Summary
IRES elements are highly structured RNA sequences that function to recruit ribosomes for the initiation of translation. In contrast to the canonical cap binding, ribosome-scanning model, the mechanism of IRES-mediated translation initiation is not well understood. IRES elements, first discovered in viral RNA genomes, were subsequently found in a subset of cellular RNAs as well. Interestingly, these cellular IRES-containing mRNAs appear to play important roles during conditions of cellular stress, development, and disease (e.g., cancer). It has been shown for viral IRESes that some require specific IRES trans-acting factors (ITAFs), while others require few if any additional proteins and can bind ribosomes directly. Current studies are aimed at elucidating the mechanism of IRES-mediated translation initiation and features that may be common or differ greatly among cellular and viral IRESes. This review will explore IRES elements as important RNA structures that function in both cellular and viral RNA translation and the significance of these structures in providing an alternative mechanism of eukaryotic translation initiation.
Keywords: internal ribosome entry site (IRES), cap-independent translation, eukaryotic translation, IRES trans-acting factors (ITAFs), RNA secondary structure
Introduction
Eukaryotic cells have many strategies to regulate the translation of mRNA for the synthesis of protein. Beyond the canonical cap-dependent model of cap recognition and ribosomal scanning, an alternative method of cap-independent translation was described first for picornaviruses and subsequently for a growing subset of cellular mRNAs. In this 5′ end-independent mechanism of translation initiation, ribosomes are recruited to the mRNA by an internal ribosome entry site (IRES). IRESes are RNA structural elements that function in the binding of ribosomes to the mRNA. IRES elements mediate translation of viral mRNAs and modulate translation of cellular mRNAs during development, stress, and disease, suggesting that these RNA structures are significant for directing translation in cellular contexts in which cap-dependent translation is down-regulated. This review will examine the mechanisms of translation initiation and the differences between cap-dependent and IRES-directed translation. Structural features of IRES elements and the identification of specific ITAFs will then be discussed. Cellular and viral IRESes will be individually explored, including ITAFs important for stimulation of translation, mechanistic possibilities for the recruitment of ribosomes by viral IRESes, and caveats to the identification of cellular IRESes. Finally, the significance of an alternative mechanism of translation initiation will be examined.
Mechanisms of translation initiation
Features of cap-dependent translation initiation
Most translational regulation occurs at the initiation stage; therefore emphasis has been placed on studying the molecular details of conventional and alternative mechanisms of translation initiation; for reviews, see [1,2]. Translation of cellular mRNAs most often occurs via a cap-dependent mechanism of initiation. As shown in Figure 1A, cellular mRNAs contain a 7-methyl guanosine cap at the 5′ end of the RNA, and this cap structure is recognized by the eukaryotic initiation factor 4F (eIF4F) cap-binding complex. The eIF4F complex consists of the initiation factors 4A, 4G, and 4E and recruits the ribosome to the mRNA for translation initiation. eIF4E is the cap-binding component of the complex, while eIF4G is a scaffolding protein that binds eIF4E, eIF4A and the mRNA. eIF4A is an ATP-dependent helicase responsible for unwinding the RNA secondary and tertiary structure during translation. This helicase activity is stimulated by the associated factor, eIF4B. The 40S ribosomal subunit binds a protein complex that consists of eIF1, eIF2-GTP-Met-tRNA (i.e., the ternary complex), eIF3, and eIF5. The assembled protein complex, termed the 43S pre-initiation complex, binds the mRNA at the cap structure via interaction of a central domain of eIF4G with eIF3. This mRNA-bound complex is referred to as the 48S complex. The bound pre-initiation complex then scans along the RNA until an AUG start codon is recognized in a favorable context [3]. When a proper start codon is encountered, GTP is hydrolyzed to GDP in the presence of eIF5, and several of the initiation factors dissociate. The large ribosomal subunit then joins the small subunit to generate an elongation-competent 80S ribosome. At this point protein synthesis begins, and initiation factors are recycled for subsequent rounds of initiation. The cellular protein poly(A)-binding protein (PABP), which binds the 3′ poly(A) tracts of cellular RNAs and also interacts with eIF4G, allows for circularization of the mRNA and provides a context for multiple rounds of translation initiation. Some cellular conditions such as stress or viral infection can result in a down-regulation of cap-dependent translation, often by interfering with initiation factors that play an important role in the cap-dependent mechanism of initiation; this topic will be discussed in a later section.
Figure 1. Recruitment of the 43S pre-initiation complex in the cap-dependent and cap-independent mechanisms of translation initiation.
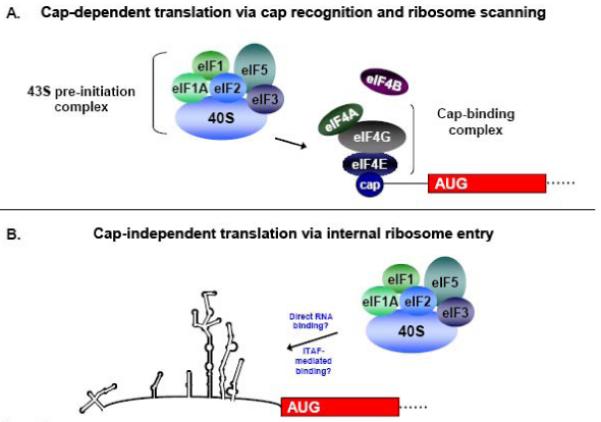
(A) In cap-dependent translation, the eIF4F cap-binding complex recognizes and binds to the 5′ cap structure of the mRNA. Following cap binding, the 43S complex scans the mRNA until an AUG is encountered in a favorable context. After GTP hydrolysis and 60S subunit joining the ribosome is now elongation-competent, and protein synthesis begins. (B) In the cap-independent mechanism of initiation, the 43S pre-initiation complex associates with RNA sequences in the IRES either directly or in conjunction with canonical or non-canonical initiation factors to facilitate initiation at the appropriate AUG start codon. Non-canonical factors are indicated as IRES trans-acting factors, or ITAFs. (Figure adapted from Semler and Waterman, 2008 [100]).
Features of cap-independent translation initiation
In the cap-independent mechanism of translation initiation, ribosomes are recruited to the RNA via an unknown mechanism. The 40S ribosomal subunit recognizes an RNA sequence, structure, or ribonucleoprotein complex within the 5′ noncoding region (5′ NCR) of the RNA, and initiation at the authentic start codon can occur several hundred nucleotides downstream from the 5′ end of the RNA. This alternative form of translation initiation does not require a 5′ cap as a site of assembly for initiation factors, and cap-recognition of the 40S ribosomal subunit via the intact eIF4F cap binding complex does not occur. In addition, the RNA may be highly structured in nature, and thus ribosomes may not be able to successfully scan through the noncoding region to reach the authentic initiation site (see Figure 1B). The cap-independent mechanism of initiation therefore involves features that are distinct from the canonical cap-binding, ribosome scanning model. Since this mechanism of translation initiation has been observed in cells during conditions such as development, stress, or disease, there is likely a significant biological relevance for this type of translation in the cell. Taken together, these factors highlight the inherent differences between cap-dependent and cap-independent translation initiation.
Overview of Internal Ribosome Entry Site (IRES) Elements and RNA Binding Proteins
IRES elements
Internal initiation of translation was first observed for viral RNAs during picornavirus infection. The uncapped viral RNA genomes of poliovirus (PV) and encephalomyocarditis virus (EMCV) were found to contain sequences in their 5′ NCRs that mediate efficient translation in eukaryotic cells via the internal binding of ribosomes [4-6]. These RNA elements were termed internal ribosome entry sites, or IRESes. During nearly all picornavirus infections, cap-dependent translation is shut down and the viral RNA utilizes IRES-mediated translation initiation to direct the synthesis of viral proteins. The details of virus-induced down-regulation of cap-dependent translation will be discussed later in more detail.
IRES trans-activating factors (ITAFs)
IRES elements typically consist of long, highly-structured RNA sequences that function to recruit ribosomes to the RNA, although the mechanism of recruitment has not been fully defined and may vary among different IRES elements. For example, cricket paralysis virus (CrPV), a member of the family Dicistroviridae, does not require any of the canonical eIFs for translation initiation and appears to recruit ribosomes directly via the secondary and tertiary structures of the IRES RNA [7-9]. Other IRES elements require at least a subset of eIFs as well as certain RNA binding proteins to facilitate IRES-mediated translation [10-15]. These non-canonical factors are termed IRES trans-activating factors, or ITAFs. Several ITAFs have been previously shown to bind IRES elements and function in IRES mediated translation. Most notably, polypyrimidine tract binding protein (PTB, also hnRNP I), poly(rC) binding protein 2 (PCBP2, also hnRNP E), hnRNP C1/C2, hnRNP D, upstream of n-ras (unr), ITAF45, and the lupus autoantigen (La) have all been implicated in IRES-mediated translation [16-23]. Of particular interest is the identification of numerous hnRNPs (heterogeneous nuclear ribonucleoproteins) as ITAFs, since these RNA binding proteins play roles in pre-mRNA processing, mRNA export, localization, stability, and translation [24].
Functional classification of IRES elements
There is no overall pattern regarding which RNA binding proteins are required for IRES-mediated translation of cellular or viral IRESes, although the requirement of any or all of these ITAFs contributes to the characterization of IRES functional classes. Also contributing to the idea of different functional classes of IRES elements is the fact that different IRESes have varying efficiencies based on the cell type or context in which translation occurs [25]. This cell-type specificity may be attributed to the availability of cellular factors required for translation of a particular IRES. Importantly, there is a lack of similarity in primary sequence among cellular IRESes. Classification of cellular IRESes would therefore require a large number of groups to account for the different cellular contexts and cellular proteins that may contribute to translation of the RNA. Viral IRES elements may share more features of translation regulation, and have been categorized based on their structure and requirements for ITAFs. The broad classification of IRES elements will be discussed further in the following sections.
