Abstract
The helicase-primase complex from herpes simplex virus-1 contains three subunits, UL5, UL52, and UL8. We generated each of the potential two subunit complexes, UL5/UL52, UL5/UL8, and UL52/UL8, and used a series of kinetic and photocrosslinking studies to provide further insights into the roles of each subunit in DNA binding and primer synthesis. UL8 increases the rate of primer synthesis by UL5/UL52 by increasing the rate of primer initiation (2 NTPs → pppNpN), the rate-limiting step in primer synthesis. The UL5/UL8 complex lacked any detectable catalytic activity (DNA dependent ATPase, primase, or RNA polymerase using a RNA primer:template and NTPs as substrates), but could still bind DNA, indicating that UL52 must provide some key amino acids needed for helicase function. The UL52/UL8 complex lacked detectable DNA dependent ATPase activity and could not synthesize primers on single-stranded DNA. However, it exhibited robust RNA polymerase activity using a RNA primer:template and NTPs as substrate. Thus, UL52 must contain the entire primase active site needed for phosphodiester bond formation, while UL5 minimally contributes amino acids needed for initiation of primer synthesis. Photocrosslinking experiments using single-stranded templates containing 5-iodouracil either before, in, or after the canonical 3′-GPyPy (Py = T or C) initiation site for primer synthesis showed that only UL5 crosslinked to the DNA. This occurred for both the UL5/UL52 and UL5/UL52/UL8 complexes, and whether or not the reactions contained NTPs. Photocrosslinking of a RNA primer:template, the product of primer synthesis, containing 5-iodouracil in the template generated the same apparent crosslinked species.
Keywords: Replication, kinetics, NTPs, polymerase, photocrosslinking
Herpes simplex virus-1 primase-helicase is one of the major virally encoded complexes essential for herpes DNA replication (1). The helicase-primase serves two essential functions during herpes replication. The helicase activity tracks along the lagging strand and unwinds the double-stranded DNA in front of the leading strand DNA polymerase, leaving in its wake single-stranded templates both for the leading strand polymerase and for primase-coupled polymerase activity on the lagging strand. The primase activity synthesizes short RNA primers for a DNA polymerase to further elongate, thereby providing a means of initiating new DNA strands.
The helicase-primase complex consists of 3 subunits (UL5, UL8, and UL52) and exhibits 3 activities — primase, helicase, and DNA-dependent ATPase activity (2, 3). This last activity provides the energy necessary for unwinding double-stranded DNA. The UL5/UL52 complex is the minimal subcomplex that has been shown to either synthesize primers on single-stranded DNA or unwind double-stranded DNA (4, 5). The UL5 protein contains 7 motifs conserved among superfamily I helicases, including the Walker A and B motifs involved in nucleotide and metal ion binding (6). The UL52 subunit contains the conserved DhD (where h is a hydrophobic amino acid) motif found in most nucleotide polymerases and that likely helps chelate a pair of divalent metal ions (7), and a zinc-binding motif found in most primases (8, 9).
Even though both UL5 and UL52 contain distinct helicase and primase motifs, respectively, neither subunit has demonstrated catalytic activity in the absence of the other (4, 10, 11). Moreover, the two activities demonstrate a complex relationship such that they are kinetically and mechanistically entangled. Weller and colleagues showed that eliminating helicase activity by mutating the conserved helicase motifs in UL5 can stimulate primase activity of the resulting complex (12). Additionally, some mutations of this Zn+2-binding motif in UL52 primarily inhibit primase activity, whereas others abrogate both primase and helicase activity (8, 13).
The UL8 subunit possesses no known catalytic function, but has several proposed functions as an essential herpes DNA replication protein (10, 14-17). On single-stranded DNA, the UL5/UL8/UL52 complex has greater activity than the UL5/UL52 complex. Furthermore, coating single-stranded DNA with the herpes single-stranded DNA binding protein ICP8 blocks primase activity by the UL5/UL52 complex, whereas the UL5/UL8/UL52 complex demonstrates robust activity. This effect of UL8 likely involves specific interactions with ICP8 since coating the DNA with E. coli single-stranded DNA binding protein blocks primer synthesis by both the UL5/UL52 and UL5/UL8/UL52 complexes. In addition, we found that UL8 enhances primase-coupled polymerase activity (i.e., primase-catalyzed primer synthesis followed by polymerase-catalyzed dNTP polymerization) by increasing primer utilization (18).
Herpes primase can initiate primer synthesis (i.e., synthesize the dinucleotide pppNpN from 2 NTPs) at any pair of template pyrimidines. However, the synthesis of primers >3 nucleotides long requires that primer initiation occurs at a 3′-GPyPy-5′ sequence where the G is cryptic (non-coding) and Py is either pyrimidine (19).1 The presence of a single 3′-GPyPy-5′ in a longer sequence eliminates initiation at any other pair of pyrimidines showing that the cryptic G directs where primase binds. After initiating primer synthesis, the efficiency with which the enzyme further elongates the dinucleotide into longer products varies tremendously depending upon the sequences surrounding the initiation site. How the surrounding template sequence alters primer synthesis remains unclear, however.
We used a combination of photocrosslinking and kinetic assays to explore DNA binding and primer synthesis by the three subunit primase-helicase complex as well as each potential two protein subcomplex. While the UL5/UL8 complex lacks detectable DNA-dependent NTPase activity, it crosslinks to single-stranded DNA and RNA primer:DNA templates in the absence of UL52. The UL52/UL8 complex catalyzes phosphodiester bond formation using a RNA primer:DNA template, but cannot initiate primer synthesis de novo. The UL8 lacks detectable DNA binding activity, but enhances primase activity by stimulating dinucleotide synthesis.
Experimental Procedures
Reagents
Unlabeled NTPs and dNTPs were purchased from Sigma and radiolabeled NTPs and dNTPs from Perkin Elmer. Synthetic DNA oligonucleotides of defined sequence were obtained from Oligos, etc. or BioSearch Technologies, Inc.; RNA oligonucleotides were obtained from Dharmacon. Oligonucleotide concentrations were determined spectrally and are reported in terms of 5′-termini. All other reagents were of the highest purity available.
Protein Purification
HSV-1 helicase-primase subcomplexes (UL5/UL8 and UL52/UL8), HSV-1 holoenzyme (UL5/UL52/UL8), and UL8 were all expressed in baculovirus-infected SF9 cells by infecting cells with the appropriate baculoviruses and grown at the Tissue Culture Core Facility at the University of Colorado Health Sciences Center . In each case, the UL8 subunit was His8-tagged such that the complexes could be purified using Ni-NTA affinity chromatography as previously described (19). The UL5/UL52 subcomplex was purified as previously described (8). All purified enzymes were identified and analyzed by SDS gel electrophoresis, followed by Western blot using monoclonal antibodies for each subunit. The antibodies were generously provided by Dr. I. Robert Lehman (Stanford University).
