Abstract
Protein localization and dynamics both play an important role in cell signal transduction. Although biochemical studies have elucidated many details about the chain of events in signal cascades, the poor temporal resolution and absence of spatial localization in these conventional techniques make it difficult to determine the “where and when” of protein interactions. Over the past decade, imaging technologies and biological tools have developed to a point where many fundamental questions about protein activities can now be addressed at the molecular level in living cells, revealing spatio-temporal information that is not provided by traditional biochemical assays. In this review, we illustrate the power of emerging fluorescence microscopy techniques to capture and quantify protein dynamics.
Spatio-temporal Aspects of Protein Behavior
Complex cellular processes are governed by signal transduction, which in turn is controlled by protein-protein interactions at the plasma membrane and along the signaling cascade. While biochemical techniques have been used for decades to determine the order of protein interactions along signaling pathways, a current thrust in cell biology is to understand the role of protein dynamics in signal transduction. For example, a membrane receptor tyrosine kinase is in constant motion as it diffuses on the plasma membrane, interacting with other proteins and membrane microdomains. Ligand binding may lead to a change in diffusive behavior and is often the trigger for receptor homo- or hetero-oligomerization, which initiates receptor phosphorylation and cytoplasmic adaptor protein recruitment. The final step is eventual removal of the activated receptor from the plasma membrane via endocytosis, after which the receptor may be recycled or continue along the endosomal pathway to degradation. Despite this common picture of protein events, there are many details in this sequence that remain unclear. The low spatial and temporal resolution of traditional biochemical techniques cannot reveal the spatial distribution and dynamic aspects of these processes. Recent developments in fluorescence microscopy are allowing us to probe protein behavior in real-time, such that we can directly visualize and quantify signal transduction events within the living cell.
Classic techniques for identifying protein-protein interactions
Evidence for direct protein interactions is often first established using biochemical methods (immunoprecipitation, pull-down assays) or genetic approaches (2-hybrid and complementation assays). To confirm the existence of protein complexes in cells, cell biologists often rely on conventional fluorescence imaging techniques for detecting colocalization or engineer pairs of fluorescent proteins for more sophisticated FRET analysis. However, these methods have their shortcomings. Traditional colocalization of two or more fluorophores in wide-field or confocal fluorescence imaging demonstrates whether labeled proteins are in the same general sub-cellular location, but cannot directly detect complex formation because resolution is limited to >250 nm. FRET (Box 1) can reliably report protein interactions[1], since proteins must be within 1–10 nm of each other of each other for energy transfer to occur, but suffers from complications that can lead to false negative results. For example, the distance between labeled components in large multi-protein complexes may be too far for FRET to occur. Therefore, a lack of energy transfer is not necessarily evidence for a lack of protein complex formation. Typically, FRET is limited to the measurement of interactions between two protein species, but creative approaches can expand this capability. For example, Grant and colleagues recently introduced four different fluorescent species into the same cell to permit simultaneous FRET measurements of two distinct FRET pairs[2]. Two-color cross-correlation FCS (Box 1) is a valuable alternative method for detecting protein interactions, with the limitations that this technique provides measurements only at a fixed location and is suitable over a narrow range of protein concentrations. Recent advances in fluorescence microscopy techniques (Box 1), such as image correlation microscopy, super-resolution techniques and FRET imaging, are providing new ways to address questions in cell biology. These creative approaches measure the average behavior of an ensemble of proteins and are developing at a fast pace.
Box 1: Comparison of fluorescence microscopy techniques used to measure protein-protein interactions and dynamics.
Here we compare and contrast some common fluorescent microscopy techniques that can monitor protein-protein interactions and dynamics. It is important to consider the advantages and limitations (Table I) of each, as well as the accessible spatio-temporal scales (Figure I), when deciding which technique is best suited to address a particular biological question.
Fluorescence Recovery After Photobleaching (FRAP)
Fluorophores in a small region of interest are photobleached with a short burst of intense laser excitation. As non-bleached molecules diffuse into the bleached region, fluorescence intensity is recovered and a diffusion constant can be calculated from the fluorescence recovery time.
Förster Resonance Energy Transfer (FRET)
Non-radiative transfer of energy from a lower wavelength (donor) to a higher wavelength chromophore (acceptor) that is dependent on the distance between them. This can be observed by a decrease in donor emission, a decrease in donor lifetime or an increase in the emission of the acceptor. FRET between like molecules can also be achieved (homo-FRET), with the added advantage of using a single fluorphore.
Single Particle Tracking (SPT)
Sparse labeling of proteins in cells allows for direct tracking of individual protein trajectories, which can be analyzed to determine diffusion constants and the type of motion (free, restricted, immobile).
Fluorescence Correlation Spectroscopy (FCS)
A small focal volume is defined with a focused laser beam and a confocal pinhole or by two-photon excitation. Intensity fluctuations are generated as fluorescently-tagged proteins diffuse in and out of the focal volume. Diffusion constant can be determined by fitting theoretical models to the autocorrelation of the intensity trace.
Image Correlation Spectroscopy (ICS)
Changes in fluorescence intensity across an image are used to calculate the spatial correlation function. Number density and aggregation state are determined by fitting models to the correlation function. Images can be acquired by confocal or TIRF microscopy. Variations of this technique include Temporal ICS (measures dynamics and number density) and Spatiotemporal ICS (returns velocity maps). These methods evaluate changes over time using confocal time series data.
Raster Image Correlation Spectroscopy (RICS)
RICS uses the time information inherent in a confocal image, created by the raster scanning of the laser across the field of view. This measures interactions on the order of µs-ms. Calculation of the correlation function using this time information can generate diffusion maps across the image.
Number and Brightness (N&B)
N&B is based upon pixel-by-pixel analysis of images in a confocal time series, with a focus on intensity fluctuations that reflect diffusion of molecules in and out of a pixel. It is possible to determine the brightness of an individual molecule and then estimate the number of molecules per pixel.
Cross-correlation (cc)
Variation on each of the above (FCCS, ICCS, ccRICS, ccN&B), based on two-color imaging. Quantification of coincident (spatial and/or temporal) fluctuations between the two channels provides information on fraction of proteins interacting, dynamics and stoichiometry of complexes.
Stimulated Emission Depletion (STED)
Combines a standard Gaussian excitation beam with a doughnut-shaped beam that depletes emission from the outside ring of the excitation spot, resulting in emission only from the central (~40 nm) region.
