally equilibrated stirred solution of apo-MnSOD [25]. Both the protein sample and the metal stock solution were equilibrated at 37°C for 7 min before addition of the CoCl2 solution. Fluorescence emission intensity was recorded at a sampling frequency of 1 s−1 for up to 2 h after addition of the metal ion. The kinetic timecourses were imported into the data analysis program Scientist (Micromath Research, St. Louis, MO) and fit to an exponential decay process with a linear time-dependent term.
Analytical methods
The concentration of the purified protein was determined by optical absorption measurements, using the published molar extinction coefficient (e280 nm = 8.66×104 M−1cm−1 (E. coli MnSOD) [31]; 1.81×105 M−1cm−1 (human MnSOD) [32]; 1.46×105 M−1cm−1 (T. thermophilus HB8 MnSOD) [33]). Optical absorption spectra were recorded using a Varian Instruments Cary 5000 UV-visible-NIR absorption spectrometer. Superoxide dismutase activity was measured with the xanthine oxidase/cytochrome c inhibition assay [34]. Metal analyses were performed using a Varian Instruments SpectrAA Model 20B atomic absorption spectrometer equipped with a GTA 96 graphite furnace. Protein homogeneity was routinely evaluated using SDS-PAGE.
Quantitation of free thiols was performed by reaction of recombinant human Apo-MnSOD with Ellman’s reagent [5,5′-dithiobis(2-nitrobenzoic acid), (DTNB)] with or without 6 M guanidinium HCl, and the sample was placed in boiling water for 2 min when denaturant was used. The formation of colored product was monitored at 412 nm according to the published procedure [35]. L-cysteine was used as calibration standard.
Protein quaternary structure was determined by size exclusion chromatography (SEC) over Sephacryl 100-HR column (1.5 × 100 cm). For measurement of SOD apoprotein quaternary structure, the elution buffer contained 1 mM EDTA and the metal-free character of the sample was verified by fluorimetric metal uptake after gel filtration to remove EDTA.
3. Results
3.1 Preparation of human MnSOD apoprotein
Recombinant human MnSOD was expressed in the tightly regulated arabinose-inducible BL21-AI expression host for high level protein production. Human apo-MnSOD was prepared from the as-isolated, fully metallated protein using a modification of procedures previously developed for preparation of the dimeric, prokaryotic E. coli apo-MnSOD [23,30], forming the soluble metal-free product in 60–70% yield, based on the total protein used in the preparation. This yield reflects losses due to formation of protein precipitates during refolding and concentration steps. Precipitates were routinely removed by centrifugation. The apoprotein formed in this way lacks manganese (0.00 ±0.01 mol Mn/mol monomer) and is devoid of superoxide dismutase activity. As expected, the optical absorption spectrum of the apoprotein lacks any significant absorption in the visible region of the spectrum or light scattering from protein aggregates (Fig. 1, trace 2). Size exclusion chromatography shows that the apoprotein is homogeneous and has the same quaternary structure as the native tetrameric human MnSOD. Under non-denaturing conditions, thiol quantitation on the apoprotein detects 1.05±0.01 free SH groups per monomer, while 2.00±0.02 SH can be detected under denaturing conditions (6 M guanidinium HCl, heat).
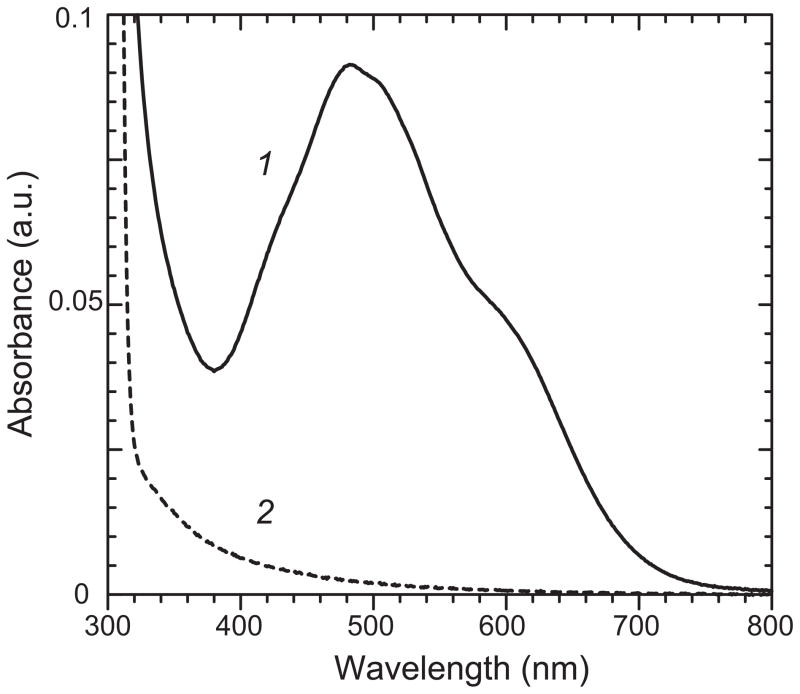
Figure 1.