Primase Assay
Primase assays (10 uL) were performed as previously described and typically contained 10-15 μM single-stranded DNA template, 0.4-1 mM [α-32P]NTPs, 50 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 5% glycerol, 1 mM DTT, and 0.1 mg/mL bovine serum albumin (20). Reactions were initiated by adding 50-100 nM helicase-primase and quenched after 30 min at 37°C.
Primer-Template Elongation
Assays to measure NTP polymerization onto RNA primer:template contained a 5′-[32P]-primer:template and NTPs and were performed as previously described (21). Products were separated by denaturing gel electrophoresis (20% acrylamide, 7.5 M urea) and analyzed by phosphorimagery (Molecular Dynamics, Inc.).
DNA-dependent ATPase
Assays to measure the DNA-dependent ATPase contained T20GTCCT19 and were performed as previously described (20).
Crosslinking Reactions
DNA templates containing 5-iodouracil were 5′-[32P] radiolabeled using [α-32P]ATP and T4 polynucleotide kinase as previously described (22, 23). Samples (usually 20-50 μL) typically contained 0.1-2 μM protein, 0.04-2 μM [32P]-labeled DNA, 50 mM Tris-HCl, pH 8.0 and 1 mM DTT. Samples were lased at 325 nm for 10-30 min using a Coherent laser as previously described (24). Products were separated by denaturing gel electrophoresis (7 or 8% polyacrylamide) and analyzed by phosphorimagery. Crosslinking to a RNA:[32P]template was performed analogously.
Western blotting of the crosslinked proteins
In order to isolate crosslinked protein and remove uncrosslinked protein from the photolysis reactions, the 5′-[32P]-templates used for photocrosslinking now contained a biotinylated uracil at the 3′-end. Reactions typically contained 1 μM primase and 1 μM [32P]DNA in a volume of 200 μL. After photolysis, the buffer was brought to 50 mM Tris-HCl, pH ~7.5, 100 mM DTT, 2% SDS and 10% glycerol. The samples were added to treptavidin-agarose beads (400 μL) pre-equilibrated with 500 uL of 50 mM Tris-HCl, pH 6.8, 100 mM DTT, 2% SDS and 10% glycerol. These mixtures were incubated for 30 min at room temperature followed by an overnight incubation at 4°C with gentle mixing on a nutator. The beads were then washed with 7- to 10 × 1 mL of the above buffer. After removal of the wash, the beads were in the wash buffer to elute the biotinylated DNA and subjected to denaturing polyacrylamide gel electrophoresis as described above. Western blotting was performed as described previously (23, 25).
RESULTS
In order to clarify the roles of each subunit in the heterotrimeric helicase-primase complex, we purified each possible dimeric complex and compared its properties to the ternary holoenzyme. As described below, this was facilitated by using baculovirus expressions systems to generate each complex followed by either classical purification (UL5/UL52 complex) or affinity chromatography (UL5/UL8 and UL52/UL8 complexes).
The UL5/UL52 complex and stimulation of primase activity by UL8
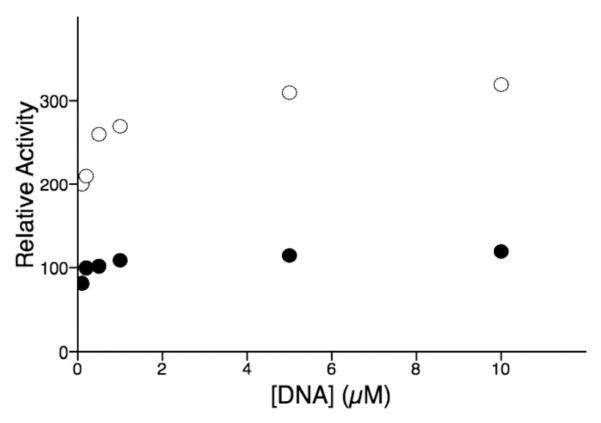
We initially examined how UL8 stimulates primase activity of the UL5/UL52 complex. Comparing primase activity on several different templates containing a single primer initiation site showed that the extent of stimulation varied from almost no stimulation to 4-fold (Table 2). The greatest stimulation occurred on a template that only supports short primer synthesis due to the lack of a 3′-GPyPy-5′ start site (Py = pyrimidine), while UL8 had smaller effects on templates that contained a canonical start site. Stimulation did not change as the DNA concentration varied (Figure 1), consistent with the low KM for DNA (19) and demonstrating that UL8 does not dramatically alter binding of UL5/UL52 to DNA. Since previous studies have shown that initiation (dinucleotide synthesis) limits the overall rate of primer synthesis and the greatest stimulation occurred on templates that support almost exclusively dinucleotide synthesis, UL8 stimulates primer synthesis via increased initiation.
Table 2.
Effects of UL8 on total primer synthesis using the noted single-stranded template in assays containing 400 μM NTPs.
| DNA | Fold stimulation by UL8 |
|---|---|
| T20GTCCT19 | 2.4 |
| (TCTG)15 | 1.9 |
| (TCTA)15 | 4.0 |
| (TC)30 | 4.5 |
| A20GTCA20 | 1.2 |
Figure 1.
Effect of increasing DNA concentrations on the rate of primer synthesis by the UL5/UL52 (λ) and UL5/UL52/UL8 ([ring2]) complexes.
In addition to synthesizing primers de novo on single-stranded templates, primase also can directly elongate a RNA primer:template via NTP polymerization (21). This latter reaction avoids the initiation step and only requires phosphodiester bond formation. UL5/UL52 and UL5/UL52/UL8 polymerized CTP onto DNA (Table 1) with catalytic efficiencies (kcat/KM) of 0.302 ±0.002 hr−1 mM CTP−1 and 0.43 ±0.01 hr−1 mM CTP−1, respectively. These similar efficiencies indicate that the loss of UL8 does not significantly affect polymerization of a single NTP onto a primer:template.
Table 1.