Localization microscopy
Intermittency in fluorescence (blinking, binding/unbinding, photoactivation/photoswitching) allows for isolated fluorophores to be localized one at a time, building up an image with ~10 nm localization accuracy. Examples include STORM[59], PALM[60], FPALM[61], and PAINT[62].
Another classic technique to probe protein dynamics in cells is single particle tracking (SPT; Box 1). SPT reveals behavior at the molecular level by tracking the motion of individual proteins, teasing out details not distinguished in methods that measure the average behavior of populations of molecules. Here, too, there have been significant recent technological advances. In the past, SPT relied on large, highly multivalent colloidal gold probes[3], which introduced complications associated with crosslinking of the molecule(s) being tracked, or relied on easily photobleached fluorochromes that limited tracking to very short time scales[4]. The introduction of improved fluorescent probes, such as the bright and photostable quantum dots (QDs; Box 2), greatly increases the duration of fluorescence-based SPT[4–6]. Another advantage of QDs is their broad excitation spectrum that allows for simultaneous excitation of many spectrally distinct QDs with a single wavelength. Combinations of multi-color probes and multi-spectral imaging allow for tracking of multiple protein species simultaneously, providing a reference frame for protein motion.
Box 2: Quantum Dots.
There are a range of fluorescent labels that can be used for visualizing proteins, including organic dyes and fluorescent proteins[69]. Quantum Dots (QDs) are a relatively new fluorescent probe composed of a semi-conducting nanocrystal core surrounded by a passivation shell and a water-soluble polymer coating[70]. A detailed comparison between QDs and organic fluorophores has recently been presented by Resch-Genger[71]. Here we briefly outline the common advantages and disadvantages of QDs.
Advantages
Photostability
QDs are photostable over minutes and even hours[4]. One caveat is that, with high excitation power, QDs can photodegrade resulting in a blue-shift of emission and eventual loss of signal[72].
Brightness
The large extinction coefficient and high quantum yield of QDs results in high brightness.
Flexible conjugation schemes
A variety of QD conjugation schemes are possible and many different coatings for QD conjugation are commercially available[70].
Broad excitation spectrum
QDs can be excited in a continuum of wavelengths below their emission spectrum, with absorption increasing towards the UV. This makes it possible to simultaneously excite many spectrally distinct QDs (or QDs plus GFPs or other dyes) with the same excitation wavelength.
Narrow emission spectrum
Their narrow emission spectra facilitate filter-based separation of spectrally distinct QDs.
Multiple colors
QDs are available in a spectrum of emission profiles.
Electron dense
The nanocrystal core can be imaged by electron microscopy[73].
Disadvantages*
Size
Typically 10–20 nm in diameter, much larger than an organic fluorophore, may cause steric interference with protein function.
Multi-valent
The multi-valency of QDs can be a complication when monovalent labeling is required. Methods have been developed to optimize 1:1 stoichiometry[74, 75].
Blinking
Intermittency of emission is a property of most QDs. This can cause complications in single QD tracking, such that when the QD is “off” the molecule can be lost. To minimize this complication, several single QD tracking algorithms have been developed[4, 5, 16]. Additionally, two groups have reported synthesis of non-blinking QDs[76, 77].
*These perceived disadvantages of QDs have been exploited by researches in particular experimental designs. For example, the blinking of QDs can be used for super-resolution imaging[78] and the multi-valency can be useful in situations where crosslinking of proteins is required[79].
Emerging fluorescence microscopy techniques can quantify protein behavior in the living cell and thus enable new and important discoveries that could not have been gained through biochemical techniques. In this review, we highlight some examples of how advanced fluorescent microscopy techniques have provided new insight into longstanding biological questions. In particular, we focus on two main areas of cell biology study: the influence of membrane microdomains on protein behavior and the quantification of protein-protein interactions. We also describe some technologies that we predict will have an increasing impact in cell biology.
Domains and corrals restrict membrane protein movements
Understanding the influence of membrane architecture on protein function is a major theme in membrane biology[7–9]. SPT has provided evidence for nanometer-sized membrane domains that restrict the lateral diffusion of membrane constituents. Along with biochemical fractionation techniques, these observations contributed importantly to several alternative hypotheses of membrane microdomain organization, such as lipid rafts[10], protein islands[11] and actin corrals[12, 13].
Elucidating the contributions of microdomains in restricting protein diffusion is needed to fully determine how these restrictions may limit receptors accessibility and govern signaling processes. For example, is membrane organization dominated by large scale segregations of proteins within the lipid sea (i.e., the protein island hypothesis)?[11] Alternatively, is membrane architecture primarily driven by phase separation of lipids and their associated proteins (i.e., the lipid raft hypothesis)?[10] How does the cortical cytoskeleton interact with proteins to create “confinement zones” (i.e., actin corrals or picket fence hypothesis where proteins collide with the cytoskeletal fence or transmembrane proteins bound to the cytoskeleton)?[14] Evidence from multicolor imaging and long-term SPT suggest that microdomains provide a plausible explanation to explain the restricted diffusion of proteins. Microscopy has also shown that this partitioning of proteins on the plasma membrane into confinement zones can be transient, where proteins transition from confined to free diffusion or between adjacent confinement zones (hop diffusion).
Diffusional trapping (Protein Islands)
In a paradigm-shifting study, Douglass and Vale used a combination of TIRF (total internal reflection fluorescence) microscopy, confocal imaging, FRAP and SPT to provide evidence that discrete microdomains in the T cell membrane transiently trap diffusing signaling proteins[15]. At the outset the authors observed that mRFP-tagged CD2, a non-raft transmembrane protein, was highly immobile. Moreover, the red fluorescent CD2 population could be imaged as large, relatively stable clusters at the adherent surface of T cells imaged in TIRF. In contrast, important signaling molecules such as the transmembrane adaptor LAT, the tyrosine phosphatase CD45 and the tyrosine kinase Lck displayed both highly mobile and immobile fractions. GFP fused to the amino terminal of Lck (Lck10-GFP), which results in dual acylation, served as a "lipid raft" marker. Contrary to the prediction that raft associated proteins would have a low diffusion coefficient, Lck10-GFP demonstrated both fast average diffusion and a very small immobile fraction. After acquiring a fixed image for CD2 fluorescence, the team switched to SPT mode to track GFP-tagged signaling proteins. By use of an overlay approach, they were able to evaluate the diffusional properties of single GFP-tagged proteins relative to the more stable CD2 regions. This provided the first concrete evidence for diffusional trapping, since both LAT and Lck were observed to have restricted mobility within CD2-defined microdomains. Abrupt changes in LAT and Lck mobility could often be seen, which correlated strongly with entry and exit from CD2 domains. This work was possible through creative use of multicolor imaging in SPT and ensemble modes, where the latter provided a reference frame for the individual molecules being tracked.
