Abstract
Background
Studies of viral load-related persistence of human papillomavirus (HPV) infection are rare, with inconsistent results reported.
Methods
Study subjects were 741 and 289 women who were positive for HPV16 and HPV18, respectively, at enrollment into in the ASCUS-LSIL Triage Study and who returned one or more times for HPV testing during a biannual 2-year follow-up. Baseline HPV16 and HPV18 copies per nanogram of cellular DNA were measured by real-time polymerase chain reaction.
Results
Women with, compared to without, persistent infection at month 6 had higher viral load at enrollment (P<0.001 for HPV16; P=0.01 for HPV18). The association of per 1 log10-unit increase in viral load with the first 6-month persistence of HPV16 or HPV18 was statistically significant among women with multiple types at enrollment (OR=1.53, 95% CI, 1.29–1.82 for HPV16; OR=1.35, 95% CI, 1.09–1.68 for HPV18) but not among those with mono-type infections (test for interaction between viral load and coinfection: P=0.002 for HPV16; P=0.34 for HPV18). Among women who continued to be positive at month 6, 12, or 18, persisting for another 6 months was unrelated to baseline viral load.
Conclusion
Higher viral load of prevalent HPV16 or HPV18 infection was associated with short- but not long-term persistence.
Keywords: Human Papillomavirus, Viral Load, Persistence
INTRODUCTION
Although infection with human papillomavirus (HPV) has been established as a necessary cause of cervical cancer [1], it is also commonly found in healthy individuals. Most HPV infections are transient and result in no discernible cervical lesion in cross-sectional screening [2–4]. As shown in a population-based study in Costa Rica [5], approximately 25% of women with single prevalent HPV infections had concurrent abnormalities of cervical cytology. Given that a persistent infection with oncogenic HPV types is a prerequisite to the development of cervical cancer [6–11], knowing determinants that are associated with viral persistence may help with improvement of programs for cervical cancer control. Since HPV DNA load reflects the productivity of viral replication, whether the level of viral DNA is able to predict the likelihood of persistence deserves consideration.
The association of higher HPV DNA load with persistence of the infection has been reported by some studies [12–17], but not others [18, 19]. These studies are, however, often limited by small sample size, use of only two timepoint measurements to define viral persistence, and a few were limited by using a semi-quantitative approach to measure DNA of a group of HPV types. It is unclear if the association of HPV DNA load with persistence of the infection differs with increasing time from the initial positive test, i.e., how persistence is defined.
In this study, we evaluated the association between baseline HPV16 and HPV18 DNA load and type-specific persistence of the infection during a 2-year follow-up among women who participated in the Atypical Squamous Cells of Undetermined Significance (ASC-US) and Low-Grade Squamous Intraepithelial Lesion (LSIL) Triage Study (ALTS).
MATERIALS AND METHODS
Study Subjects
Study subjects were women enrolled in ALTS, a large-scale randomized clinical trial designed to compare strategies for management of women with a referral Pap of ASC-US or LSIL. A detailed description of the ALTS design and study population has been reported elsewhere [20, 21]. Briefly, between January 1997 and December 1998, 5060 women with a Pap of ASC-US or LSIL in the previous 6 months were enrolled and randomly assigned to one of three trial arms. All participants underwent an entry procedure at enrollment including interview, Pap smear, and testing for HPV DNA. Three trial arms differed in their criteria for referral to colposcopy and biopsy at enrollment. Regardless of the study arms, these women returned at 6-month intervals over 2 years for cervical cytology and HPV testing. During follow-up, women were re-referred for colposcopy and biopsy if cytologic evidence of high-grade squamous intraepithelial lesion (HSIL) was detected. At exit, all women with persistent low-grade lesions or HPV-positive ASC-US were also referred to colposcopy. Women with biopsy-confirmed cervical intraepithelial neoplasia grade 2 or higher (≥CIN2) received appropriate treatment immediately. The ALTS protocol was approved by the institutional review boards at the National Cancer Institute and at each of the four clinical centers involved in the trial.
ALTS participants were eligible for this study if they had HPV16 and/or HPV18 DNA detected in their enrollment cervical samples by polymerase chain reaction (PCR)-based reverse line blot assay [22]. PCR amplicons were subjected to reverse line blot hybridization for detection of 27 types (6/11/16/18/26/31/33/35/39/40/42/45/51/52/53/54/55/56/57/58/59/66/68/73/82/83/84) [23]. During the trial, the typing capacity of the reverse line blot was expended from detection of 27 types to 38 types by additionally including 11 types (61/62/64/67/69/70/71/72/81/85/91). Of 1071 eligible women (759 positive for HPV16, 258 positive for HPV18, and 54 positive for both), 19 (11 HPV16-positive and 8 HPV18-positive) were excluded because of a lack of samples for viral quantification. We additionally excluded 76 women, including one whose enrollment sample was positive for HPV18 but negative for cellular DNA and 75 (61 HPV16-positve and 14 HPV18-positive) who did not provide any follow-up visit. This left 741 women with baseline HPV16 infection and 289 with HPV18 infection in the analyses. Baseline HPV16 or HPV18 DNA load was similar between women with and without a follow-up (data not shown). Data on HPV typing, cervical cytology and histology, and characteristics of study subjects were obtained from the ALTS database. The protocol for this study was approved by the institutional review board at the University of Washington.