DNA sequences used
| T20GTCCT19 | 3′-TTTTTTTTTTTTTTTTTTTTGTCCTTTTTTTTTTTTTTTTTTT |
| T20GCCCGAT14 | 3′-TTTTTTTTTTTTTTTTTTTTGCCCGATTTTTTTTTTTTTT |
| (TC)30 | 3′-TCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTC |
| (TCTG)13 | 3′-TCTGTCTGTCTGTCTGTCTGTCTGTCTGTCTGTCTGTCTGTCTGTCTGTCTG |
| (TCTA)13 | 3′-TCTATCTATCTATCTATCTATCTATCTATCTATCTATCTATCTATCTATCTA |
| C20GTCCA19 | 3′-CCCCCCCCCCCCCCCCCCCCGTCCAAAAAAAAAAAAAAAAAAA |
| DNA1 | 3′-TTTTTTTTTTTTTTTXTTTTGTCCTTTTTTTTTTTTTTTTTTT |
| DNA2 | 3′-TTTTTTTTTTTTTTTTTTTTGXCCTTTTTTTTTTTTTTTTTTT |
| DNA3 | 3′-TTTTTTTTTTTTTTTTTTTTGTCCXTTTTTTTTTTTTTTTTTT |
| DNA1A | 3′-TTTTTTTTTTTTTTTXTTTTATCCTTTTTTTTTTTTTTTTTTT |
| DNA2A | 3′-TTTTTTTTTTTTTTTTTTTTAXCCTTTTTTTTTTTTTTTTTTT |
| DNA3A | 3′-TTTTTTTTTTTTTTTXTTTTATCCXTTTTTTTTTTTTTTTTTT |
| DNA1B | 3′-TBTTTTTTTTTTTTTXTTTTGTCCTTTTTTTTTTTTTTTTTTT |
| DNA2B | 3′-TBTTTTTTTTTTTTTTTTTTGXCCTTTTTTTTTTTTTTTTTTT |
| DNA3B | 3′-TBTTTTTTTTTTTTTTTTTTGTCCXTTTTTTTTTTTTTTTTTT |
| P/T1 | 5′-AAAAAACAGGA 3′-TTTTTTTTTTTTTTTXTTTTGTCCTTTTTTTTTTTTTTTTTTT |
| P/T2 | 5′-AAAAAACAGGA 3′-TTTTTTTTTTTTTTTTTTTTGXCCTTTTTTTTTTTTTTTTTTT |
| P/T3 | 5′-AAAAAACAGGA 3′-TTTTTTTTTTTTTTTXTTTTGTCCXTTTTTTTTTTTTTTTTTT |
| P/T4 | 5′-AAAAAACGGG 3′-TTTTTTTTTTTTTTTTTTTTGCCCGATTTTTTTTTTTTTT |
| P/T5 | 5′-AAAAAACGGG 3′-TTTTTTTTTTTTTTTTTTTTGCCCAGTTTTTTTTTTTTTT |
| DNAG | 5′- GGGGUAAA 3′-GCCCCATTTGTAATGCATGC-5′ |
X is 5-iodouracil and B is biotinylated deoxyuridine. All primer strands are RNA and all template strands are DNA.
DNA binding by the UL5/UL52/UL8 complex
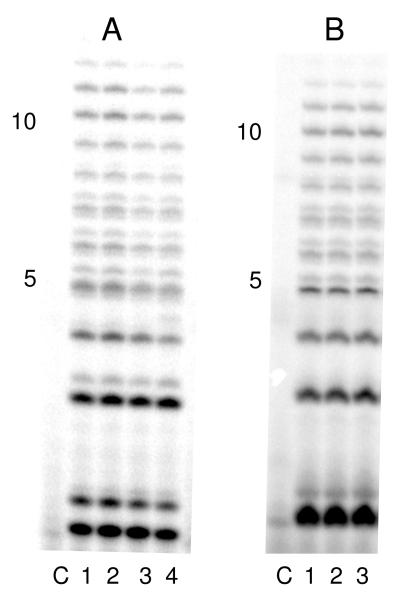
To better understand how helicase-primase interacts with DNA and any potential role of UL8, we measured photocrosslinking of 3 DNAs containing a photoactivateable crosslinking agent that structurally resembles thymine, 5-iodouracil. The 5-iodouracil was located before, within, or just after the 3′-GPyPy-5′ trinucleotide initiation site (Table 1). Primase assays showed that the 5-iodouracil minimally impacted primer synthesis compared to the identical templates containing thymine at each position both in terms of rate and the size distribution of products (Figure 2, and data not shown).
Figure 2.
Primase activity on both normal and modified templates. Assays were performed as described under Experimental Procedures and contained; Panel A in lane (1) T20GTCCT19; (2) DNA1 (3) DNA2 (4) DNA3 and (C) no enzyme, and; Panel B in lane: (1) T20GTCCT19; (2) DNA1; (3) DNA1B, and; (C) no enzyme. Primer length is noted on the side of each gel.
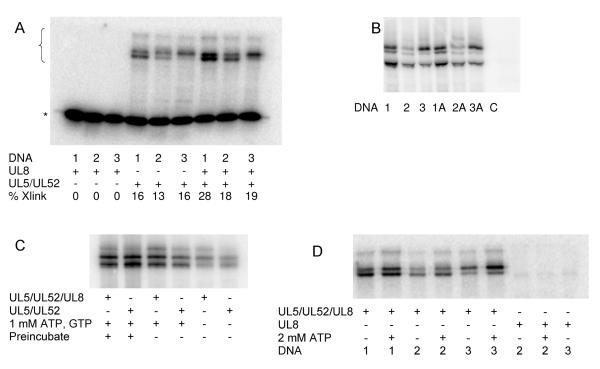
Photolysis of the helicase-primase complex and 5′-[32P]-labeled DNA1, DNA2, or DNA3 at 325 nm gave a series of crosslinked species. A typical gel of the crosslinked species is shown in Figure 3A. All 3 DNA templates gave similar products, although the relative intensities of the major bands changed depending on the position of the 5-iodouracil in the template sequence. The maximum yield of crosslinking varied by 2- to 3-fold among the 3 DNA templates. Reactions containing 1.4 μM UL5/L52/UL8 and 200 nM 5′-[32P]-DNA1, -DNA2, and -DNA3 gave 25, 9, and 11% crosslinking of the DNA after 30 min of photolysis, respectively. Control experiments showed that omitting photolysis or omitting protein eliminated crosslinking.
Figure 3.
(Panel A) Photocrosslinking of DNA1, DNA2, or DNA3 to UL8, UL5/UL52, or UL5/UL52/UL8. Photocrosslnking was performed as described under Experimental Procedures in reactions containing 280 nM of the noted protein and 40 nM of the noted 5′-[32P]-labeled DNA. Crosslinked products are noted with the bracket and the unreacted DNA is noted with a *. “% Xlink” gives the percent of the DNA crosslinked. (Panel B) Effect of eliminating the canonical 3′-GPyPy-5′ initiation site for primer synthesis. The gel shows the products of UL5/UL52/UL8 (840 nM) crosslinking to templates (120 nM) containing or lacking the 3′-GPyPy-5′ start site. Each reaction contained the noted DNA. C = No protein. (Panel C) Effect of NTPs on crosslinking. Reactions contained 100 nM DNA2, and 350 nM of the noted enzyme and NTPs. In the first two lanes, the enzyme, NTPs, and DNA were preincubated for 20 min to allow primer synthesis prior to photolysis. (Panel D) Effect of one NTP (ATP) on crosslinking. Reactions contained either 280 nM UL5/UL52/UL8 or 2.8 μM UL8 and 40 nM of the indicated DNA (5′-[32P]-labeled) in the presence or absence of 2 mM ATP.
DNA-protein crosslinking did not require the cryptic G in the 3′-GPyPy5′ initiation site (Figure 3B). Crosslinking of templates identical to DNA1, DNA2, and DNA3, except that the cryptic G was replaced with A (i.e., DNA1A, DNA2A, and DNA3A, Table 1), gave similar major crosslinked products as did the templates containing the cryptic G. Thus, crosslinking of the helicase-primase to DNA does not require an initiation site, although the cryptic G may subtly affect how the protein binds to the DNA.