Viral IRESes
Picornavirus IRES-mediated translation
IRES elements were first discovered in the genomes of picornavirus RNAs and have since been demonstrated in many additional families of viruses; refer to Table 1. Picornaviruses cause a range of diseases in humans including poliomyelitis, the common cold, myocarditis, and hepatitis. Members of this family include poliovirus (PV), coxsackievirus B3 (CVB3), human rhinovirus (HRV), hepatitis A virus (HAV), foot-and-mouth disease virus (FMDV), encephalomyocarditis virus (EMCV), Theiler’s murine encephalomyocarditis virus (TMEV), enterovirus 71 (EV71), and porcine teschovirus serotype 1 (PTV-1). Picornaviruses contain a ∼7.0 kb - 8.5 kb positive-sense, single-stranded RNA genome. The genome consists of a single open-reading frame which is translated to generate a polyprotein that is proteolytically processed. Genomic RNAs lack a 5′ cap structure and direct translation initiation via an IRES in the 5′ NCR. Picornaviruses are cytoplasmic viruses, and both viral translation and RNA replication occur in the cytoplasm of infected cells. Due to their limited coding capacity, these viruses have evolved to utilize host cell factors along with encoded viral factors and RNA secondary structures to drive viral RNA replication and translation. Viral proteinases cleave several cellular proteins, including eIF4G, to down-regulate host cell translation during infection by poliovirus, human rhinovirus, or coxsackievirus [26-28]. Other work has shown that PABP is cleaved by poliovirus and coxsackievirus proteinases and that this cleavage correlates with host cell translation shut-off [29,30]. Hepatitis A virus, however, is unique among picornaviruses in that it does not cleave translation initiation factors, and in fact requires an intact eIF4G for viral translation [31].
Table 1. Examples of viral IRES elements.
| Virus Family | Virus | Known ITAFs |
|---|---|---|
| DNA viruses | ||
| Herpesvirus | Kaposi-sarcoma-associated herpesvirus (KSHV) | |
| Polyomavirus | Simian vacuolating virus 40 (SV40) | |
| Nimavirus | White spot syndrome virus (WSSV) | |
| RNA viruses | ||
| Picornaviruses | Poliovirus (PV) | PTB, PCBP2, PCBP1, La, unr SRp20 |
| Coxsackievirus B3 (CVB3) | PCBP2, La | |
| Enterovirus 71 (EV71) | ||
| Hepatitis A virus (HAV) | PTB, PCBP2 | |
| Human rhinovirus (HRV) | PTB, PCBP2, La, unr hnRNP A1 |
|
| Foot-and-mouth disease virus (FMDV) | PTB, ITAF45, La | |
| Encephalomyocarditis cirus (EMCV) | PTB, La | |
| Theiler’s murine encephalomyelitis virus (TMEV) | PTB | |
| Porcine teschovirus serotype 1 (PTV-1) | ||
| Potyviruses | Cowpea mosaic virus (CPMV) Tobacco etch virus (TEV) |
|
| Flaviviruses | Hepatitis C virus (HCV) | PTB, PCBP2, La, hnRNP D, hnRNP L |
| Bovine viral diarrhea virus (BVDV) Classical swine fever virus (CSFV) |
||
| Dicistroviruses | Cricket paralysis virus (CrPV) Drosophila C virus (DCV) Plautia stali intestine virus (PSIV) |
|
| Retroviruses | Human T-call lymphotropic virus 1 (HTLV-1) | PTB, La |
| Moloney murine leukenia virus (MoMuLV) Rous sarcoma virus (RSV) Simian immunodeficiency virus (SIV) |
||
| Human immunodeficiency virus type 1 (HIV) | La | |
Some of the IRES elements identified in viral RNAs are shown, separated into DNA and RNA viruses, and including the virus family. Also shown are the known cellular IRES trans-acting factors (ITAFs) that function in translation for each particular viral IRES. Note that only non-canonical ITAFs are listed; this list does not include any canonical eukaryotic translation initiation factors (eIFs). Data compiled from [82,112,131,132].
Picornaviruses have been grouped into four major categories based on IRES structural similarity and ITAF requirements for translation. Enteroviruses (PV and CVB3) and rhinoviruses (HRV) contain Type I IRES elements (structure is shown in Figure 2A). By convention, Type I IRES secondary structures are denoted by roman numerals. For these IRESes, A/C-rich sequences are found in stem-loop IV and the apical loop of stem-loop V, and a pyrimidine-rich region is located in the first loop of stem-loop VI [32]. Aphthoviruses (FMDV) and cardioviruses (EMCV and TMEV) contain Type II IRES elements (see Figure 2B). By convention letters are used to designate RNA secondary structure elements for these IRESes. The Type II 5′ NCRs contain twelve stem-loop structures, A-L, and the IRES elements of FMDV and EMCV are contained within stem-loops D-L. For the Type II IRESes, A/C-rich sequences are found in the two loops of stem-loop I, and an A-rich loop is found in stem-loop K. A pyrimidine-rich region is located beyond stem-loop L.
Figure 2. Structural features of Type I and Type II picornavirus IRESes.
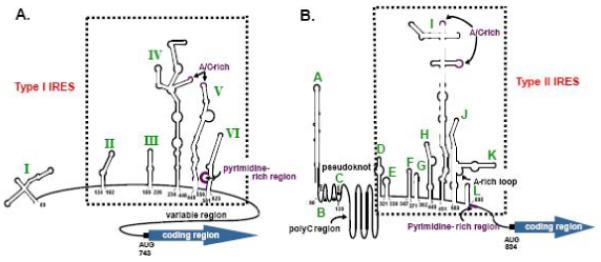
RNA secondary structures of the poliovirus (A) and encephalomyocarditis virus (B) 5′ NCRs based on chemical and enzymatic structure probing. The poliovirus IRES consists of stem-loops II-VI (numbering refers to PV type 1), the EMCV IRES consists of stem-loops D-L (numbering refers to EMCV-R strain), and each is boxed. Also shown in both IRES structures are the conserved A/C-rich loops, the pyrimidine-rich region, and the authentic AUG start codon. For the EMCV IRES, a region predicted to form pseudoknots is indicated as well as a long poly(C) tract. (Figure modified from Stewart and Semler, 1997 [32]).
Although not shown here, the Type III IRES element of HAV consists of stem-loops IIIa-V. The stem-loop structures in the HAV IRES somewhat resemble the structures of Type II IRESes; however, the functional requirements for Type III IRES activity are different from those of the Type I or Type II classes of IRESes. Notably, the HAV IRES requires an intact eIF4F complex for its function, and it has been recently shown to be inhibited by endogenous levels of La [31,33-35].
The Type IV IRESes (also not shown), such as the one encoded in the PTV-1 genomic RNA, are termed “HCV-like” IRESes. Their structural elements lack some characteristics that are common to the other types of picornavirus IRESes. The PTV-1 IRES sequence lacks any significant polypyrimidine tract and is neither stimulated nor inhibited by co-expression of an enterovirus 2A proteinase [36]. This IRES was determined to be 270 nucleotides in length [37,38], which is smaller than other previously characterized picornavirus IRES elements. There is evidence for direct interaction between the PTV-1 IRES and eIF3 as well as 40S ribosomal subunits [37], which is consistent with the similarity of Type IV IRESes to the HCV IRES. Further evidence for the similarity of HCV and PTV-1 comes from studies that inhibit the function the eIF4A. Blocking the activity of eIF4A (via dominant negative mutants or by hippuristanol treatment) also blocks the activity of the EMCV and PV IRES elements, while the activity of the HCV and PTV-1 IRES elements is largely unaffected [39-41].
The picornavirus IRESes have varying efficiencies in different cell types or in different cellular contexts; for example, EMCV translates much more efficiently than poliovirus in HeLa cell cytoplasmic extracts that have been depleted of cellular protein PCBP2, known to be required for Type I but not Type II IRES translation [21]. EMCV, in fact, requires very few ITAFs for IRES-mediated translation [42]. This observation and others have supported the idea that IRES activity differs among the viral IRESes as a result of variation in secondary and tertiary RNA structures as well as differences in ITAF requirements for stimulation of cap-independent translation.
RNA structures and ITAFs that function in picornavirus translation
IRES sequences and structures are the binding sites for canonical and non-canonical translation factors, but the way in which these elements function to assemble initiation complexes is still not clear. As shown in Figure 2A, the 5′ NCRs of enterovirus and rhinovirus RNAs are predicted to contain six stem-loop structures, with stem-loop I (or the cloverleaf) at the very 5′ end of the RNA, and the IRES consisting of stem-loops II-VI [32]. Stem-loop IV is particularly important for the IRES functions of enteroviruses and rhinoviruses, and a three nucleotide insertion in this stem-loop results in a non-infectious poliovirus genome [43].
Several cellular proteins are known to interact with the 5′ NCRs of picornaviruses, and a subset of these has been shown to have an effect on viral translation (refer to Table 1). Two cellular proteins that stimulate picornavirus IRES-mediated translation are La and PTB [10,11,17,44,45]. It was initially observed that viral RNA translation for poliovirus was restricted in rabbit reticulate lysate but was stimulated by the addition of HeLa cell cytoplasmic extracts [46,47]. This observation suggested that IRES-mediated translation required ITAFs that are present or more abundant in HeLa cell extracts and absent or in lower abundance in reticulocyte lysate. Indeed, the La protein was shown to stimulate authentic poliovirus translation in vitro when added to translation reactions containing rabbit reticulocyte lysate. La is an RNA binding protein that has many targets of both low- and high-affinity and it has been reported to have differential effects on picornavirus translation [35,48,49]. However, Costa-Mattioli and colleagues reported cell culture studies that provided additional evidence for the role of La in stimulating poliovirus IRES-mediated translation [11].