Ehrensperger and colleagues used similar SPT techniques to show that diffusional trapping occurs in neuronal cells, suggesting that plasma membranes of most cells harbor microdomains that restrict lateral movement of proteins[16]. In this study, motion of the neurotransmitter receptor, GlyR, was tracked as it traveled in and out of gephyrin clusters. While the gephyrin clusters were again marked with a chimeric gephyrin protein fused to the fluorescent protein Venus, the researchers chose to tag the GlyR with QDs. The use of QDs provided a higher signal/noise ratio and longer imaging times for tracking single GlyR molecules, compared to the GFP-tagged proteins in the Douglas and Vale study. This approach enabled the authors to extract the kinetic parameters that govern the equilibrium between GlyR exchanging in and out of gephryin clusters. Sophisticated mathematical analysis of QD-GlyR motion suggested that at least two subpopulation of receptors coexist within gephryin clusters and support novel hypotheses regarding multiple association states of protein complexes.
Actin corrals
By simultaneously tracking the dynamics of GFP-tagged actin and the trajectories of QD-labeled FcεRI, our group was the first to directly visualize actin “corralling” of membrane proteins[5] (Figure 1). This was a particularly important aspect of the study since, unlike CD2 or gephyrin clusters, the actin cytoskeleton is highly dynamic and the structural network is markedly altered on the order of seconds. Data were acquired both in TIRF, to image events at the adherent surface of mast cells, and in confocal mode, to image receptors and actin at the top of the cell. The use of QDs (Box 2) was imperative to provide long trajectories (>30s) without photobleaching so that spatial proximity and diffusional behavior of receptors relative to actin reorganization could be readily captured. This work demonstrated that membrane proteins can be transiently trapped within an actin-defined region of the membrane. Furthermore, as the cytoskeleton rearranges, the receptors can slip through temporary openings and move to new regions. Mathematical analysis of FcεRI trajectories as they approach actin revealed that the receptor does not interact with actin. Instead, actin acts as a physical barrier to protein diffusion.
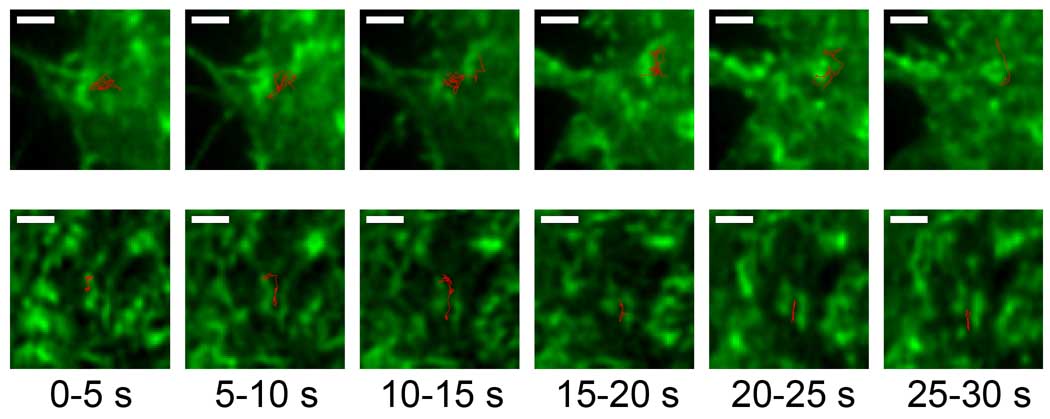
Figure 1. SPT reveals dynamic actin corrals.
Trajectories (red) of single QD655-IgE bound FcεRI can be tracked with respect to the underlying actin (green) in RBL cells expressing GFP-actin. The actin structures are dynamic on the timescale of ~1–10 seconds. Shown are two trajectories (red) broken into 5 second intervals. The actin image is the mean value of the 5 second interval. Scale bar = 1 µm. Unpublished images courtesy of Keith Lidke and similar to those found in Andrews et al[5].
Lipid Rafts
The apparent small size and dynamic nature of lipid rafts have generally precluded their detection with light microscopy. However, recent results from Eggeling and colleagues using STED (Box 1) have provided convincing evidence for lipid raft involvement in membrane compartmentalization[17]. The enhanced lateral resolution of STED (down to ~40 nm) makes it possible to probe very small areas of the membrane – regions on the order predicted for lipid raft size - and is ideal to create a smaller focal volume for FCS analysis. Results showed that fluorescently-labeled GPI-anchor proteins, as well as sphingolipids, are briefly trapped in nanometer sized (<20 nm) domains that are cholesterol dependent. An important feature of this paper is the comparison of single molecule traces acquired by STED with those acquired by confocal imaging, demonstrating that only the STED approach could capture the heterogeneous diffusion of sphingomyelin.
Clustering and oligomerization are measurable processes
An important goal in cell biology is to monitor the ebb and flow of molecular complexes that tune the overall responses of the cell. For example, what are the lifetimes of protein complexes formed during signaling and where do these productive protein interactions occur? The techniques discussed so far are powerful in their ability to obtain dynamic information from single molecules and relate individual protein behavior to its environment. Techniques that measure the behavior of a population of molecules at once can also be used to determine the location and time scale of protein interactions. Such techniques include FRAP, FRET and fluorescence correlation methods (Box 1).
Protein Oligomerization
As previously introduced, FRET is one approach used to detect close proximity between proteins. This technique has been successfully applied by many researchers to determine dynamic changes in protein-protein interactions. In one recent example, Liou and colleagues combined information from traditional FRET and FRAP to examine the oligomerization and translocation of STIM1, an endoplasmic reticulum (ER) resident whose role is to detect depleted calcium stores and stimulate store-operated calcium entry[18]. After triggering release of Ca2+ from ER stores, the authors observed a marked increase in energy transfer between CFP- and YFP-STIM1, consistent with protein aggregation. Live cell imaging also revealed that STIM1 translocates within the ER to form clusters near the plasma membrane (PM). This ER to PM translocation was apparently limited to locally available STIM1, based upon the very slow recovery of YFP-STIM1 after photobleaching. Together, the data suggest that signaling is spatially localized to a subpopulation of STIM1 located within a region of ~2 µm near putative ER-PM junctions.
