Abstract
Members of the genus Malassezia are commensal fungi found on the skin of both human and domestic animals and are associated with skin diseases including dandruff/seborrhoic dermatitis, pityriasis versicolor and atopic eczema (AE) in humans. In this study we have characterized the cell-wall carbohydrates of M. sympodialis, one of the species most frequently isolated from both AE patients and healthy individuals. Cells were grown in liquid Dixon media at 32 °C, harvested and processed using a standard Fehling's precipitation methodology for isolation of mannan and a standard base/acid extraction for (1→3)-β-D-glucans. Using these classic extraction methods we were unable to isolate precipitable mannan or insoluble (1→3)-β-D-glucan. However, acidification and addition of methanol to the remaining Fehling's-treated sample resulted in a very clean precipitate. This material was characterized by GPC-MALLS, 1D and 2D NMR and GC–MS for monomer-type and linkage-type composition. We determined that trace amounts of both mannan and branched (1→3, 1→6)-β-D glucan were present in the recovered precipitate, but not linear (1→3)-β-D-glucan. Surprisingly, NMR analysis indicated that (1→6)-β-D-glucan was the major carbohydrate component isolated from M. sympodialis cell wall. GC–MS linkage analysis confirmed the (1→6)-β-D-glucan structure. Based on these studies we have determined that the M. sympodialis cell wall contains (1→6)-β-D-glucan as the major carbohydrate component along with trace amounts of mannan and (1→3, 1→6)-β-D-glucan. In addition, these data indicate that modification of the classic mannan isolation methodology may be useful in the simultaneous isolation of both mannan and (1→6)-β-D-glucan from other fungi.
Keywords: (1→6)-β-D-Glucan, Malassezia sympodialis cell wall, Carbohydrate extraction
1. Introduction
The fungal cell wall is a complex structure composed primarily of carbohydrate molecules and lipids as well as some cell-wall associated proteins. In general, the fungal cell wall consists of branched (1→3, 1→6)-β-D-glucan that is linked to chitin via a (1→4)-β-linkage.1 (1→3)-β-glucan tends to be the predominant polymer isolated from these fungi; however, some fungal species express (1→3)-α-glucan as well.2-4 In addition to the presence of the glucans and chitin, N- and O-linked mannoproteins are found in the cell wall anchored by a glycosyl phosphatidylinositol linkage.5
Malassezia species are associated with a number of dermatological disorders including dandruff/seborrhoic dermatitis and, pityriasis versicolor.6 Malassezia yeasts have also been shown to act as allergens in atopic eczema (AE), and thirteen allergens have been cloned, characterized and produced as recombinant proteins from Malassezia species.7 Currently there are thirteen recognized species of Malassezia that have been isolated from healthy and diseased human and animal skin.6,8 Members of the Malassezia genus are lipo-dependent yeasts since the majority of them require lipids for growth. One species, M. pachydermatis is the only non-lipid-dependent lipophilic yeast.6 The cells of Malassezia species are small, unipolar budding yeasts with an unusual spiral pattern of plasma membrane grooves along the inner cell wall. The cell wall is a bilayered lipid rich structure (10-fold greater than seen with Candida albicans and Saccharomyces cerevisiae9-11) surrounded by a lipid capsule. Little is known about the cell-wall composition of Malassezia species other than from a few studies on the cell's ultrastructure.12,13 To date, no compositional analysis has been performed in order to characterize the structural components of the cell wall.
Carbohydrate polymers are not only important structural components of the fungal cell wall; they are also critically important determinants in the recognition of the fungus by the innate immune system.14-18 Very little is known about the cell-wall carbohydrates of M. sympodialis. To address this deficit in our knowledge, we characterized the cell-wall carbohydrates of M. sympodialis. Initial studies focused on the isolation of mannan and (1→3)-β-D-glucan using classical extraction methodologies.19-21 In the course of our experiments we were unable to isolate mannan or (1→3)-β-D-glucans using these methods; however, a slight modification to the mannan isolation method resulted in a novel and unexpected observation. Using the modified protocol we describe the isolation and characterization of (1→6)-β-D-glucan from M. sympodialis cell wall. Furthermore, this glucan appears to be the major carbohydrate component of the cell wall. We also detected mannan and (1→3, 1→ 6)-β-D-glucan using the modified protocol, but they were present only in trace amounts.