Subsequent studies then led to the identification of PTB as a stimulatory ITAF for poliovirus and rhinovirus [50]. Another cellular protein, unr, interacts with the human rhinovirus IRES and this interaction was found to be necessary for efficient HRV translation [12]. PCBP2 is a cellular RNA binding protein that is known to bind the 5′ NCR of several picornavirus RNAs. PCBP2 functions in viral translation for Type I IRESes but not for Type II IRESes, as mentioned previously. Poliovirus requires the binding of PCBP2 to stem-loop IV in the IRES and also requires the interaction of PCBP2 with cellular splicing factor SRp20, although the role this interaction plays in viral translation is not completely understood [51]. In addition to several non-canonical factors, canonical translation initiation factors have also been shown previously to interact with picornavirus IRES elements [13,52]. One example is eIF4B (see Figure 3A), which binds to poliovirus and FMDV IRES elements and is associated with 48S and 80S complexes [14,53].
Figure 3. The FMDV and HCV IRESes have differential ITAF requirements for the initiation of cap-independent translation.
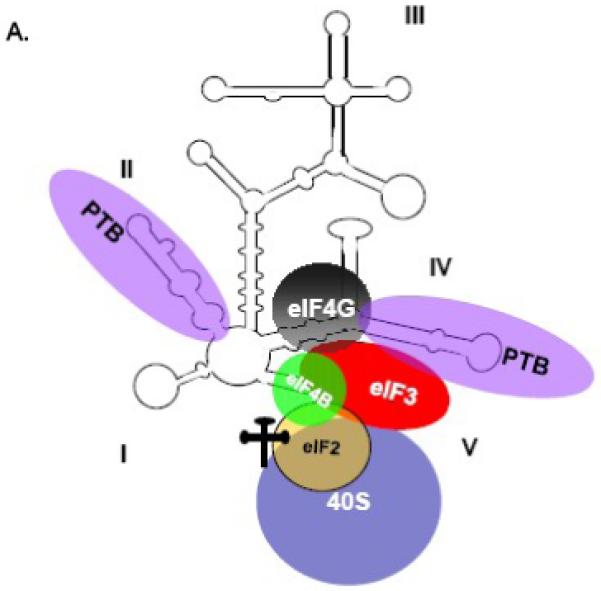
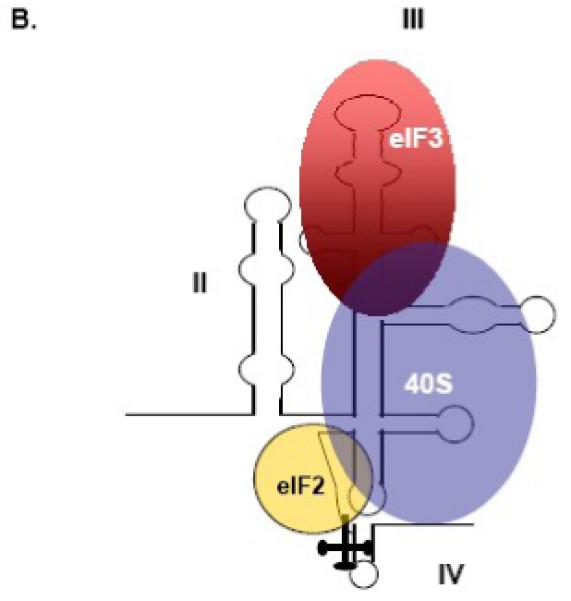
(A) RNA-protein interactions that function in FMDV translation. A schematic representation of the FMDV IRES is shown, and domain numbering is taken from Lopez de Quinto and Martinez-Salas, 2000 [135]. FMDV requires binding of canonical and non-canonical factors for IRES-mediated translation. The PTB binding sites were described by Luz and Beck and by Kolupaeva and colleagues [136,137]. The binding sites for eIF4G and eIF4B were taken from Lopez de Quinto and Martinez-Salas, 2000 [135]. (B) RNA-protein interactions that function in HCV translation. A schematic representation of the HCV IRES is shown, and domain numbering is taken from the work of Honda and colleagues [138]. HCV requires few canonical initiation factors for IRES-directed translation; shown are eIF3, eIF2 and the 40S ribosomal subunit. The eIF3 and 40S subunit binding sites have been described [71,139,140]. (Figure was redrawn from Martinez-Salas et al., 2001, with permission [112]).
As shown in Figure 2B, the 5′ NCRs of cardioviruses and aphthoviruses are much longer than those found in enterovirus RNAs [54]. Both EMCV and FMDV encode a Leader (L) protein, a feature of only cardio- and aphthoviruses. The L protein functions as a proteinase for FMDV [55,56], but not for EMCV; for review, see [57]. FMDV contains a hairpin structure termed S instead of a cloverleaf, and encodes a long poly(C) tract following the S region [58]. Additional motifs are observed in sequences upstream of the coding region for FMDV, including a region predicted to form multiple pseudoknots and the cis-acting replication element (cre) [59,60]. In addition, the FMDV genomic RNA contains two in-frame AUG triplets to initiate translation, resulting in two forms of the Leader (L) proteinase denoted Lab and Lb [61,62].
Some structural similarities exist among IRESes of different classes. As one example, a recognition motif for RNase P has been found in the EMCV IRES, but its exact position has not yet been defined [63]. This feature was shown previously for the HCV IRES [64]. However, some differences have been observed among IRESes of the same classification. Although EMCV and FMDV are similar in regard to RNA structural elements, they have different requirements for ITAF association. One case highlighting this difference is the requirement for ITAF45 between FMDV and EMCV. FMDV requires ITAF45 (also known as Ebp1) for IRES activity, while EMCV is not sensitive to depletion of ITAF45 [65,66]. Although there are similarities among IRES elements in terms of structures and ITAF requirements, there are also specific differences between IRESes - even among those of the same classification. The lack of complete conservation of structural elements among viral IRESes and the differences among the viral IRESes in ITAF requirements have made stringent classification difficult.
Ribosome recruitment during picornavirus infection
Several mechanistic possibilities have been developed to describe how IRES-mediated translation initiation may occur in a picornavirus-infected cell. Ribosomes may be recruited directly via interaction with identified ITAFs (e.g., PCBP2) bound to the viral RNA. It is also possible that ITAF binding to the RNA may stabilize the secondary or tertiary structure and allow for direct or indirect recruitment of ribosomes. An additional possibility is that ribosomes may be directly or indirectly recruited to the RNA via a larger protein complex, including identified ITAFs as well as yet unidentified factors and canonical translation initiation factors. For poliovirus, it was discovered that PCBP2, which binds the viral RNA, also interacts with cellular protein SRp20 and that these two proteins are required for viral translation [51]. Therefore it is possible that PCBP2 binding to the RNA stabilizes the secondary or tertiary structure of the RNA, but it is additionally possible that PCBP2 interacts (directly or indirectly) with other ITAFs and/or canonical translation initiation factors to recruit the ribosome to the RNA for the initiation of translation. These and other possible methods of ribosome recruitment are currently being explored to better define the mechanism of initiation for picornavirus RNA.
Other families of viral IRESes: Flaviviridae and Dicistroviridae
Flaviviridae and Dicistroviridae are two families of viruses that contain members with IRES elements (refer to Table 1), but these viruses direct translation in a way that is unique and different from the picornaviruses; for review, see [67,68]. The genomic RNA of the flavivirus hepatitis C virus (HCV) contains a ∼300 nucleotide-long IRES, while the dicistrovirus cricket paralysis virus (CrPV) genome has a ∼200 nucleotide-long intergenic region (IGR) known to contain an IRES [67]. The minimal set of factors required for HCV 48S complex formation was determined by in vitro reconstitution assays, utilizing purified translation components [69,70]. Noteworthy is that 48S complex assembly has not yet been reconstituted in vitro for the picornaviruses, likely owing to the requirement of additional unknown non-canonical factors for IRES-mediated translation. As depicted in Figure 3B, the 40S ribosomal subunit binds directly and specifically to the HCV IRES without the involvement of initiation factors (for comparison to the FMDV IRES, refer to Figure 3A), and places the initiation codon in the immediate vicinity of the P site [67]. The eIF2-ternary complex can bind the IRES-bound 40S subunit and induce a slight rearrangement of the initiation codon and the coding region. eIF3 was additionally found to bind the HCV IRES directly, and this binding is thought to stabilize the 48S complex, as it is required for 60S subunit joining and IRES function but not for 48S complex assembly [70-72]. It is speculated that eIF5 and eIF5B may play a role in 60S subunit joining. This method of initiation is remarkable in that it does not require the eIF4F complex, eIF4B, or hydrolysis of ATP. HCV translation is thus insensitive to eIF4G cleavage. It should be noted that in vitro studies have implicated a non-canonical factor, the La autoantigen, in HCV IRES-mediated translation [11,73]. A recent report has also implicated La in HCV IRES activity in cultured cells [74]. In addition, hnRNP D has also been recently shown to modulate HCV IRES-dependent translation [23].
Even more striking than HCV is CrPV, which can initiate translation of viral RNA without initiation factors or initiator tRNA [67,75-77]. The genome contains two large open reading frames, the first encoding the nonstructural proteins and the second encoding the capsid proteins. The intergenic region (IGR) of 190 nucleotides between these two coding regions contains an IRES element [75]. Initiation does not require an AUG codon, and initiation begins instead at a GCU codon. Translation of CrPV RNA is actually inhibited by the addition of eIF2 in vitro and is enhanced in cells when the amount of active ternary complex is reduced [75,78]. It has been demonstrated that the ternary complex and the CrPV IGR IRES compete for a common binding site on the 40S subunit [79]. Also noteworthy is that CrPV RNA can bind both 40S subunits and pre-assembled 80S ribosomes directly [79].
