Abstract
Genus Macaca (Cercopithecidae: Papionini) is one of the most successful primate radiations. Despite previous studies on morphology and mitochondrial DNA analysis, a number of issues regarding the details of macaque evolution remain unsolved. Alu elements are a class of non-autonomous retroposons belonging to short interspersed elements that are specific to the primate lineage. Because retroposon insertions show very little homoplasy, and because the ancestral state (absence of the SINE) is known, Alu elements are useful genetic markers and have been utilized for analyzing primate phylogenentic relationships and human population genetic relationships. Using PCR display methodology, 298 new Alu insertions have been identified from ten species of macaques. Together with 60 loci reported previously, a total of 358 loci are used to infer the phylogenetic relationships of genus Macaca. With regard to earlier unresolved issues on the macaque evolution, the topology of our tree suggests that: 1) genus Macaca contains four monophyletic species groups; 2) within the Asian macaques, the silenus group diverged first, and members of the sinica and fascicularis groups share a common ancestor; 3) Macaca arctoides are classified in the sinica group. Our results provide a robust molecular phylogeny for genus Macaca with stronger statistical support than previous studies. The present study also illustrates that SINE-based approaches are a powerful tool in primate phylogenetic studies and can be used to successfully resolve evolutionary relationships between taxa at scales from the ordinal level to closely related species within one genus.
Keywords: Macaque, Primate, Phylogeny, Retroposons
1. Introduction
The Old World monkey genus Macaca (Cercopithecidae: Papionini) represents one of the most successful radiations of anthropoid primates. About 20-22 (differs depending on the definition of different authors) species of macaques have been recognized in this genus. They are widely distributed in southern and eastern Asia, with the exception of the Barbary macaque in northern Africa (Fooden, 1982). While the earliest known fossil macaques date to around 5.5 million years ago in North Africa and Europe, they are more recent in Asia (Delson et al., 2000). Molecular estimates based on complete mitochondrial genomes and calibrated with several reasonably well accepted fossil divergent times suggest the divergence of the macaques from other members of the tribe Papionini approximately 9-10 million years ago (Raaum et al., 2005). They probably entered Eurasia via northeast Africa ~5 mya. Subsequently, the Asian macaque lineage separated into three or four species groups less than 3 mya (Tosi et al., 2003). Therefore, the radiation of this genus has taken place relatively recently, within the last 5 million years, and yet the number of species that has emerged is unequalled by any other group of primates. With such a variety of forms and species that differ in morphology and ecology, macaques represent a most alluring group for the study of species radiation and evolution. An accurate phylogeny of the extant taxa of macaques is critical, not only to our understanding of the evolutionary history of the genus, but to our general understanding of primate radiations.
Species of Macaca have been variously separated into several species groups according to different authors (Fooden, 1976; Delson, 1980; Fa, 1989; Groves, 2001). On the basis of male genitalia morphology, Fooden (1976) classified the macaques into four species groups: the silenus-sylvanus group, including M. sylvanus, M. silenus, M. nemestrina and Sulawesi macaques; the fascicularis group, including M. fascicularis, M. mulatta, M. fuscata, and M. cyclopis; the sinica group, including M. sinica, M. radiata, M. assamensis and M. thibetana; and the arctoides group, including M. arctoides. Delson (1980) modified this classification by placing M. arctoides as a member of the sinica group and removing M. sylvanus from silenus group to form a sister taxon to all of the Asian groups. Groves (2001) divided the genus into six species group with Sulawesi macaques in their own group and a separated the mulatta group as a monophyletic group. Despite the discrepancy in the species group definitions, a consensus has been reached on the existence of at least three groups, including the silenus group, the sinica group and the fascicularis group (Delson, 1980; Fooden and Lanyon, 1989; Hayasaka et al., 1996). The taxonomic positions of M. sylvanus, M. arctoides and Sulawesi macaques are still debated as to whether they belong to a monophyletic group that includes other extant macaques, or they should be classified in their own group as a sister to all other macaques.
Previously, macaque systematics and evolution was largely based on morphological characters, such as the male and female reproductive organs (Fooden, 1976; Fooden, 1980; Fooden and Lanyon, 1989), or features of the dentition and cranium (Delson, 1980). Sources of molecular data for the reconstruction of macaque phylogeny have included allozymes (Melnick and Kidd, 1985; Fa, 1989), mtDNA sequences (Zhang and Shi, 1993; Li and Zhang, 2005; Smith et al., 2007) and nuclear markers (Deinard and Smith, 2001; Tosi et al., 2002; Tosi et al., 2003). Despite the general consensus, the phylogenetic relationships among the different species and species groups have not been conclusive. A number of issues remain to be clarified. First, the number of species groups in the genus Macaca, especially in the Asian macaques is not constant; Secondly, the phylogenetic relationship among the Asian species groups has been problematic. Whether they represent different monophyletic assemblages sharing a trichotomous relationship (Delson, 1980; Tosi et al., 2000), or there is a bifurcation between the silenus group progenitor and a M. fascicularis-like taxon, with the latter representing the probable common ancestor to all non-silenus group Asian macaques (Morales and Melnick, 1998; Tosi et al., 2003). Thirdly, nearly all of previous studies propose a different phylogenetic position of M. arctoides. It is either ascribed to its own species group (Fooden, 1976; Hoelzer et al., 1992) or falling into the sinica group (Delson, 1980; Tosi et al., 2000), or being classified in the fascicularis group (Hayasaka et al., 1996; Morales and Melnick, 1998).
SINEs (short interspersed elements) are a class of non-autonomous mobile elements that insert into a genome via RNA intermediates and are usually <500bp in length. These elements not only play an important role in the shaping of the genome, but are also useful markers in phylogenetic studies. Compared to other molecular markers, SINE insertions have several important advantages making them particularly promising in systematic and evolutionary studies. Shedlock et al. (2000) and Ray et al. (2006) reviewed those advantages in detail. Briefly, SINEs are nearly homoplasy-free markers. The presence of an element in an individual is thought to represent a unique evolutionary event, sharing identity by descent (Batzer et al., 1991; Perna et al., 1992; Murata et al., 1993; Batzer et al., 1994; Stoneking et al., 1997; Batzer and Deininger, 2002). Thus, individuals found sharing a SINE at an orthologous position do so because of common ancestry. Second, the insertion of a SINE is usually unidirectional with the absence of the insertion being the ancestral state. They are relatively stable genetic markers and precise removal of SINEs is extremely rare once they have integrated within the genome (Edwards and Gibbs, 1992; van de Lagemaat et al., 2005). Third, SINEs are easy to genotype using only PCR-based assays as compared to other genetic markers such as SNPs (single nucleotide polymorphisms), or longer nuclear or mtDNA sequences. Since early investigations of fish phylogeny and population biology using SINE-based methodology (Kido et al., 1991; Murata et al., 1993; Takahashi et al., 1998), numerous studies using SINEs as phylogenetic and population genetic characters in a wider variety of taxa have been published (Hamdi et al., 1999; Shedlock and Okada, 2000; Bamshad et al., 2003; Watkins et al., 2003; Roos et al., 2004; Ray et al., 2005; Chakraborty et al., 2007; Xing et al., 2007a; Xing et al., 2007b). These studies have produced very successful results in solving controversial phylogenetic relationships that have not been resolved using traditional molecular markers.