2. Results
2.1. M. sympodialis has trace amounts of extractable mannan in its cell wall
We employed a classic mannan extraction protocol modified from Kocourek and Ballou22, and Shibata et al 23 (Fig. 1). Using this procedure we were unable to obtain any precipitable mannan, even with a large amount of starting material (10 g). In subsequent experiments, we cultured M. sympodialis in large volume (15 L of media) and were still unable to isolate mannan from a 65-g cell pellet. Since we were unable to fractionate mannan by traditional means, we treated the remaining mixture of Fehling's-soluble sample with an equal volume of methanol to precipitate any carbohydrate that might be present in the solution; however, this approach did not result in a reasonable yield. We discovered that addition of an 8:1 solution of methanol–acetic acid in equal volume to the Fehling's-soluble sample resulted in a precipitate that was sufficiently pure for NMR analysis.
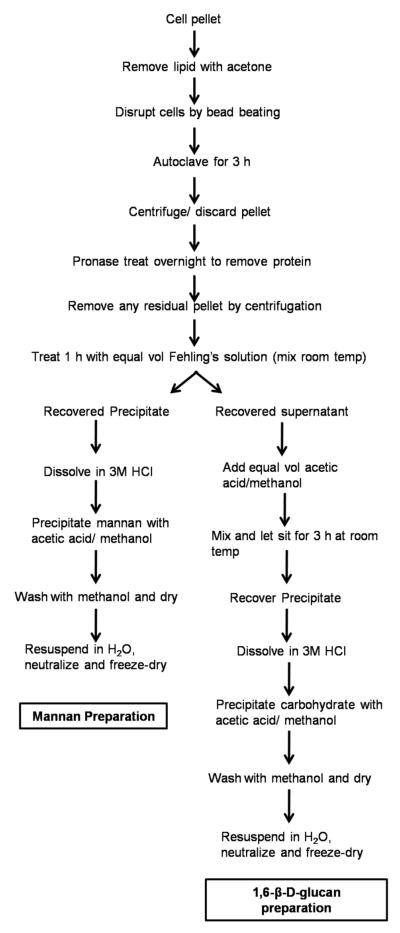
Figure 1.
General schematic for mannan and (1→6)-β-D-glucan isolation from fungi.
2.2. Isolation of material using a classic glucan extraction method
We also attempted to isolate (1→3)-β-D-glucan from M. sympodialis using a classic extraction method.19-21 Surprisingly, no (1→3)-β-D-glucan was recovered, but we were able to isolate small quantities of a (1→6)-β-D-glucan using this extraction methodology.
2.3. NMR analysis of (1→6)-β-D-glucan in the acid precipitated material
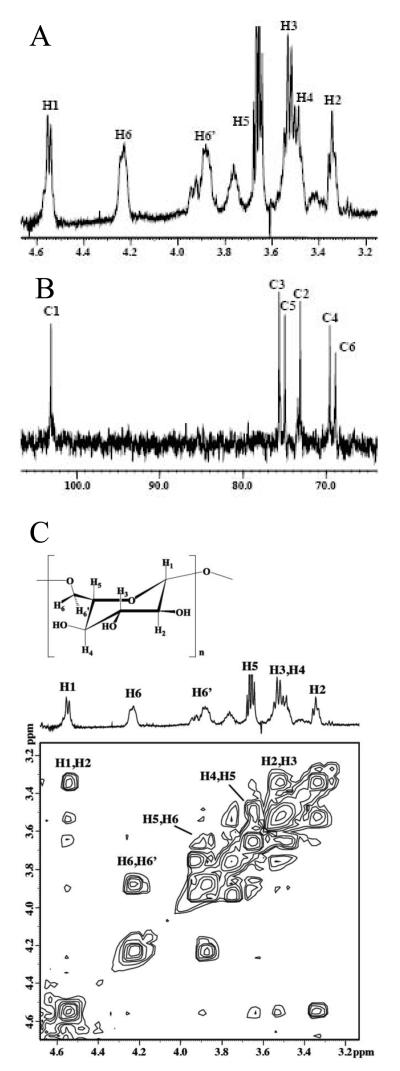
1D 1H and 13C NMR spectra (Fig. 2A and B) and a 2D COSY (Fig. 2C) NMR spectrum of the material precipitated by acetic acid–methanol treatment of the Fehling's-soluble sample mixture was consistent with similar NMR spectral evidence for a (1→6)-β-d-glucan isolated from Actinobacillus suis.27 1H and 13C NMR chemical shift assignments for this acid-precipitated glucan were compared to the isolate from A. suis (Table 1) to confirm our identification of this glucan as a (1→6)-β-d-glucan. The resonance assigned to H5 is overlapped by the methylene proton resonance from residual ethanol. The presence of more than one conformation about the (1→6)-glycosidic linkage, initially reported by Monteiro and co-workers27, is evident in this work as well. Several resonances for a lesser conformation are assigned as follows: the resonance at 4.55 ppm is assigned to anomeric proton H1 overlapped by the anomeric proton doublet resonance from the major conformation, while resonances at 3.94 and 3.76 ppm are assigned to the methylene proton resonances H6 and H6′, respectively. The small multiple resonances between 3.34 and 3.50 ppm may also be associated with other protons on the glucosyl ring in a lesser conformation.