Robia and colleagues have recently modified the FRET approach to determine protein exchange within an oligomer[19]. Förster transfer recovery (FTR) is a clever combination of FRET and FRAP in which the acceptor is photobleached and the recovery of energy transfer (rather than just intensity) in the bleached region is monitored over time. From the kinetics of the energy transfer recovery, one can determine the exchange rate between bleached and unbleached molecules in a complex. The authors used this technique with YFP- and CFP-fusion proteins to determine that phospholamban exchanges quickly in regulatory complexes with the sarcoplasmic reticulum Ca2+-ATPase (SERCA) but forms relatively stable homo-oligomers.
In the case of homo-interactions, the need for labeling with two different fluorophores is not ideal. Since the same principles that govern FRET between chemically different donor and acceptor fluorophores also applies to energy transfer between like fluorophores (i.e. GFP to GFP), the application of homo-FRET to live cell imaging makes it possible to quantify homo-clusters using a single class of fluorescent tag[20, 21]. Steady state and time-resolved fluorescence anisotropy imaging enables sensitive measurement of homo-FRET, based upon the rapid depolarization of the fluorescence. Importantly, homo-FRET can report the number of fluorophores in a complex or cluster. This approach was pioneered by Sharma et al, who focused on the clustering of GPI-anchor proteins (GPI-APs)[22]. This study showed that GPI-APs are found as both monomers and in nanoscale (<5nm), cholesterol-sensitive clusters on the plasma membrane. More recently, Bader et al[23] employed time-resolved anisotropy measurements in a specially equipped confocal microscope to spatially resolve differences in protein aggregation state across the cellular landscape (Figure 2). Using this instrument, they were able to distinguish that GPI-APs form small clusters on the plasma membrane, yet remain monomeric when residents of internal organelles.
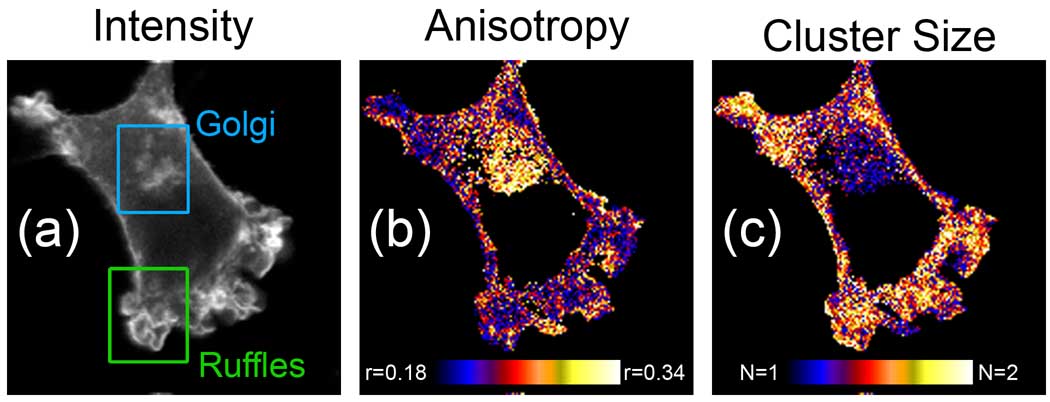
Figure 2. Homo-FRET imaging provides a map of protein aggregation.
(a) Intensity, (b) anisotropy (r) and (c) cluster size (N) images of NIH3T3 cell expressing GPI-GFP. Homo-FRET imaging reveals that GPI-GFP is found as small clusters in ruffles on the plasma membrane, but remains monomeric in the Golgi. Images courtesy of Arjen Bader and Hans Gerritsen and reproduced with permission from Bader et al[23].
Ligand-induced behavior
When used for multi-color imaging, SPT can also report protein-protein interactions[5, 24, 25]. While in the Jovin group, Lidke directly conjugated EGF to QDs to facilitate tracking of ligand-bound EGFR/erbB1[26]. The bright, photostable QD probes made it possible to directly observe the process of receptor internalization and, by use of cells stably transfected with erbB2-YFP or erbB3-mCitrine, to quantify difference in co-internalization of heterodimers. In a subsequent study, the use of EGF-QDs permitted detection of rapid retrograde erbB1 transport down cellular filopodia prior to endocytosis[24]. By combining single QD tracking and FRAP of GFP-actin, retrograde transport was shown to be coupled to actin flow rate and not dependent on a motor protein. This study was amongst the first to exploit the capabilities of two-color single QD tracking, establishing the stability of erbB1 homodimers bound to EGF-QD525 and EGF-QD605 and demonstrating that dimerization was a prerequisite for retrograde transport.
Mapping protein diffusion and aggregation
Another rapidly developing family of techniques involves the use of fluorescence correlation methods. FCS was first used to measure binding interactions in solution[27]. Cell biologists quickly adapted this technique to measure protein diffusion and aggregation state in living cells. As examples, FCS methods have been used to determine the aggregation state of membrane proteins[28], interactions between membrane proteins and downstream signaling partners[29] and the stoichiometry of protein complexes[30]. An inherent limitation of FCS is that measurements are recorded at a single position in the cell, which is defined at the beginning of the experiment. Image correlation techniques overcome this limitation by providing mobility and aggregation state data along with spatial information, generating maps of protein dynamics and interactions across a cell[31]. Importantly, these new methods can be performed using standard confocal laser scanning or TIRF microscopes that are widely available in academic core facilities. Image correlation methods have been applied to a wide variety of biological problems, such as measuring actin-integrin interactions by velocity mapping[32] and determining integrin aggregation state in adhesion organization[33]. Improvements in quantitation have been made, such as new analytical methods for calculating the fraction of interacting proteins from two-color ICCS data[34]. The Wiseman group makes available programs for analyzing data from many ICS techniques (http://wiseman-group.mcgill.ca/).