Cellular IRESes
Identification of IRES elements in cellular mRNAs
While first discovered in the RNA genomes of picornaviruses, IRES elements have also been identified in a subset of cellular mRNAs (Table 2). The mRNA encoding BiP was the first cellular mRNA reported to harbor an IRES [80]. The generally-accepted mechanism of translation initiation put forth by Kozak relies on cap recognition and ribosomal scanning [81]. Under certain cellular conditions this mechanism is compromised, and some mRNAs utilize an alternative form of initiation directed by IRES-mediated recruitment of ribosomes to the RNA. It has been difficult to identify functional classes of cellular IRESes because there is little primary sequence or major structural similarities among them, and many have varying efficiencies in different cell types. While some viral IRESes may have common contexts of synthesis and translation in the cytoplasm of normally cycling cells (leading to structural/functional classifications), a larger number of cellular contexts that require differential regulation would lead to a very large number of IRES classes with many different sequence and structural components [82]. Therefore, cellular IRESes have often been discussed in terms of the conditions that induce their activity or the conditions that result from their activity.
Table 2. Examples of cellular IRES elements.
| Gene | Cellular conditions for translation | Known ITAFs |
|---|---|---|
| Apaf-1 | Apoptosis | PTB, unr, DAP5 |
| XIAP | Apoptosis | La, hnRNP C1/C2, DAP5 |
| c-myc | Apoptosis, development, genotoxic stress, cell cycle | PCBP2, PCBP1, hnRNP C1/C2, hnRNP K, DAP5, IRP, unr, YB-1, GRSF-1, PSF, P54nrb |
| DAP5 | Apoptosis | DAP5 |
| Reaper | Apoptosis, heat shock | |
| Hsp70 | Apoptosis, heat shock | |
| Bcl-2 | Apoptosis | |
| HIAP2/c-IAP1 | Apoptosis, ER stress | DAP5 |
| Antennapedia | Development | |
| Ultrabithorax | Development | |
| ODC | Cell cycle | |
| PITSLRE | Cell cycle | unr |
| hnRNP A/B | Cell cycle | |
| Hairless | Cell cycle | |
| Notch2 | Cell cycle | |
| IGF-II | Cell Cycle | |
| VEGF | Hypoxia | PTB |
| HIF-1α | Hypoxia | |
| Cat-1 | ER stress, hypoxia | |
| Bip | Heat shock | La, NSAP1 |
| BAG-1 | Heat shock | |
| FGF-2 | Tissue/cell specific | hnRNP A1 |
| FGF-1 | Tissue/cell specific | |
| Kv1.4 | Tissue/cell specific | |
| LEF-1 | Oncogenesis | |
Some of the IRES elements that have been identified in cellular mRNAs are shown, including known cellular ITAFs that function in translation for each particular IRES. Note that only non-canonical ITAFs are listed; this list does not include any canonical eukaryotic translation initiation factors (eIFs). Cellular mRNAs listed are roughly grouped by the cellular condition that induces translation of the particular IRES. Apaf-1, apoptotic protease activating factor 1; XIAP, X-linked inhibitor of apoptosis protein; DAP5, death-associated protein 5; Bcl-2, B-cell CLL/lymphoma 2; HIAP2/c-IAP1, inhibitor of apoptosis protein; ODC, ornithine decarboxylase; IGF-II, insulin-like growth factor 2; VEGF, vascular endothelial growth factor; HIF-1α, hypoxia-inducible factor 1; Cat-1, cationic amino acid transporter 1; BiP, immunoglobulin heavy chain-binding protein; BAG-1, BCL2-associated athanogene 1; FGF, fibroblast growth factor; LEF-1, lymphoid enhancer-binding factor 1. Data compiled from [82,112,131-134].
Secondary structures have been derived for several cellular IRESes utilizing enzymatic and chemical structure probing from transcripts, including c-Myc [83], Apaf-1 [84], Kv1.4 [85], and FGF-2 [86], among others. A common Y structure has been predicted for cellular IRESes based on the computational comparison of several orthologs of the BiP and FGF-2 5′ NCRs [87]. As shown in Figure 4, Le and Maizel compared the common structural motif found in picornavirus [88], HCV, and pestivirus IRESes to the proposed structural model for the human BiP IRES. Similarities were observed between the proposed BiP IRES structural elements and those of picornaviruses (compare Figure 4A and 4B). The common feature is a Y stem-loop structure, which is followed by another stem-loop structure upstream of the authentic AUG start codon [87]. A higher-order structure similar to the conserved structural motif found in the IRES elements of BiP mRNAs was also observed in the IRES element of FGF-2 mRNA. A sequence alignment of the 5′ NCRs of other BiP and FGF family members revealed more details of the common Y-type structure in cellular mRNAs. In this structure, stem A contains an internal loop of eight to 13 base pairs (refer to Figure 4). Stems B and C contain four to eight base pairs and a hairpin loop that contains three to eight unpaired bases. Stem A is often connected to stem C; stem D is variable in length and is only a few nucleotides upstream of the authentic AUG initiator codon [87].
Figure 4. Schematic representation of the common structural core in IRES elements of cellular mRNAs (A) and IRES elements of picornavirus, HCV, and pestivirus (B).
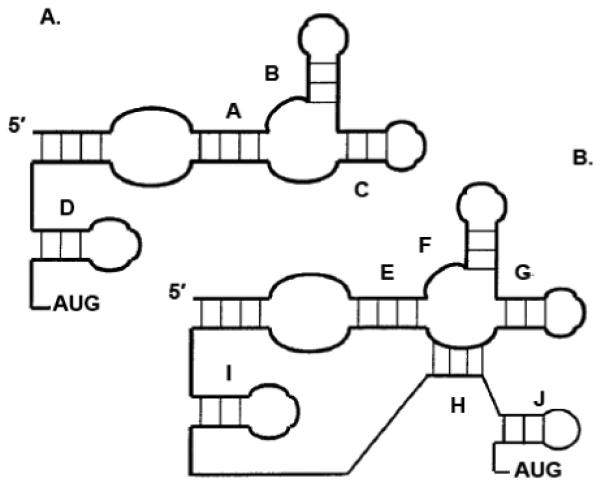
Stems A and E are shown as including an internal loop. The potential unpaired nucleotides in stems B-D and stems F-J are not shown. Stems A-D in the structural core of the cellular IRES correspond to stems E-I/J in the viral IRES core. (Figure reproduced and modified from Le and Maizel, 1997, with permission [87]).
The Y motif has been used for computer-based identification of putative IRESes in cellular mRNAs [82]. In these studies on putative cellular IRESes, only some IRESes show any preference for preservation of structure in order to retain complete activity. This would suggest that there is not a requirement for an overall structure for cellular IRESes, in contrast to what is seen in some viral IRESes [82]. It has been proposed that cellular IRESes are not well-defined in their overall structure but may be dependent upon short motifs and ITAFs for their function [89]. Indeed, the minimal length of an RNA for IRES activity has been investigated; in one study a nine-nucleotide segment in the 5′ leader of the mRNA encoding the Gtx homeodomain protein was reported to have IRES activity [90]. The activity directed by the nine-nucleotide Gtx mRNA sequence was later described as potentially due to ribosomal shunting [91]. Ribosomal shunting has been defined as a cap-dependent, scanning-independent mechanism of translation initiation in which the initiation complex bypasses specific stretches of nucleotides upstream of the initiator AUG [92,93].
Functional data and ITAFs important for cellular IRES activity
Identification of many cellular IRES elements has been based on the demonstration of the function of these elements - their ability to direct internal initiation of translation. The most common test for IRES activity is a mammalian cell transfection assay utilizing dicistronic reporter constructs. Expression of a second, downstream open reading frame is dependent on IRES sequences directing internal ribosome entry and translation initiation. Although it is an important first step in the determination of whether an RNA sequence has IRES activity, this approach has been criticized because mRNAs generated from transfected dicistronic plasmid DNAs generally have a much higher IRES activity than when the corresponding RNAs are transfected [94]. The difference in expression levels may be interpreted as an artifact of DNA transfection assays that leads to reporter expression that is independent of IRES function [95-98]. Despite this criticism, it is clear that IRES-mediated translation does occur for certain cellular mRNAs [99]. One proposed explanation for the difference between the translation of reporter mRNAs delivered via transfection of plasmid DNAs into the nucleus versus translation of reporter mRNAs transfected into the cytoplasm of cells is that IRES-containing cellular mRNAs require a ‘nuclear experience,’ which does not occur efficiently following RNA transfections [18,25,100]. Indeed, one recent study has re-examined the nuclear transcription of BACE1 mRNA and compared this to cytoplasmic transcription or RNA transfection [101]. The results suggested that when the mRNA is transcribed in the nucleus, translation occurs via a shunting mechanism. This is in contrast to conclusions drawn from previous studies by other groups, in which the mRNA was transcribed in the cytoplasm or when RNAs were transfected into cells (or translated in a cell-free system). Although it did not specifically address IRES-mediated translation, this study provides evidence suggesting that the site of transcription affects translation efficiency. The synthesis of mRNAs in the nucleus and subsequent transport to the cytoplasm may allow for the assembly of specific (and possibly unique) RNP complexes on individual mRNAs, an idea that is depicted in Figure 5 [100]. In this way, classes of cellular IRESes may emerge based on the sub-cellular site of IRES synthesis.