Figure 2.
(A) 1H and (B) 13C NMR spectra and (C) 2D COSY NMR spectrum of (1→6)-β-d-glucan isolated from M. sympodialis.
Table 1.
1H and 13C NMR chemical shift assignments (in ppm) for (1→6)-β-d-glucan isolated from M. sympodialis compared to assignments for a similar glucan from A. suis.
2.4. Monosaccharide composition and linkage analysis
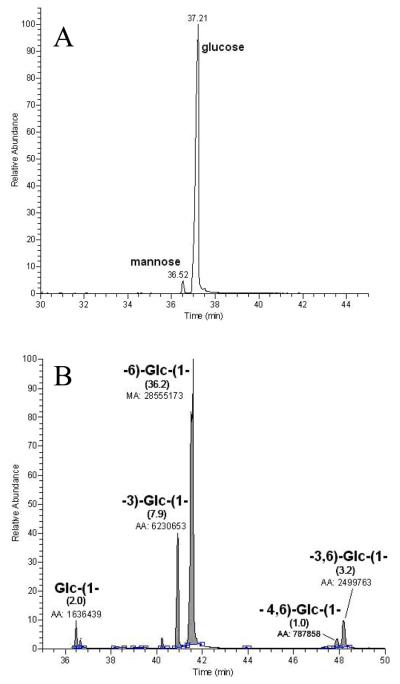
Monosaccharide composition (Fig. 3A) analysis carried out on the preparation precipitated by acetic acid–methanol treatment of the Fehling's-soluble sample mixture revealed that this material was dominantly composed of glucose (Glc) with accompanying minor traces mannose (Man). A linkage-type analysis performed on the same material showed that Glc was mainly present as a 6-substituted unit (Fig. 3B). Glc substituted at the 3-position was the other mono-substituted residue detected. Albeit in small amounts, two branch points were also observed in the form of 4,6- and 3,6-disubstituted Glc units. These analyses showed that this material contained as the predominant structural feature a (1→6)-β-d-glucan. The fact that we observed 3-substituted glucose, 3,6- and 4,6-disubstituted glucose (branch points) indicated that the 6-linked glucan may carry a limited number of branching points containing in part 3-substituted glucose, or/and another independent polysaccharide motif composed of these three minor units (with or without 6-substituted glucoses) may be present in this M. sympodialis preparation.
Figure 3.
Monosaccharide composition and linkage analysis. (A) GC–MS profile of the monosaccharide composition analysis showing glucose as the main component in this M. sympodialis preparation; and (B) GC–MS profile of the monosaccharide linkage analysis, with respective area counts, showing that a linear 6-linked glucan is the dominant structural unit. Although in minor amounts, the detection of 3-substituted glucose, 3,6- and 4,6-disubstituted glucose (branch points) indicates that the 6-linked glucan may contain a limited number of branching points carrying in part 3-substituted glucose or/and another independent polysaccharide motif composed of these three minor units (with or without 6-substituted glucoses) may be present in this preparation.
2.5. Molecular weight distribution of the M. sympodialis (1→6)-β-D-glucan
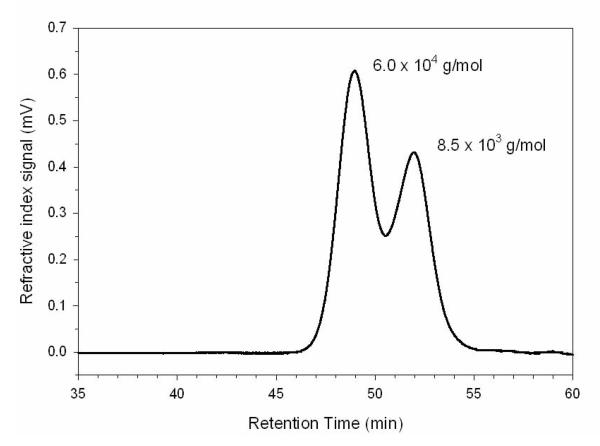
GPC–MALLS analysis demonstrated that the (1→6)-β-D-glucan had a bimodal polymer distribution. Two peaks corresponding to molecular weights of 6.0 × 104 and 8.5 × 103 g/mol were observed (Fig. 4). The average molecular weight for the entire polymer distribution, i.e. both peaks, was 2.7 × 104 g/mol (Fig. 4). Each peak was harvested by fractionation and analyzed by NMR spectroscopy. NMR analysis confirmed that both peaks were (1→6)-β-D-glucan (data not shown).