Recently, ccRICS and ccN&B analysis (Box 1) have emerged as promising new methods. Both are based on cross-correlating fluorescence in two-color pairs of images acquired by laser scanning confocal microscopy. Digman et al have used ccN&B to evaluate the exchange of focal adhesion kinase (FAK), paxillin and vinculin within adhesion complexes of fibroblasts[35]. Based upon the amplitude of the correlated fluctuations, it was possible to determine the stoichiometry of proteins in large aggregates that dissociated from adhesions as they disassembled. This group has also used ccRICS to create localized "brightness" maps, reflecting the dynamics of these proteins as they enter or leave adhesion structures[36]. As a concluding remark for this section, we note that attempts to capture the dynamics of FAK and vinculin binding to Pax by FRET methods were unsuccessful[36]. Clearly, the choice of analytical technique for measuring specific protein-protein interactions must sometimes be a trial and error process.
On the forefront
The examples described above demonstrate the ability of fluorescence microscopy to obtain information about biological processes that could not have been gained with conventional techniques. However, many other biological questions still exist that will require new technology, including: 1) While tracking multiple protein species simultaneously with multi-color imaging is useful, is there a way to see more than the typical 2 colors? 2) Can we track protein motion in three dimensions, permitting observations of proteins restricted by junctional complexes in polarized cells or moving through the interior of the cell? 3) Can we combine super-resolution with high temporal resolution to monitor dynamics at the nanometer scale? In the following sections, we describe applications of both established and new techniques that demonstrate these challenging questions can be addressed by the cell biologist.
How can we see more?
The ability to track the motion of specific proteins with respect to a reference (other proteins or lipids) using two-color imaging has already proven to provide much more information than single color imaging. In the quest to resolve more proteins, multiplex or multi-color imaging holds much promise. However, filter-based imaging systems are typically limited to 2–4 different fluorophores due to limitations in excitation sources and overlapping emission spectra of conventional dyes. New generation hyperspectral microscopes will greatly increase the number of labels that can be imaged simultaneously, such as the confocal imaging system developed by Sinclair et al. While several commercial hyperspectral microscopes are available (i.e. Zeiss META), this new instrument acquires the full emission spectrum at each sampled point with an exceptional 1–3 nanometer spectral resolution[37]. When combined with sophisticated analysis routines, even closely overlapping fluorophores can be distinguished[38]. Spectral imaging can also improve FRET measurements[39, 40].
Tracking protein motion in 3D
Traditional imaging techniques are limited to a 2D focal plane. Several groups are developing 3D tracking systems based on different approaches[41–45]. Ober and colleagues have designed an instrument that allows for simultaneous imaging in two focal planes: one at the membrane surface by TIRF and the second inside the cytoplasm by epifluorescence. Using this microscope, they have monitored protein endocytosis, recycling and exocytosis in real-time[43, 46]. Werner and colleagues have taken a different approach by developing a microscope that “locks onto” a single QD probe and tracks its motion in x, y, and z by moving the microscope stage to always keep the QD in focus[42, 47] (Figure 3). Manipulation of the excitation or emission light can also provide information in the z-dimension. In astigmatic imaging, a cylindrical lens is used to introduce an xy asymmetry in the fluorphore emission that is related to z-position[41]. This technique has recently been combined with STORM to generate super-resolution 3D images of the microtubule network[48] and in single QD tracking to study intracellular transport[49]. Pavani et al have demonstrated the ability to localize molecules in the z-dimension using a special shaped double-helix point spread function[50]. Hagen et al have recently developed a Programmable Array Microscope (PAM), that combines structured illumination and detection to produce video rate optical sectioning with photobleaching in arbitrary regions of interest, and used the PAM to measure diffusion of the membrane protein erbB3[51].
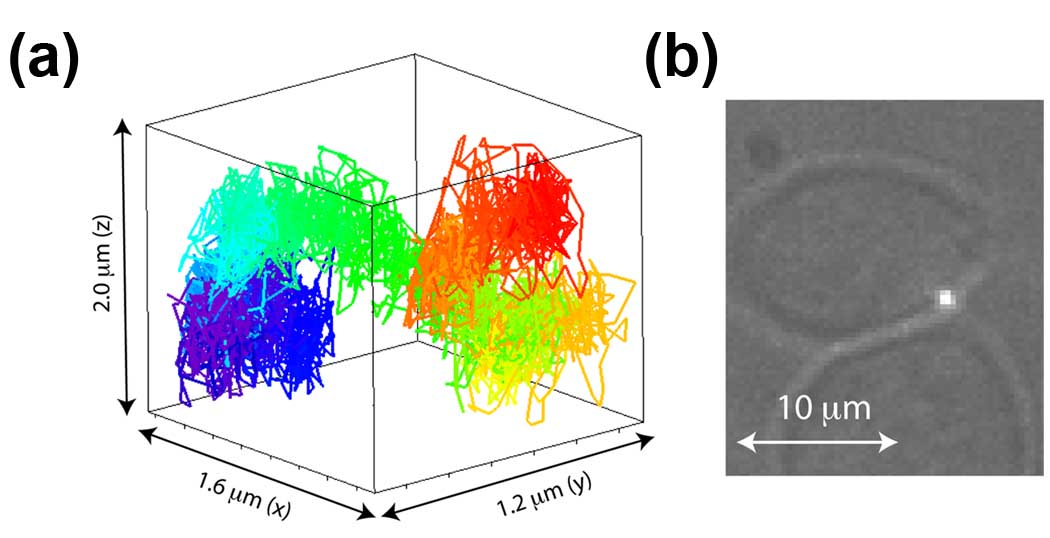
Figure 3. 3D tracking of membrane protein motion.
(a) 3D trajectory of QD-IgE-FcεRI diffusion along the side of the plasma membrane for 40 s. Trajectories are color coded as a function of time from start (red) to finish (violet) following the rainbow (ROYGBIV) scheme. (b) Transmission image of cell overlayed with the QD emission (white spot) to show the location of the tracked receptor. Unpublished figure courtesy of James Werner and similar to those found in Wells et al[47].
Capturing protein dynamics at super-resolution
Recently, a number of super-resolution imaging techniques have been developed that can “break” the diffraction limit of the light microscope and provide 100 nm or better resolution with light microscopy[52]. For example, new single molecule localization techniques are powerful ways to map protein localization, but initially relied on the sequential localization of individual fluorophores in fixed cells and required minutes to hours to generate super-resolution images. The field is now focused on bridging dynamic and super-resolution measurements through the use of novel, photoactivable probes in live cells. Techniques like sptPALM[53] and live-cell PALM[54], discussed in the article by Lippincott-Schwartz in this issue, are examples of this emerging technology. Live imaging with STED has also been performed to image dendrite spine or organelle motion with rates up to 1 frame every 10 s[55, 56]. Structured Illumination Microscopy (SIM) is another super-resolution technique that can increase lateral resolution over conventional microscopes. In SIM a sample is illuminated with a series of patterned light and computational analysis reconstructs the super-resolution images from high frequency information encoded in Moiré fringes[57] and has recently captured kinesin and microtubule dynamics with 100 nm resolution[58].