Alu elements are primate-specific SINEs with a full-length of ~300bp long. They are the most abundant SINEs in primates and comprise ~10% of primate genomes (Lander et al., 2001; Gibbs et al., 2007; Han et al., 2007). Alu elements have been extensively used in the study of primate phylogenetic relationships, including the human, chimpanzee and gorilla trichotomy (Salem et al., 2003), Old World monkey (Xing et al., 2005; Xing et al., 2007a) and New World monkey (Ray et al., 2005; Osterholz et al., 2009) phylogenies, the tarsier affiliation (Schmitz et al., 2001) and the strepsirrhine phylogeny (Roos et al., 2004). However, to date, no mobile element based phylogenetic analysis has been conducted to resolve the relationships within the macaques. In this study, PCR display methodology was employed to identify potentially informative Alu insertions in the genomes of ten macaque species and 298 new Alu insertions were identified. Together with 60 previously reported Alu insertions (Xing et al., 2005; Han et al., 2007), a total of 358 Alu insertions were used to reconstruct a robust phylogeny of the genus Macaca.
2. Materials and Methods
2.1. PCR display methodology and genotyping
The PCR display methodology used in this analysis was described in detail by Ray et al. (2005) and Xing et al. (2005). In summary, genomic DNAs (~500 ng) were digested using an NdeI, restriction endonuclease, as recommended by the manufacturer (New England Biolabs, Beverly, MA). The digested genomic DNAs were then ligated with double stranded linkers (Table 1) and amplified by using the LNP primer and a rhesus Alu specific primer (Alu YdI or Alu YbI) in order to obtain partial Alu sequences with the accompanying flanking unique sequences from each template. A second round amplification was performed using the LNP primer and a second nested rhesus Alu specific primer (Alu YdII or Alu YbII) to increase the specificity of the amplicons. The Alu specific primers were designed according to the consensus sequences of the rhesus Alu Yb and Alu Yd subfamilies (Han et al., 2007). All primers used for PCR display are listed in Table 1. Seven macaque species were subjected to this methodology to identify potentially informative Alu insertions, including M. sylvanus, M. silenus, M. nigra, M. arctoides, M. thibetana, M. radiata, and M. fascicularis. Informative Alu insertions from four other species, M. mulatta, M. fuscata and M. nemestrina and Papio hamadryas were obtained by testing primers reported previously by Han et al. (2007) and Xing et al. (2005).
Table 1.
Linkers and primers sequences used for PCR display methodology
| Name | Sequence (5'-3') |
|---|---|
| Linkers | |
| Top | TAGAAGGAGAGGACGCTGTCTGTCGAAGG |
| Bottom | GAGCGAATTCGTCAACATAGCATTTCTGTCCTCTCCTTC |
| Primers | |
| LNPa | GAATTCGTCAACATAGCATTTCT |
| Alu YdIb | GACGGAGTCTCGCTGTGTCT |
| Alu YdIIb | GTTTCACCATGTTAGCCAGGATA |
| Alu YbIb | GACGGAGTCTCACGCTGTT |
| Alu YbIIb | AAGCTCCGCCTCCCGGGTTCC |
linker primer
Alu-specific primer
All oligonucleotide primer pairs were initially tested for amplification on the rhesus macaque (M. mulatta) DNA templates using temperature gradient PCR (48-60 °C) to determine the most appropriate annealing temperature. The primers were then tested on a macaque phylogenetic panel that was composed of DNA samples from ten macaque species and P. hamadryas, which was used as an outgroup (Table 2). All loci that were successfully amplified were screened on the macaque panel to identify the informative loci. For those taxa for which limited amounts of genomic DNA was available, a whole genome pre-amplification protocol was performed following the protocol for the GenomiPhi genome amplification kit (Amersham, Sunnyvale, CA). The genome amplified products were then used as templates for allele specific PCR analysis.
Table 2.
DNA samples of all species examined in this study
| Species Names | Common Names | Origin | ID number |
|---|---|---|---|
| Macaca mulatta | Rhesus macaque | SFBRa | 17573BRNY |
| Macaca nemestrina | Pig-tailed macaque | Coriellb | NG08452 |
| Macaca nigra | Celebes crested macaque | Coriellb | NG07101 |
| Macaca arctoides | Stump-tailed macaque | M. Rocchic | N/A |
| Macaca sylvanus | Barbary macaque | M. Rocchic | N/A |
| Macaca fascicularis | Crab-eating macaque | H.-S. Kimd | 1155(♀) |
| Macaca radiata | Bonnet macaque | H.-S. Kimd | 2831(♂) |
| Macaca thibetana | Tibetan macaque | B. Yuee | TM1 |
| Papio hamadryas | Hamadryas baboon | T. Disotellf | BE97089(♂) |
| Macaca silenus | Lion-tailed macaque | SDZIg | OR1890 |
| Macaca fuscata | Japanese macaque | IPBIRh | PR00782 |
DNA samples from Dr. Jeffrey Rogers, Southwest Foundation for Biomedical Research, P.O. Box 760549, San Antonio, TX 78245, USA
Coriell Institute for Medical Research, 403 Haddon Avenue, Camden, NJ 08103, USA
DNA samples from Dr. Mariano Rocchi, Primate Cytogenetics, Dip. di Genetica e Microbiologia, Via Amendola 165/A 70126 Bari, Italy
DNA sample from Dr. Heui-Soo Kim, Pusan National University, Busan 609-735, Korea
DNA sample from Sichuan Key Laboratory of Conservation Biology on Endangered Wildlife, Chengdu 610064, China
DNA samples from Dr. Todd Disotell, Department of Anthropology, New York University, New York, NY 10003, USA
San Diego Zoo's Institute for Conservation Research, San Diego, California 92112, USA
Integrated Primate Biomaterials and Information Resource, 403 Haddon Avenue, Camden, NJ 08103, USA
2.2. PCR and DNA sequencing
PCR amplification for each locus was performed in a 25μl reaction mixture containing 25ng of genomic DNA, 200 nM of each oligonucleotide primer, 200mM dNTPs, in 50mM KCl, 1.5mM MgCl2, 10mM Tris-HCl (pH 8.4), and 2.5U Taq DNA polymerase. PCR reaction conditions were as follows: an initial denaturation step of 94 °C for 2 min, followed by 32 cycles of denaturation at 94 °C for 30 sec, annealing at indicated annealing temperature for 30 sec, extension at 72 °C for 1 min and 30 sec, followed by a final extension step at 72 °C for 5 min. PCR products were run on a 2% agarose gel with 0.25 μg ethidium bromide and visualized using UV fluorescence (Bio-Rad, Hercules, CA). Detailed information on each locus including primer sequences, annealing temperature, PCR product sizes, chromosomal locations and amplification results are available on the Batzer Laboratory Web site (http://batzerlab.lsu.edu) under supplemental data.
