Abstract
Purpose
To identify the pulse sequence and acquisition parameters that result in the most accurate and repeatable measurements of glutamate (Glu) concentration in the brain at 3T.
Materials and Methods
Simulations were performed to compare the accuracy and repeatability of eleven pulse sequences and acquisition parameters, within four general classes (PRESS, STEAM, Car-Purcell PRESS (CPRESS) and TE averaged PRESS (JPRESS)), the majority of which were previously suggested as optimal for Glu detection. Three of the simulated acquisitions were implemented in a clinical scanner, and measures of repeatability in vivo were compared to their simulated values.
Results
Good agreement was demonstrated between simulated and experimentally determined measures of repeatability. Among the acquisitions considered, a CPRESS sequence with minimal echo time, together with, possibly, a short TE PRESS sequence, result in the most repeatable Glu measurements, while slightly overestimating the Glu concentration. Excellent accuracy is demonstrated by the simulations for a JPRESS sequence, at the expense of lower repeatability than optimal PRESS or CPRESS sequences.
Conclusion
Further proof of concept is presented towards validation of a simulation approach to understand pulse sequence performance in measuring the concentration of a given metabolite. Improved Glu measurement repeatability is predicted for CPRESS and demonstrated .
Keywords: Simulation, spectroscopy, glutamate, 1H MRS
Introduction
Glutamate (Glu) is the most abundant excitatory neurotransmitter in the mammalian nervous system 1. Transmitter glutamate is compartmentalized (and highly concentrated) in synaptic vesicles within the neurons 1; nerve impulses trigger the release of Glu from the pre-synaptic cell in the synaptic space, where it binds to postsynaptic receptors. Following the neuro-transmission process, Glu is internalized into astrocytes, where it is converted to glutamine (Gln) 1. Gln leaves the astrocytes, is taken up by neurons, where glutaminase regenerates Glu and completes the well-known Glu/Gln cycle.
Disruptions in Glu levels have been reported in a multitude of pathological conditions of the brain, including multiple sclerosis 2, schizophrenia 3, HIV 4, cerebral ischemia and hepatic encelopathy 5. Accurate, non-invasive measurements of brain Glu concentration may, therefore, offer a valuable tool for diagnosing and monitoring these pathologic conditions. Although Glu concentration in the normal brain reaches as high as 12mM 6, accurate determination of its concentration in vivo, though 1H MRS, is very difficult. A study surveying existing literature reports indicates average coefficients of variation (CV’s), expressed as standard deviation divided by the mean, in excess of 10% 7. This poor measurement repeatability is mainly due to the strong coupling between the five protons providing the observable signal 6.
Many attempts have been made to tailor acquisition strategies for optimal Glu detection 8–10. It is not trivial, however, to assess theoretically if such tailored approaches, which usually aim at selectively increasing the signal of interest or selectively decreasing the overlapping signals, offer improved measurement performance. Proposed methods typically also have undesired side effects: increasing the signal from the metabolite of interest usually results in higher background signals (which potentially leads to less reliable fitting); decreasing the signal overlapping the metabolite of interest usually also results in a reduction of the signal of interest, and therefore reduced measurement repeatability 11. While data acquired in vivo would offer the ultimate proof of the usefulness of a given approach, it is cumbersome to acquire in vivo data resulting in statistically relevant comparisons, in particular when multiple pulse sequences or acquisition parameters are tested. Yield computations can provide insight into measurement performance of pulse sequences 12. The yield factors, however, do not generally consider T2 decay, do not account for the acquisition signal to noise ratio (SNR)-- significant in determining measurement repeatability 11-- and do not result in the calculation of the true parameters of interest: measurement accuracy and repeatability.
A simulation approach, recently documented and used for identifying the best pulse sequence for myo-inositol measurement 13, was used in this work to identify the optimal strategy for enhanced Glu detection. Eleven acquisitions pertaining to four classes (PRESS, STEAM, Carr-Purcell PRESS and TE averaged PRESS sequence), the majority of which were proposed in the past as improved choices for Glu detection, were included in this comparative study. Three of the simulated pulse sequences were implemented in a clinical scanner, and five volunteers were scanned with each of these three sequences, three times in a row for each sequence, in a single scanning session. Excellent agreement between repeatability measures resulting from simulations and the ones measured in vivo further validate the simulation approach as a useful tool for identifying the best pulse sequence or acquisition parameters for enhanced detection of a given metabolite. More repeatable Glu detection was predicted by simulations, and demonstrated in vivo with a Carr Purcell PRESS sequence.
