Abstract
The enhanced sensitivity Trofile assay was used to re-test co-receptor usage at study screening and entry for the 118 ACTG A5211 treatment-experienced subjects who had CCR5-tropic (R5) virus by the original Trofile assay at study screening. Among 90 vicriviroc recipients, a significantly (P<0.001) greater mean reduction in HIV-1 RNA was observed in 72 subjects with R5 virus versus 15 subjects reclassified with dual/mixed-tropic viruses at screening: −1.11 vs. −0.09 (day 14), −1.91 vs. −0.57 (week 24) log10 copies/mL, respectively. Results suggest that the enhanced sensitivity assay is a better screening tool for determining patient eligibility for CCR5 antagonist therapy.
Keywords: Enhanced Trofile, CCR5, CXCR4, vicriviroc
INTRODUCTION
HIV entry into target cells requires 3 steps: viral attachment to the CD4 receptor, viral attachment to a chemokine receptor (CCR5 or CXCR4), and viral-cell membrane fusion [1]. CCR5 antagonists are designed to prevent HIV entry into target cells by blocking a functional interaction with the CCR5 co-receptor [2]. The Trofile HIV co-receptor tropism assay (Monogram Biosciences, South San Francisco, CA) determines whether a patient’s viral population uses the CCR5 or CXCR4 co-receptor exclusively (R5 or X4 virus, respectively), or consists of a population of dual- (R5X4) or mixed-tropic viruses (dual/mixed [DM] virus) [3]. This assay is useful for selecting appropriate patients for treatment with CCR5 antagonists.
The original tropism assay was validated to detect minority X4 variants at 10% and 5% of a population with 100% and 85% sensitivity, respectively, using mixtures of plasmids carrying different HIV-1 envelopes [3]. Low level CXCR4-using (X4 or dual-tropic) variants below the detection limit of the original assay are better identified by an enhanced version of the assay validated to detect as little as 0.3% X4 variants with 100% sensitivity [4, 5, 6].
Vicriviroc is a CCR5 antagonist that potently inhibits R5 HIV-1 in vitro and in vivo [7]. We previously reported that in ACTG A5211, 10 vicriviroc recipients with DM virus detected at study entry had significantly reduced virologic responses compared to 71 vicriviroc recipients with R5 virus at entry as determined by the original tropism assay [8]. We recently used the enhanced sensitivity tropism assay to re-test the screening and entry samples from individuals enrolled in ACTG A5211. The study’s key virologic and immunologic endpoints were reanalyzed based on enhanced assay tropism classifications to determine if the enhanced tropism assay would better identify patients who may benefit from CCR5 antagonist therapy.
SUBJECTS AND METHODS
The design of ACTG A5211 has been described in detail elsewhere [8]. In brief, the study was a double-blind, randomized study of vicriviroc in treatment-experienced subjects with 48-week follow-up. Within 6 weeks prior to study entry, subjects were screened for R5 virus by the original Trofile assay (Monogram Biosciences). At study entry, eligible subjects were randomly assigned to one of three doses of vicriviroc (5, 10, or 15 mg QD; Schering-Plough Research Institute), or placebo, added to their failing background antiretroviral regimen that contained ritonavir (100-800 mg/day). After 14 days, subjects continued their randomized, double-blind study treatment and started a new optimized antiretroviral regimen that contained ritonavir.
The original tropism assay was used in determining viral co-receptor usage for ACTG A5211 participants at study screening, entry, day 14, weeks 8, 24, and 48, and if applicable, at a confirmatory virologic failure which was defined as a confirmed plasma HIV-1 RNA level of <1 log10 copies/mL (Ultra Sensitive Roche Amplicor HIV-1 Monitor assay) below the baseline level at or after 16 weeks. Patient envelope expression vectors generated from study screening and entry samples of subjects enrolled in ACTG A5211 were re-tested using the enhanced sensitivity Trofile assay (Monogram Biosciences) [6]. Written informed consent was obtained from study participants. Human experimentation guidelines of the U.S. Department of Health and Human Services were followed in the conduct of this research.