Bright future for quantitative imaging
The fluorescence imaging techniques described here are capable of capturing biochemical and biophysical events in the living cell. These emerging techniques provide the opportunity to examine cellular events on unprecedented scales in time and space. With up-and-coming technologies poised for innovative biological applications, the cell biologist will be able to address questions that have previously been experimentally inaccessible. Importantly, many of these techniques can be performed using commercially available instrumentation. The trend is clear: fluorescence microscopy will play an increasingly important role in cell biology, shaping the way cell biologists approach questions and providing quantitative information that compliments and extends traditional biochemical techniques.
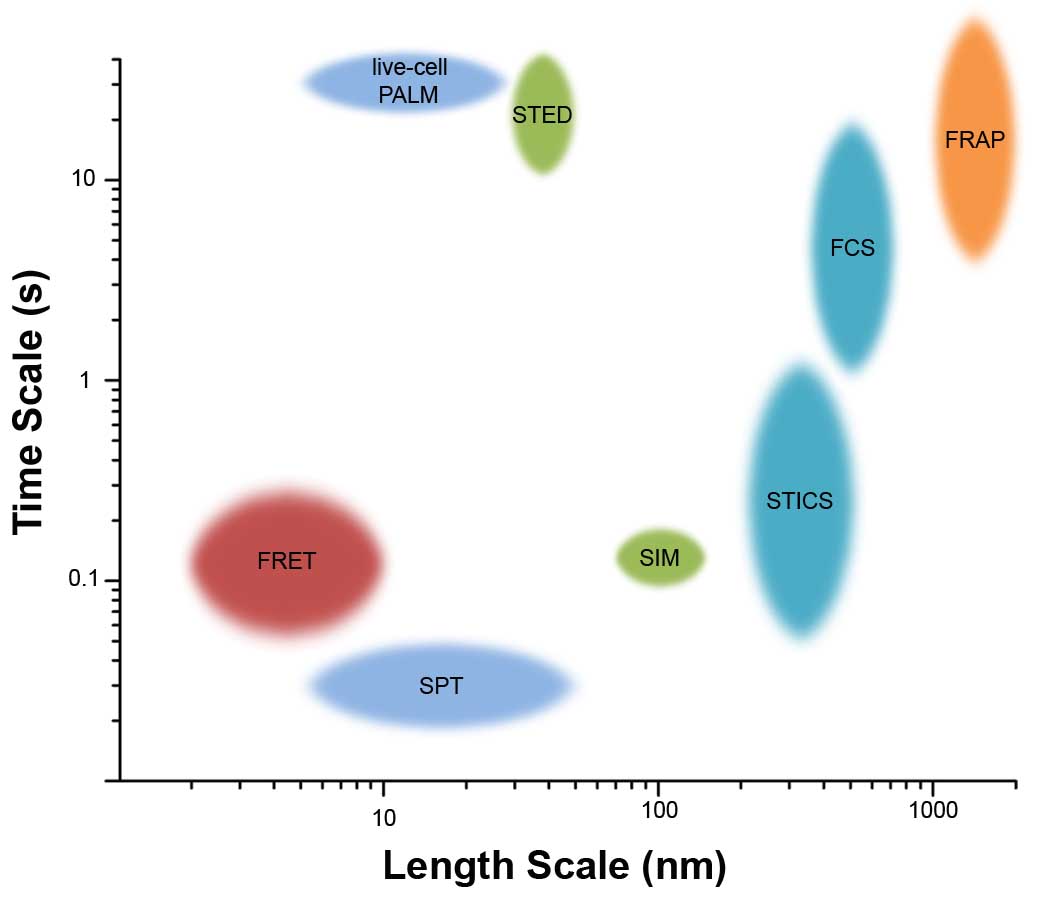
Box 1 Figure 1. Comparison of length and time scales accessible by fluorescence microscopy techniques.
Length scales refer to resolution (STED, SIM), localization accuracy (PALM, SPT), distance over which interactions can be detected (FRET), or the limiting size of the measurement field (FCS, ICS, FRAP). The time scale refers to the amount of time to complete one measurement, representing the maximum rate at which dynamic changes in the sample can be detected. In most cases, slower events and longer length scales can also be detected. Size of oval approximates the typical range of length and time scales in live cell imaging. Similar techniques are grouped by color.
Box 1 Table 1.