Materials and Methods
The simulation approach employed here is thoroughly detailed in a recent publication 13. For ease of reading, the essential features are also detailed here; for more details, the reader is referred to the initial article 13. The response of 14 metabolites to a pulse sequence is computed using the GAMMA set of libraries 14, using previously published values for chemical shift and J-coupling values 6, in a manner identical with the one described elsewhere 15. The metabolites included in our simulations, listed here together with their concentrations are N-acetyl aspartate (NAA) [12mM], phospho-choline (Cho) [2mM], creatine (Cr) [7mM], phospho-creatine (PCr) [3mM], Glu [10mM], Gln [4.5mM], Tau [1.2mM], mI [6mM], lactate (Lac) [0.4mM], Gly [0.7mM], aspartate (Asp) [1.2mM], alanine (Ala) [0.8mM], gamma amino butyric acid GABA [1.6mM] and guanidine (Gua) [0.2mM].
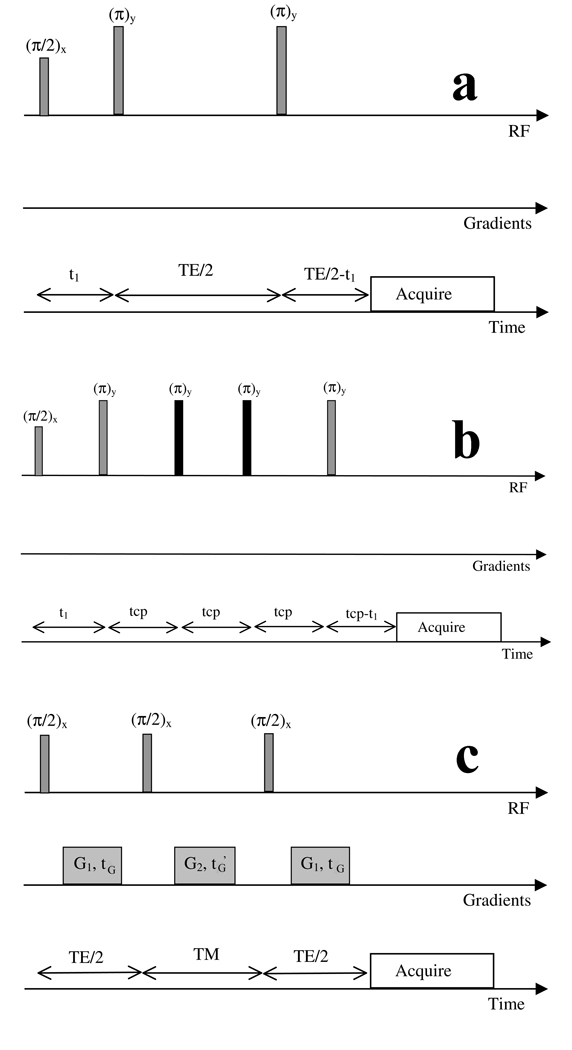
The diagrams and timing denominations of the separate types of acquisitions considered in our simulations (PRESS, CPRESS and STEAM pulse sequences) are depicted in Figure 1a, b and c, respectively. For TE averaged PRESS (JPRESS), a number of PRESS acquisitions were simulated/acquired with increasing echo times, and the resulting spectra were averaged together. Eleven separate pulse sequences were considered within these 4 classes as following:
PRESS, TE=35ms, t1=7ms (this is our standard clinical pulse sequence)
PRESS, TE=15ms, t1=3.8ms (this is the close to the potential minimum timing achievable for a PRESS sequence on a 3T scanner)
PRESS, TE=45ms, t1=7ms (this is a sequence whose timing is close to the one proposed in the past as optimal for Glu detection 16)
PRESS, TE=80ms, t1=7ms (this is a sequence whose timing is close to the one proposed in the past as optimal for Glu detection 17)
PRESS, TE=144ms, t1=7ms (this is a standard long TE sequence, which should allow almost complete decay of MM signals)
STEAM, TE=5ms, TM=5ms. This is close to the minimum timing achievable on a clinical scanner 18, providing maximum yield for the Glu signal
STEAM, TE=72ms, TM=6ms (this is the timing of the pulse sequence found through simulations to offer the best signal to background ratio for the Glu signal 10)
JPRESS, t1=9.6ms, 128 equally spaced TE steps between 35ms and 352.5ms (this is one of the JPRESS acquisition schemes considered in the past for optimized Glu detection 8)
JPRESS, t1=9.6ms, 64 TE steps between 35ms and 192.5ms (this has also been used in the past as potentially optimal for Glu detection 4)
CPRESS with 2 additional refocusing pulses (CPRESS 2), TE=45ms, tcp=11.2ms, t1=5.8ms (this is a minimum TE CPRESS sequence achievable on our clinical scanner, proposed in the past for optimized detection of coupled metabolites 19,20)
CPRESS with 4 additional refocusing pulses (CPRESS 4), TE=67ms, tcp=11.2ms, t1=5.8ms (this is a similar acquisition with the previous one, additionally allowing stronger decay of MM signals)
Figure 1.