To confirm the phylogenetic distribution of Alu insertions on the macaque panel, filled and empty loci from selected taxa were chosen for sequencing analysis. In addition, when the PCR amplification patterns were different from that suggested by the majority of genetic systems analyzed, representative PCR products were selected for DNA sequence analysis to identify the source of the disparity. Individual PCR products were cloned and sequenced as described previously (Ray et al., 2005). Sequences for loci identified experimentally were aligned against the orthologous rhesus macaque genome sequence (rheMac2; http://genome.brc.mcw.edu/cgibin/hgBlat) obtained via the BLAST-like alignment tool (BLAT) search. The DNA sequences generated during this project have been deposited in GenBank under accession numbers FJ978786-FJ979008.
2.3. Phylogenetic analysis
Alu insertion loci were included in phylogenetic analysis if clearly distinguishable products (either empty or filled) were amplified from at least eight of the eleven taxa in our panel. Primer pairs that generated multiple paralogous fragments across the panel were excluded from our analysis. Six examples of gel electrophoresis patterns of amplification results are shown in Fig. 1. Phylogenetic analysis was performed as described by Xing et al. (2005) by implementing an exhaustive search via the PAUP*4.0b10 software (Swofford, 2003) using Dollo parsimony analysis and designating P. hamadryas as an outgroup taxon. For a few loci containing adjacent independent insertion events (see Discussion), the independent insertions were treated as independent markers. In total, 10,000 bootstrap replicates were performed on the data. A statistical test for evaluating SINE insertions based on a likelihood model (Waddell et al., 2001) was also employed to assess the statistical significance of each branch of the resulting tree.
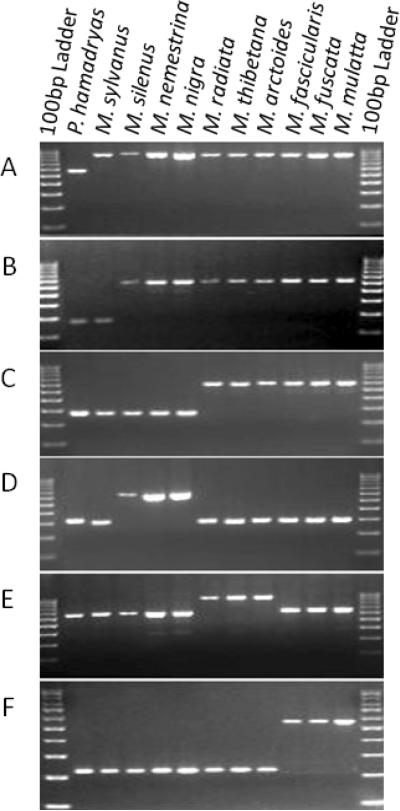
Figure 1. PCR amplification analysis of Alu insertion polymorphisms in Macaca.
Six gel electrophoresis results on Macaca species are shown with P. hamadryas as an outgroup.
A: Amplification of locus Han1_b4, an Alu insertion specific to macaques.
B: Amplification of locus Mni_d1t_60, an Alu insertion shared by all Asian macaques.
C: Amplification of locus YRb2_13, an Alu insertion clustering the sinica group and the fascicularis group.
D: Amplification of locus MS_b1_155, an Alu insertion specific to the silenus group.
E: Amplification of locus MA_b1b_72, an Alu insertion specific to the sinica group.
F. Amplification of locus MacYb5_25, an Alu insertion specific to the fascicularis group.
3. Results
3.1. Phylogeny of the macaques
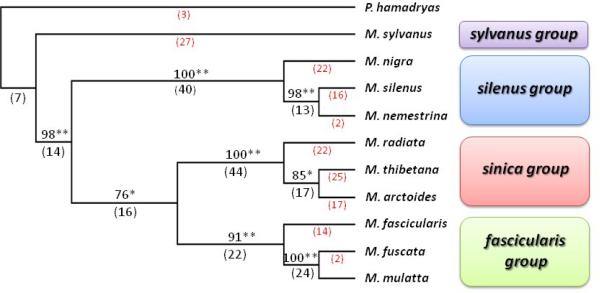
In total, 345 Alu insertion loci were found to be potentially useful for phylogenetic analysis. Of these, 285 loci were obtained from different macaque taxa by PCR display methodology while the remaining 60 loci were recovered from three macaque taxa previously reported by Han et al. (2007) and Xing et al. (2005). In addition, sequencing verification showed that twelve of the 345 loci contained two adjacent independent Alu insertion events, and one locus contained three adjacent independent Alu insertion events. At these loci, two or three different Alu elements have inserted independently in the genomes of different species that were within sufficiently close genomic proximity that they be amplified by a single set of PCR primers. These loci were treated as independent markers for phylogenetic analysis. Thus, a total of 358 loci were identified from 11 taxa and used for phylogenetic analysis. Among them, 197 loci were found to be parsimony informative, and generated a single most parsimonious tree (Fig. 2; consistency index (CI) =0.7732; homoplasy index (HI) =0.2268; retention index (RI) =0.7125). The likelihood test for every branch was significant at the 0.001 level and 0.05 level as illustrated in Fig. 2.
Figure 2. Macaque phylogenetic relationships derived from 358 Alu insertion polymorphisms.
The amplification patterns of the Alu insertions were used to construct a dollo parsimony tree of macaque phylogenetic relationships using P. hamadryas as an outgroup and PAUP*4.0b10 program. The numbers above the branches indicate the percentage of bootstrap replicates (10,000 iterations) producing trees that including that node. The Numbers below the branches indicate the numbers of unambiguous insertions supporting each node. The numbers in red are insertions that are present in only one species. These loci are not parsimony informative. The significance level of each node is indicated by either * (P<0.05) or ** (P<0.001).
A total of seven insertions were present in all Macaca species but not in the P. hamadryas, supporting the monophyly of the genus Macaca. The topology of our tree clearly defines four distinct species groups, sylvanus, silenus, sinica and fascicularis groups within the genus. This result is congruent with the classification of Delson (1980) and several previous molecular phylogenetic studies (Tosi et al., 2000; Li and Zhang, 2005), whereas our results provide much stronger statistical support for each species group. Thirty-four Alu insertions are found only in M. sylvanus thus supporting a sylvanus group, with the only African member of the genus as a sister clade to all Asian macaques. Fourteen unambiguous insertions support all Asian species clustering together into a monophyletic assemblage. Within the Asian macaques, three groups that have been previously established are divided into two assemblages: the silenus group and the proto-sinica-fascicularis group. The silenus group consisting of M. silenus, M. nemestrina and M. nigra forms a monophyletic clade, which is strongly supported by 40 Alu insertions. This clade is joined as a sister clade by the proto-sinica-fascicularis assemblage, which is supported by 16 Alu insertions. The proto-sinica-fascicularis assemblage then experienced a radiation to the extant sinica and fascicularis groups.