Quantification of HPV16 and HPV18 DNA Load
HPV16 and HPV18 E7 copy number and cellular DNA amount in enrollment cervical swab samples were measured by multiplex real-time PCR, as described previously [24, 25]. Briefly, the assay was set up in a reaction volume of 25 μL with the TaqMan Universal PCR Master Mix kit (Applied Biosystems, Foster City, CA). Amplification was carried out on the Applied Biosystems 7900 HT. Two logarithmic-phase five-point standard curves were implemented in each set of real-time PCR assays; one for HPV16 or HPV18 DNA and the other for cellular DNA. Each sample was assayed in triplicate. The viral load was normalized to the input amount of cellular DNA and expressed as E7 copy number per nanogram of cellular DNA.
E7 DNA was undetectable by real-time PCR in 58 HPV16-positive and 21 HPV18-positve samples. Considering that the negative result might be due to a tiny amount of viral DNA, a value of one viral copy per nanogram of cellular DNA was assigned to each sample. Similar results were obtained when these samples were excluded from the analysis (data not shown).
Statistical Analyses
The normalized HPV16 and HPV18 DNA load was log10-transformed; the mean value of the triplicate measurements was used for analyses.
A linear regression model [26] was used to compare log10-transformed HPV16 or HPV18 DNA load at enrollment between women who remained positive and those who became negative at each follow-up visit while adjusting for cervical cytology at enrollment, current smoking status, and coinfection with other HPV types. Coinfection was defined based on testing results of 27 HPV types for 52% of the enrollment samples and 38 HPV types for 48%. In ALTS, the events of becoming viral DNA-negative were ascertained by 6-month intervals. A woman was eligible for analysis of HPV status at a given visit if she had type-specific HPV DNA detected at all scheduled previous visits. For example, a woman with a baseline HPV16 infection was eligible for analysis of HPV16 status at month 6 visit; if she continued to be HPV16-positive at month 6 she would be eligible for the analysis at month 12. Although this analysis is straightforward, it excluded all visits after an initial missing one.
A second analysis was performed to evaluate odds ratios (ORs) and 95% confidence intervals (CIs) using a logistic regression model for the association of baseline viral load (to be fitted as a continuous covariate) with risk of being positive at various follow-up visits. The ORs were adjusted for coinfection with other types, current smoking status, and cervical cytology at enrollment. The significance of the interaction between viral load and time (visit) was assessed using a likelihood ratio test. For illustrative purposes the probability of remaining positive at various follow-up visits were plotted for the 25, 50, or 75 percentile of baseline HPV16 (or HPV18) DNA load. We also examined coinfection with other types, current smoking status, or cervical cytology at enrollment to determine if they modified the effect of baseline viral load in this model.
In ALTS, ≥CIN2 was histologically confirmed in 377 (50.9%) of 741 initially HPV16-positive women (253 at enrollment and 124 during follow-up) and 90 (31.1%) of 289 initially HPV18-positive women (54 at enrollment and 36 during follow-up). As reported elsewhere [27–31], risks of ≥CIN2 varied with HPV DNA load; treatment of ≥CIN2 altered HPV persistence. To determine whether the estimates of persistence by viral load would be distorted by presence of ≥CIN2, we performed parallel analyses in which women with ≥CIN2 were censored at the time of initial diagnosis. The results remained similar; for simplicity, these results were not presented.
We used student t test to compare viral load by age at enrollment, race, current use of hormonal contraceptives, lifetime number of sex partners, current smoking status, coinfection with other types, and HPV variant. Differences in viral load by number of visits followed and by baseline cervical cytology were tested by one way ANOVA. Among women with multiple HPV types at enrollment, the proportions of coinfection with the same species type between women with and without detectable HPV16 or HPV18 at month 6 were compared by chi-square test. The types co-infected were phylogenetically classified as non-HPV16 alpha-9 species (i.e., HPV16-related, including HPV31/33/35/52/58/67) for analysis of HPV16 and non-HPV18 alpha-7 species (i.e., HPV18-related, including HPV39/45/59/68/70/85) for analysis of HPV18. All statistical tests were at the 5% two-sided significance level.
RESULTS
The present study included 741 women with HPV16 and 289 women with HPV18 infection at enrollment. Forty-one of them were positive for both HPV16 and HPV18. The mean (SD) value of log10-transformed HPV16 E7 copy number per 1 nanogram of cellular DNA was 2.82 (1.33), 2.81 (1.34), 2.86 (1.29) and 2.76 (1.37) for 72, 74, 196, and 399 women who provided 1, 2, 3, and 4 follow-up visits, respectively (p=0.87). The corresponding values of viral load for those with HPV18 infection were 4.34 (1.20), 3.79 (1.68), 3.85 (1.68), and 3.79 (1.67) for 20, 33, 72, and 164 women, respectively (p=0.39). As shown in Table 1, within the study population, baseline HPV16 DNA load differed significantly by age, race, current smoking status, coinfection with other HPV types, and cervical cytology at enrollment. Baseline HPV18 DNA load differed significantly by current smoking status, coinfection with other HPV types, and cervical cytology at enrollment.
Table 1.