Wilcoxon rank sum test and Fisher’s exact test were used to compare subjects’ baseline characteristics. Changes in HIV-1 RNA (log10 copies/mL) were analyzed using linear regression for censored data to account for values below the limit of assay quantification (50 copies/ml) and adjusting for baseline log10 HIV-1 RNA level and each of three stratification factors used in the randomization (prior enfuvirtide use, anticipated enfuvirtide use, and CD4 count <50 cells/mm3). The proportions of subjects achieving various HIV-1 RNA thresholds were compared using Fisher’s exact test. An intent-to-treat approach was used. Analysis of changes in CD4 count followed the same approach as for the analysis of changes in log10 HIV-1 RNA. All tests were two-sided. P<0.05 was considered statistically significant. The analyses were performed using SAS software (version 9.1, SAS Institute; Cary, NC).
RESULTS
ACTG A5211 enrolled 118 subjects with R5 virus at study screening by the original tropism assay [8]. Among subjects with tropism results available by the enhanced sensitivity assay, 89/114 (78%) had R5 virus and 25/114 (22%) were reclassified with DM virus at study screening, and 85/111 (77%) subjects had R5 virus and 26/111 (23%) had DM virus at study entry; samples were unavailable for 4 subjects at study screening and 7 at study entry. At study entry, all 12 subjects with DM virus detected by the original assay [8] also had DM virus detected by the enhanced assay. Baseline characteristics, including age, sex, race/ethnicity, screening HIV-1 RNA level, screening CD4 count, nadir CD4 count, and screening genotypic and phenotypic susceptibility scores [9, 10, 11], were not significantly different between subjects with R5 virus at study screening and entry and those with DM virus at either study screening or entry by the enhanced assay (data not shown).
The association of co-receptor usage by the enhanced assay at study screening with a change in co-receptor tropism results by the original assay at study entry and on study was assessed among the 90 subjects randomized to receive vicriviroc (Table 1). Fifteen vicriviroc recipients were reclassified with DM virus at study screening by the enhanced assay and would have been excluded from the trial if the enhanced assay had been used to determine eligibility. Among these 15, 12 (80%) had DM/X4 viruses detected by the original assay by week 24 and would have been appropriately excluded. Of the other 3 subjects, 2 experienced virologic failure at week 16 and 32, respectively, and the third discontinued study at week 11 due to severe debilitation. Conversely, the original tropism assay detected DM/X4 viruses in 23/90 (26%) vicriviroc recipients by week 24 (Table 1). Of these 23 subjects, 12 (52%) had DM virus detected by the enhanced assay at study screening.
Table 1.
Comparison of co-receptor tropism according to the original and enhanced sensitivity tropism assay for ACTG A5211 subjects randomized to receive vicriviroc.
| Frequency (row percent) | Tropism by the original assay between study entry and week 24 |
|||
|---|---|---|---|---|
| R5 | DM or X4 | Total | ||
| Tropism by the enhanced sensitivity assay at study screening |
R5 | 61 (85%) | 11 (15%) | 72 |
| DM | 3 (20%) | 12 (80%) | 15 | |
| Missing | 3 (100%) | 0 (0%) | 3 | |
| Total | 67 | 23* | 90 | |
Of these 23 subjects, DM or X4 viruses were first detected at study entry for 10 subjects, at week 2 for 8, at week 8 for 2, and between week 19 and 24 for 3 subjects.
Eight subjects had DM virus at study screening but only R5 virus at study entry by the enhanced assay. Four of these 8 subjects were randomized to receive vicriviroc. During study follow-up, 2 of these 4 vicriviroc recipients were found to have DM virus by the original assay (week 2). The other 2 had R5 virus by the original assay while on study.