Classes of pulse sequences and their timings used in our simulations. a) PRESS pulse sequence b) Carr Purcell (CPRESS) pulse sequence with two extra refocusing pulses (4 extra refocusing pulses were also considered, by inserting two additional tcp intervals prior to the final z slice select pulse) c) STEAM sequence. TE represents the echo time in all sequences, t1 the time interval between the first and second excitation pulses, tG and tG’ gradient timings, TM mixing time for STEAM, tcp time between refocusing pulses in the Carr Purcell sequence. The action of the slice select/crusher gradients in sequences a) and b) (not displayed) was not simulated; the action of the gradients shown in c) was simulated indirectly, as described in ref. 15.
Separate pseudo-codes were written using the GAMMA set of libraries for each class of pulse sequence. Similar to 13, the volume selective pulses depicted in the pulse sequences of Figure 1 are replaced in our simulations with idealized hard pulses. The only gradients simulated in this work are the G1 gradients following the first and third pulse of the STEAM sequence, and the G2 gradients applied during the TM period of the STEAM sequence (Figure 1c). The action of these gradients was simulated indirectly (therefore the strength of these gradients, G1 and G2, and their duration, tG and tG’, is irrelevant), in a manner identical to the one described in 15. For the CPRESS acquisitions, the two additional refocusing pulses (depicted in black in Figure 1b) were implemented in the scanner as quadratic phase 180 degree pulses, of 2.01ms duration and 2500Hz bandwidth 21.
Relaxation was not explicitly included in the simulations. A penalty factor of exp(-TE/T2), however, multiplied all time-domain data, to account for signal loss in longer TE sequences. Here T2 is the transverse relaxation time; consistent with published literature reports at 3T 22,23, the T2 values used for all the metabolites and all the pulse sequences apart from CPRESS were 250ms. For lack of a better reference documenting in vivo MM T2 value at 3T for PRESS and CPRESS, as well as metabolite T2 CPRESS values, 1.5T data 20 were used to choose PRESS MM’s T2’s of 35ms and CPRESS MM T2’s of 75ms. The same 1.5T report 20 was used to extract CPRESS metabolite T2’s (375ms). While MM T2’s can only decrease with field strength, a “worst case scenario” was considered throughout this work as far as MM T2’s are concerned. To verify that we have not accidentally favored the CPRESS sequence by choosing longer metabolite T2’s than for the other sequences, a second set of simulations were conducted for CPRESS with equal relaxation times (metabolite and MM’s) as for the other sequences.
For each of the pulse sequences considered, and each metabolite, simulations were initially run, and resulting spectra were line-broadened to 3Hz for construction of LCModel basis set spectra. Subsequently, a set of spectra line-broadened to 7 Hz was generated, providing more realistic simulations for in vivo acquisitions. The 14 resulting spectra were added together with weights corresponding to their in vivo concentrations. Additionally, a residual water signal, and a simulated MM signal (identical to the one presented in 13, and designed to be similar to MM signals acquired in vivo under similar conditions 20) were added to the simulated brain spectrum. The absolute scaling of the MM signals was such as the ratio of the NAA peak to the 1.45ppm MM signal was ~5 for the PRESS, TE=35ms pulse sequence, somewhat typical of an in vivo acquisition.