Baseline HPV16 or HPV18 DNA load by characteristics of study subjects
| Log10 HPV16 E7 copies per 1 nanogram of cellular DNA |
Log10 HPV18 E7 copies per 1 nanogram of cellular DNA |
|||||
|---|---|---|---|---|---|---|
| Variables | No. | Mean (SD) | P-value | No. | Mean (SD) | P-value |
| Age at study entry (years) | ||||||
| 18–24 | 467 | 2.86 (1.33) | 0.09 | 188 | 3.87 (1.58) | 0.75 |
| ≥25 | 274 | 2.69 (1.36) | 101 | 3.80 (1.76) | ||
| Race a | ||||||
| White | 535 | 2.87 (1.31) | 0.03 | 175 | 3.86 (1.66) | 0.87 |
| Nonwhite | 201 | 2.62 (1.41) | 113 | 3.82 (1.63) | ||
| Current use of hormonal contraceptives b | ||||||
| No | 372 | 2.76 (1.42) | 0.42 | 153 | 3.90 (1.79) | 0.56 |
| Yes | 363 | 2.84 (1.26) | 134 | 3.78 (1.47) | ||
| No. of life time male sex partners c | ||||||
| 0–5 | 363 | 2.80 (1.38) | 0.93 | 143 | 3.81 (1.60) | 0.68 |
| ≥6 | 367 | 2.79 (1.32) | 141 | 3.89 (1.69) | ||
| Current smoking | ||||||
| No | 408 | 2.68 (1.33) | 0.01 | 174 | 3.67 (1.61) | 0.03 |
| Yes | 333 | 2.94 (1.34) | 115 | 4.11 (1.67) | ||
| Coinfection with other HPV types | ||||||
| No | 218 | 2.93 (1.31) | 0.09 | 72 | 4.34 (1.63) | <0.001 |
| Yes | 523 | 2.74 (1.35) | 217 | 3.68 (1.62) | ||
| HPV variant d | ||||||
| European | 572 | 2.97 (1.21) | 0.41 | 107 | 4.26 (1.38) | 0.26 |
| Non-European | 130 | 2.88 (1.18) | 144 | 4.05 (1.52) | ||
| Cytology at enrollment e | ||||||
| Within normal limits | 140 | 1.76 (1.31) | <0.001 | 58 | 2.79 (1.72) | <0.001 |
| ASC-US | 208 | 2.76 (1.24) | 81 | 3.88 (1.48) | ||
| LSIL | 248 | 3.10 (1.26) | 118 | 4.19 (1.54) | ||
| HSIL | 140 | 3.39 (1.03) | 32 | 4.38 (1.47) | ||
NOTE. Excluded were 5 HPV16-positive women and one HPV18-positive woman who did not provide race information. A category of nonwhite includes African American, American Indian/Alaskan, or Asian/Pacific Islander women.
Excluded were 6 HPV16-positive women and 2 HPV18-positive women who did not provide information on use of hormonal contraceptives.
Excluded was 11 HPV16-positive woman and 5 HPV18-positive women who did not provide information of number of lifetime sex partners.
Excluded were 39 HPV16-positive women and 38 HPV18-positive whose enrollment samples were insufficient for variant characterization.
Excluded were 5 HPV16-positive women whose enrollment samples were inadequate for cytologic diagnosis.
Table 2 shows differences in viral load at enrollment between women with and without persistence of the infection at consecutive visits. Women who remained type-specific positive as compared to those who became negative at month 6 had a significantly higher baseline HPV16 (P<0.001) or HPV18 DNA load (P=0.01). Among those who remained HPV16- or HPV18-positive at month 6, there was no appreciable difference in baseline viral load by type-specific positivity at the subsequent follow-up visit at month 12. Similarly, subsequent persistence of infections still present at month 12 or month 18 was not predicted by viral load at enrollment.
Table 2.
Baseline HPV16 or HPV18 DNA load between women who continued to be type-specific positive and those who became negative at each of follow-up visits, starting with 741 HPV16-positive and 289 HPV18-positive, respectively
| Log10 HPV16 E7 copies per 1 nanogram of cellular DNA in women | Log10 HPV18 E7 copies per 1 nanogram of cellular DNA in women | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Follow-up visit | not seen No. | remaining positive |
becoming negative |
not seen No. | remaining positive |
becoming negative |
||||||
| No. | Mean (SD) | No. | Mean (SD) | P-value a | No. | Mean (SD) | No. | Mean (SD) | P-value a | |||
| month 6 | 106 | 279 | 3.00 (1.21) | 356 | 2.61 (1.41) | <0.001 | 44 | 102 | 4.07 (1.49) | 143 | 3.56 (1.79) | 0.01 |
| month 12 | 51 | 138 | 2.94 (1.24) | 90 | 3.19 (1.16) | 0.28 | 16 | 52 | 3.98 (1.57) | 34 | 4.14 (1.19) | 0.55 |
| month 18 | 19 | 82 | 3.01 (1.15) | 37 | 2.69 (1.40) | 0.24 | 2 | 34 | 4.17 (1.40) | 16 | 3.78 (1.91) | 0.76 |
| month 24 | 11 | 48 | 3.04 (1.22) | 23 | 3.10 (1.10) | 0.76 | 2 | 23 | 4.06 (1.35) | 9 | 4.53 (1.68) | 0.34 |
NOTE. Adjusted for cervical cytology (within normal limits, ASC-US, LSIL or HSIL), current smoking status (yes or no), and coinfection with other types at enrollment (yes or no)
We next assessed the association between a 1 log10 unit increase in baseline viral load and type-specific positivity at various follow-up visits. The interaction between the follow-up visit number and baseline viral load was statistically significant (likelihood ratio test: p=0.05 for women with HPV16; p=0.04 for those with HPV18); it was included in the logistic model. With adjustment for current smoking status, coinfection with other types, and cervical cytology at enrollment, the ORs for the association of per 1 log10 unit increase in baseline viral load with HPV16 positivity at month 6, 12, 18 and 24 were 1.35 (95% CI, 1.18–1.54), 1.14 (95% CI, 0.99–1.30), 1.12 (95% CI, 0.96–1.30), and 1.03 (95% CI, 0.89–1.20), respectively. The ORs for the association with HPV18 positivity at month 6, 12, 18 and 24 were 1.19 (95% CI, 1.00–1.40), 1.13 (95% CI, 0.94–1.37), 1.02 (95% CI, 0.85–1.21), and 0.97 (95% CI, 0.82–1.15), respectively. The associations remained similar when visits subsequent to the initial negative test were excluded (data not shown). We also tested the model with treating the intercurrent negative visit as being positive; the results remained similar (data not shown).