Figure 4.
GPC–MALLS analysis of M. sympodialis (1→6)-β-d-glucan revealed a bimodal polymer distribution. The first peak has an Mw of 6.0 × 104 g/mol, while the second peak has an Mw of 8.5 × 103 g/mol. The average molecular weight for the entire polymer distribution, i.e., both peaks, is 2.7 × 104 g/mol.
2.6. Staining of cell-wall components
Since we were unable to isolate any significant amount of mannan or (1→3)-β-D-glucan by our extraction methods, we grew M. sympodialis cells for 72 hours in Dixon media and subjected them to staining with ConA–FITC for mannan, and used an anti- (1→3)-β-D-glucan antibody labeled with FITC and an aniline blue stain specific for (1→ 3)-β-D-glucan as a means of indirect measurement of these components in the cell wall. For ConA–FITC staining, there was little-to-no fluorescence in many cells with very few showing specific staining, indicating that there was very little mannan present in the cell wall (data not shown). Staining with either the anti-(1→3)-β-D-glucan antibody or an-line blue resulted in no visible fluorescence for (1→3)-β-d-glucan (data not shown). When taken together, these data confirm our studies using the extraction methodologies.
3. Discussion
Cell walls of fungi play an important role in maintaining cell morphology as well as a role in modulating adherence to host and non-biological surfaces (i.e., biofilms). From the host point of view, the fungal cell wall is an important target for pathogen recognition. For example the surface receptor dectin-1 recognizes the (1→3)-β-D-glucan present in fungal cell walls.14,15 Mice genetically altered to lack this receptor have an increased susceptibility to fungal infections.16 Another C-type lectin receptor of interest besides dectin-1 is Mincle, which is expressed on antigen-presenting cells, and recently was shown to be an activating receptor for Malassezia.28
In this study we have identified a major structural component of the cell wall from M. sympodialis. The majority of Malassezia species require lipid supplementation for growth except for M. pacydermatis.6 Ultrastructural studies on Malassezia spp. have demonstrated that the cell wall appears to be bilayered and lipid rich with a surrounding lipid capsule.12,13 Our initial foray into characterizing the M. sympodialis cell-wall components originally focused on the mannan component of the cell wall. Using a classical methodology for mannan extraction, we were unable to isolate any mannan material from this organism. Though modification of our protocol did demonstrate that there were trace amounts of mannan present in the cell wall, it may be that the mannan present in the cell wall is of simple structure such as O-linked mannan (3–5 mannose residues). One possible reason for this reduced mannan presence could be due to the surrounding lipid capsule. Mannosylated proteins on the cell-wall surface of other fungi such as C. albicans are known to play roles in cell aggregation, and adherence to surfaces and may be immunological targets for host cell recognition.29-31 The presence of the lipid capsule may allow for the organism to dispense with this feature common to other fungi and serve a protective function (for immune evasion) similar to the capsule of Cryptococcus.32,33
An unexpected observation made during this study is that M. sympodialis apparently lacks any (1→3)-β-D-glucan. Using classical extraction methods we were unable to isolate (1→3)-β-D-glucan.19-21 This result goes against the current dogma of fungal cell-wall structure, since (1→3)-β-D-glucan is normally a major part of a fungal cell wall. One possible explanation for our inability to isolate (1→3)-β-D-glucan from this organism could be that the molecule is too small for classical extraction techniques or is readily degraded under these conditions. Attempts to confirm this by staining with an anti-(1→3)-β-d-glucan antibody demonstrated little to no fluorescence when staining was performed in the presence of nonspecific competitors such as serum and bovine serum albumin (data not shown). When taken together these data make a strong case that M. sympodialis does not contain substantive amounts of (1→3)-β-D-glucan.