Figure 5. The nuclear versus cytoplasmic experience of an IRES-containing mRNA.
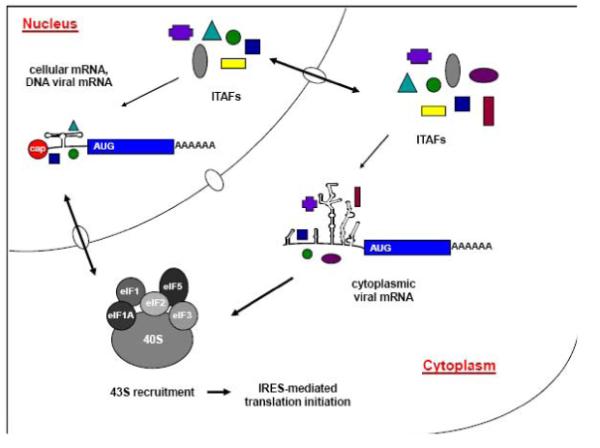
Cellular mRNAs and viral mRNAs from DNA genomes that contain IRES elements are first transcribed in the nucleus and subsequently exported to the cytoplasm for translation. RNP complexes may associate with these IRES elements in the nucleus and remain associated through export into the cytoplasm, where additional ITAFs may also associate with the IRES to recruit ribosomes for translation initiation. RNA genomes from RNA viruses that contain IRES elements are translated in the cytoplasm of eukaryotic cells, and the RNA does not go through a “nuclear experience.” ITAFs (colored shapes) bind different IRES elements in the nucleus and/or the cytoplasm, generating common or unique RNP complexes between cellular and viral IRESes. These distinct, functional RNP complexes may be formed on specific IRES elements based on the site of synthesis of the IRES. Shown in the figure are the sites of IRES synthesis in a eukaryotic cell, nuclear and cytoplasmic ITAFs, and the 43S initiation complex that is recruited for the subsequent initiation of IRES-mediated translation. (Figure redrawn from Semler and Waterman, 2008 [100]).
Additional caveats have been described for the identification of IRES function for some cellular mRNAs [95,97,102]. These include concerns that tested putative IRES sequences may actually function as cryptic promoters, splice sites, or sequences that modulate cleavage by RNases. To address these issues, in part, it is important to look for promoter activity in the NCR, reinitiation of the ribosome on the second open reading frame, aberrant splicing, mRNA cleavage, and inconsistent values of the dual luciferase reporter gene construct [82]. Re-evaluation of some mRNAs, including the 5′ NCRs of PDGF, PIM-1, and p27kip1, has suggested that they may not encode IRES elements as initially thought [103-105].
Even with the shortcomings noted above, IRES-mediated translation occurs for a subset of cellular mRNAs. One well-studied cellular IRES is the c-myc IRES element [106]. Proteins belonging to the Myc family have a role in proliferation and apoptosis, and up-regulated Myc expression has been associated with tumorigenesis [107-109]. The c-myc mRNA has a cap structure at its 5′ end, and it can be translated via both cap-dependent and cap-independent mechanisms [25]. The RNA is translated via its IRES element in various cell types with different efficiencies, a topic that was introduced earlier in this review. Thus, it is likely that the variation in abundance of specific ITAFs contributes to this difference in translation efficiency. Several ITAFs have been identified that stimulate c-myc translation, including PTB, PCBP2, IRP, unr, DAP5, YB-1, GRSF-1, PSF, p54nrb [110-112]. Specific ITAFs likely function to stimulate c-myc IRES-driven translation during situations when cap-dependent translation in compromised.
Another identified cellular IRES is in the 5′ NCR of the lymphoid enhancer factor-1 (LEF-1) mRNA [94]. LEF-1 is a LEF/TCF transcription factor that mediates WNT signal control of cell cycling and differentiation. LEF-1 regulated genes are involved in cell growth in mitotically active cells. WNT signaling has a profound effect on cell growth, and numerous types of cancers (e.g., colon cancer) are a result of overactive WNT signaling. The LEF-1 mRNA is translated to generate a full-length (oncogenic) form of the protein as well as a truncated (growth-suppressive) form [113]. The shorter LEF-1 mRNA, transcribed from a second promoter in the second intron of the gene, generates the smaller inhibitory LEF-1 protein that competes with β-catenin for binding to WNT target genes. It was demonstrated that the full-length form of the LEF-1 mRNA (but not the truncated form) contains an IRES element that directs its translation. It is thought that the full length LEF-1 mRNAs are not translated as efficiently in cells under normal steady-state conditions and are likely to be subject to greater regulation [94]. Under conditions of mis-regulation in cells (e.g., neoplastic transformation, where cells are also rapidly dividing and likely under stress) the full-length LEF-1 mRNA is transcribed, while the truncated inhibitory form is not. Translation of full-length LEF-1 mRNAs then remains constant, and is not subject to the suppression of cap-dependent translation that occurs during neoplastic transformation.
Additional ITAFs have been identified for several cellular IRESes, including the X-linked inhibitor of apoptosis IRES (hnRNP C, La autoantigen), Apaf-1 IRES (PTB, unr), and VEGF IRES (PTB), among others (refer to Table 2). Interestingly, nucleo-cytoplasmic shuttling protein hnRNP A1 has been implicated in the IRES-mediated translation of both human rhinovirus type 2 (HRV-2) RNA and human apoptotic peptidase activating factor 1 (apaf-1) mRNA [114]. hnRNP A1 re-localizes from the nucleus to the cytoplasm during HRV-2 infection or after irradiation with ultraviolet light; re-localization enhances HRV-2 translation during infection, but inhibits apaf-1 IRES-mediated translation following ultraviolet irradiation. This suggests that IRES elements are regulated differentially under varying physiological conditions, a topic that will be investigated further in the following section. While cellular IRESes are continuing to be identified and added to the growing list of IRES-containing mRNAs, ITAFs are also currently being investigated for these IRESes. There does not appear to be one or more “universal” ITAFs for IRES-mediated translation in eukaryotic cells. However, PTB has been shown to function in both viral and cellular IRES translation [115-117]. In addition, there is emerging evidence that many ITAFs regulating cellular IRES translation are nucleo-cytoplasmic shuttling proteins [20,118].
Requirement for an alternative mechanism of translation initiation
Cellular conditions that provide for preferential translation of IRES-containing mRNAs
While the focus of many studies on the mechanisms of IRES-mediated translation has been on the interactions between the IRES RNA and initiation factors or yet unidentified, non-canonical ITAFs, it is important to take a broader look at the conditions that invoke such a requirement for an alternative form of translation initiation. Within the eukaryotic cell, the environment and the context that it provides for mRNA translation is not static; there are conditions under which the cellular environment is dramatically altered from that of steady state. Some of these conditions include stress (e.g., heat shock, hypoxia, nutrient deprivation), cellular growth and differentiation, apoptosis, and disease. Under several of these conditions, cap-dependent translation is down-regulated, and there is an inherent need for translation of certain mRNAs to direct the cell either to recovery or to apoptosis. It has been proposed that IRES-mediated translation provides the cell with a high degree of spatial and temporal control often required to mount an appropriate stress response [118].
One example of an IRES-containing mRNA that is associated with translating polysomes during hypoxic stress, which also corresponds to the environment present during tumor growth, is HIF-1α [119]. The mechanism of cap-dependent translation shut-down has not been completely defined during hypoxia; however in the prostate cancer cell line PC-3, under hypoxic conditions the HIF-1α mRNA appears to remain associated with the active translation machinery [119]. As mentioned in the previous section, the LEF-1 mRNA contains an IRES and translation of this RNA is important for the regulation of genes involved in cell growth during mitosis, a time in which most cellular cap-dependent translation is inhibited. Mis-regulation of LEF-1 translation (i.e., continued translation of full-length LEF-1) is linked to a cancer phenotype. Also mentioned previously, up-regulation of the c-myc mRNA has been implicated in some cancers. It is clear then that IRES-mediated translation is subject to regulation as a result of the particular cellular environment. Taken together, this evidence would implicate IRES-containing mRNAs as important players with specific roles during certain conditions in the cell. These particular cellular conditions then dictate the requirement for IRES-mediated translation, the suppression of IRES-mediated translation, or the mis-regulation of this type of translation and potential causative environment for disease.
IRES elements are also important for directing translation at specific stages of the cell cycle. During mitosis canonical cap-dependent translation is impaired, although certain cell cycle regulatory proteins must still be expressed. For example, ornithine decarboxylase (ODC) is a key enzyme in the biosynthesis of polyamines, which are proposed to affect chromatin and mitotic spindle organization; thus, maintaining an abundance of polyamines is essential for cells during mitosis. ODC expression peaks at both G1/S and G2/M [120]. There is a general augmentation of protein synthesis at G1/S; however, ODC expression at G2/M is in stark contrast to the strong inhibition of protein synthesis as cells enter mitosis. It is thought that ODC translation is generally cap-dependent, which correlates well with its expression at G1/S. Importantly, ODC has also been shown to contain an IRES in its 5′ NCR that functions exclusively during G2/M to mediate cap-independent translation of ODC during a time of overall suppression of cap-dependent translation [121].
The PITSLRE protein kinases are related to the cyclin-dependent kinases and also have an important role in cell cycle progression. Two PITSLRE isoforms (p110PITSLRE and p58PITSLRE) are generated from a single transcript; p110 PITSLRE is expressed during all phases of the cell cycle, but the p58 PITSLRE isoform is expressed predominately at G2/M. It has been demonstrated that the p110 PITSLRE protein is produced via a cap-dependent mechanism of translation, while the p58PITSLRE isoform is translated in a cap-independent manner [122]. This IRES element is the first IRES identified in the mRNA coding region and is utilized during G2/M (when cap-dependent translation is suppressed) to produce the p58 PITSLRE isoform. The PITSLRE IRES is therefore controlled by yet-unidentified cell cycle-specific factors. Taken together, these examples suggest an importance for IRES-mediated translation in cell cycle progression and control, as well as the possibility of cell cycle-dependent ITAFs functioning to stimulate translation directed by IRES elements during specific stages of the cell cycle. It is likely that during transitions between different cell cycle phases, both cap-dependent and IRES-mediated translation occur simultaneously (although perhaps only transiently).