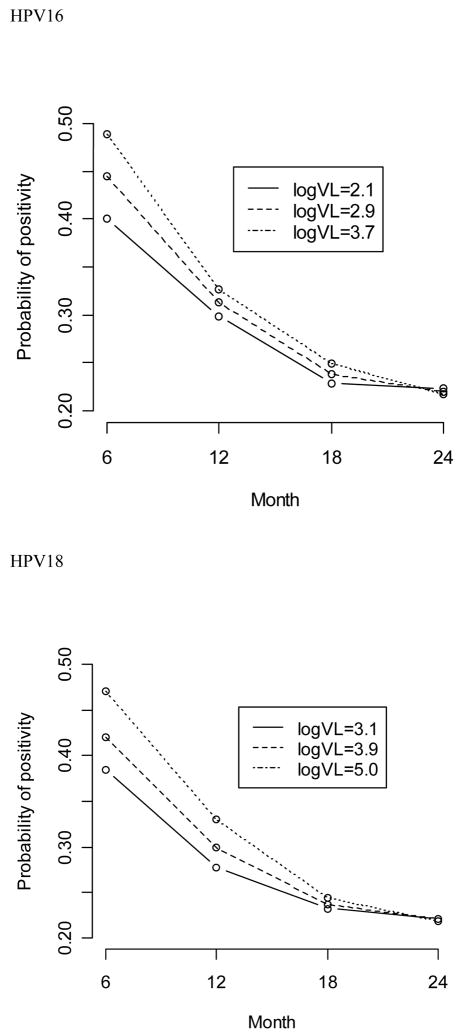
Figure 1 illustrates probabilities of remaining type-specific positive at various follow-up visits by baseline viral load. The values of 25, 50, and 75 percentile of HPV16 or HPV18 DNA load were chosen to represent low-, medium-, and high-level of viral load, respectively. Irrespective of the viral load at enrollment, probabilities of positivity dropped dramatically overtime. Consistent with the estimates of risk association, probabilities of type-specific positivity at month 6 differed substantially by baseline viral load but the differences diminished quickly as increasing of follow-up time and disappeared at month 18 and 24 visits.
Figure 1.
Probability of HPV16 or HPV18 positivity at month 6, 12, 18, or 24 visits among women with values of 25 (solid line), 50 (dashed line), or 75 (dot line) percentile of log10-transformed baseline HPV16 (log10 viral loads of 2.1, 2.9 and 3.7, respectively) or HPV18 DNA load (log10 viral loads of 3.1, 3.9, 5.0, respectively)
Given a significant association between the higher viral load at enrollment and type-specific positivity at month 6, we further examined whether the association was modified by viral load-related factors. As shown in Table 3, in contrast to a null association of baseline viral load with type-specific positivity at month 6 among women with mono-type infection at enrollment, the association among those with multiples HPV types was statistically significant (OR = 1.53, 95% CI, 1.29–1.82 for women with baseline HPV16 infections; OR = 1.35, 95% CI, 1.09–1.68 for those with baseline HPV18 infections). The interaction between the viral load and baseline coinfection with other HPV types was statistically significant for women with HPV16 at enrollment (P = 0.002) but not for those with HPV18 (P = 0.34). The magnitude of the risk association was not meaningfully different by current smoking status or cervical cytology at enrollment. Among women with multiple HPV types at enrollment, coinfection with HPV16-related types from the alpha-9 species was detected in 76 (39.4%) of 193 women with and 116 (45.1%) of 257 women without detectable HPV16 DNA at month 6 (P = 0.22). Baseline coinfection with HPV18-related types from the alpha-7 species was detected in 22 (28.9%) of 76 women with and 40 (36.7%) of 109 women without detectable HPV18 DNA at month 6 (P = 0.27).
Table 3.