Since we were unable to isolate mannan by classical means from M. sympodialis, the remaining Fehling's supernatant mixture was treated with an equal volume of methanol–acetic acid. This supernatant is normally discarded, but in an attempt to identify any carbohydrates that might remain in the supernatant, we discovered that, upon addition of methanol–acetic acid to the solution, a precipitate formed that was readily recoverable. We initially, thought this might be a mannan that was soluble under the Fehling's precipitation conditions and therefore proceeded to isolate the material. Upon recovery we noted that the majority of the material recovered was water soluble. To our surprise, NMR spectroscopy and GC–MS analysis revealed that the material was predominantly (1→6)-β-D-glucan with trace amounts of mannan. A small amount of (1→6)-β-D-glucan was also isolated using the classical methodology for (1→3)-β-d-glucan extraction (data not shown), which tends to confirm that M. sympodialis cell wall expresses (1→6)-β-d-glucan. We also detected trace amounts of (1→3, 1→6)-β-d-glucan. Our isolation methodology differed significantly from the method used by Iorio et al34 which might indicate that the molecular size of the glucan that we observed is possibly more biologically relevant than what they have reported for C. albicans. In fact we have recently used this same procedure to isolate β-(1→6)-d-glucan from C. albicans ourselves and report a significantly larger molecule than what they observed (M.D.K, D.W.L, and D.L.W manuscript in preparation). Traditionally the isolation of (1→6)-β-d-glucan and (1→3)-β-d-glucan is performed using a hot alkaline extraction with several steps for enzymatic digest to further increase the purity of the insoluble material in which the remaining material is generally the (1→3)-β-d-glucan. The (1→6)-β-d-glucan is generally considered to be alkali soluble and therefore should be released from the cell wall upon hot alkali treatment. The report by Iorio et al34 describes the isolation of (1→6)-β-d-glucan from the insoluble particulate after digestion with a (1→3)-β-d-glucanase. What we have shown here is that the (1→6)-β-d-glucan can also be water soluble and readily extractable using high heat and pressure for a period of time. The solubility of the molecule is probably a reflection of the size of the polymer and not necessarily the nature of the material. The (1→6)-β-d-glucan remains soluble even during the Fehling's precipitation of mannan. It isn't until slight acidification and precipitation with methanol that allows for recovery of (1→6)-β-d-glucan from the now nearly mannan-free mixture. This new method that we describe for isolation of (1→6)-β-d-glucan is dramatically simplified from the classic glucan isolation methodology and may represent a means to obtain a more biologically relevant molecule that hasn't been over-processed during the procedure for its extraction.
In conclusion, we have demonstrated that M. sympodialis cell wall contains a simple mannan, but it lacks (1→3)-β-d-glucan. Our data indicate that (1→6)-β-d-glucan as a major component of the M. sympodialis cell wall. Additionally, we have presented evidence that a modification to existing isolation methodology may be useful in the simultaneous isolation of both mannan and (1→6)-β-d-glucan from other fungi.
4. Experimental
4.1. Strain and cultivation conditions
The M. sympodialis strain used for this study was ATCC strain 42132 (American Type Culture Collection). Routine cultivation was performed on Dixon medium (3.6 % Malt Extract, 0.6 % Mycological peptone, 2 % Oxgall, 1 % Tween 60, 0.2 % Glycerol, 0.05 g Chloramphenicol) at 32 ° C.
4.2. Procedure for extraction of mannan and (1→6)-β-d-glucan
The following protocol (Fig. 1) is modified from those of Kocourek and Ballou22 and Shibata et al.23 M. sympodialis was grown in 12 L of Dixon medium for a period of 5 days at 32 °C. Cells were harvested by centrifugation (7000 × g) and washed with 200 mL of acetone to remove lipid and contaminating medium. The washed pellet was allowed to dry for 1 h, allowing for acetone to evaporate. The dry weight yield was on average 65 g of material. The pellet was then broken up using a rubber policeman and suspended in 150 mL of deionized H2O and transferred to a bead beater chamber (Biospec). Glass beads corresponding to the weight of the pellet were added to the container, and the mixture was subjected to ten 15-s pulses by bead beating on ice. The mixture was transferred to a 1-L flask and autoclaved for 3 h. The sample was allowed to cool, and then centrifuged for 15 min at 7000 × g. The supernatant was saved and the pellet was discarded. The cleared supernatant was divided in half. Sodium azide was added to a final concentration of 1 mM to each sample. One half of the sample had 100 mg of heat-treated pronase (100 mg of pronase, Roche Diagnostics dissolved in 5 mL of deionized H2O, filter sterilized through a 0.45-μm filter and heated for 30 min at 70 °C to remove any glycosidic activity) added to remove protein, and samples were incubated at 37 °C for 16–20 h. The pronase-treated and untreated samples were then centrifuged 5 min at 7000 × g, to remove any remaining particulates from the samples. The supernatant was transferred to a 250-mL beaker, and an equal volume of Fehling's solution was added to the sample with stirring for 1 h at room temperature to allow for a copper–mannan complex to precipitate. In processing the samples, one of the problems seen was poor copper–mannan complex precipitation, which required centrifugation of the solution to determine if mannan was present at all. This allowed us to recover trace amounts of material from this species. The remaining supernatant was saved for further processing (see below). The trace copper–mannan precipitate complex was dissolved in 3–5 mL of 3 M HCl. An 8:1 mixture of MeOH–HOAc (50 mL) was added, and the solution was briefly stirred to mix. The samples were then allowed to sit for several hours to allow for the carbohydrate to settle. After the precipitate had settled it was washed several times with the 8:1 MeOH–HOAc solution until all traces of color were removed. The precipitate was then subjected to several MeOH washes to reduce the acidity of the product. The precipitate was then allowed to dry thoroughly, dissolved in deionized H2O and the pH adjusted to pH 6.5–7.0. The sample was then frozen and lyophilized for storage until further use. Generally we isolated trace amounts of material from M. sympodialis using the traditional mannan extraction protocol. The amounts we obtained were negligible for further use in compositional analysis.