Following the generation of a noiseless brain spectrum for each of the pulse sequences considered, noise was added with a given standard deviation σ. The resulting spectrum had an SNR equivalent to a spectrum acquired in vivo, with 128 acquisitions of TR=2s, from a 16cc voxel (SNR, defined as the maximum signal in the spectrum minus the baseline signal divided by 2σ, equals ~40 for a PRESS, TE=35ms acquisition). While the signals of different metabolites simulated in various pulse sequences varied as a function of echo time due to evolution under J coupling and relaxation, the standard deviation of the noise added was kept constant between the different acquisition studied.
The “noised” brain spectra were then fitted using LCModel version 6.2, with no custom parametrization of the lipid and MM signals in the basis sets, and the metabolite concentrations were recorded for each run. The process was repeated 1000 times for each pulse sequence with different noise seeds. The Cramer Rao Lower Bounds (CRLB’s) reported by LCModel, as well as metabolite concentrations were saved for each individual run. Average CRLB’s, average metabolite concentration, and standard deviations were computed for each pulse sequence and metabolite of interest. Absolute scaling of the spectrum was performed such as the average NAA concentration for each particular pulse sequence was calibrated to its known level (12mM). This is equivalent to obtaining the overall calibration factor fcalib needed for LCModel using NAA phantom measurements. The difference between the smallest fcalib and the largest fcalib factor for all pulse sequences was ~ 5%. The absolute errors (defined as the average concentration resulting from the simulations minus the known concentration used in the simulations divided by the known concentration were reported for each pulse sequence/metabolite of interest).
Phantom experiments
Three 600ml spherical phantoms, containing 50mM Glu (Fluka 49449), 50mM NAA (Fluka 00920) and 100mM Gln (Fluka 49419) with their pH adjusted to 7.2, were initially scanned to insure a proper match between our simulations and the experimental data acquired using PRESS, TE=35ms.
In vivo experiments
Five volunteers (average age 36 years) were subjected to a one-hour exam, in which three acquisitions were acquired with each of the following pulse sequences: PRESS (TE=35ms), CPRESS2 (TE=45ms) and JPRESS (TE=35–192.5ms). All nine spectroscopic measurements from each exam were acquired from a 2cm × 2cm × 4cm voxel (with the largest dimension along the anterior/posterior direction) situated in the posterior cingulate gyrus, and had TR=2s, and 128 averages, and a total acquisition time of ~5 minutes. While such a large voxel is not the norm for brain MRS exams, it