Absorption spectra for Mn reconstituted recombinant human MnSOD forms. (1, solid line) Mn reconstituted recombinant human MnSOD (0.35 mM in 50 mM potassium phosphate pH 7). (2, dashed line) Human apo-MnSOD (0.35 mM in 20 mM MOPS pH 7) prepared as described in the Methods section.
3.2 In vitro reconstitution of human MnSOD
Mn-reconstituted apoprotein prepared as described in the Methods was found to contain 0.89±0.03 mol Mn/mol monomer. The superoxide dismutase activity of the in vitro reconstituted enzyme measured using the xanthine oxidase/cytochrome c reduction assay was 7240±80 U/mg, compared to 7164±48 U/mg for the as-isolated manganese-containing recombinant enzyme. Optical absorption spectra of reconstituted human MnSOD exhibits the typical Mn(III) absorption spectrum characteristic of MnSOD proteins (Fig. 1, trace 1).
3.3 In vitro metal uptake kinetics
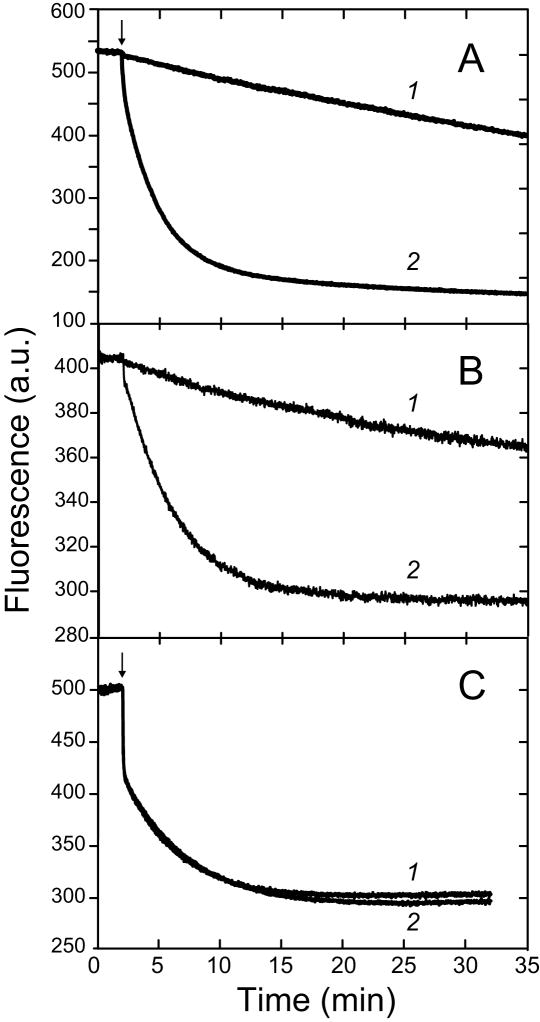
Continuous, real time fluorescence measurements of Co2+ uptake by human apo-MnSOD reveal a simple monophasic single exponential behavior (Fig. 2, A), quite distinct from the relatively complex biphasic metal uptake previously observed for the dimeric E. coli apo-MnSOD (Fig. 2, C). Monophasic metal uptake kinetics are also observed for Thermus apo-MnSOD (Fig. 2, B). The rate of metal uptake by the tetrameric apo-MnSODs is sensitive to the metal ion concentration (Fig. 2, A&B). The general expression for the metal dependence rate law is:
| (1) |
| (2) |
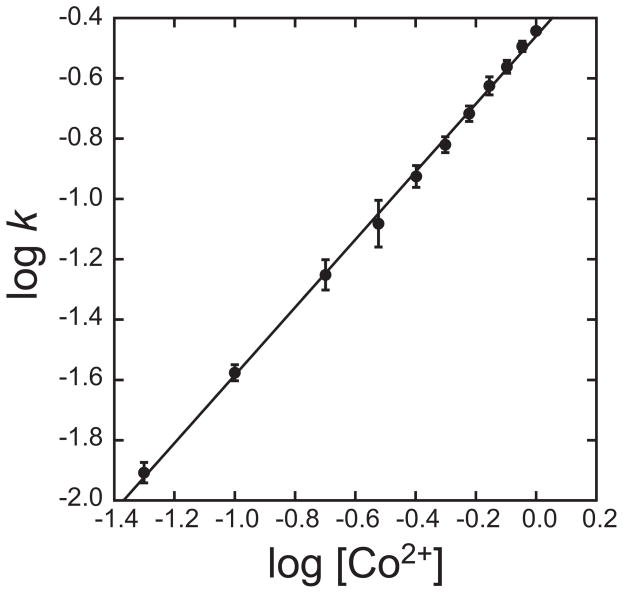
where k is the pseudo first order rate constant evaluated from the reaction timecourses. The kinetic order can be empirically evaluated from the slope of a log-log plot, as illustrated in Fig. 3 for human apo-MnSOD. Systematic variation of the metal concentration shows a first order rate dependence on Co2+, with a regression slope of 1.1±.01. In contrast, the rate of metal uptake by E. coli apo-MnSOD is independent of the metal concentration [25] (Fig. 2, C) and thus zero-order in Co2+ ion at concentrations above 10 μM, which represents a practical limit for accurately measuring the kinetics.