To determine whether cell-wall carbohydrates remained in the supernatant after the initial precipitation, the remaining Fehling's-treated supernatant was treated with an equal volume of 8:1 MeOH–HOAc, which resulted in the formation of a greenish precipitate. The precipitate was allowed to settle for several hours until the solution appeared clear. The supernatant was decanted, and the precipitate was dissolved in 8–10 mL of 3 M HCl. Again 50 mL of 8:1 MeOH–HOAc solution was added to the sample with mixing, and the precipitate was allowed to settle. As with the processing of the mannan sample, the precipitate was washed several times with 8:1 MeOH–HOAc until colorless. The precipitate was then washed several more times with MeOH. The sample was then dried, dissolved in deionized H2O and the pH adjusted to pH 6.5– 7.0. The sample was then frozen and lyophilized for storage until further use.
4.3. Procedure for the isolation of (1→3)-β-d-Glucan
We also attempted to isolate (1→3)-β-D-glucan from M. sympodialis using a classic extraction method.19-21 Briefly, lyophilized M. sympodialis was extracted with a base–acid extraction protocol, followed by an organic extraction to remove lipids.19-21
4.4. Molecular weight and polymer distribution analysis
GPC–MALLS analyses were performed essentially as described by Adams et al.24 Glucan samples were dissolved (3 mg/mL) in the mobile phase (50 mM sodium nitrite, pH 7.0). The samples were incubated for 15 min at ambient temperature, followed by sterile filtration (0.45 μm) and injection into the GPC at a concentration of 600 g in 200 L of mobile phase. The data were analyzed using Wyatt ASTRA v 5.3.4.13 software (Wyatt Technology, Santa Barbara, CA).
4.5. NMR characterization of glucan
1D and 2D gradient-selective homonuclear correlation spectroscopy (COSY) 1H NMR spectra were acquired on a JEOL Eclipse+ 600 NMR spectrometer in 5-mm OD NMR tubes operating at ambient probe temperature in D2O. Internal chemical shift reference was provided by trimethylsilyl-2,2,3,3-d4-propionic acid (TSP) at 0.0 ppm for 1H NMR and −2.80 ppm for 13C NMR. Spectral collection and processing parameters for the 1D 1H NMR spectrum were the following: 15 ppm spectral width centered at 5.0 ppm, 32,768 data points, 2048 scans, 5 s relaxation delay, 2.18 s acquisition time and exponential apodization. Spectral collection and processing parameters for the 1D 13C NMR spectrum were the following: 250 ppm spectral width centered at 110.0 ppm, 65,536 data points, 2708 scans, 3 s relaxation delay, 1.74 s acquisition time and exponential apodization. Spectral collection and processing parameters for the 2D COSY spectrum were the following: 512 points in f2 and 128 points in f1 zero-filled 4 times for a 512 × 512 data array size, 10 ppm spectral width centered at 5 ppm in both dimensions, 4 prescans, 128 scans and 1 s relaxation delay.