Since the helicase uses NTPs to power DNA unwinding and primase uses NTPs as substrates, we considered the effects of NTPs on crosslinking. Figures 3C and 3D show that adding a single NTP to allow an active helicase but not allow primer synthesis or adding two NTPs to allow both primer synthesis and an active helicase did not alter the primary crosslinked species, although the band intensities varied slightly. Likewise, incubating the enzyme with ATP and GTP to allow primer synthesis prior to photolysis also did not alter the crosslinked species.
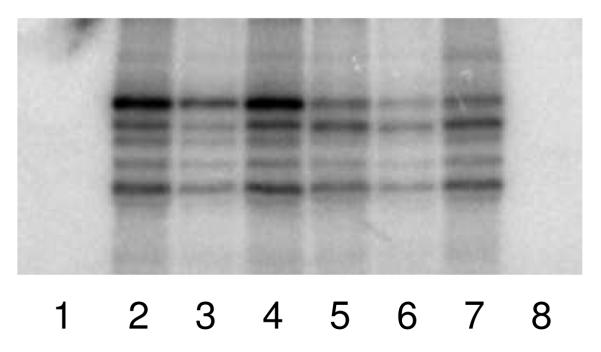
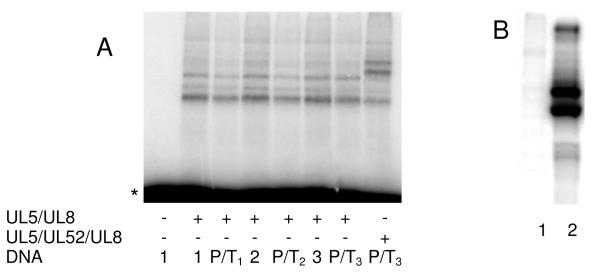
We synthesized a series of RNA primer:templates, the products of primase activity, based on DNA1-3 (i.e, P/T1-3, Table 1) and examined their ability to crosslink to UL5/UL52/UL8. Figure 4 shows that these constructs efficiently react with the protein complex, but again gave similar patterns of bands as did the single-stranded templates alone.
Figure 4.
Photocrosslinking of UL5/UL52/UL8 to primer:templates. In lane 1 and lane 8, the photolysis reactions contained DNA2 or P/T2 and no enzyme. Lanes 2-7 contained UL5/UL52/UL8 (2 μM) and either; lane 2, DNA2; lanes 3 and 4, P/T2; lane 5, DNA3, lanes 6 and 7, P/T3. The concentration of DNA was 2 μM. Reactions in lanes 3 and 6 were photolyzed for 10 min, while all other reactions were photolyzed for 20 min.
UL8 Does Not Affect Crosslinking of DNA to UL5/UL52
Previous studies using gel shift assays suggested that UL8 did not bind DNA (14, 26). Consistent with these results, photolyzing 280 nM UL8 with 40 nM of any of the DNAs containing iodouracil resulted in no detectable crosslinking products (Figure 3A). At an extremely high UL8 concentration (2.8 μM), an extremely faint band results from photolysis with DNA2 or DNA3 (Figure 3D). By comparison, crosslinking with only 280 nM UL5/UL52/UL8 gives >40-fold more crosslinked products (Figure 3D). As a second probe of UL8 crosslinking to DNA, we compared the UL5/UL52 and UL5/UL52/UL8 complexes. Figures 3A and 3C shows that both complexes gave similar crosslinking pattern under several different conditions, and Western blotting of the products with an anti-UL8 antibody gave no detectable signal at the position of the crosslinked species (data not shown). Additionally, the extent of crosslinking was similar for both UL5/UL52 and UL5/UL52/UL8 (Figure 3). Thus, UL8 does not greatly alter how the DNA binds to UL5/UL52 and by itself reacts at best extremely weakly with iodouracil in DNA.
UL5/UL8 Complex
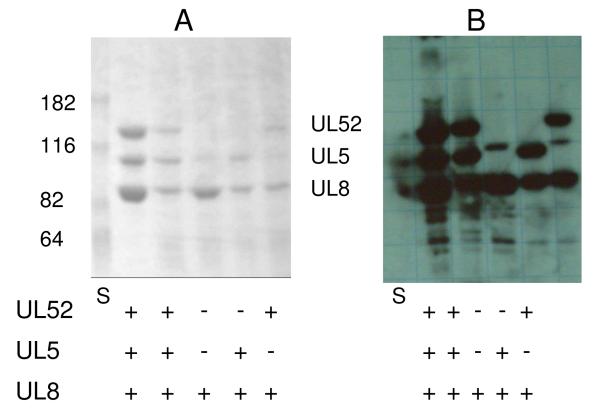
To determine if UL5 and UL8 form a stable and isolatable complex, we coinfected insect cells with baculoviruses coding for UL5 and His8-UL8, and purified the resulting proteins via Ni+2-NTA affinity chromatography. Figure 5 shows that this resulted in purification of the UL5/UL8 complex, and the yield of complex was comparable to either His8-UL8 alone or the UL5/UL52/UL8 ternary complex (0.51 mg of UL5/UL8, 0.27 mg of UL8 and 0.42 mg of UL5/UL52/UL8 from 2 × 107 cells). Thus, UL5 and UL8 stably interact and the resulting complex is readily purified.
Figure 5.
Purification of UL5/UL8 and UL52/UL8. SDS-PAGE gels (7% acrylamide) of proteins purified from SF9 insect cells infected with baculoviruses for the noted proteins and probed with either Coomassie Blue staining (Panel A) or Western blotting using a polyclonal antibody raised against UL5/UL52/UL8 (Panel B). Molecular weights for the Benchmark Pre-stained Protein Ladder from Invitrogen (lane S) and the UL5, UL52, and UL8 subunits are shown on the side of the gels.
Previous studies reported that the UL5 subunit lacks catalytic activity in the absence of UL52 (4, 5). Likewise, we found that the UL5/UL8 complex also lacked detectable catalytic activity in 3 different assays — DNA-dependent ATPase, primer synthesis on single-stranded DNA, and polymerization of NTPs onto a RNA primer template (<0.01% of the activity of UL5/UL52/UL8).
An inability to bind DNA and/or altered DNA binding by the UL5/UL8 complex could explain the lack of DNA-dependent ATPase activity by the UL5/UL8 complex. Thus, we probed DNA binding using the DNA templates containing 5-iodouracil as described above. UL5/UL8 clearly binds DNA, as evidenced by the crosslinking between DNA1-3 and UL5/UL8 (Figure 7). However, the pattern of crosslinked products varied substantially between the UL5/UL8 and UL5/UL52/UL8 complexes, indicating that UL52 affects how the helicase-primase binds DNA (Figure 7). Crosslinking of a RNA primer:template, P/T1, P/T2. or P/T3, gave identical crosslinked products as those using just single-stranded DNA (Figure 6), and the absence of UL52 also altered the crosslinking pattern of the protein with primer:templates.