Figure 2.
Metal concentration dependence of uptake by apo-MnSOD. (A) Fluorimetric metal uptake timecourses for recombinant human apo-MnSOD (25 μg/mL in 20 mM HEPES pH 8.2) at 37°C. 1, 50 μM CoCl2; 2, 1 mM CoCl2. (B) Fluorimetric metal uptake timecourses for recombinant Thermus thermophilus HB8 apo-MnSOD (50 μg/mL in 20 mM MOPS pH 7) at 65°C. 1, 100 μM CoCl2; 2, 2 mM CoCl2. (C) Fluorimetric metal uptake timecourses for Escherichia coli apo-MnSOD (50 μg/mL in 20 mM MOPS pH 7.6) at 37°C. 1, 24 μM CoCl2; 2, 240 μM CoCl2.
Figure 3.
Empirical evaluation of the reaction order for Co2+ in the uptake reaction with human apo-MnSOD. Human apo-MnSOD apoprotein (25 μg/mL in 20 mM HEPES pH 8.2) was equilibrated to 37°C and metal binding initiated by addition of CoCl2 (50 μM – 1 mM). Metal binding was monitored fluorimetrically as described in the Methods, and the experimental timecourses were analyzed to evaluate the pseudo-first order reaction rate constants. The fitted line has a slope of 1.1±0.01.
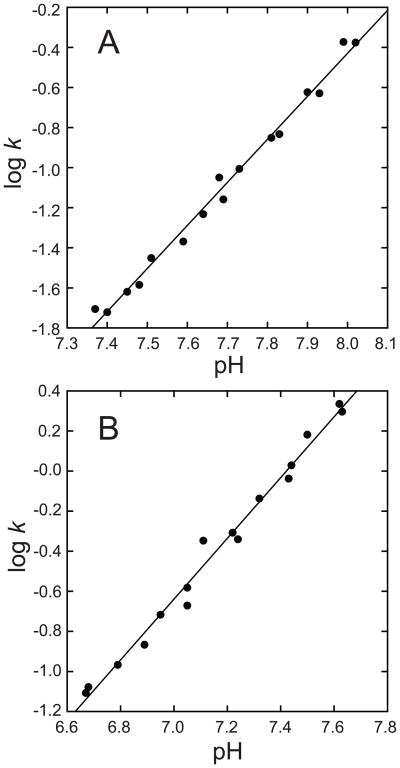
The rate of metal uptake is sensitive to both pH (Fig. 4) and temperature (Fig. 5&6) for human and Thermus apo-MnSOD. Monophasic kinetic timecourses are observed for human apo-MnSOD over the full range of physical variables (pH 7.4–8.4, temperature 30–60°C), with varying rate constants (Fig. 4, A). A plot of log k vs. pH is linear with a slope of 1.8±0.1, implying ionization of two protons in the rate determining step. Similar behavior is observed for the Thermus apo-MnSOD (Fig. 4, B), yielding a plot of log k vs. pH that is linear with a slope of 2.05±0.08.