is appropriate for a number of non-focal brain diseases. It was chosen in this work to probe the limits of in vivo Glu measurement repeatability-- as repeatability is a function of the acquisition SNR. The subjects were not repositioned between the acquisitions, and prescan was not performed between the three repeat scans of each acquisition type. All in vivo exams were performed under a protocol approved by the Institutional Review Board. Cramer Rao Lower Bounds (CRLB’s) and intra-volunteer, intra-session coefficients of variation (CV’s) were reported as measures of repeatability, and compared to the quantities predicted by simulations.
Results
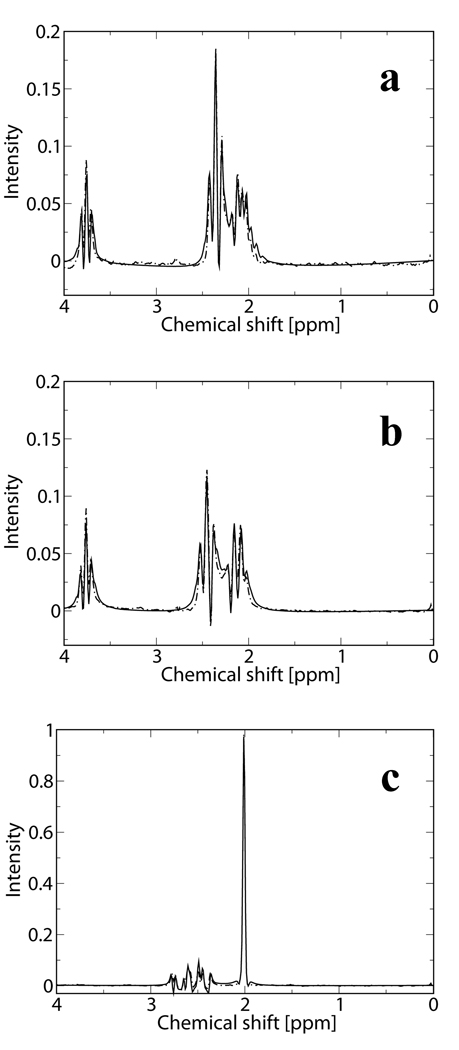
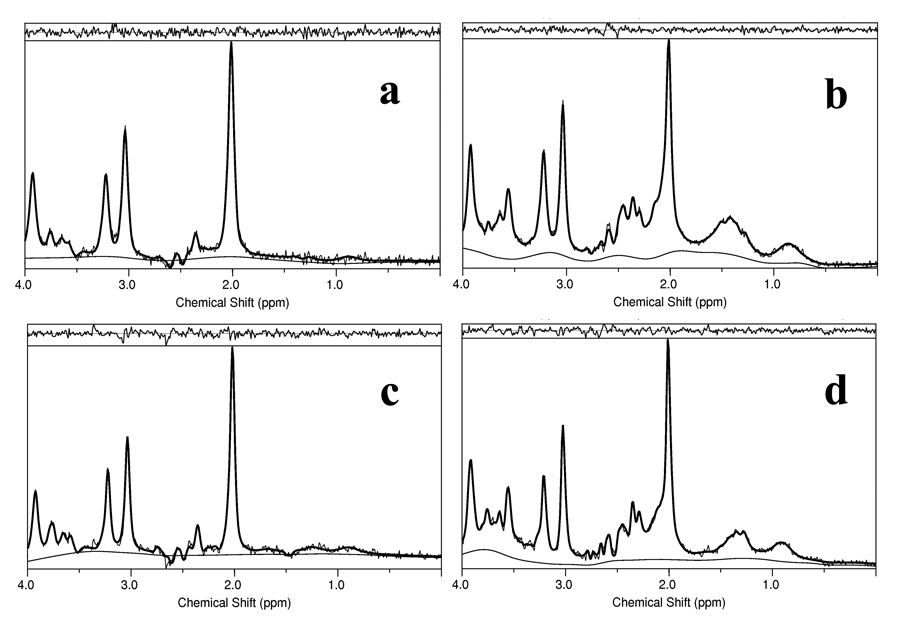
Figure 2 and Figure 3 demonstrate a validation of the performance of our simulations tools. Figure 2a represents a fit of the experimental data acquired from our 50mM Glu phantom, using PRESS (TE=35ms), (dashed line) with a simulated Glu signal using the same experimental pulse sequence parameters (continuous line). Figure 2b presents the same experimental data and fit for the Gln phantom, while Figure 2c experimental data and fit for the NAA phantom. The intensity of the three spectra is scaled by the phantoms’ concentrations. Given the slight differences between our experimental conditions and the conditions reported in 6, the agreement between the experimental data and simulations is convincing. Figures 3a and 3b, respectively, represent simulated brain spectra for the JPRESS, TE=35–192.5ms pulse sequence, and for the CPRESS2 pulse sequence, along with LCModel fits. Figures 3c and 3d display spectra acquired in vivo from a normal volunteer using the same two sequences, along with the LCModel fits. As observed from the matches between Figures 3a and 3c, and Figures 3b and 3d, respectively, there is a close resemblance between the simulated the in vivo data. Note that the varying baseline signals in the simulated brain spectra are due to the lack of any attempt to match the macromolecule resonances used in our simulations with the MM/lipid signals included in LCModel’s basis set.
Figure 2.
Validation of the performance of the simulation tools. Simulated (continuous line) and experimentally acquired data (dashed line) from a) a 50mM Glu phantom b) a 100mM Gln phantom and c) a 50mM NAA phantom using PRESS, TE=35ms. Data is scaled by the phantoms’ concentration.
Figure 3.
Simulated brain data sets and fits for a) JPRESS, TE=35–192.5ms pulse sequence b) CPRESS2, TE=45ms. In vivo data sets, acquired from a 2cm×2cm×4cm voxel situated in the posterior cingulate gyrus of normal volunteers using c) JPRESS, TE=35–192.5ms and d) CPRESS 2, TE=45ms. The residuals and the fitted baselines are also displayed.