Within the silenus group, thirteen Alu insertions support M. silenus and M. nemestrina sharing a closer relationship and suggesting an earlier divergence of the M. nigra in the group. These two species form a sister clade to M. nigra. Fourty-four Alu insertions support a monophyletic sinica group including M. radiata, M. thibetana and M. arctoides. Within the sinica group, a bifurcation is apparent between M. radiata lineage and the other two members. Fifteen Alu insertions support that M. thibetana and M. arctoides share a closer relationship than either does with M. radiata. Three species from our sample fall into the fascicularis group, linked by 22 Alu insertions. Among these species, 33 Alu insertions are unique to M. fascicularis. This clade is joined as a sister clade by an assemblage containing M. fuscata and M. mulatta, which is itself supported by 24 Alu insertions.
3.2. Incongruent PCR patterns of Alu insertions
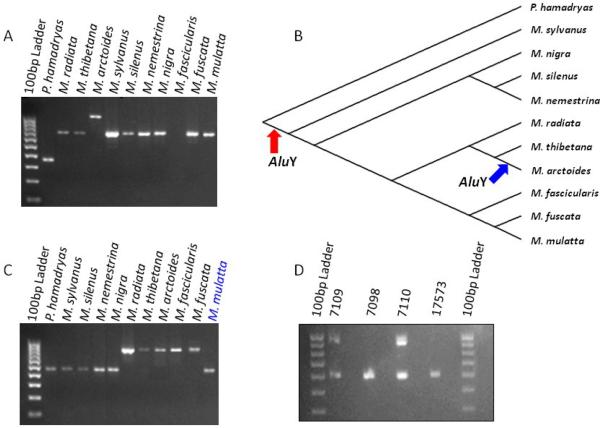
When a SINE-based phylogenetic analysis is used, several confounding events may occur to disrupt the interpretation of the tree topology (see Discussion). Although vast majority Alu insertions in our study support a single most parsimonious tree, it is noteworthy that PCR amplification patterns in several loci appeared to be incongruent with our tree (see Supplemental Table 1). To confirm the phylogenetic distribution of Alu insertion patterns and to determine the nature of the incongruent loci, we sequenced a total of 223 amplicons from 345 loci, either filled or empty (as indicated in the Supplemental Table 1). Among them, twelve loci result from adjacent independent insertion events. Locus MAb1_b_56 provides an example of adjacent independent insertions event (Fig. 3A, B). The sequencing results indicate that this locus contains two independent Alu insertions. One Alu element inserted after macaques diverged from baboons and is shared by all macaque species with no Alu element observed in P. hamadryas. The second Alu inserted specifically into M. arctoides genome, just 13-bp upstream of the first Alu element. These loci were treated as two independent markers in the analysis.
Figure 3. Potential confounding Alu insertions.
Two examples of incongruent loci are shown. The upper part of the figure shows an adjacent independent insertion event occurring at locus MAb1_b_56. Picture (A) shows an agarose gel electrophoresis result with the final tree superimposed on the (B). Two arrows (red and blue) on the tree indicate Alu element insertion events. The two Alu elements independently insert in different lineages of macaque genomes. The integration sites are separated by 13-bp. The first AluY (red arrow) is shared by all macaque species; the second AluY (blue arrow) specifically inserted in M. arctoides genome.
The bottom part of the figure shows an incomplete lineage sorting occurring at locus JH70. (C) is an agarose gel electrophoresis analysis of our macaque diversity panel. One Alu element is shared by members of sinica group and fascicularis group except for M. mulatta. (D) is the agarose gel electrophoresis result from four individual M. mulatta samples. It is polymorphic in the population of M. mulatta. ID 17573BRNY is the individual that was subjected to rhesus macaque reference genome (rheMac2).
In addition to the adjacent independent insertion events, we found nine loci that appear to contain the same Alu insertion in the sequenced taxa but support an alternative phylogeny different from the final tree (Supplemental Table 1). Several factors, including the close relationship among the macaque species under investigation, interspecific hybridation among macaque species, incomplete lineage sorting and concurrent polymorphism may have contributed to this result. Another incongruent PCR pattern found at locus JH70 is an example of incomplete lineage sorting event. At locus JH70, an Alu element identified by PCR display methodology from the M. thibetana genome, was found present in M. thibetana and all members of the sinica and fascicularis groups except M. mulatta. The amplification pattern presented at this locus is incongruent with the topology of our tree. When we amplified this locus using four rhesus macaque individuals including a sample of ID 17573BRNY, the locus are found to be polymorphic among individuals, with an Alu present as heterozygous in IDs 7109 and 7110 and absent in IDs. 7098 and 17573 (Fig. 3C, D).
4. Discussion
4.1. Phylogeny of macaque species group
Our results based on Alu elements clearly define four groups that are largely congruent with Delson's (1980) classification. Although the present study did not include any sample from Sulawesi macaques, we sampled each of the four extant species groups. There is little doubt that the Sulawesi group represents a monophyletic clade that clusters with M. nemestrina based upon mitochondrial, Y chromosome, and autosomal sequences (Tosi et al., 2003). With respect to the previous studies, our results are strongly supported not only phylogenetically, with long branches leading to each species group, but also cladistically, with much higher bootstrap values (Fig. 2).
With respect to the relationships among species groups, no strong evidence to date has been presented to support a consistent phylogenetic tree connecting the different species groups. Previous investigations have proposed scenarios regarding the macaque phylogeny based on morphology, allozymes (Cronin et al., 1980; Fooden and Lanyon, 1989); mitochondrial DNA (mtDNA) (Hoelzer et al., 1992; Hayasaka et al., 1996; Morales and Melnick, 1998; Li and Zhang, 2004; Li and Zhang, 2005); Y-chromosomal and autosomal DNA (Tosi et al., 2000; Deinard and Smith, 2001; Tosi et al., 2003) markers. However, when different genetic markers were used, conflicting hypothesis have been generated concerning several branches leading to different species groups (Tosi et al., 2000; Tosi et al., 2003). For example, two closely linked Y-chromosome markers, TSPY (testis-specific protein, Y-encoded) and SRY (sex-determining region Y-chromosome) were used by Tosi et al. (2000) to determine the phylogenetic relationship among 18 macaque species. Four main clades are depicted, but their tree failed to resolve the polytomy between the four species groups. Another study by Tosi et al. (2003) based on autosomal markers (C4 intron 9 and IRBP intron 3) only showed low levels of resolution. The bootstrap values of many branches in their tree are low. This is probably due to the low diversity of the nuclear markers they used. These markers, such as TSPY, a protein coding gene, may be under strong selection and thus may be less appropriate for the reconstruction of recent phylogenetic relationships. Therefore, more neutral and homoplasy-free nuclear markers are needed to accurately elucidate phylogenetic relationships among macaque species groups. In this study we report the first large scale Alu element based phylogenetic study of genus Macaca. Our results suggest that the first species group that diverged from the rest of clade is the sylvanus group (M. sylvanus). The Asian macaques diverged from a common ancestor with M. sylvanus, followed by a split between the silenus group members and the proto-sinica-fascicularis group. A subsequent divergence separated the monophyletic sinica group from the fascicularis group. The topology of our tree supports Delson's (1980) species group relationships based on morphology, and is generally consistent with earlier studies based on mtDNA analysis with the exception of the position of M. arctoides (Morales and Melnick, 1998; Li and Zhang, 2005).