Figure 7.
Western blots of crosslinked proteins. Photolysis reactions and separation of crosslinked protein from uncrosslinked protein was performed as described under Experimental Procedures. Reactions contained 350 nM protein and 100 nM 5′-[32P]-DNA1B. The images show the crosslinked products generated from photolysis of [32P]-DNA1B with UL5/UL52/UL8 (right side) and the results of a Western blot using a MAb against UL5 (left) and a MAb against UL52 (center).Lane 1, UL5/UL52/UL8; Lane 2, UL5/UL52; Lane 3, UL5/UL52/UL8 and 1 mM ATP and GTP; Lane 4, UL5/UL52 and 1 mM ATP and GTP; Lane 5, UL5/UL52/UL8, 1 mM ATP and GTP with a 20 min preincubation prior to photolysis, and; Lane 6, UL5/UL52, 1 mM ATP and GTP with a 20 min preincubation prior to photolysis. The lane labeled P contained UL5/UL52.
Figure 6.
Crosslinking of DNA to UL5/UL8 or UL52/UL8. Panel A. Photolysis reactions were performed as described under Experimental Procedures and contained either 500 nM UL5/UL8 or 500 nM UL5/UL52/UL8 and 500 nM of the 5′-[32P]-labeled DNA as noted. The primer:templates were labeled on the primer strand. Panel B. Photolysis reactions contained either 1 μM UL52/UL8 (lane 1) or UL5/UL52/UL8 (lane 2) and 1 μM DNA1.
UL52/UL8 Complex
To determine if the UL52/UL8 complex could be stably expressed, we coinfected insect cells with baculoviruses encoding UL52 and His8-UL8 and purified the resulting proteins via Ni+2-NTA affinity chromatography. Figure 5 shows that the purified complex contains both UL52 and UL8, and the yield of UL52/UL8 is comparable to the yield of either UL8 alone or the ternary UL5/UL52/UL8 complex (0.43 mg of UL52/UL8, 0.27 mg of UL8 and 0.42 mg of UL5/UL52/UL8 from 2 × 107 cells.). Thus, UL52 and UL8 form a stable complex.
The catalytic properties of the UL52/UL8 complex were analyzed using three different assays — DNA dependent ATPase, primer synthesis on a single-stranded template, and elongation of a RNA primer-template. Consistent with previous work indicating that UL5 contains the ATPase activity and primase activity requires UL52 and UL5 (4-6), the UL52/UL8 complex lacked detectable ATPase activity and could not synthesize primers on single-stranded templates (<0.01% of UL5/UL52/UL8). However, UL52/UL8 did elongate RNA primer-templates via polymerization of the next required NTP. UL52/UL8 elongated primer-templates around 20- to 25-fold less efficiently than UL5/UL52/UL8 (kcat/KM), due both to an increased KM (NTP) and decreased kcat (Table 3). Importantly, these results indicate that UL52 contains all of the amino acids required for phosphodiester bond formation.
Table 3.
Comparison of RNA primer:template elongation by UL5/UL52/UL8 and UL52/UL8.
| Protein | Primer-template | kcat (hr−1) |
KM (NTP) (μM) |
kcat/KM (hr−1 μM−1) |
|---|---|---|---|---|
| UL5/UL52/UL8 | P/T4 | 8.1±0.1 | 64±4 | 0.13 |
| UL52/UL8 | P/T4 | 2.3±0.3 | 450±180 | 0.0051 |
| UL5/UL52/UL8 | P/T5 | 7.3±0.3 | 190±30 | 0.038 |
| UL52/UL8 | P/T5 | 0.58±0.04 | 280±50 | 0.0021 |
UL52/UL8 must bind a RNA primer-template in order to catalyze phosphodiester bond formation. However, the inability of this complex to synthesize a primer on single-stranded DNA might have resulted from an inability to bind single stranded DNA. To exclude this possibility and test to what extent DNA sequence affects the affinity of UL52/UL8 for DNA, we measured the effect of adding single-stranded DNA to assays that measured elongation of a primer-template. If UL52/UL8 could not bind single-stranded DNA, then adding single-stranded DNA to these assays should have no effect. However, the single-stranded DNA did inhibit primer-template elongation, indicating that UL52/UL8 does bind single stranded DNA. Single-stranded DNA inhibits elongation of primer-templates by both UL5/UL52/UL8 and UL52/UL8 with similar potency (Table 4), suggesting that the loss of UL5 does not greatly affect binding of single-stranded DNA to UL52. Additionally, the presence or absence of a canonical GPyPy initiation sequence did not affect binding of the single-stranded DNAs, indicating that this feature is not a major determinant of DNA binding by UL52.
Table 4.
Inhibition of primer-template elongation by single-stranded DNAs.
| Single-stranded DNA | Enzyme | IC60 (μM) |
|---|---|---|
| T20GC3AGT14 | UL5/UL52/UL8 | 2.9±0.6 |
| T20GC3AGT14 | UL52/UL8 | 2.3±0.5 |
| T20GC3GAT14 | UL5/UL52/UL8 | 3.5±0.5 |
| T20GC3GAT14 | UL52/UL8 | 1.6±0.9 |
| (TTC)20 | UL52/UL8 | 1.6±0.2 |
| (CCT)20 | UL52/UL8 | 1.3±0.5 |
We next asked if we could detect crosslinking of DNA to UL52 within the context of the UL52/UL8 complex. However, even with high concentrations of DNA and UL52/UL8, at most trace amounts of DNA crosslinked (Figure 6B, compare crosslinking to UL52/UL8 and UL5/UL52/UL8).
DNA Crosslinks to UL5
We determined the subunit(s) of the UL5/UL52/UL8 complex to which DNA crosslinked by Western blotting. Since UL5, UL8 and UL52 all have similar molecular masses (99, 80, and 114 kDa, respectively) and covalently bound DNA could affect the electrophoretic mobility of the crosslinked products, we first separated crosslinked protein from native protein. To accomplish this, the DNA templates for these studies contained both a 5-iodouracil and a biotin near its 3′-terminus (DNA1B — DNA3B, Table 1). Control experiments showed that the biotin did not affect primase activity (Figure 2). In order to eliminate uncrosslinked protein prior to Western blotting, the photolyzed primase-helicase complex was first denatured, the biotinylated DNA and any covalently bound proteins were captured onto streptavidin agarose, and the beads washed to remove uncrosslinked proteins. Finally, the products bound to the beads were analyzed by denaturing polyacrylamide gel electrophoresis and phosphorimagery. This purification procedure efficiently captured any protein that crosslinked to the DNA as evidenced by; i) analysis of the material that did not bind to the column showed that it lacked any detectable crosslinked species — only trace amounts of [32P] that comigrated with the input [32P]DNA, and; ii) the relative amounts of each crosslinked species captured by the beads were the same as the relative amounts prior to capture.