Figure 4.
pH sensitivity of metal uptake by tetrameric apo-MnSODs. Fluorimetric metal binding timecourses were recorded for Co2+ binding to human apo-MnSOD (A) or Thermus thermophilus apo-MnSOD (B) over a range of pH values and the pseudo first-order rate constant was evaluated by regression analysis in Scientist. The fitted lines have slopes (A) 1.8±0.1, and (B) 2.05±0.08.
Figure 5.
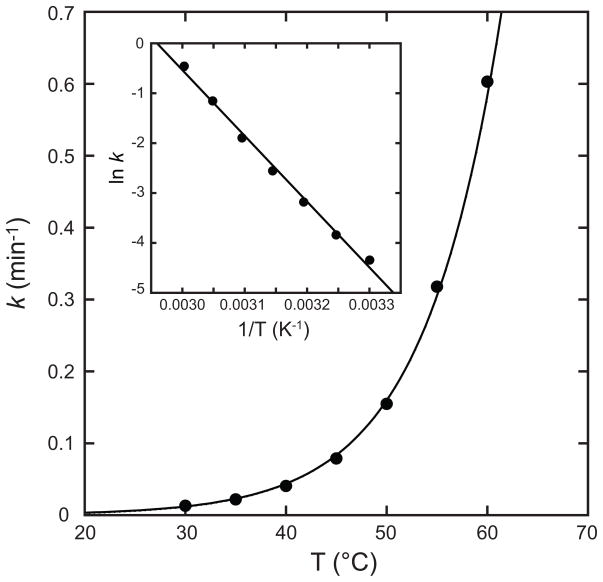
Temperature dependence of metal uptake by human apo-MnSOD. Human apo-MnSOD apoprotein (25 μg/mL in 20 mM HEPES pH 8.2) was equilibrated to the target temperature and metal binding initiated by addition of CoCl2 (to 0.1 mM final concentration). Metal binding was monitored fluorimetrically as described in the Methods. A regression line fitting the data is shown. Inset Arrhenius plot of temperature-dependent rate data. A regression line fitting the data is shown (Ea = 110 kJ/mol; ln A = 39).
Figure 6.
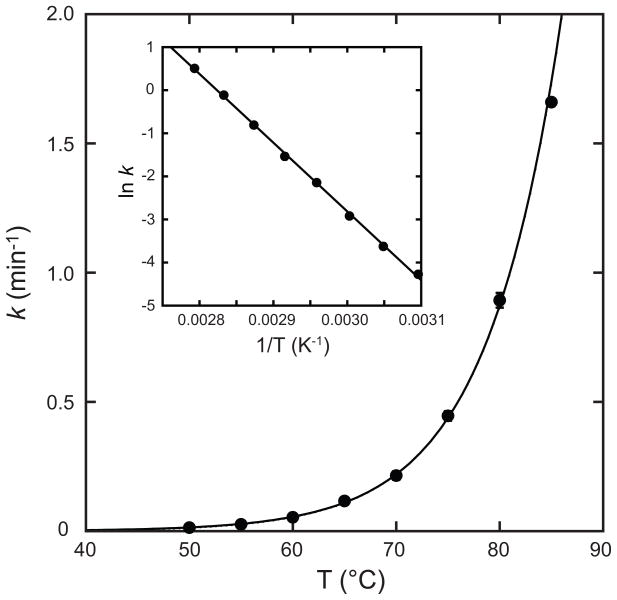
Temperature dependence of metal uptake by Thermus thermophilus HB8 apo-MnSOD. T. thermophilus apo-MnSOD apoprotein (50 μg/mL in 20 mM MOPS pH 7.6) was equilibrated to the target temperature and metal binding initiated by addition of CoCl2 (to 0.1 mM final concentration). Metal binding was monitored fluorimetrically as described in the Methods. A regression line fitting the data is shown. Inset Arrhenius plot of temperature-dependent rate data. A regression line fitting the data is shown (Ea = 133 kJ/mol; ln A = 20).