Table 1 presents the coefficient of variation (% CV), absolute error, and the CRLB’s for the Glu concentration for all 11 acquisitions considered. The simulation results for the overlapping NAA resonance and Glx (glutamate and glutamine) signals-- sometimes quantified instead Glu-- is also displayed in this table. Note that the absolute error for NAA was not added, as the absolute errors for the other metabolites were scaled with respect to the average NAA signal for each sequence (as it is typical when absolute scaling is done in vivo). Data displayed in normal font represent simulation results; data displayed in bold, however, represent the intra-volunteer, intra-session %CV’s, and the average CRLB’s from the in vivo acquisitions. Note a few very remarkable results from this table:
Not all pulse sequences that were suggested in the past as potential candidates for improved Glu detection result in a positive outcome (note, eg, the poor results of the optimized STEAM sequence) 10. This is because long TE, coupled with a penalty factor of 2 of the STEAM sequence results in a spectrum SNR which negatively impacts repeatability.
As also previously experimentally observed 16, a standard, clinical PRESS, TE=35ms tends to result in measurements of larger Glu concentrations than longer TE sequences. It is only through simulations, however, that one can decide that long TE sequences assess Glu concentrations appropriately, while a TE=35ms results in an overestimation of this metabolite’s concentration.
Optimized Glu detection does not imply optimized Glx detection. Although these results were not verified experimentally (being outside the scope of this work), they suggest, somewhat surprisingly, that at least at this field strength and SNR level, the best choice for best Glx detection is a long TE PRESS sequence (TE=144ms).
Most importantly, however, the simulations results of Table 1 give a ranking of pulse sequences to be used for improved Glu detection. Not only that the trend suggest by the simulated data (normal font) is matched well by the in vivo data (bold font) for the 3 sequences tested in vivo, but the absolute value of the CRLB’s and CV’s resulting from the simulation are also matched well in vivo, confirming the good performance for the simulation approach.
Table 1.
Simulations (black) and in vivo results (bold black) for coefficients of variation (%CV’s) and Cramer Rao Lower Bounds (CRLB’s) for glutamate (Glu), glutamate+ glutamine (Glx) and N-acetyl aspartate (NAA) concentrations obtained from fitting simulated brain spectra and in vivo data acquired at 3T. In vivo data was acquired from a 16cc voxel in the posterior cingulate gyrus of 5 healthy subjects, scanned 3 times each.
| Pulse Sequence | % CV Glu | CRLB Glu [%] |
abs error Glu [%] |
% CV Glx |
CRLB Glx [%] |
abs error Glx [%] |
%CV NAA |
CRLB NAA [%] |
|---|---|---|---|---|---|---|---|---|
| PRESS | ||||||||
| TE=35ms | 4.00 (4.3) | 6.50 (7.4) | 14.8 | 4.3 | 6.1 | 16.3 | 1.3 | 2.0 |
| TE=15ms | 2.9 | 4.9 | 2.7 | 2.8 | 4.7 | 8.8 | 1.4 | 2.0 |
| TE=45ms | 5.9 | 8.4 | 7.4 | 5.3 | 7.4 | 7.4 | 1.3 | 2.0 |
| TE=80ms | 5.1 | 7.8 | −2.2 | 4.7 | 6.9 | −2.4 | 1.5 | 2.1 |
| TE=144ms | 5.1 | 7.0 | −5.0 | 2.6 | 3.3 | −7.2 | 1.7 | 2.0 |
| STEAM | ||||||||
| TE/TM=5/5ms | 4.1 | 6.8 | −1.0 | 3.4 | 5.2 | 9.9 | 1.6 | 3.0 |
| TE/TM=72/6ms | 9.6 | 13.0 | 6.7 | 8.0 | 10.7 | 10.1 | 2.0 | 2.9 |
| JPRESS | ||||||||
| TE=35–355ms | 8.5 | 11.4 | −6.6 | 11.0 | 13.8 | −29.2 | 1.6 | 1.1 |
| TE=35–190ms | 5.7 (5.2) | 8.0 (8.8) | −0.2 | 5.8 | 7.6 | −4.1 | 1.7 | 1.9 |
| Carr Purcell echo train | ||||||||
| CPRESS 2 (TE=45ms) | 3.3 (3.8) | 5.4 (6.3) | 10.8 | 3.8 | 5.9 | 12.6 | 1.3 | 2.0 |
| CPRESS 4 (TE=67ms) | 4.5 | 6.4 | 16.5 | 5.9 | 7.7 | 13.4 | 1.3 | 2.0 |
Levene’s homogeneity of variance test was performed for the three pulse sequences tested in vivo in order to verify if that the improvement in repeatability predicted by the simulations is statistically significant. For the 1000 data points available from each of the simulation runs, CPRESS2 offers smaller measurement variance (p value for Levene test p<10−3) than PRESS, TE=35. At the same time, JPRESS (TE=35–192.5ms) results in CV’s that are significantly higher than the variance of the measurements with PRESS, TE=35ms. This confirms that, among the pulse sequences considered and tested in vivo, a Carr Purcell PRESS sequence, may offer the most repeatable Glu measurements, at the expense of a slight overestimation of the Glu concentration. Additionally, according to the simulation results, should accuracy of a given acquisition be the deciding factor in choosing an acquisition approach for a given investigator, it is the JPRESS acquisition that offers exquisite accuracy, at the expense of lower repeatability.