In contrast to the earlier studies of Morales and Melnick (1998) and Tosi et al. (2003), which indicated that the fascicularis group is a paraphyletic assemblage giving rise to all non-silenus groups including the sinica group, our results show a sister relationship between the fascicularis group and the sinica group. Hayasaka's (1996) results suggest a closer relationship between these two Asian groups, while the topology in that study showed a monophyletic fascicularis species group derived from a polyphyletic sinica group assemblage. The phylogenetic relationships of the fascicularis and sinica groups depicted in our tree are in agreement with Li's (2005), which was based on mtDNA analysis. Overall, our results are consistent with the following scenario: after the macaques entered Asia, they diverged into a silenus group and a proto-fascicularis-sinica group. All members of the fascicularis and the sinica groups were derived from a common ancestor, followed by an expansion and radiation into the two monophyletic extant species groups. In addition, within the fascicularis group, our results indicate that the M. fascicularis lineage was first to diverge from a common ancestor, after which the other members of the group (M. mulatta and M. fuscata) diverged. The evolutionary relationship among fascicularis group members is generally consistent with previous hypothesis based on morphology as well as mtDNA data. (Fooden and Lanyon, 1989; Smith et al., 2007).
4.2. The phylogenetic position of M. arctoides
The phylogenetic position of M. arctoides, a taxon unique in reproductive behavior and morphology, is one of the most hotly debated questions in the macaque phylogeny. According to morphological analysis, Delson (1980) suggests that M. arctoides should be included in the sinica group because the modifications of the glans penis in M. arctoides represent an extreme of the sagittate form already present in the sinica group. Moreover, the extensive similarities: such as allozyme frequencies (Cronin et al., 1980; Fooden and Lanyon, 1989; Fooden, 1990), hair growth patterns (Fooden, 1988; Fooden, 1990; Inagaki, 1996), and craniofacial structure (Delson, 1980; Fooden, 1990), that are found between M. arctoides and the sinica group members suggest that M. arctoides is closely related to the sinica group, specifically to M. thibetana.
Compared to the consistent results based on morphology, the molecular data with respect to the position of M. arctoides, are less concordant. Hoelzer et al. (1992) suggested a sister relationship of M. arctoides with the sinica group based on mtDNA restriction sites. However, it appears that M. arctoides fell outside the rest of the sinica group because they only included M. nemestrina as an outgroup in this phylogenetic analysis. Nearly all subsequent studies based on mtDNA data indicate that M. arctoides is more closely associated with the fascicularis group than the sinica group and should be classified into the fascicularis group (Hayasaka et al., 1996; Tanaka and Takenaka, 1996; Morales and Melnick, 1998; Tosi et al., 2003; Li and Zhang, 2005). By contrast, macaque phylogenetic analyses based on nuclear DNA markers, including Y-chromosomal and autosomal genes, consistently agree with morphological studies in assigning M. arctoides to the sinica group (Tosi et al., 2000; Deinard and Smith, 2001; Tosi et al., 2003).
One of the most distinct differences between our tree and previous mtDNA phylogenetic trees is the position of M. arctoides. The Alu insertion data strongly supports (with a bootstrap value of 100%) the morphological studies and nuclear DNA studies arguing a monophyletic sinica group including M. arctoides. With respect to other species groups, 44 Alu insertions are shared by the three members within the sinica group. This is to date the most robust support placing M. arctoides within the sinica group. Furthermore, our results define a clear position of M. arctoides within the sinica group. It is neither a primitive sister taxon (Purvis, 1995; Chakraborty et al., 2007), nor a paraphyletic clade to all other sinica members (Tosi et al., 2000; Tosi et al., 2003). M. arctoides exhibits a closer relationship to M. thibetana than to M. radiata.
The unusual discrepancy about the position of M. arctoides between the mtDNA and the nuclear DNA topologies has been noted by previous studies. After investigating and comparing the position of M. arctoides in the Y-chromosomal DNA tree, mitochondrial DNA tree and autosomal DNA tree derived from the same of macaque individuals, Tosi et al. (2000, 2003) suggested a possible hybrid origin of M. arctoides. They concluded that extensive hybridization between proto-M. assamensis/thibetana and proto-M. fascicularis in a Pleistocene forest refugium may have given rise to a unique entity that is M. arctoides. However, they also mentioned that this hybrid hypothesis awaits an extensive survey of autosomal markers.
Our results are consistent with a particular type of hybrid origin for M. arctoides. The majority of Alu insertions examined here support a close relationship between M. arctoides and the silica group. If the hybrid origin of M. arctoides were the result of population fusion consisting of equal contributions from an early fascicularis species and an early silica species, then we would expect approximately half of the Alu insertions to link M. arctoides to the silica group, and half to link it to the fascicularis group. We find that the clear majority of insertions (44 Alu insertions) associate M. arctoides with the sinica group, while only two Alu insertions (GROUP6&7_17 and YdJXRh20) link M. arctoides to the fascicularis clade. However, if M. arctoides were the result of male-mediated gene flow from an early silica group species into an early fascicularis group species, then we might expect the nuclear genome of M. arctoides to consist predominantly (but not entirely) of alleles derived from the silica group lineage. Male-mediated gene flow from a silica group species into a population of an early fascicularis group species would be expected to alter the nuclear genome, but this new hybrid population would retain the mtDNA from its fascicularis lineage matrilineal ancestor. An origin for M. arctoides based on sex-biased genetic contributions (male nuclear input primarily from silica and female mtDNA input primarily from fascicularis) might be expected to produce a new species with a nuclear genome that carries more Alu insertions derived from the silica lineage than the other, while its mtDNA would be more closely related to fascicularis group species. Gene flow between closely related species of macaques such as M. mulatta and M. fascicularis has previously been implicated to explain the high rate of overlap of SNPs from these species (Street et al., 2007). Our data are consistent with this scenario, and with the discussion presented by Tosi et al. (2003), though of course there may be other possible scenarios that are also plausible.
4.3. SINEs in phylogenetic studies
Due to their essentially homoplasy-free nature and known ancestral state, SINE-based methods have been shown to be a powerful tool in phylogenetic studies. This approach has been used to successfully resolve the phylogenetic relationships of primates among different suborders (Roos et al., 2004), families (Salem et al., 2003; Ray et al., 2005; Xing et al., 2005) and genera (Xing et al., 2007a). Using this approach, we reconstruct a robust phylogeny of the macaques that is generally consistent with previous morphological and molecular studies, but has stronger statistical support. Our study indicates that Alu based methods can be successfully adapted to lower taxonomic levels (for instance, within a genus) to resolve relationships of closely related species whose radiation and speciation have occurred very recently.