Western blot analysis of the crosslinked products using antibodies to UL5 and UL52 showed that all of the major crosslinked species contained UL5, with no detectable UL52 (Figure 7, crosslinking with DNA1B). As noted previously, we were unable to detect crosslinking of DNA to UL8. The Western blot also shows only trace amounts of uncrosslinked protein, demonstrating the effectiveness of the purification. Similarly, we generated biotinylated versions of DNA2B and DNA3B, and found that all of the crosslinked species contained only UL5 as detected by Western blotting (not shown).
The relative abilities of the anti-UL5 and anti-UL52 antibodies to detect UL5 and UL52, respectively, were compared to provide a limit of detection. Western blots with different amounts of UL5 and UL52 loaded onto the gel showed that the anti-UL52 antibody could detect ≤0.75 fmol of protein under the conditions used, while the anti-UL5 antibody could detect ≤0.075 fmol of protein (data not shown). The amount of sample loaded onto the gel for Western blotting contained approximately 37 fmol of crosslinked protein for the samples containing UL5/UL52/UL8, and 20 fmol crosslinked protein for the samples containing UL5/UL52. The lack of detectable signal from the Western blot with the UL52 antibody in combination with the data showing that all of the crosslinked species contain UL5 indicate that only UL5 reacts significantly with the DNA, even though the primase active site resides within UL52.
Discussion
Herpes primase-helicase consists of 3 subunits whose precise roles in primase and helicase activity have not been well defined. Here, we used a combination of kinetic and DNA crosslinking approaches to better define the roles and catalytic properties of each subunit.
All three potential two-protein subcomplexes of the ternary UL5/UL52/UL8 helicase-primase complex can be readily expressed and purified using a baculovirus expression system. The yields of UL5/UL8 and UL52/UL8 were comparable to the yields of UL8 alone or the UL5/UL52/UL8 complex, indicating that the absence of one subunit does not significantly impede expression and folding of either UL5 or UL52 in the presence of UL8. Purification of UL5/UL52 cannot be directly compared to these other complexes since it employs a very different purification protocol (8).
The ability to purify the UL5/UL8, UL52/UL8 and UL5/UL52 complexes indicates that each subunit of the primase-helicase complex binds to the other two subunits with sufficiently strong interactions to allow purification of each complex. Thus, UL8 binds to a remarkable number of viral proteins, including UL5, UL52, ICP8, UL9, and UL30 (15, 16, 27, 28). Stow and colleagues used UL8 deletion mutants to identify regions important for the interaction with UL5 and UL52, and found that residues 340-470 of UL8 likely play critical roles (28). It is unknown, however, if all of the protein-protein interactions can occur simultaneously and if different interactors share a common binding domain on UL8.
Previous studies have suggested that the primase and helicase/DNA-dependent ATPase activities require both UL5 and UL52, although these studies did not explicitly examine purified UL5 or UL52 (3, 4). Consistent with helicase/ATPase requiring both UL5 and UL52, the UL5/UL8 complex showed no detectable DNA-dependent ATPase activity. The UL5/UL8 and UL5/UL52/UL8 complexes gave distinctly different crosslinking patterns to DNA, indicating that UL52 alters the binding of DNA to UL5/UL8 as compared to UL5/UL52/UL8. How binding changes, however, cannot be derived from these data. Potentially, this altered DNA binding could account for the lack of ATPase activity. Consistent with this idea, mutations in the Zn+2 binding domain of UL52, a domain that likely binds DNA, also affect helicase activity (8). It should be noted that the changes in crosslinking pattern do not rule out the possibility that UL52 also contributes catalytically essential residues for helicase/DNA-dependent ATPase activity.
Primer synthesis on single-stranded DNA, a reaction that requires rate-limiting dinucleotide synthesis followed by polymerization of additional NTPs, also requires both UL5 and UL52. However, if one only considers the phosphodiester bond forming reaction (i.e., polymerization of a NTP onto a primer:template), then UL52 alone suffices. UL52 must, therefore, contain the entire primase active site for NTP polymerization, but lacks some critical residues for initiation. Potential roles of UL5 during initiation include proper positioning of the DNA, perhaps including an interaction with the cryptic template G, or binding the NTP that becomes the 5′-terminal nucleotide of the primer. The active sites of DNA polymerases and primases typically contain 3 critical aspartates that chelate two essential divalent metal ions (29-33), suggesting that the herpes primase active site will also contain 3 essential aspartates. Two of the critical aspartates, D628 and D630 have been identified while the identity of the third remains unknown (7). The ability of UL52 to catalyze phosphodiester bond formation indicates that the third aspartate also resides within UL52. Even though the complex analyzed contains UL8, UL8 cannot contain the third aspartate since primase activity does not require UL8.
This phenomenon of the catalytic subunit of primase catalyzing phosphodiester bond formation but not dinucleotide synthesis provides another commonality between herpes and human primase. Human primase also consists of two subunits, p58 and p49 (9). p49 contains the active site and can elongate primer-templates by itself, but cannot initiate primer synthesis without p58 (34). p49 and UL52 also exhibit mild sequence similarity, but more significantly, appear to discriminate between right and wrong NTPs and between different sugars using virtually identical mechanisms (21, 35-37).
In all of the crosslinking studies, we only detected crosslinking to UL5, even in the UL52/UL8 complex lacking UL5. This occurs even though UL52 can bind primer-templates and single-stranded DNA within the context of both the UL5/UL52/UL8 and UL52/UL8 complexes, as evidenced by our kinetic studies. Thus, the absence of crosslinking to UL52 within the context of the UL5/UL52 (±UL8) complex likely did not result from the DNA primarily binding to UL5. Rather, these data indicate that the DNA binding domain(s) in UL52 lack amino acids to which 5-iodouracil can crosslink when probed using single-stranded DNA and primer:templates. Upon photoactivation, 5-iodouracil has only been observed to crosslink with Phe, Tyr, His and Met (38). Thus, the DNA binding domain of UL52 likely lacks these amino acids in near proximity and/or in the appropriate orientation that would allow crosslinking to 5-iodouracil.
Regardless of the location of the 5-iodoracil in the template, the same primary crosslinked species resulted upon photolysis of the UL5/UL52 and UL5/UL52/UL8 complexes. The multiple species generated indicate that several different amino acids in UL5 can react with the photoactivated 5-iodouracil, and the apparently identical species observed with each template indicate that the same set of amino acids always reacts. This occurred whether or not (1) the reactions contained NTPs, (2) when the 5-iodouracil was contained within a single-stranded template with or without the cryptic G needed for efficient synthesis of long primers, and (3) when the 5-iodouracil was in a primer-template. The relative amount of each crosslinked species did, however, vary somewhat as the aforementioned parameters were varied. These results suggest that when bound in the UL5 DNA binding site(s), the different conditions do not change the amino acids with which the 5-iodouracil can react, although the accessibility of these amino acids may vary. Perhaps the simplest mechanism to account for this result is that the DNA can slide within the DNA binding domain. Additionally, the DNA binding domain appears to bind both single-stranded DNA and a RNA-DNA duplex since both nucleic acids give the same crosslinked species.