The exponential temperature dependence of metal uptake rate for human apo-MnSOD (Fig. 5) was analyzed using the Arrhenius equation
| (3) |
where k is the pseudo first order rate constant, Ea is the activation energy, R the gas constant, T the absolute temperature and A the Arrhenius pre-exponential factor. The Arrenhius plot (Fig. 5, Inset) yields an estimates of the activation barrier for metal uptake (Ea = 110 kJ/mol). Similar results are obtained for the temperature dependence of Co2+ uptake kinetics for Thermus thermophilus apo-MnSOD (Fig. 6), although metal uptake is associated with a higher activation barrier for the thermophilic protein (Ea = 133 kJ/mol). For comparison, the activation barrier estimated by Arrhenius analysis of the temperature dependence for the slow phase of metal uptake is nearly twice as large (Ea = 240 kJ/mol [25]), suggesting a different metallation mechanism may be effective in that case.
4. Discussion
Metal delivery is an essential step in the functional maturation of every metalloenzyme that, for most proteins, is just beginning to be investigated [36]. In principle, metal binding in vivo may occur at any point during protein biosynthesis, from the nascent polypeptide emerging from the ribosome to the fully folded, assembled apoprotein, complicating analysis of the biological metallation processes. In vitro studies on metal binding by the isolated apoprotein, on the other hand, can provide insight into the role of protein structural features in forming the metal complex and may be investigated over a relatively wide range of well-defined experimental conditions.
The method used to prepare human apo-MnSOD for these studies is based on the denaturation-chelation-refolding method previously used to prepare the E. coli MnSOD apoprotein [23,30], but modified to address the thiol oxidation sensitivity and special refolding requirements of the human enzyme, which contains two cysteine residues in each polypeptide chain. The human apo-MnSOD prepared by this protocol contains the full complement of sulfhydryl groups. Metal-free human apo-MnSOD is a colorless (Fig. 1, trace 2), soluble protein, and size exclusion chromatography demonstrates that the refolded protein retains the tetrameric structure of the native enzyme. Under appropriate conditions, the apoprotein takes up Mn forming the fully metallated, active holoenzyme product, with the characteristic absorption spectrum of the Mn3+-containing enzyme (Fig. 1, trace 1), as previously reported for the as-isolated protein [37].
Previous studies on the dimeric MnSOD from E. coli have revealed a surprising conformational gating requirement for metal binding, reflected in biphasic kinetics, zero-order metal ion dependence, and distinctive temperature and pH sensitivities [25,26]. Fluorimetric metal uptake analysis of human and Thermus apo-MnSODs (Fig. 2, A&B) shows that there are significant differences between the tetrameric and dimeric proteins. The monophasic kinetics and sensitivity to metal ion concentration indicate that the metal binding reaction is not gated for human or Thermus apo-MnSOD. Still, as previously found for the E. coli apo-MnSOD, the metal binding reaction is a thermally activated process. There is also a requirement for proton ionization in the rate determining step for all three proteins, although the pH dependence implies two protons are ionized in metal binding by the tetrameric apoproteins (Fig. 4), while a single proton ionization is found for the slow phase of the E. coli metal uptake reaction [26]. The temperature dependence of the metal uptake reactions of both human and Thermus apo-MnSODs reflect a simple Arrhenius behavior (Fig. 5&6), unlike the temperature-dependent structural transition observed for the E. coli protein [26].
These results demonstrate that there are both differences and significant similarities in the in vitro metal uptake behavior displayed by apo-MnSOD proteins. For all three proteins compared here, metal binding occurs through a thermally activated mechanism, with the activation barrier being higher for the thermophilic protein (T. thermophilus MnSOD) than for the human homolog. The most obvious contrasts, on the other hand, are associated with the absence of a conformational gating mechanism for the human and Thermus proteins. Since Thermus apo-MnSOD can be isolated directly as apoprotein from the expression host, the differences that we observe are unlikely to be a consequence of the denaturation/refolding cycle used to prepare the human apo-MnSOD. Also, Thermus apo-MnSOD provides a control for potential thiol interference in the binding reaction, since that protein lacks cysteine residues.