Comparison of fluorescence microscopy techniques
| Method | Measurables | Advantages | Limitations |
|---|---|---|---|
| FRAP [8, 9] |
|
|
|
| FRET [1, 18, 19] |
|
|
|
| Homo-FRET [21, 63] |
|
|
|
| SPT [5, 8, 9, 24, 25] |
|
|
|
| FCS/FCCS [28–30, 64, 65] |
|
|
|
| ICS/ICCS [31] |
|
|
|
| RICS/ccRICS [31, 36, 66] |
|
|
|
| N&B/ccN&B [35, 67] |
|
|
|
| STED [52, 55,56, 68] |
|
|
|
| Localization Microscopy [48, 52, 53, 59–61] |
|
|
|
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jares-Erijman EA, Jovin TM. FRET imaging. Nat Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 2.Grant DM, et al. Multiplexed FRET to image multiple signaling events in live cells. Biophys J. 2008;95:L69–L71. doi: 10.1529/biophysj.108.139204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saxton MJ, Jacobson K. Single-particle tracking: applications to membrane. Annu Rev Biophys Biomol Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- 4.Dahan M, et al. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science. 2003;302:442–445. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- 5.Andrews NL, et al. Actin restricts FcepsilonRI diffusion and facilitates antigen induced receptor immobilization. Nat Cell Biol. 2008;10:955–963. doi: 10.1038/ncb1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang YP, et al. Tracking bio-molecules in live cells using quantum dots. J Biophotonics. 2008;1:287–298. doi: 10.1002/jbio.200810029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day CA, Kenworthy AK. Tracking microdomain dynamics in cell membranes. Biochim Biophys Acta. 2009;1788:245–253. doi: 10.1016/j.bbamem.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marguet D, et al. Dynamics in the plasma membrane: how to combine fluidity and order. Embo J. 2006;25:3446–3457. doi: 10.1038/sj.emboj.7601204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owen DM, et al. Quantitative Microscopy: Protein Dynamics and Membrane Organisation. Traffic. 2009 doi: 10.1111/j.1600-0854.2009.00908.x. [DOI] [PubMed] [Google Scholar]
- 10.Pike LJ. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J Lipid Res. 2006;47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Lillemeier BF, et al. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc Natl Acad Sci U S A. 2006;103:18992–18997. doi: 10.1073/pnas.0609009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kusumi A, et al. Paradigm shift of the plasma membrane concept from the two dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu Rev Biophys Biomol Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 13.Kusumi A, et al. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys J. 1993;65:2021–2040. doi: 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujiwara T, et al. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J Cell Biol. 2002;157:1071–1081. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrensperger MV, et al. Multiple association states between glycine receptors and gephyrin identified by SPT analysis. Biophys J. 2007;92:3706–3718. doi: 10.1529/biophysj.106.095596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eggeling C, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 18.Liou J, et al. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci U S A. 2007;104:9301–9306. doi: 10.1073/pnas.0702866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robia SL, et al. Forster transfer recovery reveals that phospholamban exchanges slowly from pentamers but rapidly from the SERCA regulatory complex. Circ Res. 2007;101:1123–1129. doi: 10.1161/CIRCRESAHA.107.159947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levitt JA, et al. Fluorescence lifetime and polarization-resolved imaging in cell biology. Curr Opin Biotechnol. 2009;20:28–36. doi: 10.1016/j.copbio.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Tramier M, Coppey-Moisan M. Fluorescence anisotropy imaging microscopy for homo-FRET in living cells. Methods Cell Biol. 2008;85:395–414. doi: 10.1016/S0091-679X(08)85017-0. [DOI] [PubMed] [Google Scholar]
- 22.Sharma P, et al. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–589. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- 23.Bader AN, et al. Imaging of protein cluster sizes by means of confocal time-gated fluorescence anisotropy microscopy. Optics Express. 2007;15:6934–6945. doi: 10.1364/oe.15.006934. [DOI] [PubMed] [Google Scholar]
- 24.Lidke DS, et al. Reaching out for signals: filopodia sense EGF and respond by directed retrograde transport of activated receptors. Journal of Cell Biology. 2005;170:619–626. doi: 10.1083/jcb.200503140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roullier V, et al. High-Affinity Labeling and Tracking of Individual Histidine-Tagged Proteins in Live Cells Using Ni2+ Tris-nitrilotriacetic Acid Quantum Dot Conjugates. Nano Letters. 2009;9:1228–1234. doi: 10.1021/nl9001298. [DOI] [PubMed] [Google Scholar]
- 26.Lidke DS, et al. Quantum dot ligands provide new insights into erbB/HER receptor511 mediated signal transduction. Nature Biotechnology. 2004;22:198–203. doi: 10.1038/nbt929. [DOI] [PubMed] [Google Scholar]
- 27.Magde D, et al. Thermodynamic Fluctuations in a Reacting System - Measurement by Fluorescence Correlation Spectroscopy. Physical Review Letters. 1972;29:705–708. [Google Scholar]
- 28.Saffarian S, et al. Oligomerization of the EGF receptor investigated by live cell fluorescence intensity distribution analysis. Biophysical Journal. 2007;93:1021–1031. doi: 10.1529/biophysj.107.105494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larson DR, et al. Temporally resolved interactions between antigen-stimulated IgE receptors and Lyn kinase on living cells. Journal of Cell Biology. 2005;171:527–536. doi: 10.1083/jcb.200503110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Muller JD. Determining the stoichiometry of protein heterocomplexes in living cells with fluorescence fluctuation spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3147–3152. doi: 10.1073/pnas.0606557104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolin DL, Wiseman PW. Advances in image correlation spectroscopy: Measuring number densities, aggregation states, and dynamics of fluorescently labeled macromolecules in cells. Cell Biochemistry and Biophysics. 2007;49:141–164. doi: 10.1007/s12013-007-9000-5. [DOI] [PubMed] [Google Scholar]
- 32.Brown CM, et al. Probing the integrin-actin linkage using high-resolution protein velocity mapping. Journal of Cell Science. 2006;119:5204–5214. doi: 10.1242/jcs.03321. [DOI] [PubMed] [Google Scholar]
- 33.Wiseman PW, et al. Spatial mapping of integrin interactions and dynamics during cell migration by Image Correlation Microscopy. Journal of Cell Science. 2004;117:5521–5534. doi: 10.1242/jcs.01416. [DOI] [PubMed] [Google Scholar]
- 34.Comeau JWD, et al. Accurate measurements of protein interactions in cells via improved spatial image cross-correlation spectroscopy. Molecular Biosystems. 2008;4:672–685. doi: 10.1039/b719826d. [DOI] [PubMed] [Google Scholar]
- 35.Digman MA, et al. Stoichiometry of molecular complexes at adhesions in living cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2170–2175. doi: 10.1073/pnas.0806036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Digman MA, et al. Detecting protein complexes in living cells from laser scanning confocal image sequences by the cross correlation raster image spectroscopy method. Biophys J. 2009;96:707–716. doi: 10.1016/j.bpj.2008.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinclair MB, et al. Design, construction, characterization, and application of a hyperspectral microarray scanner. Applied Optics. 2004;43:2079–2088. doi: 10.1364/ao.43.002079. [DOI] [PubMed] [Google Scholar]