Weller and coworkers reported that within the context of the primase-helicase complex, UL52 can crosslink to a forked DNA substrate containing 5-iodouracil (12). This raises the possibility that when engaged at a forked junction, the DNA binding domain in UL52 either undergoes a structural rearrangement that exposes reactive amino acids or that a forked DNA binds in a different portion of the UL52 DNA binding domain than either single-stranded DNA or a primer:template.
UL8 is a key component of the herpes replication apparatus, interacting with multiple proteins and stimulating the catalytically active UL5/UL52 subcomplex. Comparing primer synthesis by the UL5/UL52 and UL5/UL52/UL8 complexes showed that stimulation resulted from increased rates of primer initiation. Consistent with UL8 primarily affecting initiation, UL8 had little effect on primer:template elongation by UL5/UL52. UL8 did not greatly affect binding of UL5/UL52 to DNA, as evidenced by similar extents of stimulation at different DNA concentrations and the similar crosslinking patterns to DNA. Previous electromobility shift assays suggested that the UL5/UL52/UL8 complex binds DNA only slightly tighter than does the UL5/UL52 complex (12), consistent with the crosslinking studies showing (Figure 3). UL8 also did not alter the length of products synthesized on single-stranded DNA, indicating that it does not affect the ability of primase to count. Thus, the effects of UL8 on primer synthesis appear directed solely to increasing either a conformational change or phosphodiester bond formation associated with dinucleotide synthesis. Consistent with this conclusion, UL8 did not affect the ability of UL5/UL52 to polymerize a single NTP onto a RNA primer:template.
While both UL5 and UL52 contain DNA binding domains, their location and how they interact remain unclear. Weller and colleagues have shown that the primase and helicase activities exhibit a remarkably complex interdependence (12). How UL5 and UL52 communicate mechanistically remains unclear, but interacting DNA binding domains in the two proteins would help explain this interdependence. Indeed, a contiguous DNA binding domain would allow for direct information transfer between UL5 and UL52.
Acknowledgements
We are especially grateful to Dr. Tad Koch at the University of Colorado for the use of his laser for some of the crosslinking studies.
Footnotes
This work was supported by NIH grant AI059764 (R.D.K.).
- DTT
- dithiothreitol
- EDTA
- ethylene diamine tetraacetic acid
- Py
- pyrimidine
- Tris-HCl
- Tris(hydroxymethyl)aminomethane, HCl salt
References
- 1.Lehman IR, Boehmer PE. Replication of Herpes Simplex Virus DNA. J. Biol. Chem. 1999;274:28059–28062. doi: 10.1074/jbc.274.40.28059. [DOI] [PubMed] [Google Scholar]
- 2.Crute JJ, Lehman IR. Herpes Simplex Virus-1 Helicase-Primase: Physical and Catalytic Properties. J. Biol. Chem. 1991;266:4484–4488. [PubMed] [Google Scholar]
- 3.Dodson MS, Crute JJ, Bruckner RC, Lehman IR. Overexpression and Assembly of the Herpes Simplex Virus Type I Helicase-Primase in Insect Cells. J. Biol. Chem. 1989;264:20835–20838. [PubMed] [Google Scholar]
- 4.Dodson MS, Lehman IR. Association of DNA helicase and Primase Activities with a Subassembly of the Herpes Simplex Virus 1 Helicase-Primase Composed of the UL5 and UL52 Gene Products. Proc. Natl. Acad. Sci. USA. 1991;88:1105–1109. doi: 10.1073/pnas.88.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calder JM, Stow ND. Herpes simplex Virus Helicase-Primase: the UL8 Protein Is not Required for DNA-Dependent ATPase and DNA Helicase Activities. Nuc. Ac. Res. 1990;18:3573–3578. doi: 10.1093/nar/18.12.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu LA, Weller SK. The Six Conserved Helicase Motifs of the UL5 Gene Product, a Component of the Herpes Simplex Virus Type I Helicase-Primase, Are Essential for its Function. J. Virol. 1992;66:469–479. doi: 10.1128/jvi.66.1.469-479.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dracheva S, Koonin EV, Crute JJ. Identification of the Primase Active Site of the Herpes Simplex Virus Type I Helicase-primase. J. Biol. Chem. 1995;270:14148–14153. doi: 10.1074/jbc.270.23.14148. [DOI] [PubMed] [Google Scholar]
- 8.Biswas N, Weller SK. A Mutation in the C-terminal Putative Zn2+ Finger Motif of UL52 Severely Affects the Biochemical Activities of the HSV-1 Helicase-Primase Subcomplex. J. Biol. Chem. 1999;274:8068–8076. doi: 10.1074/jbc.274.12.8068. [DOI] [PubMed] [Google Scholar]
- 9.Frick DN, Richardson CC. DNA Primases. Annu. Rev. Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 10.Carmichael EP, Weller SK. Herpes Simplex Virus Type 1 DNA Synthesis Requires the Product of the UL8 Gene: Isolation and Characterization of an ICP6::lacZ Insertion Mutation. J. Virol. 1989;63:591–599. doi: 10.1128/jvi.63.2.591-599.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parry ME, Stow ND, Marsden HS. Purification and Properties of the Herpes Simplex Virus Type 1 UL8 Protein. J. Gen. Vir. 1993;74:607–612. doi: 10.1099/0022-1317-74-4-607. [DOI] [PubMed] [Google Scholar]
- 12.Biswas N, Weller SK. The UL5 and UL52 Subunits of the Herpes Simplex Virus Type 1 Helicase-Primase Subcomplex Exhibit a Complex Interdependence for DNA Binding. J. Biol. Chem. 2001;276:17610–17619. doi: 10.1074/jbc.M010107200. [DOI] [PubMed] [Google Scholar]
- 13.Graves-Woodward KL, Gottlieb J, Challberg MD, Weller SK. Biochemical Analyses of Mutations in the HSV-1 Helicase-Primase That Alter ATP Hydrolysis, DNA Unwinding, and Coupling Between Hydrolysis and Unwinding. J. Biol. Chem. 1997;272:4623–4630. doi: 10.1074/jbc.272.7.4623. [DOI] [PubMed] [Google Scholar]
- 14.Falkenberg M, Bushnell DA, Elias P, Lehman IR. The UL8 Subunit of the Heterotrimeric Herpes Simplex Virus Type 1 Helicase-Primase Is Required for the Unwinding of Single Stand DNA-binding Protein (ICP8)-coated DNA Substrates. J. Biol. Chem. 1997;272:22766–22770. doi: 10.1074/jbc.272.36.22766. [DOI] [PubMed] [Google Scholar]