Additional control of IRES-mediated translation may be exerted by microRNAs (miRs), short single-stranded regulatory RNA elements that have been extensively studied in recent years. In experiments carried out with the EMCV IRES, miRs were unable to inhibit viral translation [123]. In contrast, the HCV and CrPV IRESes are sensitive to miRs [124,125]. It was then proposed that cellular IRESes were likely regulated by miRs, since cellular IRESes are typically found in critical growth regulatory genes that would be subject to miR regulation [126]. It has been determined that the vascular endothelial growth factor (VEGF) mRNA, which encodes a growth and survival factor for endothelial cells, contains two separate IRES elements (A and B) [127]. One study has shown that a specific miRNA, miR-16, inhibits IRES-mediated translation from IRES-B of the VEGF mRNA [126]. This provides evidence that cellular IRESes may serve as targets for regulation via miRNAs in the cell.
Cellular conditions generated by viral infection that provide for preferential translation initiation of viral RNA
Viruses have evolved to take advantage of the host cell and its machinery while at the same time preventing competition for resources by down-regulating normal cellular processes. This is certainly true for IRES-containing viruses, as exemplified by the picornaviruses. These positive-strand RNA viruses can be immediately translated in the cytoplasm of infected cells by an IRES-mediated mechanism. Picornaviruses not only utilize an alternative mechanism of translation, they also efficiently down-regulate host cell cap-dependent translation. As noted earlier, enteroviruses and rhinoviruses encode proteinases that cleave cellular proteins eIF4G and PABP, effectively preventing initiation and re-initiation for capped mRNAs [26-30,128]. Picornaviruses can then presumably utilize available canonical initiation factors, ribosomes, and additional non-canonical factors to selectively translate their own genomic RNA. EMCV employs a different method of down-regulating cap-dependent translation and does not cleave eIF4G; instead it activates the dephosphorylation of eIF4E-binding protein 1 (4E-BP1), which then activates 4E-BP1 for binding of eIF4E and sequestration from the functional eIF4F complex [129].
Viral infections also generate conditions of stress in the infected cell. This has been recently described for poliovirus-infected cells, in which the viral infection induces the formation of cytoplasmic stress granules [130]. Stress granule formation may alter the availability of cellular proteins for viral RNA translation, or it may actually function to concentrate proteins necessary for translation (or other viral processes) to specific sub-cellular regions. The significance and function of stress granules during poliovirus infection has yet to be determined, but one can speculate that this is a natural cellular response to an external stress on the cell that then serves either directly, as a cellular attempt to aid itself in thwarting the viral infection, or perhaps indirectly, to aid the virus in the course of infection. This finding may provide insight and promote further study into the significance of alternative ways to bridge the translation machinery to cellular mRNAs for the initiation of protein synthesis.
Acknowledgments
We are grateful to Andrea Cathcart and Amanda Chase for critical comments on the manuscript. Research described from the authors’ laboratory is supported by Public Health Service grant AI 26765 from the National Institutes of Health. K.D.F. is a predoctoral trainee of Public Health Service training grant AI 07319.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005;33:1231. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- 2.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984;12:857. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelletier J, Kaplan G, Racaniello VR, Sonenberg N. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol. Cell Biol. 1988;8:1103. doi: 10.1128/mcb.8.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 7.Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol. 2006;13:1092. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 8.Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 2002;324:889. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 10.Kaminski A, Hunt SL, Patton JG, Jackson RJ. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA. 1995;1:924. [PMC free article] [PubMed] [Google Scholar]
- 11.Costa-Mattioli M, Svitkin Y, Sonenberg N. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell Biol. 2004;24:6861. doi: 10.1128/MCB.24.15.6861-6870.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunt SL, Hsuan JJ, Totty N, Jackson RJ. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 1999;13:437. doi: 10.1101/gad.13.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pestova TV, Shatsky IN, Hellen CU. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell Biol. 1996;16:6870. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ochs K, Rust RC, Niepmann M. Translation initiation factor eIF4B interacts with a picornavirus internal ribosome entry site in both 48S and 80S initiation complexes independently of initiator AUG location. J. Virol. 1999;73:7505. doi: 10.1128/jvi.73.9.7505-7514.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walter BL, Parsley TB, Ehrenfeld E, Semler BL. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 2002;76:12008. doi: 10.1128/JVI.76.23.12008-12022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell SA, Spriggs KA, Bushell M, Evans JR, Stoneley M, Le Quesne JP, Spriggs RV, Willis AE. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes Dev. 2005;19:1556. doi: 10.1101/gad.339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meerovitch K, Svitkin YV, Lee HS, Lejbkowicz F, Kenan DJ, Chan EK, Agol VI, Keene JD, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 1993;67:3798. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holcik M, Gordon BW, Korneluk RG. The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol. Cell Biol. 2003;23:280. doi: 10.1128/MCB.23.1.280-288.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sella O, Gerlitz G, Le SY, Elroy-Stein O. Differentiation-induced internal translation of c-sis mRNA: analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein. Mol. Cell Biol. 1999;19:5429. doi: 10.1128/mcb.19.8.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schepens B, Tinton SA, Bruynooghe Y, Parthoens E, Haegman M, Beyaert R, Cornelis S. A role for hnRNP C1/C2 and Unr in internal initiation of translation during mitosis. EMBO J. 2007;26:158. doi: 10.1038/sj.emboj.7601468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walter BL, Nguyen JH, Ehrenfeld E, Semler BL. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA. 1999;5:1570. doi: 10.1017/s1355838299991483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blyn LB, Towner JS, Semler BL, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 1997;71:6243. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paek KY, Kim CS, Park SM, Kim JH, Jang SK. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J. Virol. 2008;82:12082. doi: 10.1128/JVI.01405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 25.Stoneley M, Subkhankulova T, Le Quesne JP, Coldwell MJ, Jopling CL, Belsham GJ, Willis AE. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 2000;28:687. doi: 10.1093/nar/28.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 1982;257:14806. [PubMed] [Google Scholar]
- 27.Krausslich HG, Nicklin MJ, Toyoda H, Etchison D, Wimmer E. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. J. Virol. 1987;61:2711. doi: 10.1128/jvi.61.9.2711-2718.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd RE, Jense HG, Ehrenfeld E. Restriction of translation of capped mRNA in vitro as a model for poliovirus-induced inhibition of host cell protein synthesis: relationship to p220 cleavage. J. Virol. 1987;61:2480. doi: 10.1128/jvi.61.8.2480-2488.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joachims M, Van Breugel PC, Lloyd RE. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 1999;73:718. doi: 10.1128/jvi.73.1.718-727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerekatte V, Keiper BD, Badorff C, Cai A, Knowlton KU, Rhoads RE. Cleavage of Poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J. Virol. 1999;73:709. doi: 10.1128/jvi.73.1.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borman AM, Kean KM. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology. 1997;237:129. doi: 10.1006/viro.1997.8761. [DOI] [PubMed] [Google Scholar]
- 32.Stewart SR, Semler BL. RNA determinants of picornavirus cap-independent translation initiation. Semin. Virol. 1997;8:242. [Google Scholar]
- 33.Whetter LE, Day SP, Elroy-Stein O, Brown EA, Lemon SM. Low efficiency of the 5′ nontranslated region of hepatitis A virus RNA in directing cap-independent translation in permissive monkey kidney cells. J. Virol. 1994;68:5253. doi: 10.1128/jvi.68.8.5253-5263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ali IK, McKendrick L, Morley SJ, Jackson RJ. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol. 2001;75:7854. doi: 10.1128/JVI.75.17.7854-7863.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cordes S, Kusov Y, Heise T, Gauss-Muller V. La autoantigen suppresses IRES-dependent translation of the hepatitis A virus. Biochem. Biophys. Res. Commun. 2008;368:1014. doi: 10.1016/j.bbrc.2008.01.163. [DOI] [PubMed] [Google Scholar]