As previous studies have mentioned, there are three events that may lead to confounding results and should be treated with caution when using a SINE-based phylogenetic analysis (Ray et al., 2006). These are adjacent independent insertions, hybridization between species and incomplete lineage sorting. For the adjacent independent insertion cases, different Alu elements insert independently in close genomic proximity in different species. Therefore, genotyping using agarose gel electrophoresis of the loci at which these events occurred can be interpreted as homoplasy. In our study, when an anomalous pattern was observed compared to the overall tree, we sequenced the locus to determine if the independent insertion events had occurred. Eleven loci containing two independent Alu insertions (one example has been shown in Fig. 3) and one locus containing three independent Alu insertions were recovered in our study. After verification by automated DNA sequencing, locus Mfab1_T_8 contains three independent insertions. No Alu element is found in P. hamadryas at this locus. The first Alu element is shared by all macaques, while two additional different Alu elements inserted independently into the M. nemestrina and M. fascicularis genomes after the first Alu insertion occurred. By sequencing the informative loci, especially those with incongruent PCR amplification patterns, confounding results caused by adjacent independent insertions can be eliminated. It is noteworthy that because not every PCR product from each locus in specific species has been verified by sequencing in this study, a small number of the genotypes ascertained by PCR may still contain unidentified homoplasious information.
Introgressions via hybridization present yet another scenario leading to the disruption of interpretations of SINE-based cladogram (Churakov et al., 2009). The taxa under investigation are closely related species with radiation and speciation occurring very rapidly. In addition, the geographic distribution of these closely related macaques often overlap. Therefore, it is likely that interspecific hybridization may have occurred during the evolutionary history of these species. Hybridization among macaque species has been noted in the wild as well in captivity (Champoux et al., 1994; Evans et al., 2001). This hybridization has been observed between M. mulatta and M. fascicularis (Tosi et al., 2002). This may be one reason why our tree produced a somewhat lower consistency index and higher homoplasy index (CI=0.7732, HI=0.2268) as compared with previous phylogenetic studies based on Alu elements in less recently diverged taxa of primates (CI=0.983, HI=0.017 in Old World monkeys; CI=1 in platyrrhine; CI=0.873, HI=0.183 in guenons) (Ray et al., 2005; Xing et al., 2005; Xing et al., 2007a). As mentioned above, four Alu insertions are shared between M. thibetana, M. arctoides and M. fascicularis. This suggests that hybridization events among them could have happened. These hybridization events would have contributed to the somewhat lower index values observed between the sinica group and fascicularis group in our study.
Finally, incomplete lineage sorting is another possible cause of incongruent Alu insertion patterns. Incomplete lineage sorting is mainly caused by the presence of a polymorphic insertion in the ancestral species that alternatively becomes fixed or extinct in the genomes of descendent species. In addition, precise deletion of retroposons mediated by recombination between identical target site duplications flanking the elements could mimic incomplete lineage sorting in primate evolution but it is considered a very rare event (Edwards and Gibbs, 1992; van de Lagemaat et al., 2005). Several studies reported that incomplete lineage sorting can be particularly problematic when the taxa investigated have undergone rapid bursts of speciation (reviewed in Shedlock and Okada, 2000; Ray et al., 2006; Churakov et al., 2009). Molecular estimates suggest the divergence of the macaques from other members of the tribe Papionini approximately 9-10 million years ago (Raaum et al., 2005), and the earliest fossil macaques in Asia are less than ~5.5 mya (Delson et al., 2000). This means that broad geographic radiation and multiple speciation events occurred within only 5 million years to create the three Asian species groups containing more than 20 extant species. MtDNA data further suggests a divergence of the silenus group from the common ancestor of all other Asian species at ~4.9 mya, and a subsequent bifurcation between the fascicularis and sinica group ancestors at ~3.2 mya (Tosi et al., 2003). Therefore, the rapid speciation that occurred in the ancestral macaque populations give many Alu elements reported in the present study the potential to be polymorphic during the subsequent speciation events. These insertions will eventually fix in or be lost from the genomes of descendent species and thereby give rise to incongruent insertion patterns. In particular, more caution is warranted when a locus is polymorphic in the reference genome. One such incongruent PCR pattern found in our study resulted from a locus that is still polymorphic in the M. mulatta population (Locus JH70, Fig. 3C, D). In our study the reference genome (rheMac2) is used to target new Alu insertion by comparing PCR display ascertained sequences to the reference genome which is derived from only one rhesus macaque individual (ID number 17573BRNY, Table 2). However, the divergence of M. mulatta is estimated to have occurred from a M. fascicularis-like ancestor only ~2.5 mya (Delson, 1980; Morales and Melnick, 1998) or ~1.2 mya (Tosi et al., 2003). Due to its recent speciation, some of the Alu insertion loci may still be polymorphic in the population of M. mulatta, which may contributed to the incongruent phylogenetic patterns of some Alu insertions. However, we could not rule out that this polymorphic locus occurred by a precise Alu deletion even though this process is quite rare.
Conclusion
This study represents the first large-scale application of SINEs to study macaque phylogeny. A total 358 loci, including 298 newly identified loci and 60 loci collected from previously studies, were used to construct a robust hypothesis of macaque phylogeny. The main findings of our Alu element-based study can be distilled to the following: 1) Phylogenies of genus Macaca generally support Delson's (1980) revision of Fooden's (1976) species groups with much stronger statistical support. 2) After the silenus group diverged from other Asian macaques, the sinica and the fascicularis groups originate from a common ancestor followed by a final radiation and speciation to a monophyletic sinica group and a monophyletic fascicularis group. 3) Forty-four Alu insertions support a placement of M. arctoides within the sinica group. 4) Alu element-based approaches can be utilized to resolve phylogenetic relationships even among closely related species. Basic precautions and careful interpretation can limit the potential impact of incongruent events in Alu element-based phylogenetic studies.
Supplementary Material
Acknowledgements
We thank T. Meyer for his useful comments during preparation of the manuscript, J. A. Walker for her help throughout this project, and Dr. M. Rocchi for DNA samples. This research was supported by National Basic Research Program of China (973 project: 2007CB411605), National Science Foundation grant BCS-0218338 (M.A.B.), and National Institutes of Health Grant RO1 GM59290 (M.A.B.).