4.6. Monosaccharide composition and linkage analysis
Monosaccharide composition analysis was performed by the alditol acetate method.25 The glycosyl hydrolysis were carried out with 4 M CF3CO2H at 105 °C for 5 h, followed by reduction in H2O with NaBD4 overnight at room temperature, and subsequent acetylation by Ac2O at 100 °C for 2 h. The alditol acetate derivatives were analyzed by GC using a Varian 3400 gas chromatograph equipped with a 30-m DB-17 capillary column [210 °C (30 min), then 240 °C at 2 °C/min], and by GC–MS in the electron-impact and chemical-ionization modes on a ThermoFinnigan PolarisQ instrument. Sugar linkage-type analysis was performed by the methylation procedure (NaOH–Me2SO–CH3I) and with characterization of the permethylated alditol acetate derivatives by GC–MS in the electron-impact mode (DB-17 column, isothermal at 190 °C for 60 min).26
4.7. Fluorescent staining of the M. sympodialis cell wall
Cells were grown in 20 mL of Dixon medium for 72 h at 32 °C. To stain the cell wall, the capsule was removed by briefly washing the cells with 50 mL of acetone. The cell pellet was harvested by centrifugation and suspended in 20 mL of deionized H2O. To stain for the presence of mannan we used a ConA–FITC-labeled probe (Sigma-Aldrich). A stock solution was prepared (1 mg/mL) and diluted 1:500 for staining. To stain for (1→3)-μ-D-glucan we used two different approaches. First, we used an aniline blue fluorchrome (Biosupplies, Australia) that was prepared and used according to manufacturer's instructions. A second approach was using an anti-(1→3)-μ-D-glucan IgG antibody (Biosupplies Australia). The antibody was reconstituted as a 1 mg/mL stock solution in 10 mM phosphate buffer, pH 7.2. The antibody was labeled using the Zenon mouse IgG labeling kit (Molecular Probes) according to manufacturer's instructions. For all staining procedures, the cells were blocked with a solution containing 0.5% bovine serum albumin, 1 mM EDTA to reduce nonspecific antibody–antigen interactions. The antibody was incubated with the cells for 30 min at room temperature and then washed to remove unbound antibody. For all staining methods the cells were mounted on the slide with ProLong Antifade kit (Molecular Probes) according to manufacturer's instructions. The cells were examined for fluorescence under an Axiovert S 100 microscope (Zeiss) and photographed.
Acknowledgements
Part of this study was supported by ETSU faculty startup funding to M.D.K. and National Institutes of Health grant NIH GM53552 to D. L.W. C. S. and A. S. were supported by grants from the Swedish Research Council, the Swedish Asthma and Allergy Association's Research Foundation, and through the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and the Karolinska Institutet. Y.H.C. and M.A.M. were supported by the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Latge JP. Mol. Microbiol. 2007;66:279–290. doi: 10.1111/j.1365-2958.2007.05872.x. [DOI] [PubMed] [Google Scholar]
- 2.Anas A, Lowman DW, Williams DL, Millen S, Pai SS, Sajeevan TP, Philip R, Singh ISB. Aquacult. Res. 2009 Published Online: Apr 29 2009. [Google Scholar]
- 3.Hogan LH, Klein BS. Infect. Immun. 1994;62:3543–3546. doi: 10.1128/iai.62.8.3543-3546.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klimpel KR, Goldman WE. Infect. Immun. 1998;56:2997–3000. doi: 10.1128/iai.56.11.2997-3000.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaffin WL, López-Ribot JL, Casanova M, Gozalbo D, Martínez JP. Microbiol. Mol. Biol. Rev. 1998;62:130–80. doi: 10.1128/mmbr.62.1.130-180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashbee HR, Evans EGV. Clin. Microbiol. Rev. 2002;15:21–57. doi: 10.1128/CMR.15.1.21-57.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheynius A, Crameri C. In: Malassezia and the Skin. Velegraki A, Mayser P, Boekhout T, editors. Springer Verlag; Heidelberg: 2009. in press. [Google Scholar]
- 8.Cabañes FJ, Theelen B, Castellá G, Boekhout T. FEMS Yeast Res. 2007;7:1064–1076. doi: 10.1111/j.1567-1364.2007.00217.x. [DOI] [PubMed] [Google Scholar]
- 9.Hechemy KE, Vanderwyk RW. Bacteriolog. Proc. 1968;68:42. [Google Scholar]