- 36.Kaku Y, Chard LS, Inoue T, Belsham GJ. Unique characteristics of a picornavirus internal ribosome entry site from the porcine teschovirus-1 talfan. J. Virol. 2002;76:11721. doi: 10.1128/JVI.76.22.11721-11728.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pisarev AV, Chard LS, Kaku Y, Johns HL, Shatsky IN, Belsham GJ. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 2004;78:4487. doi: 10.1128/JVI.78.9.4487-4497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belsham GJ, Nielsen I, Normann P, Royall E, Roberts LO. Monocistronic mRNAs containing defective hepatitis C virus-like picornavirus internal ribosome entry site elements in their 5′ untranslated regions are efficiently translated in cells by a cap-dependent mechanism. RNA. 2008;14:1671. doi: 10.1261/rna.1039708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7:382. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chard LS, Kaku Y, Jones B, Nayak A, Belsham GJ. Functional analyses of RNA structures shared between the internal ribosome entry sites of hepatitis C virus and the picornavirus porcine teschovirus 1 Talfan. J. Virol. 2006;80:1271. doi: 10.1128/JVI.80.3.1271-1279.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2006;2:213. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- 42.Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell Biol. 1996;16:6859. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trono D, Andino R, Baltimore D. An RNA sequence of hundreds of nucleotides at the 5′ end of poliovirus RNA is involved in allowing viral protein synthesis. J. Virol. 1988;62:2291. doi: 10.1128/jvi.62.7.2291-2299.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hellen CU, Witherell GW, Schmid M, Shin SH, Pestova TV, Gil A, Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. U. S. A. 1993;90:7642. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borman A, Howell MT, Patton JG, Jackson RJ. The involvement of a spliceosome component in internal initiation of human rhinovirus RNA translation. J. Gen. Virol. 1993;74:1775. doi: 10.1099/0022-1317-74-9-1775. [DOI] [PubMed] [Google Scholar]
- 46.Brown BA, Ehrenfeld E. Translation of poliovirus RNA in vitro: changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology. 1979;97:396. doi: 10.1016/0042-6822(79)90350-7. [DOI] [PubMed] [Google Scholar]
- 47.Dorner AJ, Semler BL, Jackson RJ, Hanecak R, Duprey E, Wimmer E. In vitro translation of poliovirus RNA: utilization of internal initiation sites in reticulocyte lysate. J. Virol. 1984;50:507. doi: 10.1128/jvi.50.2.507-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Svitkin YV, Meerovitch K, Lee HS, Dholakia JN, Kenan DJ, Agol VI, Sonenberg N. Internal translation initiation on poliovirus RNA: further characterization of La function in poliovirus translation in vitro. J. Virol. 1994;68:1544. doi: 10.1128/jvi.68.3.1544-1550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheung P, Zhang M, Yuan J, Chau D, Yanagawa B, McManus B, Yang D. Specific interactions of HeLa cell proteins with Coxsackievirus B3 RNA: La autoantigen binds differentially to multiple sites within the 5′ untranslated region. Virus Res. 2002;90:23. doi: 10.1016/s0168-1702(02)00138-7. [DOI] [PubMed] [Google Scholar]
- 50.Hunt SL, Jackson RJ. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA. 1999;5:344. doi: 10.1017/s1355838299981414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bedard KM, Daijogo S, Semler BL. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007;26:459. doi: 10.1038/sj.emboj.7601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de BS, Yu Y, Unbehaun A, Pestova TV, Hellen CU. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9197. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ochs K, Saleh L, Bassili G, Sonntag VH, Zeller A, Niepmann M. Interaction of translation initiation factor eIF4B with the poliovirus internal ribosome entry site. J. Virol. 2002;76:2113. doi: 10.1128/jvi.76.5.2113-2122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fernandez-Miragall O, Quinto SL, Martinez-Salas E. Relevance of RNA structure for the activity of picornavirus IRES elements. Virus Res. 2009;139:172. doi: 10.1016/j.virusres.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 55.Strebel K, Beck E. A second protease of foot-and-mouth disease virus. J. Virol. 1986;58:893. doi: 10.1128/jvi.58.3.893-899.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Devaney MA, Vakharia VN, Lloyd RE, Ehrenfeld E, Grubman MJ. Leader protein of foot-and-mouth disease virus is required for cleavage of the p220 component of the cap-binding protein complex. J. Virol. 1988;62:4407. doi: 10.1128/jvi.62.11.4407-4409.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skern T, Hampolz B, Guarne A, Fita I, Bergmann E, Jens P, James MNG. Structure and Function of Picornavirus Proteinases. In: Semler BL, Wimmer E, editors. Molecular biology of picornaviruses. ASM Press; Washington, D.C.: 2002. pp. 199–212. [Google Scholar]
- 58.Rowlands DJ, Harris TJ, Brown F. More precise location of the polycytidylic acid tract in foot and mouth disease virus RNA. J. Virol. 1978;26:335. doi: 10.1128/jvi.26.2.335-343.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Escarmis C, Dopazo J, Davila M, Palma EL, Domingo E. Large deletions in the 5′-untranslated region of foot-and-mouth disease virus of serotype C. Virus Res. 1995;35:155. doi: 10.1016/0168-1702(94)00091-p. [DOI] [PubMed] [Google Scholar]
- 60.Mason PW, Bezborodova SV, Henry TM. Identification and characterization of a cis-acting replication element (cre) adjacent to the internal ribosome entry site of foot- and-mouth disease virus. J. Virol. 2002;76:9686. doi: 10.1128/JVI.76.19.9686-9694.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Belsham GJ. Dual initiation sites of protein synthesis on foot-and-mouth disease virus RNA are selected following internal entry and scanning of ribosomes in vivo. EMBO J. 1992;11:1105. doi: 10.1002/j.1460-2075.1992.tb05150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lopez de Quinto S, Martinez-Salas E. Involvement of the aphthovirus RNA region located between the two functional AUGs in start codon selection. Virology. 1999;255:324. doi: 10.1006/viro.1999.9598. [DOI] [PubMed] [Google Scholar]
- 63.Lyons AJ, Robertson HD. Detection of tRNA-like structure through RNase P cleavage of viral internal ribosome entry site RNAs near the AUG start triplet. J. Biol. Chem. 2003;278:26844. doi: 10.1074/jbc.M304052200. [DOI] [PubMed] [Google Scholar]
- 64.Nadal A, Martell M, Lytle JR, Lyons AJ, Robertson HD, Cabot B, Esteban JI, Esteban R, Guardia J, Gomez J. Specific cleavage of hepatitis C virus RNA genome by human RNase P. J. Biol. Chem. 2002;277:30606. doi: 10.1074/jbc.M203595200. [DOI] [PubMed] [Google Scholar]
- 65.Monie TP, Perrin AJ, Birtley JR, Sweeney TR, Karakasiliotis I, Chaudhry Y, Roberts LO, Matthews S, Goodfellow IG, Curry S. Structural insights into the transcriptional and translational roles of Ebp1. EMBO J. 2007;26:3936. doi: 10.1038/sj.emboj.7601817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CU. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028. [PMC free article] [PubMed] [Google Scholar]
- 67.Pisarev AV, Shirokikh NE, Hellen CU. Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. C. R. Biol. 2005;328:589. doi: 10.1016/j.crvi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 68.Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol. 2007;5:29. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 69.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 1998;72:4775. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Odreman-Macchioli FE, Tisminetzky SG, Zotti M, Baralle FE, Buratti E. Influence of correct secondary and tertiary RNA folding on the binding of cellular factors to the HCV IRES. Nucleic Acids Res. 2000;28:875. doi: 10.1093/nar/28.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc. Natl. Acad. Sci. U. S. A. 1997;94:2249. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Romero V, Fellows E, Jenne DE, Andrade F. Cleavage of La protein by granzyme H induces cytoplasmic translocation and interferes with La-mediated HCV-IRES translational activity. Cell Death. Differ. 2009;16:340. doi: 10.1038/cdd.2008.165. [DOI] [PubMed] [Google Scholar]
- 75.Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 76.Jan E, Kinzy TG, Sarnow P. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15410. doi: 10.1073/pnas.2535183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pestova TV, Hellen CU. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thompson SR, Gulyas KD, Sarnow P. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12972. doi: 10.1073/pnas.241286698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pestova TV, Lomakin IB, Hellen CU. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Macejak DG, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 81.Kozak M. The scanning model for translation: an update. J. Cell Biol. 1989;108:229. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA. 2006;12:1755. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Le Quesne JP, Stoneley M, Fraser GA, Willis AE. Derivation of a structural model for the c-myc IRES. J. Mol. Biol. 2001;310:111. doi: 10.1006/jmbi.2001.4745. [DOI] [PubMed] [Google Scholar]
- 84.Mitchell SA, Spriggs KA, Coldwell MJ, Jackson RJ, Willis AE. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell. 2003;11:757. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 85.Jang GM, Leong LE, Hoang LT, Wang PH, Gutman GA, Semler BL. Structurally distinct elements mediate internal ribosome entry within the 5′-noncoding region of a voltage-gated potassium channel mRNA. J. Biol. Chem. 2004;279:47419. doi: 10.1074/jbc.M405885200. [DOI] [PubMed] [Google Scholar]
- 86.Bonnal S, Schaeffer C, Creancier L, Clamens S, Moine H, Prats AC, Vagner S. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 2003;278:39330. doi: 10.1074/jbc.M305580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Le SY, Maizel JV., Jr. A common RNA structural motif involved in the internal initiation of translation of cellular mRNAs. Nucleic Acids Res. 1997;25:362. doi: 10.1093/nar/25.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pilipenko EV, Blinov VM, Chernov BK, Dmitrieva TM, Agol VI. Conservation of the secondary structure elements of the 5′-untranslated region of ca. Nucleic Acids Res. 1989;17:5701. doi: 10.1093/nar/17.14.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baird SD, Lewis SM, Turcotte M, Holcik M. A search for structurally similar cellular internal ribosome entry sites. Nucleic Acids Res. 2007;35:4664. doi: 10.1093/nar/gkm483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chappell SA, Edelman GM, Mauro VP. Biochemical and functional analysis of a 9-nt RNA sequence that affects translation efficiency in eukaryotic cells. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9590. doi: 10.1073/pnas.0308759101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chappell SA, Dresios J, Edelman GM, Mauro VP. Ribosomal shunting mediated by a translational enhancer element that base pairs to 18S rRNA. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9488. doi: 10.1073/pnas.0603597103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yueh A, Schneider RJ. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 1996;10:1557. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]