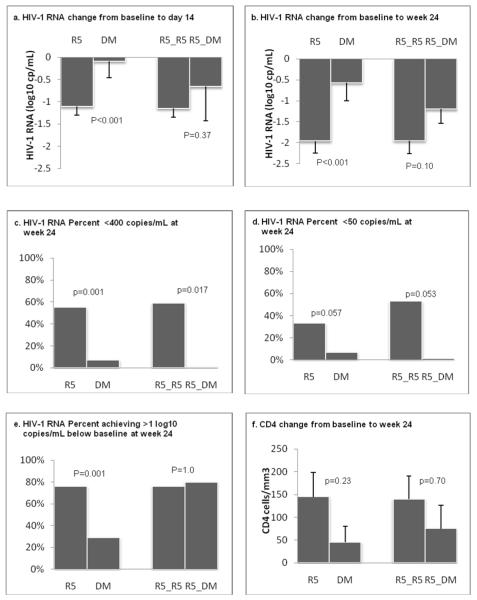
Among the subjects randomized to receive vicriviroc, the mean reductions in HIV-1 RNA from baseline to day 14 and week 24 were significantly greater for the 72 subjects with R5 virus (R5 virus group) than for the 15 subjects with DM virus (DM virus group) by the enhanced assay at study screening: −1.11 versus −0.09 log10 copies/mL, respectively, at day 14 (P<0.001, Figure 1a) and −1.91 versus −0.57 log10 copies/mL, respectively, at week 24 (P<0.001, Figure 1b). Thirty-six (55%) subjects in the R5 virus group achieved HIV-1 RNA < 400 copies/mL at week 24. In contrast, only one subject (7%) in the DM virus group achieved HIV-1 RNA < 400 copies/mL (P=0.001, Figure 1c); this subject had R5 virus detected by the enhanced sensitivity assay at study entry. Similar trends were seen from a comparison of the R5 and DM virus groups with respect to proportions with <50 copies/mL (33% vs. 7%, P=0.057) at week 24 (Figure 1d) and proportions with declines of ≥1 log10 copies/mL (76% vs. 29%, P=0.001, Figure 1e). Subgroup analysis showed that subjects with R5 virus at screening and entry by the enhanced tropism assay (R5_R5 group; N=64) had greater reductions in plasma HIV-1 RNA levels at day 14 (−1.15 log10 copies/mL) and week 24 (−1.95 log10 copies/mL) as compared to those with R5 virus at screening but DM virus at entry (R5_DM group; N=5; −0.66 log10 copies/mL at day 14 and −1.20 log10 copies/mL at week 24, Figure 1a,b), but these differences were not statistically significant. Four subjects (80%) in the R5_DM group achieved >1 log10 copies/mL reduction in viral load at week 24, but no subjects in this group attained <50 or <400 copies/mL (Figure 1c,d,e).
Figure 1. Virologic and immunologic responses according to co-receptor usage by the enhanced sensitivity tropism assay among subjects randomized to receive vicriviroc.
Three vicriviroc dose arms combined. Intent-to-treat approach; last observation carried forward following virologic failure or study discontinuation. N= 15, 72, 5, 64 for subjects who had DM virus at screening (DM virus group), R5 virus at screening (R5 virus group), R5 virus at screening but DM virus at entry (R5_DM virus group), and R5 virus at both screening and entry (R5_R5 virus group), respectively, for the day 14 results; N=14, 66, 5, and 58 for the same 4 groups, respectively for the week 24 results; Seven subjects in the 5-mg group who increased their dose to 15 mg prior to week 24 were not included in the week 24 analysis. The 15 subjects in the DM virus group include 4 subjects who had R5 virus by the enhanced sensitivity assay at study entry. P-values were from linear regression analysis adjusted for study stratification factors (see Methods), and baseline log10 HIV-1 RNA (for change in HIV-1 RNA) or CD4 count (for change in CD4 count). Vertical bars on the graph represent lower bounds of 95% confidence intervals.
The mean changes in CD4 count (cells/mm3) at week 24 were +145 (R5 virus group) and +45 (DM virus group); in the subgroup analysis, they were +140 (R5_R5 virus group) and +75 (R5_DM virus group). These differences were not statistically significant in an analysis adjusted for baseline CD4 count and study stratification factors (Figure 1f).