Odds ratios (ORs) and 95% confidence intervals (CIs) for the association of a 1 log10 increase in baseline HPV16 or HPV18 DNA load with type-specific positivity at month 6 visit, stratified by coinfection with other types, current smoking status, or cervical cytology at enrollment
| Baseline HPV16 DNA load in women | Baseline HPV18 DNA load in women | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| remaining positive |
becoming negative |
remaining positive |
becoming negative |
|||||||
| Stratum | No. | Mean (SD) | No. | Mean (SD) | OR(95% CI)a | No. | Mean (SD) | No. | Mean (SD) | OR (95% CI)a |
| Coinfection | ||||||||||
| No | 86 | 2.89 (1.39) | 99 | 3.04 (1.29) | 1.04 (0.79–1.36) | 26 | 4.46 (1.31) | 34 | 4.19 (1.97) | 1.08 (0.76–1.53) |
| Yes | 193 | 3.05 (1.12) | 257 | 2.44 (1.42) | 1.53 (1.29–1.82) | 76 | 3.94 (1.54) | 109 | 3.37 (1.69) | 1.35 (1.09–1.68) |
| Current smoking | ||||||||||
| No | 150 | 2.87 (1.24) | 203 | 2.58 (1.38) | 1.31 (1.09–1.59) | 63 | 3.98 (1.43) | 87 | 3.34 (1.71) | 1.26 (1.00–1.59) |
| Yes | 129 | 3.16 (1.16) | 153 | 2.65 (1.46) | 1.45 (1.17–1.79) | 39 | 4.22 (1.60) | 56 | 3.91 (1.87) | 1.38 (0.99–1.91) |
| Cervical cytology b | ||||||||||
| Within normal limits | 54 | 2.08 (1.20) | 64 | 1.38 (1.30) | 1.52 (1.11–2.07) | 20 | 3.23 (1.45) | 30 | 2.24 (1.79) | 1.75 (1.11–2.77) |
| ASC-US | 79 | 2.98 (1.16) | 101 | 2.56 (1.23) | 1.35 (1.04–1.75) | 27 | 4.14 (1.18) | 40 | 3.56 (1.74) | 1.30 (0.91–1.86) |
| LSIL | 103 | 3.28 (1.01) | 105 | 2.90 (1.40) | 1.32 (1.04–1.69) | 51 | 4.27 (1.54) | 50 | 4.11 (1.59) | 1.11 (0.87–1.43) |
| HSIL | 42 | 3.60 (1.02) | 82 | 3.27 (1.09) | 1.37 (0.94–1.99) | 4 | 5.35 (1.54) | 23 | 4.12 (1.42) | |
NOTE. Associating per 1 log10 unit increase in baseline HPV16 or HPV18 DNA load with type-specific positivity at month 6, adjusted for all variables listed in the table except for the stratifier
Excluded were 5 HPV16-positive women whose enrollment samples were inadequate for cytologic diagnosis. Because of the limited number of women with HSIL who remained HPV18-positive at month 6, women with HSIL were grouped with those with LSIL for analysis of risk association.
DISCUSSION
In this 2-year longitudinal study of women with HPV16 and/or HPV18 infection at enrolment, we found that baseline HPV16 or HPV18 DNA load was significantly higher among those with, compared to without, a persistent infection in the first 6-month follow-up. However, for women who remained viral DNA-positive at month 6, 12, or 18 visits, the probabilities of persisting for another 6 months did not differ appreciably by HPV16 or HPV18 DNA load at enrollment. This suggests that higher viral load of the prevalent infection predicted greater short- but not long-term persistence.
The loss of strength in the association over time cannot be explained by loss-to-follow-up or the length of follow-up, because the viral load was equivalent between women with and without follow-up and by the number of follow-up visits. Ascertainment bias was not an issue because measurement of viral load was performed without information on viral persistence. A substantial number of ALTS participants received a therapeutic procedure for ≥CIN2. In view of the association between viral load and risk of ≥CIN2 [27–29] and between ≥CIN2 treatment and viral clearance [30, 31], a diagnosis of ≥CIN2 might be an intermediate step of the pathway from viral load to persistence. Therefore, we performed parallel analyses by censoring ≥CIN2 cases at the time of initial diagnosis rather than adjusting for a diagnosis of ≥CIN2. The comparable results between the analyses with and without censoring suggested that it is unlikely that the loss of strength in the association overtime was biased by ≥CIN2 treatment.
Studies of the association between HPV DNA load and persistence are rare, with inconsistent findings reported [12–19]. None of the previous studies except for the one by van Duin el al. [15] examined risk association overtime. In that study, cervical samples assayed were a mixture of entry and follow-up samples which were divided into 3 time groups, i.e., 3–12 months, 13–24 months, and 25–59 months before the end of follow-up. Viral clearance was defined as no HPV16 DNA in the last follow-up sample or absence of HPV16 DNA in at least 2 consecutive samples. They found a significant association of lower median HPV16 DNA load with viral clearance in the group of samples taken 3–12 months before the end of the study but not in other two groups of samples. Although the study by Duin et al. was limited by a small sample size and lack of individual viral load normalization, the findings somewhat concur in our results.
The mechanism for the association of higher baseline HPV16 or HPV18 DNA load with short-but not long-term persistence is not clear. One possibility is that a subset of prevalent infections was close to the end of the natural course of HPV infection. The viral load of these infections would be likely to decreasing with time, thereby reducing the viral load average for women without a positive detection at month 6. Alternatively, low viral load infections might indicate in some cases contamination by sexual partners or other viral states destined to produce short-term viral DNA positivity.
However, it is difficult to see how to explain our finding that the association of higher viral load with short-term persistence was seen among women with coinfection of other HPV types but not among those with mono-type infection, especially for HPV16. It is possible that some coinfection-related factors play a role in the association of viral load with short-term persistence. As suggested by reduced rates of HPV clearance in individuals with HIV infection and/or low CD4 count [32–36], the host’s cellular immune response is thought to be critical to clearance of the infection. In a recent study [37], we observed statistically significant associations between lower HPV16 DNA load and coinfection with HPV16-related types and between lower HPV18 DNA load and coinfection with HPV18-related types. Given that closely related HPV types may share some conserved T cell epitopes to elicit cross-reactive immune responses [38, 39], the coinfection-associated reduction of viral load was presumed to be related to cross-reactivity of cellular immune response. Accordingly, the cellular immunity induced by other HPV types may help with clearance of the infection by targeting the infected cells and/or limiting the virus for viral DNA replication or production of viral progeny.