Abbreviations
- mya
million years ago
- SINE
short interspersed element
- SNPs
single nucleotide polymorphisms
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bamshad MJ, Wooding S, Watkins WS, Ostler CT, Batzer MA, Jorde LB. Human population genetic structure and inference of group membership. Am J Hum Genet. 2003;72:578–589. doi: 10.1086/368061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Batzer MA, Gudi VA, Mena JC, Foltz DW, Herrera RJ, Deininger PL. Amplification dynamics of human-specific (HS) Alu family members. Nucleic Acids Res. 1991;19:3619–3623. doi: 10.1093/nar/19.13.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Stoneking M, Alegria-Hartman M, Bazan H, Kass DH, Shaikh TH, Novick GE, Ioannou PA, Scheer WD, Herrera RJ, Deininger PL. African origin of human-specific polymorphic Alu insertions. Proc Natl Acad Sci U S A. 1994;91:12288–12292. doi: 10.1073/pnas.91.25.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty D, Ramakrishnan U, Panor J, Mishra C, Sinha A. Phylogenetic relationships and morphometric affinities of the Arunachal macaque Macaca munzala, a newly described primate from Arunachal Pradesh, northeastern India. Mol Phylogenet Evol. 2007;44:838–849. doi: 10.1016/j.ympev.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Champoux M, Suomi SJ, Schneider ML. Temperament differences between captive Indian and Chinese-Indian hybrid rhesus macaque neonates. Lab Anim Sci. 1994;44:351–357. [PubMed] [Google Scholar]
- Churakov G, Kriegs JO, Baertsch R, Zemann A, Brosius J, Schmitz J. Mosaic retroposon insertion patterns in placental mammals. Genome Res. 2009;19:868–875. doi: 10.1101/gr.090647.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin JE, Cann R, Sarich VM. Molecular evolution and systematics of the genus Macaca. In: Lindburg DG, editor. The macaques: Studies in ecology behavior and evolution. Van Nostrand Reinhold Co.; New York: 1980. pp. 31–51. [Google Scholar]
- Deinard A, Smith DG. Phylogenetic relationships among the macaques: evidence from the nuclear locus NRAMP1. J Hum Evol. 2001;41:45–59. doi: 10.1006/jhev.2001.0480. [DOI] [PubMed] [Google Scholar]
- Delson E. Fossil macaques phyletic relationships and a scenario of development. In: Lindburg DG, editor. The macaques: Studies in ecology behavior and evolution. Van Nostrand Reinhold Co.; New York: 1980. pp. 10–30. [Google Scholar]
- Delson E, Tattersall I, Van Couvering JA, Brooks AS. Cercopithecidae. In: Delson E, Tattersall I, Van Couvering JA, Brooks AS, editors. Encyclopedia of Human Evolution and Prehistroy. 2nd ed. Garland; New York: 2000. pp. 166–171. [Google Scholar]
- Edwards MC, Gibbs RA. A human dimorphism resulting from loss of an Alu. Genomics. 1992;14:590–597. doi: 10.1016/s0888-7543(05)80156-9. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Supriatna J, Melnick DJ. Hybridization and population genetics of two macaque species in Sulawesi, Indonesia. Evolution. 2001;55:1686–1702. doi: 10.1111/j.0014-3820.2001.tb00688.x. [DOI] [PubMed] [Google Scholar]
- Fa JE. The genus Macaca: A review of taxonomy and evolution. Mammal Rev. 1989;19:45–81. [Google Scholar]
- Fooden J. Provisional classifications and key to living species of macaques (primates: Macaca) Folia Primatol (Basel) 1976;25:225–236. doi: 10.1159/000155715. [DOI] [PubMed] [Google Scholar]
- Fooden J. Classification and distribution of living macaques. In: Lindburg DG, editor. The macaques: Studies in ecology behavior and evolution. Van Nostrand Reinhold Co.; New York: 1980. pp. 1–9. [Google Scholar]
- Fooden J. Taxonomy and evolution of the sinica group of macaques: 3. species and subspecies accounts of Macaca assamensis. Fieldiana Zool. 1982;10:1–52. [Google Scholar]
- Fooden J. Taxonomy and evolution of the sinica group of macaques: 6. Interspecific comparisons and synthesis. Fieldiana Zool. 1988;45:1–44. [Google Scholar]
- Fooden J. The bear macaque, Macaca arctoides: a systematic review. J Hum Evol. 1990;19:607–686. [Google Scholar]
- Fooden J, Lanyon SM. Blood protein allele frequencies and phylogenetic relationships in Macaca: A review. Am J Primatol. 1989;17:209–241. doi: 10.1002/ajp.1350170304. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Groves CP. Primate Taxonomy. Smithsonian Institution Press; Washington DC: 2001. [Google Scholar]
- Hamdi H, Nishio H, Zielinski R, Dugaiczyk A. Origin and phylogenetic distribution of Alu DNA repeats: irreversible events in the evolution of primates. J Mol Biol. 1999;289:861–871. doi: 10.1006/jmbi.1999.2797. [DOI] [PubMed] [Google Scholar]
- Han K, Konkel MK, Xing J, Wang H, Lee J, Meyer TJ, Huang CT, Sandifer E, Hebert K, Barnes EW, Hubley R, Miller W, Smit AF, Ullmer B, Batzer MA. Mobile DNA in Old World monkeys: a glimpse through the rhesus macaque genome. Science. 2007;316:238–240. doi: 10.1126/science.1139462. [DOI] [PubMed] [Google Scholar]
- Hayasaka K, Fujii K, Horai S. Molecular phylogeny of macaques: implications of nucleotide sequences from an 896-base pair region of mitochondrial DNA. Mol Biol Evol. 1996;13:1044–1053. doi: 10.1093/oxfordjournals.molbev.a025655. [DOI] [PubMed] [Google Scholar]
- Hoelzer GA, Hoelzer MA, Melnick DJ. The evolutionary history of the sinicagroup of macaque monkeys as revealed by mtDNA restriction site analysis. Mol Phylogenet Evol. 1992;1:215–222. doi: 10.1016/1055-7903(92)90018-c. [DOI] [PubMed] [Google Scholar]
- Inagaki H. Some hair characteristics of Macaca monkeys and an attempt to group them based on those features. In: Shotaka T, Wada K, editors. Variations in the Asian Macaques. Tokai University Press; Tokyo: 1996. pp. 89–96. [Google Scholar]
- Kido Y, Aono M, Yamaki T, Matsumoto K, Murata S, Saneyoshi M, Okada N. Shaping and reshaping of salmonid genomes by amplification of tRNA-derived retroposons during evolution. Proc Natl Acad Sci U S A. 1991;88:2326–2330. doi: 10.1073/pnas.88.6.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Li QQ, Zhang YP. A Molecular Phylogeny of Macaca Based on Mitochondrial Control Region Sequences. Zool Res. 2004;25:385–390. [Google Scholar]
- Li QQ, Zhang YP. Phylogenetic relationships of the macaques (Cercopithecidae: Macaca), inferred from mitochondrial DNA sequences. Biochem Genet. 2005;43:375–386. doi: 10.1007/s10528-005-6777-z. [DOI] [PubMed] [Google Scholar]
- Melnick DJ, Kidd KK. Genetic and evolutionary relationships among Asian macaques. Int J Primatol. 1985;6:123–160. [Google Scholar]
- Morales JC, Melnick DJ. Phylogenetic relationships of the macaques (Cercopithecidae: Macaca), as revealed by high resolution restriction site mapping of mitochondrial ribosomal genes. J Hum Evol. 1998;34:1–23. doi: 10.1006/jhev.1997.0171. [DOI] [PubMed] [Google Scholar]