DISCUSSION
In this study, samples from ACTG A5211 subjects who had R5 virus at study screening as determined by the original tropism assay were retested using an enhanced sensitivity assay. Subjects reclassified with DM virus at study screening by the enhanced assay had suboptimal virologic responses to vicriviroc compared to those with R5 virus by the enhanced assay. Only one vicriviroc recipient who had DM virus at study screening, but R5 virus detected at study entry by the enhanced assay, successfully responded to vicriviroc (achieving HIV-1 RNA < 400 copies/mL at week 24). Therefore the enhanced sensitivity tropism assay identified a subset of treatment-experienced patients who were more likely to achieve suppression of HIV-1 viremia with an antiretroviral regimen containing a CCR5 antagonist, at least through 24 weeks.
A maximum time period of 6 weeks between study screening and entry visits was allowed for A5211. Differences in co-receptor usage between these visits were observed for some patients with the original and enhanced assays. Differences in tropism calls during this time frame could represent (1) the presence of low levels of DM virus fluctuating around the limit of detection of each assay; (2) very low levels of DM virus at both screening and entry which is only amplified at one time point due to the stochastic nature of polymerase chain reaction; (3) a more discernable change in relative levels of DM virus between screening and entry due to natural fluctuations in viral populations or adherence to the failing antiretroviral regimen.
Interestingly, a small number (5) of vicriviroc recipients with R5 virus at screening and DM at entry by the enhanced assay had an intermediate viral load reduction from baseline to day 14 and week 24, compared to subjects with R5 virus at screen and entry, the significance of which could not be ascertained due to the small sample size. However these patients did not achieve <400 or <50 HIV RNA copies/mL at week 24. Therefore, the presence of presumably very low-level DM virus around the detection limit of the enhanced assay also portends reduced CCR5 antagonist efficacy.
Limitations of this study include that it represents a retrospective analysis of ACTG A5211 participants who had R5 virus as determined by the original assay at study screening. Some analyses may have insufficient power due to relatively small sample size. For example, we saw a clinically relevant, but not statistically significant improvement in immunologic responses at week 24 for subjects with R5 virus as compared to those with DM virus by the enhanced assay as evaluated at study screening.
Our main study findings in this treatment-experienced cohort are supported by a similar reanalysis of the MERIT trial of the CCR5 antagonist maraviroc versus efavirenz (both in combination with co-formulated zidovudine and lamivudine) in treatment-naïve individuals with R5 virus at screen by the original tropism assay [12]. In the original study analysis, virologic response to maraviroc did not reach a pre-specified non-inferiority threshold at 48 weeks. Reanalysis of MERIT samples with the enhanced tropism assay revealed that 15% of the study subjects had DM virus at screening. Exclusion of these subjects in a retrospective analysis showed that maraviroc would have achieved non-inferiority to efavirenz in subjects with R5 virus at screening by the enhanced tropism assay.
In summary, the enhanced sensitivity co-receptor tropism assay identified A5211 subjects with DM virus below the detection limit of the original assay at study screening. These subjects experienced significantly inferior virologic responses to the CCR5 inhibitor vicriviroc. Therefore, these results, along with those from reanalysis of the MERIT trial, suggest that the enhanced tropism assay would be a better screening tool for determining the eligibility of HIV-1-infected patients for CCR5 antagonist therapy.
ADDIONAL MEMBERS OF THE AIDS CLINICAL TRIALS GROUP A5211 TEAM AND RESEARCH STAFF AND MONOGRAM BIOSCIENCES PERSONNEL
Other protocol team members
Robert Gross, MD, MSCE, Scott Hammer, MD, Martin Hirsch, MD, and Andrew Zolopa, MD (Co-Investigators); Catherine Godfrey, MD and Carla Pettinelli MD, PhD (Co-Medical Officers); Beatrice Kallungal, BS (Clinical Trials Specialists); David Clifford, MD (Co-Investigator, Protocol Neurologist); Mary Dobson, BS (Laboratory Data Coordinator); Antoine Simmons BS, MT (Laboratory Technologists); Valery Hughes, NP (Field Representative); Ana Martinez, RPh (Protocol Pharmacist); Susan Owens, RN, MS (Data Manager); and Jim Smith (Community Representative).