Given the above interpretation, one may expect a rapid clearance in women with, compared to without, coinfection of other HPV types. However, previous studies of natural history of HPV infection demonstrated either no [40] or positive association [41, 42] between coinfection and persistence. The positive association was explained by Trottier et al. [42] that women with multiple types might have immune-responded poorly to the virus and consequently permitted persistence of the infection. Interestingly, we observed here, among women with multiple HPV types, a slightly lower proportion of coinfection with the same species types in those with, compared to without, detectable type-specific positivity at month 6. This difference, although not statistically significant, somewhat corroborates a previous report that the extent of cellular immune response to HPV16 L1 VLP vaccine correlated with phylogenetic distance of HPV types [43]. Thus, the results of this study extended the previous interpretation and raised an intriguing possibility that the association of viral load with short-term persistence among women with coinfection may be in part attributable to differences in cross-reactivity of cellular immune response induced by the same versus other species types.
It should be pointed out that a lack of the association between HPV16 or HPV18 DNA load and short-term persistence among women with mono-type infections appeared dissimilar to a resent report from the ALTS participants who had single infections (Manuscript by Maucort-Boulch et al. under revision at Int J Cancer). They found that persistence was associated with higher viral load. In that study, however, the exposure was semi-quantitative measurement of viral load by Hybrid Capture and the endpoint was average type-specific persistence of a group of high-risk types. Although the estimate of viral load by real-time PCR versus Hybrid Capture may be comparable for samples with a single type of HPV, the range of types summarized as an outcome in that study extended to 14 types that included HPV16 and HPV18. This may in part explain the discrepancy of the results between two studies.
Several study limitations should be addressed. First, women included in this study were those who were positive for HPV16 and/or HPV18 at enrollment. Because we did not observe the time of acquisition, we were unable to define the time from initial infection to regression. By selecting participants with prevalent HPV infection, we might have biased our study population towards those with a prolonged positive duration. However, the influence of this is likely to be non-differential. Second, we were unable to assess impacts of change of viral load on persistence of the infections because in this study only baseline viral load was measured. Although this does not affect the validity of the findings, data on type-specific positivity subsequent to the viral load at each of follow-up visits would further provide a dynamic view of the risk association. Third, HPV positivity in ALTS was detected by interval. The disappearance and occurrence within the interval remain undetermined. Such a misclassification may lead to an overestimate of the length of viral persistence. It is unknown whether this would be differentially related to viral load. Lastly, because of the PCR’s detection limit, whether the undetectable HPV DNA is due to a clearance of the infection or a latent infection in basal cells can not be determined.
In summary, our data indicated that the persistence of type-specific infection in the first 6 months after enrollment was significantly associated with baseline HPV16 or HPV18 DNA load. Among women who continued to be positive at month 6, 12, or 18 visits, persisting for another 6 months was unrelated to baseline viral load. The association of higher viral load with short-term persistence among women with multiple HPV types but not among those with mono-type infections suggests a potential role of coinfection-related cross-reactivity of cellular immune response in clearance of the infection.
Acknowledgments
The work was supported by Public Health Service grant CA 84396 to L.F.X.
This study was part of the project ancillary to the ALTS clinical trial but does not represent the ALTS Group. The authors would like to thank the ALTS Group for providing the biological specimens and HPV typing results and Dr Kathrin Jansen for providing the HPV16 and HPV18 DNA standard.
Footnotes
Potential conflicts of interest: L.A.K. receives research funds from Merck Research Laboratories. Other authors have no commercial or other associations that might pose a conflict of interest.
References
- 1.Munoz N, Bosch FX, de Sanjose S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–27. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 2.Nobbenhuis MA, Helmerhorst TJ, van den Brule AJ, et al. Cytological regression and clearance of high-risk human papillomavirus in women with an abnormal cervical smear. Lancet. 2001;358:1782–3. doi: 10.1016/S0140-6736(01)06809-X. [DOI] [PubMed] [Google Scholar]
- 3.Franco EL, Villa LL, Sobrinho JP, et al. Epidemiology of acquisition and clearance of cervical human papillomavirus infection in women from a high-risk area for cervical cancer. J Infect Dis. 1999;180:1415–23. doi: 10.1086/315086. [DOI] [PubMed] [Google Scholar]