We have shown that, as previously found for the dimeric E. coli apo-MnSOD, refolded tetrameric human and Thermus apo-MnSODs are able to bind exogenous metal ions to form native, metallated complexes. However, there are significant differences in the metal uptake behavior for dimeric and tetrameric apo-proteins, most dramatically expressed in the simple single phase metal uptake kinetics and metal concentration sensitivity observed for the latter, which may relate to the distinct quaternary structures of these proteins. In particular, the tetrameric organization of the human and Thermus apo-MnSODs may constrain motions on the subunit interface, a region that has been shown to be important for metal uptake in the corresponding E. coli apoprotein [26], accounting for their altered metal binding behavior. The zero-order metal ion dependence observed for the dimeric E. coli apo-MnSOD metal uptake suggests that in that case, metal interactions do not contribute to the rate limiting step, whereas the behavior of the tetrameric apo-MnSODs suggests that metal entry into the active site contributes significantly to the activation barrier. This implies that there are distinct rate limiting steps for metal uptake in the dimeric and tetrameric proteins. The doubling of the subunit composition (dimer/tetramer) may also be reflected in the doubling of the proton ionization requirement for the metal uptake by the tetrameric apo-MnSODs, implying cooperative subunit interactions within the tetramer. The most striking difference, however, is the metal concentration dependence of these reactions. The intracellular concentration of Mn2+ has been estimated to be in the nM to μM range [38], which would be predicted to result in extremely slow (hours timescale) metallation of tetrameric apo-SODs at neutral pH, based on their sensitivity to metal concentration in vitro. The observation that metal uptake by the fully folded, tetrameric apo-MnSODs requires relatively high (approaching mM range) concentrations of metal ion to proceed at significant rates (compared to the low μM range concentrations that are sufficient for metallation of the dimeric E. coli protein) may indicate a chaperone requirement or co-translational metal insertion mechanism for the tetrameric enzymes in vivo.
Acknowledgments
This work was supported by the National Institutes of Health (GM 042680 to J.W.W.) and by a grant from the Oregon Health and Science University Medical Research Foundation.
Footnotes
This work was supported by the National Institutes of Health (GM42680 to J.W.W.) and the Oregon Medical Research Foundation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fridovich I. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 2.Whittaker JW. Biochem Soc Trans. 2003;31:1318–1321. doi: 10.1042/bst0311318. [DOI] [PubMed] [Google Scholar]
- 3.Carlioz A, Touati D. EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCord JM, Edeas MA. Biomed Pharmacother. 2005;59:139–142. doi: 10.1016/j.biopha.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Lebovitz RM, Zhang H, Vogel H, Cartwright J, Jr, Dionne L, Lu N, Huang S, Matzuk MM. Proc Natl Acad Sci USA. 1996;93:9782–9787. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duttaroy A, Paul A, Kundu M, Belton A. Genetics. 2003;165:2295–2299. doi: 10.1093/genetics/165.4.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ose DE, Fridovich I. J Biol Chem. 1979;194:360–364. doi: 10.1016/0003-9861(79)90628-3. [DOI] [PubMed] [Google Scholar]
- 8.Yamakura F, Kobayashi K, Ue H, Konno M. Eur J Biochem. 1995;227:700–706. doi: 10.1111/j.1432-1033.1995.tb20191.x. [DOI] [PubMed] [Google Scholar]
- 9.Parker MW, Blake CC, Barra D, Bossa F, Schinina ME, Bannister WH, Bannister JV. Protein Eng. 1987;1:393–397. doi: 10.1093/protein/1.5.393. [DOI] [PubMed] [Google Scholar]