Monogram Biosciences
Wei Huang, MB, Linda Kiss, PhD, Jeff Larson, PhD, Yang Liu, MB, Neil Parkin, PhD, Chris Petropoulos, PhD, Lan Trinh, BS, Jeannette Whitcomb, PhD, Terri Wrin, BS, and the Monogram Biosciences Clinical Reference Laboratory.
Pharmaceutical Supporters
Schering-Plough Research Institute provided study drug, funds to support patient screening, and specialized laboratory assays.
AIDS Clinical Trials Group (ACTG) Study Sites (listed in order of patient accrual)
Jon Gothing, RN (Brigham and Women’s Hospital, Boston), Betsy Adams, RN, BSN, ACRN (Boston Medical Center), Lynn Bubley, RN, Mary Albrecht, MD (Beth Israel-Deaconess Medical Center); Pat Cain, RN, Jane Norris, PA-C (Stanford); Christine Hurley, RN, Carol Greisberger, RN (Rochester); Sharon Riddler, MD, MPH, Nancy Mantz, MSN, CRNP (University of Pittsburgh), Joseph Timpone, MD, Ioulia Vvedenskaya (Georgetown); Todd Stroberg, RN, Glenn Sturge, BS (Cornell); Margie Vasquez, RN, Judith Aberg, MD (New York University); Turkia Akaki, MD, Clifford Gunthel, MD (Emory); Sheila Dunaway, MD, Ann C. Collier, MD (University of Washington); Michael Morgan, FNP, Fred Nicotera (Vanderbilt); Beverly Putnam, MSN, John Koeppe (University of Colorado); Benigno Rodriguez, MD, Patricia Walton, BSN, RN (Case Western Reserve University), Mary Wild, RN and Ann Conrad, RN (MetroHealth Center, Cleveland); Judith Currier, MD, Maria Palmer, PA-C (University of California, Los Angeles); Mark Rodriguez, RN, BSN, David B. Clifford, MD (Washington University, St. Louis); Michael F. Para, MD, Barbara Ehrgott, RN (Ohio State); Allan S. Tenorio, MD, Beverly E. Sha, MD (Rush University Medical Center, Chicago); Donna Mildvan, MD, Manuel Revuelta, MD (Beth Israel Hospital, New York); Karen T. Tashima, MD, Helen Sousa, LPN (Miriam Hospital, Providence); Kristine Patterson, MD, Susan Richard, PA (University of North Carolina, Chapel Hill); Jody Lawrence, MD, Michele Downing, RN (University of California, San Francisco); Jeffery L. Meier, MD, Barbara Ann Wiley, BSN, RN (University of Iowa); William A. O’Brien, MD, MS, Cheryl Mogridge, RN (University of Texas, Galveston); Julie Hoffman, RN, Linda Meixner, RN (University of California; San Diego); Lorna Nagamine, RN, Scott Souza, PharmD (University of Hawaii at Manoa); Wayne Wagner, RN, MSW, Pablo Tebas, MD (University of Pennsylvania); Beth Zwickl, RN, MSN, Mitchell Goldman, MD (Wishard Hospital, Indianapolis).
Acknowledgments
We thank the study participants.
Financial support: Sponsored by the AIDS Clinical Trial Group (ACTG)/NIAID/NIH (AI-68636) and Statistical Data Management Center (AI68634), Cornell CTU (AI-69419), Cornell CTSC (RR024996), K24 AI51966 (Gulick), and K23 AI55038 (Wilkin), Johns Hopkins ACTU grant AI27668, PENN AIDS Clinical Trials Unit (U01-AI32783), PENN Center for AIDS Research (P30-AI45008 ), Harvard-BMC ACTU (AI27659). Tropism testing by the enhanced Trofile assay was supported by Monogram Biosciences.
Footnotes
Potential conflicts of interest
Dr. Reeves and Coakley are employed by Monogram Biosciences, Inc.
Dr. Hughes is a paid Data and Safety Monitoring Board member for Boehringer Ingelheim Corporation, Tibotec Pharmaceuticals Limited, and Virionyx Corporation Limited.