- 38.Haaland DM, et al. Hyperspectral Confocal Fluorescence Imaging: Exploring Alternative Multivariate Curve Resolution Approaches. Applied Spectroscopy. 2009;63:271–279. doi: 10.1366/000370209787598843. [DOI] [PubMed] [Google Scholar]
- 39.Forde TS, Hanley QS. Spectrally resolved frequency domain analysis of multi-fluorophore systems undergoing energy transfer. Appl Spectrosc. 2006;60:1442–1452. doi: 10.1366/000370206779321544. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Y, et al. Imaging lifetime and anisotropy spectra in the frequency domain. J Microsc. 2009;234:80–88. doi: 10.1111/j.1365-2818.2009.03145.x. [DOI] [PubMed] [Google Scholar]
- 41.Kao HP, Verkman AS. Tracking of single fluorescent particles in three dimensions: use of cylindrical optics to encode particle position. Biophys J. 1994;67:1291–1300. doi: 10.1016/S0006-3495(94)80601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lessard GA, et al. Three-dimensional tracking of individual quantum dots. Applied Physics Letters. 2007;91:224106. [Google Scholar]
- 43.Prabhat P, et al. Elucidation of intracellular recycling pathways leading to exocytosis of the Fc receptor, FcRn, by using multifocal plane microscopy. Proc Natl Acad Sci U S A. 2007;104:5889–5894. doi: 10.1073/pnas.0700337104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schutz GJ, et al. Imaging single molecules in three dimensions. Single Molecules. 2001;2:69–73. [Google Scholar]
- 45.Watanabe TM, et al. Three-dimensional nanometry of vesicle transport in living cells using dual-focus imaging optics. Biochemical and Biophysical Research Communications. 2007;359:1–7. doi: 10.1016/j.bbrc.2007.04.168. [DOI] [PubMed] [Google Scholar]
- 46.Ram S, et al. High accuracy 3D quantum dot tracking with multifocal plane microscopy for the study of fast intracellular dynamics in live cells. Biophys J. 2008;95:6025–6043. doi: 10.1529/biophysj.108.140392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wells NP, et al. Going beyond 2D: following membrane diffusion and topography in the IgE-Fc[epsilon]RI system using 3-dimensional tracking microscopy. In: Jorg E, et al., editors. SPIE; 2009. 71850Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang B, et al. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holtzer L, et al. Nanometric three-dimensional tracking of individual quantum dots in cells. Applied Physics Letters. 2007;90:053902. [Google Scholar]
- 50.Pavani SRP, et al. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2995–2999. doi: 10.1073/pnas.0900245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hagen GM, et al. Fluorescence recovery after photobleaching and photoconversion in multiple arbitrary regions of interest using a programmable array microscope. Microsc Res Tech. 2009;72:431–440. doi: 10.1002/jemt.20686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang B, et al. Super-resolution fluorescence microscopy. Annu Rev Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manley S, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nature Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 54.Shroff H, et al. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat Methods. 2008;5:417–423. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hein B, et al. Stimulated emission depletion (STED) nanoscopy of a fluorescent protein-labeled organelle inside a living cell. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14271–14276. doi: 10.1073/pnas.0807705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagerl UV, et al. Live-cell imaging of dendritic spines by STED microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18982–18987. doi: 10.1073/pnas.0810028105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heintzmann R, Ficz G. Breaking the resolution limit in light microscopy. Brief Funct Genomic Proteomic. 2006;5:289–301. doi: 10.1093/bfgp/ell036. [DOI] [PubMed] [Google Scholar]
- 58.Kner P, et al. Super-resolution video microscopy of live cells by structured illumination. Nat Methods. 2009;6:339–342. doi: 10.1038/nmeth.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rust MJ, et al. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nature Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 61.Hess ST, et al. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophysical Journal. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharonov A, Hochstrasser RM. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc Natl Acad Sci U S A. 2006;103:18911–18916. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thaler C, et al. Structural rearrangement of CaMKIIalpha catalytic domains encodes activation. Proc Natl Acad Sci U S A. 2009;106:6369–6374. doi: 10.1073/pnas.0901913106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chiantia S, et al. Fluorescence correlation spectroscopy in membrane structure elucidation. Biochim Biophys Acta. 2009;1788:225–233. doi: 10.1016/j.bbamem.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 65.Haustein E, Schwille P. Fluorescence correlation spectroscopy: novel variations of an established technique. Annu Rev Biophys Biomol Struct. 2007;36:151–169. doi: 10.1146/annurev.biophys.36.040306.132612. [DOI] [PubMed] [Google Scholar]
- 66.Digman MA, et al. Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophysical Journal. 2005;89:1317–1327. doi: 10.1529/biophysj.105.062836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Digman MA, et al. Mapping the number of molecules and brightness in the laser scanning microscope. Biophys J. 2008;94:2320–2332. doi: 10.1529/biophysj.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klar TA, et al. Stimulated emission depletion microscopy with an offset depleting beam. Applied Physics Letters. 2001;78:393–395. [Google Scholar]
- 69.Giepmans BNG, et al. Review - The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 70.Smith AM, et al. Engineering luminescent quantum dots for In vivo molecular and cellular imaging. Annals of Biomedical Engineering. 2006;34:3–14. doi: 10.1007/s10439-005-9000-9. [DOI] [PubMed] [Google Scholar]
- 71.Resch-Genger U, et al. Quantum dots versus organic dyes as fluorescent labels. Nat Methods. 2008;5:763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 72.Grecco HE, et al. Ensemble and single particle photophysical properties (Two-Photon excitation, anisotropy, FRET, lifetime, spectral conversion) of commercial quantum dots in solution and in live cells. Microscopy Research and Technique. 2004;65:169–179. doi: 10.1002/jemt.20129. [DOI] [PubMed] [Google Scholar]
- 73.Giepmans BNG, et al. Correlated light and electron microscopic imaging of multiple endogenous proteins using Quantum dots. Nature Methods. 2005;2:743–749. doi: 10.1038/nmeth791. [DOI] [PubMed] [Google Scholar]
- 74.Howarth M, et al. Monovalent, reduced-size quantum dots for imaging receptors on living cells. Nature Methods. 2008;5:397–399. doi: 10.1038/nmeth.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lidke DS, et al. Biotin-ligand complexes with streptavidin quantum dots for in vivo cell labeling of membrane receptors. Methods Mol Biol. 2007;374:69–79. doi: 10.1385/1-59745-369-2:69. [DOI] [PubMed] [Google Scholar]
- 76.Chen Y, et al. "Giant" multishell CdSe nanocrystal quantum dots with suppressed blinking. Journal of the American Chemical Society. 2008;130:5026–5027. doi: 10.1021/ja711379k. [DOI] [PubMed] [Google Scholar]
- 77.Wang X, et al. Non-blinking semiconductor nanocrystals. Nature. 2009;459:686–689. doi: 10.1038/nature08072. [DOI] [PubMed] [Google Scholar]
- 78.Lidke KA, et al. Superresolution by localization of quantum dots using blinking statistics. Optics Express. 2005;13:7052–7062. doi: 10.1364/opex.13.007052. [DOI] [PubMed] [Google Scholar]
- 79.Andrews NL, et al. Small, mobil FceRI aggregates are signaling competent. Immunity. doi: 10.1016/j.immuni.2009.06.026. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]