- 10.Jackson SM, Cooper JB. Biometals. 1998;11:159–173. doi: 10.1023/a:1009238214394. [DOI] [PubMed] [Google Scholar]
- 11.Wintjens R, Noel C, May AC, Gerbod D, Dufernez F, Capron M, Viscogliosi E, Rooman M. J Biol Chem. 2004;279:9248–9254. doi: 10.1074/jbc.M312329200. [DOI] [PubMed] [Google Scholar]
- 12.Edwards RA, Baker HM, Whittaker MM, Whittaker JW, Jameson GB, Baker EN. J Biol Inorg Chem. 1998;3:161–171. [Google Scholar]
- 13.Lah MS, Dixon MM, Pattridge KA, Stallings WC, Fee JA, Ludwig ML. Biochemistry. 1995;34:1646–1660. doi: 10.1021/bi00005a021. [DOI] [PubMed] [Google Scholar]
- 14.Ludwig ML, Metzger AL, Pattridge KA, Stallings WC. J Mol Biol. 1991;219:335–358. doi: 10.1016/0022-2836(91)90569-r. [DOI] [PubMed] [Google Scholar]
- 15.Borgstahl GE, Parge HE, Hickey MJ, Beyer WF, Jr, Hallewell RA, Tainer JA. Cell. 1992;71:107–118. doi: 10.1016/0092-8674(92)90270-m. [DOI] [PubMed] [Google Scholar]
- 16.Parker MW, Blake CC. J Mol Biol. 1988;199:649–661. doi: 10.1016/0022-2836(88)90308-7. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt M, Meier B, Parak F. J Biol Inorg Chem. 1996;1:532–541. [Google Scholar]
- 18.Stoddard BL, Howell PL, Ringe D, Petsko GA. Biochemistry. 1990;29:8885–8893. doi: 10.1021/bi00490a002. [DOI] [PubMed] [Google Scholar]
- 19.Cooper JB, McIntyre K, Badasso MO, Wood SP, Zhang Y, Garbe TR, Young D. J Mol Biol. 1995;246:531–544. doi: 10.1006/jmbi.1994.0105. [DOI] [PubMed] [Google Scholar]
- 20.Beyer WF, Jr, Fridovich I. J Biol Chem. 1991;266:303–308. [PubMed] [Google Scholar]
- 21.Whittaker JW, Whittaker MM. J Am Chem Soc. 1991;113:5528–5540. [Google Scholar]
- 22.Edwards RA, Whittaker MM, Whittaker JW, Jameson GB, Baker EN. J Am Chem Soc. 1998;120:9684–9685. [Google Scholar]
- 23.Mizuno K, Whittaker MM, Bächinger HP, Whittaker JW. J Biol Chem. 2004;279:27339–27344. doi: 10.1074/jbc.M400813200. [DOI] [PubMed] [Google Scholar]
- 24.Vance CK, Miller AF. Biochemistry. 2001;40:13079–13087. doi: 10.1021/bi0113317. [DOI] [PubMed] [Google Scholar]
- 25.Whittaker MM, Mizuno K, Bächinger HP, Whittaker JW. Biophys J. 2006;90:598–607. doi: 10.1529/biophysj.105.071308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whittaker MM, Whittaker JW. Biochemistry. 2008;47:11625–11636. doi: 10.1021/bi8015636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guzman LM, Belin D, Carson MJ, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang K, Hallewell RA, Bell GI. Nucleic Acids Res. 1987;15:7654. doi: 10.1093/nar/15.18.7654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whittaker MM, Whittaker JW. J Biol Chem. 1999;274:34751–34757. doi: 10.1074/jbc.274.49.34751. [DOI] [PubMed] [Google Scholar]
- 30.Quijano C, Hernandez-Saavedra D, Castro L, McCord JM, Freeman BA, Radi R. J Biol Chem. 2001;276:11631–11638. doi: 10.1074/jbc.M009429200. [DOI] [PubMed] [Google Scholar]
- 31.Beyer WF, Jr, Reynolds JA, Fridovich I. Biochemistry. 1989;28:4403–4409. doi: 10.1021/bi00436a042. [DOI] [PubMed] [Google Scholar]
- 32.Ho YS, Crapo JD. FEBS Lett. 1988;229:256–260. doi: 10.1016/0014-5793(88)81136-0. [DOI] [PubMed] [Google Scholar]
- 33.Sato S, Nakazawa K. J Biochem (Tokyo) 1978;83:1165–1171. doi: 10.1093/oxfordjournals.jbchem.a132007. [DOI] [PubMed] [Google Scholar]
- 34.McCord JM, Fridovich I. J Biol Chem. 1969;244:6049–55. [PubMed] [Google Scholar]
- 35.Riddles PW, Blakeley RL, Zerner B. Meth Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- 36.Culotta VC, Yang M, O’Halloran TV. Biochim Biophys Acta. 2006;1763:747–758. doi: 10.1016/j.bbamcr.2006.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lévêque VJP, Vance CK, Nick HS, Silverman DN. Biochemistry. 2001;40:10586–10591. doi: 10.1021/bi010792p. [DOI] [PubMed] [Google Scholar]
- 38.Weatherburn DC. In: Handbook on Metalloproteins. Bertini I, Sigel A, Sigel H, editors. Marcel Dekker; New York, NY: 2001. p. 197. [Google Scholar]