Dr. Flexner has served on a scientific advisory board for Schering-Plough
Dr. Greaves is employed by Schering Plough Research Institute.
Dr. Kuritzkes is a consultant to Schering-Plough and Monogram Biosciences, and has received research funding from Schering-Plough.
Dr. Gulick receives research grant support from Pfizer and Schering-Plough and has served as an ad-hoc consultant for Monogram Biosciences, Pathway, Pfizer and Schering-Plough.
All other authors: no conflicts.
References
- 1.Moore JP, Doms RW. The entry of entry inhibitors: a fusion of science and medicine. Proc Natl Acad Sci U S A. 2003;100(19):10598–602. doi: 10.1073/pnas.1932511100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clotet B. CCR5 inhibitors: promising yet challenging. J Infect Dis. 2007;196(2):178–80. doi: 10.1086/518799. [DOI] [PubMed] [Google Scholar]
- 3.Whitcomb JM, Huang W, Fransen S, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob Agents Chemother. 2007;51(2):566–75. doi: 10.1128/AAC.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reeves JD, Han D, Liu Y, Wrin T, Huang W, Coakley E, Petropoulos C, Parkin N. Enhancements to the Trofile HIV coreceptor tropism assay enable reliable detection of CXCR4-using subpopulations at less than 1%. 47th annual interscience conference on antimicrobial agents and chemotherapy; Chicago, Illinois, USA. 2007 17-20 September; 2007. 2007. Abstract H-1026. [Google Scholar]
- 5.Trinh L, Han D, Huang W, Wrin T, Larson J, Kiss L, Coakley E, Petropoulos CJ, Parkin NT, Whitcomb JM, Reeves JD. Technical validation of an enhanced sensitivity Trofile HIV co-receptor tropism assay for selecting patients for therapy with entry inhibitors targeting CCR5. Antivir Therapy. 2008;13(Suppl 3):A128. [Google Scholar]
- 6.Reeves JD, Coakley E, Petropoulos CJ, Whitcomb JM. An Enhanced-Sensitivity Trofile™ HIV Coreceptor Tropism Assay for Selecting Patients for Therapy with Entry Inhibitors Targeting CCR5. Journal of Viral Entry. 2009 (In Press) [Google Scholar]
- 7.Strizki JM, Tremblay C, Xu S, et al. Discovery and characterization of vicriviroc (SCH 417690), a CCR5 antagonist with potent activity against human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2005;49:4911–9. doi: 10.1128/AAC.49.12.4911-4919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gulick RM, Su Z, Flexner C, et al. Phase 2 study of the safety and efficacy of vicriviroc, a CCR5 inhibitor, in HIV-1-Infected, treatment-experienced patients: AIDS clinical trials group 5211. J Infect Dis. 2007;196(2):304–12. doi: 10.1086/518797. [DOI] [PubMed] [Google Scholar]
- 9.Anderson JA, Jiang H, Ding X, et al. Genotypic susceptibility scores and HIV type 1 RNA responses in treatment-experienced subjects with HIV type 1 infection. AIDS Res Hum Retroviruses. 2008;24(5):685–94. doi: 10.1089/aid.2007.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swanstrom R, Bosch RJ, Katzenstein D, et al. Weighted phenotypic susceptibility scores are predictive of the HIV-1 RNA response in protease inhibitor-experienced HIV-1-infected subjects. J Infect Dis. 2004;190(5):886–93. doi: 10.1086/422692. [DOI] [PubMed] [Google Scholar]
- 11.Coakley EP, Chappey C, Flandre P, et al. Defining lower and upper phenotypic clinical cutoffs for tipranavir, lopinavir, saquinavir and amprenavir co-administered with ritonavir within the RESIST dataset using the PhenoSense assay (Monogram Biosciences) Antivir Ther. 2006;11:S81. [Google Scholar]
- 12.Saag M, Heera J, Goodrich J, et al. Reanalysis of the MERIT study with the enhanced Trofile assay (MERIT-ES). 48th Interscience Conference on Antimicrobial Agents and Chemotherapy; Washington. 2008. poster abstract H-1232a. [Google Scholar]