- 10.Shepherd MG, Gopal PK. In: Candida and Candidamycosis. Tunbay E, Seeliger HPR, Ang O, editors. Plenum Press; New York: 1991. pp. 21–33. [Google Scholar]
- 11.Thompson E, Colvin JR. Can. J. Microbiol. 1970;16:263–265. doi: 10.1139/m70-048. [DOI] [PubMed] [Google Scholar]
- 12.David M, Gabriel M, Kopecká M. Cell. Biol. Int. 2007;31:16–23. doi: 10.1016/j.cellbi.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Mittag H. Mycoses. 1995;38:13–21. doi: 10.1111/j.1439-0507.1995.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 14.Gow NA, Netea MG, Munro CA, Ferwerda G, Bates S, Mora-Montes HM, Walker L, Jansen T, Jacobs L, Tsoni V, Brown GD, Odds FC, Van der Meer JW, Brown AJ, Kullberg BJ. J. Infect. Dis. 2007;196:1565–1571. doi: 10.1086/523110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reid DM, Gow NA, Brown GD. Curr. Opin. Immunol. 2009;21:30–37. doi: 10.1016/j.coi.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, Haynes K, Steele C, Botto M, Gordon S, Brown GD. Nat. Immunol. 2007;8:31–38. doi: 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van de Veerdonk FL, Marijnissen RJ, Kullberg BJ, Koenen HJ, Cheng SC, Joosten I, van den Berg WB, Williams DL, van der Meer JW, Joosten LA, Netea MG. Cell Host Microbe. 2009;5:329–340. doi: 10.1016/j.chom.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Werner JL, Metz AE, Horn D, Schoeb TR, Hewitt MM, Schwiebert LM, Faro-Trindade I, Brown GD, Steele C. J. Immunol. 2009;15:4938–4946. doi: 10.4049/jimmunol.0804250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowman DW, Williams DL. J. Agri. Food. Chem. 2001;49:4188–4191. doi: 10.1021/jf010435l. [DOI] [PubMed] [Google Scholar]
- 20.Lowman DW, Ferguson DA, Williams DL. Carbohydr. Res. 2003;338:1491–1496. doi: 10.1016/s0008-6215(03)00169-1. [DOI] [PubMed] [Google Scholar]
- 21.Williams DL, McNamee RB, Jones EL, Pretus HA, Ensley HE, Browder IW, Di Luzio NR. Carbohydr. Res. 1991;219:203–213. doi: 10.1016/0008-6215(91)89052-h. [DOI] [PubMed] [Google Scholar]
- 22.Kocourek J, Ballou C. J. Bacteriol. 1969;100:1175–1181. doi: 10.1128/jb.100.3.1175-1181.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shibata N, Kobayashi H, Okawa Y, Suzuki H. Eur. J. Biochem. 2003;270:2565–2575. doi: 10.1046/j.1432-1033.2003.03622.x. [DOI] [PubMed] [Google Scholar]
- 24.Adams EL, Rice PJ, Graves B, Ensley HE, Yu H, Brown GD, Gordon S, Monteiro MA, Papp-Szabo E, Lowman DW, Power TD, Wempe MF, Williams DL. J. Pharmacol. Exp. Ther. 2008;325:115–123. doi: 10.1124/jpet.107.133124. [DOI] [PubMed] [Google Scholar]
- 25.Sawardeker JS, Sloneker JH, Allene J. Anal. Chem. 1967;37:1602–1604. [Google Scholar]
- 26.Ciucanu I, Kerek F. Carbohydr. Res. 1984;131:209–217. [Google Scholar]
- 27.Monteiro MA, Slavic D, Michael F, Brisson JR, MacInnes JI, Perry MB. Carbohydr. Res. 2000;329:121–130. doi: 10.1016/s0008-6215(00)00148-8. [DOI] [PubMed] [Google Scholar]
- 28.Yamasaki S, Matsumoto M, Takeuchi O, Matsuzawa T, Ishikawa E, Sakuma M, Tateno H, Uno J, Hirabayashi J, Mikami Y, Takeda K, Akira S, Saito T. Proc. Natl. Acad. Sci. USA. 2009;106:1897–1902. doi: 10.1073/pnas.0805177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Bernardis F, Liu H, O'Mahony R, La Valle R, Bartollino S, Sandini S, Grant S, Brewis N, Tomlinson I, Basset RC, Holton J, Roitt IM, Cassone A. J. Infect. Dis. 2007;195:149–157. doi: 10.1086/509891. [DOI] [PubMed] [Google Scholar]
- 30.Pietrella D, Bistoni G, Corbucci C, Perito S, Vecchiarelli A. Cell. Microbiol. 2006;8:602–612. doi: 10.1111/j.1462-5822.2005.00651.x. [DOI] [PubMed] [Google Scholar]
- 31.Pietrella D, Lupo P, Rachini A, Sandini S, Ciervo A, Perito S, Bistoni F, Vecchiarelli A. Infect. Immun. 2008;76:4359–4367. doi: 10.1128/IAI.00669-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McFadden D, Zaragoza O, Casadevall A. Trends Microbiol. 2006;14:497–505. doi: 10.1016/j.tim.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Thomas DS, Ingham E, Bojar RA, Holland KT. FEMS Immunol. Med. Microbiol. 2008;54:203–214. doi: 10.1111/j.1574-695X.2008.00468.x. [DOI] [PubMed] [Google Scholar]
- 34.Iorio E, Torosantucci A, Bromuro C, Chiani P, Ferretti A, Giannini M, Cassone A, Podo F. Carbohydr Res. 2008;343:1050–1061. doi: 10.1016/j.carres.2008.02.020. [DOI] [PubMed] [Google Scholar]