- 15.Mardsen HS, McLean GW, Barnard EC, Francis GJ, MacEachran K, Murphy M, McVey G, Cross A, Abbotts AP, Stow ND. The Catalytic Subunit of the DNA Polymerase of Herpes Simplex Virus Type 1 Interacts Specifically with the C Terminus of the UL8 Component of the Viral Helicase-Primase Complex. J. Vir. 1997;71:6390–6397. doi: 10.1128/jvi.71.9.6390-6397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Gac N. Tanguy, Villani G, Hoffmann JS, Boehmer PE. The UL8 Subunit of the Herpes Simplex Virus Type-1 DNA Helicase-Primase Optimizes Utilization of DNA Templates Covered by the Homologous Single-Strand DNA-Binding Protein ICP8. J. Biol. Chem. 1996;271:21645–21651. doi: 10.1074/jbc.271.35.21645. [DOI] [PubMed] [Google Scholar]
- 17.Tenney DJ, Hurlburt WW, Micheletti PA, Bifano M, Hamatake RK. The UL8 Component of the Herpes Simplex Virus Helicase-primase Complex Stimulates Primer Synthesis by a Subassembly of the UL5 and UL52 Components. J. Biol. Chem. 1994;269:5030–5035. [PubMed] [Google Scholar]
- 18.Cavanaugh NA, Kuchta RD. Initiation of New DNA Strands by the Herpes Primase-Helicase Complex and either Herpes DNA Polymerase or Human DNA Polymerase alpha. J. Biol. Chem. 2009;284:1523–1532. doi: 10.1074/jbc.M805476200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramirez-Aguilar KA, Low-Nam NA, Kuchta RD. Key Role of Template Sequence for Primer Synthesis by the Herpes Simplex Virus 1 Helicase-Primase. Biochemistry. 2002;41:4569–4579. doi: 10.1021/bi026680v. [DOI] [PubMed] [Google Scholar]
- 20.Ramirez-Aguilar K, Kuchta RD. Mechanism of Primer Synthesis by the Herpes Simplex Virus 1 Helicase-Primase. Biochemistry. 2004;43:1103–1112. doi: 10.1021/bi035519x. [DOI] [PubMed] [Google Scholar]
- 21.Keller KE, Cavanaugh NA, Kuchta RD. Interaction of Herpes Primase with the Sugar of a NTP. Biochemistry. 2008;47:8977–8984. doi: 10.1021/bi8008467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuchta RD, Mizrahi V, Benkovic PA, Johnson KA, Benkovic SJ. Kinetic Mechanism of DNA Polymerase I (Klenow) Biochemistry. 1987;26:8410–8417. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Sptring Harbor Laboratories; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 24.Arezi B, Kirk BW, Copeland WC, Kuchta RD. Interactions of DNA with Human DNA Primase Monitored with Photoactivatable Cross-linking Agents: Implications for the Role of the p58 Subunit. Biochemistry. 1999;38:12899–12907. doi: 10.1021/bi9908991. [DOI] [PubMed] [Google Scholar]
- 25.Knecht DA, Randall LD. Visualization of Antigenic Proteins on Western Blots. Anal. Biochem. 1984;136:180–184. doi: 10.1016/0003-2697(84)90321-x. [DOI] [PubMed] [Google Scholar]
- 26.Gac NTL, Villani G, Hoffmann J-S, Boehmer PE. The UL8 Subunit of the Herpes Simplex Virus Type-1 DNA Helicase-Primase Optimizes Utilization of DNA Templates Covered by the Homologous Single-strand DNA-binding Protein ICP8. J. Biol. Chem. 1996;271:21645–21651. doi: 10.1074/jbc.271.35.21645. [DOI] [PubMed] [Google Scholar]
- 27.McLean GW, Abbotts AP, Parry ME, Marsden HS, Stow ND. The Herpes Simplex Virus Type 1 Origin-binding Protein Interacts Specifically with the Viral UL8 Protein. J. Gen. Vir. 1994;75:2699–2706. doi: 10.1099/0022-1317-75-10-2699. [DOI] [PubMed] [Google Scholar]
- 28.Barnard EC, Brown G, Stow ND. Deletion Mutants of the Herpes Simplex Virus Type 1 UL8 Protein: Effect on DNA Synthesis and Ability to Interact with and Influence the Intracellular Localization of the UL5 and UL52 Proteins. Virology. 1997;237:97–106. doi: 10.1006/viro.1997.8763. [DOI] [PubMed] [Google Scholar]
- 29.Steitz TA. DNA Polymerases: Structural Diversity and Common Mechanisms. J. Biol. Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Sattar AK, Wang CC, Karam JD, Konigsberg WH, Steitz TA. Crystal Structure of a Pol alpha Family Replication DNA Polymerase from Bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 31.Keck JL, Roche DD, Lynch AS, Berger JM. Structure of the RNA Polymerase Domain of E. coli Primase. Science. 2000;287:2482–2486. doi: 10.1126/science.287.5462.2482. [DOI] [PubMed] [Google Scholar]
- 32.Augustin MA, Huber R, Kaiser JT. Crystal Structure of a DNA-Dependent RNA Polymerase (DNA Primase) Nature Struct. Molec. Biol. 2001;8:57–61. doi: 10.1038/83060. [DOI] [PubMed] [Google Scholar]
- 33.Ito N, Nureki O, Shirouzu M, Yokoyama S, Hanaoka F. Crystal Structure of the Pyrococcus horikoshii DNA Primase-UTP Complex: Implications for the Mechanism of Primer Synthesis. Genes Cells. 2003;8:913–923. doi: 10.1111/j.1365-2443.2003.00693.x. [DOI] [PubMed] [Google Scholar]
- 34.Zerbe L, Kuchta RD. The p58 Subunit of Human DNA Primase Is Important for Primer Initiation, Elongation, and Counting. Biochemistry. 2002;41:4891–4900. doi: 10.1021/bi016030b. [DOI] [PubMed] [Google Scholar]
- 35.Moore CL, Zivkovic A, Engels J, Kuchta RD. Human DNA Primase Uses Watson-Crick Hydrogen Bonding Groups to Distinguish between Correct and Incorrect NTPs. Biochemistry. 2004;43:12367–12374. doi: 10.1021/bi0490791. [DOI] [PubMed] [Google Scholar]
- 36.Ramirez-Aguilar KA, Moore CL, Kuchta RD. Herpes Simplex Virus I Primase Employs Watson-Crick Hydrogen Bonding to Identify Cognate NTPs. Biochemistry. 2005;44:15585–15593. doi: 10.1021/bi0513711. [DOI] [PubMed] [Google Scholar]
- 37.Lakshminarayan MI, Koonin EV, Leipe DD, Aravind L. Origin and Evolution of the Archaeo-Eukaryotic Primase Superfamily and Related Palm-Domain Proteins: Structural Insights and New Members. Nuc. Ac. Res. 2005;33:3875–3896w. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meisenheimer KM, Koch TH. Photocross-Linking of Nucleic Acids to Associated Proteins. Crit. Rev. Biochem. Molec. Biol. 1997;32:101–140. doi: 10.3109/10409239709108550. [DOI] [PubMed] [Google Scholar]