- 4.Syrjanen S, Shabalova IP, Petrovichev N, et al. Clearance of high-risk human papillomavirus (HPV) DNA and PAP smear abnormalities in a cohort of women subjected to HPV screening in the New Independent States of the former Soviet Union (the NIS cohort study) Eur J Obstet Gynecol Reprod Biol. 2005;119:219–27. doi: 10.1016/j.ejogrb.2004.07.036. [DOI] [PubMed] [Google Scholar]
- 5.Kovacic MB, Castle PE, Herrero R, et al. Relationships of human papillomavirus type, qualitative viral load, and age with cytologic abnormality. Cancer Res. 2006;66:10112–9. doi: 10.1158/0008-5472.CAN-06-1812. [DOI] [PubMed] [Google Scholar]
- 6.Ho GY, Burk RD, Klein S, et al. Persistent genital human papillomavirus infection as a risk factor for persistent cervical dysplasia. J Natl Cancer Inst. 1995;87:1365–71. doi: 10.1093/jnci/87.18.1365. [DOI] [PubMed] [Google Scholar]
- 7.Schlecht NF, Kulaga S, Robitaille J, et al. Persistent human papillomavirus infection as a predictor of cervical intraepithelial neoplasia. Jama. 2001;286:3106–14. doi: 10.1001/jama.286.24.3106. [DOI] [PubMed] [Google Scholar]
- 8.Ylitalo N, Josefsson A, Melbye M, et al. A prospective study showing long-term infection with human papillomavirus 16 before the development of cervical carcinoma in situ. Cancer Res. 2000;60:6027–32. [PubMed] [Google Scholar]
- 9.Remmink AJ, Walboomers JM, Helmerhorst TJ, et al. The presence of persistent high-risk HPV genotypes in dysplastic cervical lesions is associated with progressive disease: natural history up to 36 months. Int J Cancer. 1995;61:306–11. doi: 10.1002/ijc.2910610305. [DOI] [PubMed] [Google Scholar]
- 10.Nobbenhuis MA, Walboomers JM, Helmerhorst TJ, et al. Relation of human papillomavirus status to cervical lesions and consequences for cervical-cancer screening: a prospective study. Lancet. 1999;354:20–5. doi: 10.1016/S0140-6736(98)12490-X. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez AC, Schiffman M, Herrero R, et al. Rapid clearance of human papillomavirus and implications for clinical focus on persistent infections. J Natl Cancer Inst. 2008;100:513–7. doi: 10.1093/jnci/djn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai CH, Chao A, Chang CJ, et al. Host and viral factors in relation to clearance of human papillomavirus infection: a cohort study in Taiwan. Int J Cancer. 2008;123:1685–92. doi: 10.1002/ijc.23679. [DOI] [PubMed] [Google Scholar]
- 13.Fontaine J, Hankins C, Money D, et al. Human papillomavirus type 16 (HPV-16) viral load and persistence of HPV-16 infection in women infected or at risk for HIV. J Clin Virol. 2008;43:307–12. doi: 10.1016/j.jcv.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 14.Dalstein V, Riethmuller D, Pretet JL, et al. Persistence and load of high-risk HPV are predictors for development of high-grade cervical lesions: a longitudinal French cohort study. Int J Cancer. 2003;106:396–403. doi: 10.1002/ijc.11222. [DOI] [PubMed] [Google Scholar]
- 15.van Duin M, Snijders PJ, Schrijnemakers HF, et al. Human papillomavirus 16 load in normal and abnormal cervical scrapes: an indicator of CIN II/III and viral clearance. Int J Cancer. 2002;98:590–5. doi: 10.1002/ijc.10232. [DOI] [PubMed] [Google Scholar]
- 16.Park JY, Lee KH, Dong SM, Kang S, Park SY, Seo SS. The association of pre-conization high-risk HPV load and the persistence of HPV infection and persistence/recurrence of cervical intraepithelial neoplasia after conization. Gynecol Oncol. 2008;108:549–54. doi: 10.1016/j.ygyno.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Song SH, Lee JK, Oh MJ, et al. Persistent HPV infection after conization in patients with negative margins. Gynecol Oncol. 2006;101:418–22. doi: 10.1016/j.ygyno.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 18.Molano M, Van den Brule A, Plummer M, et al. Determinants of clearance of human papillomavirus infections in Colombian women with normal cytology: a population-based, 5-year follow-up study. Am J Epidemiol. 2003;158:486–94. doi: 10.1093/aje/kwg171. [DOI] [PubMed] [Google Scholar]
- 19.Kulmala SM, Shabalova IP, Petrovitchev N, et al. Type-specific persistence of high-risk human papillomavirus infections in the New Independent States of the former Soviet Union Cohort Study. Cancer Epidemiol Biomarkers Prev. 2007;16:17–22. doi: 10.1158/1055-9965.EPI-06-0649. [DOI] [PubMed] [Google Scholar]
- 20.Schiffman M, Adrianza ME. ASCUS-LSIL Triage Study. Design, methods and characteristics of trial participants. Acta Cytol. 2000;44:726–42. doi: 10.1159/000328554. [DOI] [PubMed] [Google Scholar]
- 21.Results of a randomized trial on the management of cytology interpretations of atypical squamous cells of undetermined significance. Am J Obstet Gynecol. 2003;188:1383–92. doi: 10.1067/mob.2003.457. [DOI] [PubMed] [Google Scholar]