- Murata S, Takasaki N, Saitoh M, Okada N. Determination of the phylogenetic relationships among Pacific salmonids by using short interspersed elements (SINEs) as temporal landmarks of evolution. Proc Natl Acad Sci U S A. 1993;90:6995–6999. doi: 10.1073/pnas.90.15.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterholz M, Walter L, Roos C. Retropositional events consolidate the branching order among New World monkey genera. Mol Phylogenet Evol. 2009;50:507–513. doi: 10.1016/j.ympev.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Perna NT, Batzer MA, Deininger PL, Stoneking M. Alu insertion polymorphism: a new type of marker for human population studies. Hum Biol. 1992;64:641–648. [PubMed] [Google Scholar]
- Purvis A. A composite estimate of primate phylogeny. Philos Trans R Soc Lond B Biol Sci. 1995;348:405–421. doi: 10.1098/rstb.1995.0078. [DOI] [PubMed] [Google Scholar]
- Raaum RL, Sterner KN, Noviello CM, Stewart CB, Disotell TR. Catarrhine primate divergence dates estimated from complete mitochondrial genomes: concordance with fossil and nuclear DNA evidence. J Hum Evol. 2005;48:237–257. doi: 10.1016/j.jhevol.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Ray DA, Xing J, Hedges DJ, Hall MA, Laborde ME, Anders BA, White BR, Stoilova N, Fowlkes JD, Landry KE, Chemnick LG, Ryder OA, Batzer MA. Alu insertion loci and platyrrhine primate phylogeny. Mol Phylogenet Evol. 2005;35:117–126. doi: 10.1016/j.ympev.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Ray DA, Xing J, Salem AH, Batzer MA. SINEs of a nearly perfect character. Syst Biol. 2006;55:928–935. doi: 10.1080/10635150600865419. [DOI] [PubMed] [Google Scholar]
- Roos C, Schmitz J, Zischler H. Primate jumping genes elucidate strepsirrhine phylogeny. Proc Natl Acad Sci U S A. 2004;101:10650–10654. doi: 10.1073/pnas.0403852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem AH, Ray DA, Xing J, Callinan PA, Myers JS, Hedges DJ, Garber RK, Witherspoon DJ, Jorde LB, Batzer MA. Alu elements and hominid phylogenetics. Proc Natl Acad Sci U S A. 2003;100:12787–12791. doi: 10.1073/pnas.2133766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J, Ohme M, Zischler H. SINE insertions in cladistic analyses and the phylogenetic affiliations of Tarsius bancanus to other primates. Genetics. 2001;157:777–784. doi: 10.1093/genetics/157.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedlock AM, Milinkovitch MC, Okada N. SINE evolution, missing data, and the origin of whales. Syst Biol. 2000;49:808–817. doi: 10.1080/106351500750049851. [DOI] [PubMed] [Google Scholar]
- Shedlock AM, Okada N. SINE insertions: powerful tools for molecular systematics. Bioessays. 2000;22:148–160. doi: 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Smith DG, McDonough JW, George DA. Mitochondrial DNA variation within and among regional populations of longtail macaques (Macaca fascicularis) in relation to other species of the fascicularis group of macaques. Am J Primatol. 2007;69:182–198. doi: 10.1002/ajp.20337. [DOI] [PubMed] [Google Scholar]
- Stoneking M, Fontius JJ, Clifford SL, Soodyall H, Arcot SS, Saha N, Jenkins T, Tahir MA, Deininger PL, Batzer MA. Alu insertion polymorphisms and human evolution: evidence for a larger population size in Africa. Genome Res. 1997;7:1061–1071. doi: 10.1101/gr.7.11.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street SL, Kyes RC, Grant R, Ferguson B. Single nucleotide polymorphisms (SNPs) are highly conserved in rhesus (Macaca mulatta) and cynomolgus (Macaca fascicularis) macaques. BMC Genomics. 2007;8:480. doi: 10.1186/1471-2164-8-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*:Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4.0b10 Sinauer Associates; Sunderland, Massachusetts: 2003. [Google Scholar]
- Takahashi K, Terai Y, Nishida M, Okada N. A novel family of short interspersed repetitive elements (SINEs) from cichlids: the patterns of insertion of SINEs at orthologous loci support the proposed monophyly of four major groups of cichlid fishes in Lake Tanganyika. Mol Biol Evol. 1998;15:391–407. doi: 10.1093/oxfordjournals.molbev.a025936. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Takenaka O. Phylogenetic relationship of the genus Macaca inferred from DNA sequence. International symposium: evolution of Asian primates; Freude and Kyoto University Primate Research Institute, Inuyama, Aichi, Japan. 1996. p. 10. [Google Scholar]
- Tosi AJ, Morales JC, Melnick DJ. Comparison of Y chromosome and mtDNA phylogenies leads to unique inferences of macaque evolutionary history. Mol Phylogenet Evol. 2000;17:133–144. doi: 10.1006/mpev.2000.0834. [DOI] [PubMed] [Google Scholar]
- Tosi AJ, Morales JC, Melnick DJ. Y-chromosome and mitochondrial markers in Macaca fascicularis indicate introgression with Indochinese M. mulatta and a biogeographic barrier in the Isthmus of Kra. Int J Primatol. 2002;23:161–178. [Google Scholar]
- Tosi AJ, Morales JC, Melnick DJ. Paternal, maternal, and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution. 2003;57:1419–1435. doi: 10.1111/j.0014-3820.2003.tb00349.x. [DOI] [PubMed] [Google Scholar]
- van de Lagemaat LN, Gagnier L, Medstrand P, Mager DL. Genomic deletions and precise removal of transposable elements mediated by short identical DNA segments in primates. Genome Res. 2005;15:1243–1249. doi: 10.1101/gr.3910705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell PJ, Kishino H, Ota R. A phylogenetic foundation for comparative mammalian genomics. Genome Inform. 2001;12:141–154. [PubMed] [Google Scholar]
- Watkins WS, Rogers AR, Ostler CT, Wooding S, Bamshad MJ, Brassington AM, Carroll ML, Nguyen SV, Walker JA, Prasad BV, Reddy PG, Das PK, Batzer MA, Jorde LB. Genetic variation among world populations: inferences from 100 Alu insertion polymorphisms. Genome Res. 2003;13:1607–1618. doi: 10.1101/gr.894603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Wang H, Han K, Ray DA, Huang CH, Chemnick LG, Stewart CB, Disotell TR, Ryder OA, Batzer MA. A mobile element based phylogeny of Old World monkeys. Mol Phylogenet Evol. 2005;37:872–880. doi: 10.1016/j.ympev.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Xing J, Wang H, Zhang Y, Ray DA, Tosi AJ, Disotell TR, Batzer MA. A mobile element-based evolutionary history of guenons (tribe Cercopithecini) BMC Biol. 2007a;5:5. doi: 10.1186/1741-7007-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Witherspoon DJ, Ray DA, Batzer MA, Jorde LB. Mobile DNA elements in primate and human evolution. Am J Phys Anthropol. 2007b;45(Suppl):2–19. doi: 10.1002/ajpa.20722. [DOI] [PubMed] [Google Scholar]
- Zhang YP, Shi LM. Phylogenetic relationships of macaques as inferred from restriction endonuclease analysis of mitochondrial DNA. Folia Primatol (Basel) 1993;60:7–17. doi: 10.1159/000156671. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.