- 93.Schmidt-Puchta W, Dominguez D, Lewetag D, Hohn T. Plant ribosome shunting in vitro. Nucleic Acids Res. 1997;25:2854. doi: 10.1093/nar/25.14.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jimenez J, Jang GM, Semler BL, Waterman ML. An internal ribosome entry site mediates translation of lymphoid enhancer factor-1. RNA. 2005;11:1385. doi: 10.1261/rna.7226105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 96.Van Eden ME, Byrd MP, Sherrill KW, Lloyd RE. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA. 2004;10:720. doi: 10.1261/rna.5225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kozak M. A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 2005;33:6593. doi: 10.1093/nar/gki958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bert AG, Grepin R, Vadas MA, Goodall GJ. Assessing IRES activity in the HIF-1alpha and other cellular 5′ UTRs. RNA. 2006;12:1074. doi: 10.1261/rna.2320506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schneider R, Agol VI, Andino R, Bayard F, Cavener DR, Chappell SA, Chen JJ, Darlix JL, Dasgupta A, Donze O, Duncan R, Elroy-Stein O, Farabaugh PJ, Filipowicz W, Gale M, Jr., Gehrke L, Goldman E, Groner Y, Harford JB, Hatzglou M, He B, Hellen CU, Hentze MW, Hershey J, Hershey P, Hohn T, Holcik M, Hunter CP, Igarashi K, Jackson R, Jagus R, Jefferson LS, Joshi B, Kaempfer R, Katze M, Kaufman RJ, Kiledjian M, Kimball SR, Kimchi A, Kirkegaard K, Koromilas AE, Krug RM, Kruys V, Lamphear BJ, Lemon S, Lloyd RE, Maquat LE, Martinez-Salas E, Mathews MB, Mauro VP, Miyamoto S, Mohr I, Morris DR, Moss EG, Nakashima N, Palmenberg A, Parkin NT, Pe’ery T, Pelletier J, Peltz S, Pestova TV, Pilipenko EV, Prats AC, Racaniello V, Read GS, Rhoads RE, Richter JD, Rivera-Pomar R, Rouault T, Sachs A, Sarnow P, Scheper GC, Schiff L, Schoenberg DR, Semler BL, Siddiqui A, Skern T, Sonenberg N, Tahara SM, Thomas AA, Toulme JJ, Wilusz J, Wimmer E, Witherell G, Wormington M. New ways of initiating translation in eukaryotes. Mol. Cell Biol. 2001;21:8238. doi: 10.1128/MCB.21.23.8238-8246.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Semler BL, Waterman ML. IRES-mediated pathways to polysomes: nuclear versus cytoplasmic routes. Trends Microbiol. 2008;16:1. doi: 10.1016/j.tim.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 101.Koh DC, Mauro VP. Reconciling contradictory reports regarding translation of BACE1 mRNA: initiation mechanism is altered by different expression systems. RNA Biol. 2009;6:54. doi: 10.4161/rna.6.1.7567. [DOI] [PubMed] [Google Scholar]
- 102.Kozak M. New ways of initiating translation in eukaryotes? Mol. Cell Biol. 2001;21:1899. doi: 10.1128/MCB.21.6.1899-1907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Han B, Dong Z, Zhang JT. Tight control of platelet-derived growth factor B/c-sis expression by interplay between the 5′-untranslated region sequence and the major upstream promoter. J. Biol. Chem. 2003;278:46983. doi: 10.1074/jbc.M304976200. [DOI] [PubMed] [Google Scholar]
- 104.Wang Z, Weaver M, Magnuson NS. Cryptic promoter activity in the DNA sequence corresponding to the pim-1 5′-UTR. Nucleic Acids Res. 2005;33:2248. doi: 10.1093/nar/gki523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu Z, Dong Z, Han B, Yang Y, Liu Y, Zhang JT. Regulation of expression by promoters versus internal ribosome entry site in the 5′-untranslated sequence of the human cyclin-dependent kinase inhibitor p27kip1. Nucleic Acids Res. 2005;33:3763. doi: 10.1093/nar/gki680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nanbru C, Lafon I, Audigier S, Gensac MC, Vagner S, Huez G, Prats AC. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J. Biol. Chem. 1997;272:32061. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 107.Paulin FE, West MJ, Sullivan NF, Whitney RL, Lyne L, Willis AE. Aberrant translational control of the c-myc gene in multiple myeloma. Oncogene. 1996;13:505. [PubMed] [Google Scholar]
- 108.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat. Rev. Cancer. 2002;2:764. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 109.West MJ, Stoneley M, Willis AE. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene. 1998;17:769. doi: 10.1038/sj.onc.1201990. [DOI] [PubMed] [Google Scholar]
- 110.Cobbold LC, Spriggs KA, Haines SJ, Dobbyn HC, Hayes C, de Moor CH, Lilley KS, Bushell M, Willis AE. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol. Cell Biol. 2008;28:40. doi: 10.1128/MCB.01298-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Evans JR, Mitchell SA, Spriggs KA, Ostrowski J, Bomsztyk K, Ostarek D, Willis AE. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene. 2003;22:8012. doi: 10.1038/sj.onc.1206645. [DOI] [PubMed] [Google Scholar]
- 112.Martinez-Salas E, Ramos R, Lafuente E, Lopez de QS. Functional interactions in internal translation initiation directed by viral and cellular IRES elements. J. Gen. Virol. 2001;82:973. doi: 10.1099/0022-1317-82-5-973. [DOI] [PubMed] [Google Scholar]
- 113.Hovanes K, Li TW, Munguia JE, Truong T, Milovanovic T, Lawrence MJ, Holcombe RF, Waterman ML. Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat. Genet. 2001;28:53. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- 114.Cammas A, Pileur F, Bonnal S, Lewis SM, Leveque N, Holcik M, Vagner S. Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Mol. Biol. Cell. 2007;18:5048. doi: 10.1091/mbc.E07-06-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Belsham GJ, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev. 1996;60:499. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Giraud S, Greco A, Brink M, Diaz JJ, Delafontaine P. Translation initiation of the insulin-like growth factor I receptor mRNA is mediated by an internal ribosome entry site. J. Biol. Chem. 2001;276:5668. doi: 10.1074/jbc.M005928200. [DOI] [PubMed] [Google Scholar]
- 117.Kim YK, Hahm B, Jang SK. Polypyrimidine tract-binding protein inhibits translation of bip mRNA. J. Mol. Biol. 2000;304:119. doi: 10.1006/jmbi.2000.4179. [DOI] [PubMed] [Google Scholar]
- 118.Graber TE, Holcik M. Cap-independent regulation of gene expression in apoptosis. Mol. Biosyst. 2007;3:825. doi: 10.1039/b708867a. [DOI] [PubMed] [Google Scholar]
- 119.Thomas JD, Johannes GJ. Identification of mRNAs that continue to associate with polysomes during hypoxia. RNA. 2007;13:1116. doi: 10.1261/rna.534807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fredlund JO, Johansson MC, Dahlberg E, Oredsson SM. Ornithine decarboxylase and S-adenosylmethionine decarboxylase expression during the cell cycle of Chinese hamster ovary cells. Exp. Cell Res. 1995;216:86. doi: 10.1006/excr.1995.1011. [DOI] [PubMed] [Google Scholar]
- 121.Pyronnet S, Pradayrol L, Sonenberg N. A cell cycle-dependent internal ribosome entry site. Mol. Cell. 2000;5:607. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- 122.Cornelis S, Bruynooghe Y, Denecker G, Van HS, Tinton S, Beyaert R. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell. 2000;5:597. doi: 10.1016/s1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- 123.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 2006;21:533. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 124.Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 125.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, Filipowicz W, Duchaine TF, Sonenberg N. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 126.Karaa ZS, Iacovoni JS, Bastide A, Lacazette E, Touriol C, Prats H. The VEGF IRESes are differentially susceptible to translation inhibition by miR-16. RNA. 2009;15:249. doi: 10.1261/rna.1301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huez I, Creancier L, Audigier S, Gensac MC, Prats AC, Prats H. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol. Cell Biol. 1998;18:6178. doi: 10.1128/mcb.18.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kuyumcu-Martinez NM, Joachims M, Lloyd RE. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. J. Virol. 2002;76:2062. doi: 10.1128/jvi.76.5.2062-2074.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gingras AC, Svitkin Y, Belsham GJ, Pause A, Sonenberg N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5578. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.White JP, Cardenas AM, Marissen WE, Lloyd RE. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host. Microbe. 2007;2:295. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 131.Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 132.Komar AA, Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J. Biol. Chem. 2005;280:23425. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- 133.Cho S, Park SM, Kim TD, Kim JH, Kim KT, Jang SK. BiP internal ribosomal entry site activity is controlled by heat-induced interaction of NSAP1. Mol. Cell Biol. 2007;27:368. doi: 10.1128/MCB.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bonnal S, Pileur F, Orsini C, Parker F, Pujol F, Prats AC, Vagner S. Heterogeneous nuclear ribonucleoprotein A1 is a novel internal ribosome entry site trans-acting factor that modulates alternative initiation of translation of the fibroblast growth factor 2 mRNA. J. Biol. Chem. 2005;280:4144. doi: 10.1074/jbc.M411492200. [DOI] [PubMed] [Google Scholar]
- 135.Lopez de Quinto S, Martinez-Salas E. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA. 2000;6:1380. doi: 10.1017/s1355838200000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Luz N, Beck E. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J. Virol. 1991;65:6486. doi: 10.1128/jvi.65.12.6486-6494.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kolupaeva VG, Hellen CU, Shatsky IN. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA. 1996;2:1199. [PMC free article] [PubMed] [Google Scholar]
- 138.Honda M, Beard MR, Ping LH, Lemon SM. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 1999;73:1165. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Buratti E, Tisminetzky S, Zotti M, Baralle FE. Functional analysis of the interaction between HCV 5′UTR and putative subunits of eukaryotic translation initiation factor eIF3. Nucleic Acids Res. 1998;26:3179. doi: 10.1093/nar/26.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kolupaeva VG, Pestova TV, Hellen CU. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J. Virol. 2000;74:6242. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