- 22.Gravitt PE, Peyton CL, Alessi TQ, et al. Improved amplification of genital human papillomaviruses. J Clin Microbiol. 2000;38:357–61. doi: 10.1128/jcm.38.1.357-361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gravitt PE, Peyton CL, Apple RJ, Wheeler CM. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J Clin Microbiol. 1998;36:3020–7. doi: 10.1128/jcm.36.10.3020-3027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xi LF, Kiviat NB, Galloway DA, Zhou XH, Ho J, Koutsky LA. Effect of cervical cytologic status on the association between human papillomavirus type 16 DNA load and the risk of cervical intraepithelial neoplasia grade 3. J Infect Dis. 2008;198:324–31. doi: 10.1086/589715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xi LF, Koutsky LA, Castle PE, et al. Human papillomavirus type 18 DNA load and 2-year cumulative diagnoses of cervical intraepithelial neoplasia grades 2–3. J Natl Cancer Inst. 2009;101:153–61. doi: 10.1093/jnci/djn461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slinker BK, Glantz SA. Multiple linear regression is a useful alternative to traditional analyses of variance. Am J Physiol. 1988;255:R353–67. doi: 10.1152/ajpregu.1988.255.3.R353. [DOI] [PubMed] [Google Scholar]
- 27.Ylitalo N, Sorensen P, Josefsson AM, et al. Consistent high viral load of human papillomavirus 16 and risk of cervical carcinoma in situ: a nested case-control study. Lancet. 2000;355:2194–8. doi: 10.1016/S0140-6736(00)02402-8. [DOI] [PubMed] [Google Scholar]
- 28.Josefsson AM, Magnusson PK, Ylitalo N, et al. Viral load of human papilloma virus 16 as a determinant for development of cervical carcinoma in situ: a nested case-control study. Lancet. 2000;355:2189–93. doi: 10.1016/S0140-6736(00)02401-6. [DOI] [PubMed] [Google Scholar]
- 29.Moberg M, Gustavsson I, Wilander E, Gyllensten U. High viral loads of human papillomavirus predict risk of invasive cervical carcinoma. Br J Cancer. 2005;92:891–4. doi: 10.1038/sj.bjc.6602436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fen J, Yoshinouchi M, Nakamura K, et al. Eradication of HPV post-surgical treatments, its correlation with specific types, types of surgery and the physical status. Oncol Rep. 2004;12:375–9. [PubMed] [Google Scholar]
- 31.Sarian LO, Derchain SF, Pittal Dda R, Andrade LA, Morais SS, Figueiredo PG. Human papillomavirus detection by hybrid capture II and residual or recurrent high-grade squamous cervical intraepithelial neoplasia after large loop excision of the transformation zone (LLETZ) Tumori. 2005;91:188–92. doi: 10.1177/030089160509100216. [DOI] [PubMed] [Google Scholar]
- 32.Ahdieh L, Klein RS, Burk R, et al. Prevalence, incidence, and type-specific persistence of human papillomavirus in human immunodeficiency virus (HIV)-positive and HIV-negative women. J Infect Dis. 2001;184:682–90. doi: 10.1086/323081. [DOI] [PubMed] [Google Scholar]
- 33.Sun XW, Kuhn L, Ellerbrock TV, Chiasson MA, Bush TJ, Wright TC., Jr Human papillomavirus infection in women infected with the human immunodeficiency virus. N Engl J Med. 1997;337:1343–9. doi: 10.1056/NEJM199711063371903. [DOI] [PubMed] [Google Scholar]
- 34.Moscicki AB, Ellenberg JH, Farhat S, Xu J. Persistence of human papillomavirus infection in HIV-infected and -uninfected adolescent girls: risk factors and differences, by phylogenetic type. J Infect Dis. 2004;190:37–45. doi: 10.1086/421467. [DOI] [PubMed] [Google Scholar]
- 35.Rowhani-Rahbar A, Hawes SE, Sow PS, et al. The impact of HIV status and type on the clearance of human papillomavirus infection among Senegalese women. J Infect Dis. 2007;196:887–94. doi: 10.1086/520883. [DOI] [PubMed] [Google Scholar]
- 36.Vernon SD, Reeves WC, Clancy KA, et al. A longitudinal study of human papillomavirus DNA detection in human immunodeficiency virus type 1-seropositive and -seronegative women. J Infect Dis. 1994;169:1108–12. doi: 10.1093/infdis/169.5.1108. [DOI] [PubMed] [Google Scholar]
- 37.Xi LF, Edelstein ZR, Meyers C, Ho J, Cherne SL, Schiffman M. Human Papillomavirus Types 16 and 18 DNA Load in Relation to Coexistence of Other Types, Particularly Those in the Same Species. Cancer Epidemiol Biomarkers Prev. doi: 10.1158/1055-9965.EPI-09-0482. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Combita AL, Touze A, Bousarghin L, Christensen ND, Coursaget P. Identification of two cross-neutralizing linear epitopes within the L1 major capsid protein of human papillomaviruses. J Virol. 2002;76:6480–6. doi: 10.1128/JVI.76.13.6480-6486.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCarthy C, Youde SJ, Man S. Definition of an HPV18/45 cross-reactive human T-cell epitope after DNA immunisation of HLA-A2/KB transgenic mice. Int J Cancer. 2006;118:2514–21. doi: 10.1002/ijc.21643. [DOI] [PubMed] [Google Scholar]
- 40.Liaw KL, Hildesheim A, Burk RD, et al. A prospective study of human papillomavirus (HPV) type 16 DNA detection by polymerase chain reaction and its association with acquisition and persistence of other HPV types. J Infect Dis. 2001;183:8–15. doi: 10.1086/317638. [DOI] [PubMed] [Google Scholar]
- 41.Ho GY, Bierman R, Beardsley L, Chang CJ, Burk RD. Natural history of cervicovaginal papillomavirus infection in young women. N Engl J Med. 1998;338:423–8. doi: 10.1056/NEJM199802123380703. [DOI] [PubMed] [Google Scholar]
- 42.Trottier H, Mahmud S, Prado JC, et al. Type-specific duration of human papillomavirus infection: implications for human papillomavirus screening and vaccination. J Infect Dis. 2008;197:1436–47. doi: 10.1086/587698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pinto LA, Viscidi R, Harro CD, et al. Cellular immune responses to HPV-18, -31, and -53 in healthy volunteers immunized with recombinant HPV-16 L1 virus-like particles. Virology. 2006;353:451–62. doi: 10.1016/j.virol.2006.06.021. [DOI] [PubMed] [Google Scholar]