Note that, according to our simulation, a short TE PRESS sequence may have been a better candidate for enhanced Glu detection. While a couple of reports exist documenting the use of TE’s below 20ms for single voxel PRESS acquisitions at 3T 24,25, such exams make use of more than truly clinical hardware capabilities, compromise slice profile, or allow inhomogeneous B1 across the voxels. It is estimated that B1’s in excess of 0.25G are needed for reducing TE’s below 20ms. The absence of the capability of obtaining such strong homogeneous excitation field in a clinical scanner prevented us from validating this prediction.
Discussion
A simulation approach was used to identify the best approach for detection of Glu at 3T, and validated in vivo. The results of the simulations matched data acquired in vitro and in vivo well: simulated Glu, Gln and NAA spectra approximated well spectra acquired from phantoms (Figure 2), and good match was also demonstrated between simulated brain spectra and spectra acquired in vivo with multiple pulse sequences (Figure 3). Most importantly, however, measures of repeatability resulting from simulations (CRLB’s and %CV’s) were matched well by data acquired in vivo in this study. Note that, due to the relatively low number of statistical samples, there is large variability in determining the coefficients of variation in vivo. For example that, if the data acquired in the last volunteer is removed from the computation of the coefficients of variation, the Glu CV for CPRESS 2 decreases from 3.8% to 3.2%. At the same time, removal of the last data volunteer from the pool of in vivo data results in an increase in the TE-35ms PRESS CV from 4.3% to 4.4%. CRLB’s change significantly less as a consequence of removal of this last volunteer’s data sets (6.3% to 6.2% for CPRESS 2 and 7.4% to 7.6% for PRESS, TE=35ms). Consequently, in the absence of large in vivo data sets, and in agreement with previous literature reports 26, CRLB’s are probably better measures of repeatability.
Our simulation results are also matched well by data presented in a very recent literature report aiming at experimentally comparing the performance of a small number of acquisitions16, demonstrating the capability of this simulation method for identifying the best technique for optimized metabolite detection. Such method can provide a very versatile tool in the hands of investigators embarking in clinical trial with a 1H MRS endpoint; using this approach, one can easily determine the choice of pulse sequence/ acquisition parameters to be used, and the number of subjects to be enrolled, as a function of the main requirements of the study (accuracy vs. repeatability).
While very generic, this simulation technique, implemented as described above, may be susceptible to providing erroneous answers in certain situations. One such case may be presented when excitation pulses (simulated as ideal in this work) become experimentally longer – a significant fraction of 1/J. In such cases, RF pulses and gradients may need to be explicitly simulated. While significantly slowing down the simulation time, the GAMMA libraries can easily accommodate this approach, as previously demonstrated 27. For our experimental case, in which none of the excitation pulses exceeded 5.4ms, the match between experimental and simulated data did not require this additional correction.
A second factor that may impact the results of the simulations is the shape of the MM signals. To investigate the impact of this factor, a number of simulations were run for four acquisition approaches [PRESS (TE=15ms), PRESS (TE=35ms), JPRESS (TE=35–192.5ms) and CPRESS2 (TE=45ms)], in which the overall lipid/MM signals added to the idealized brain signal were 50% higher than the ones used for the rest of the simulations. While this case is probably at the very high end of what is expected in vivo, the simulation results did not change dramatically: coefficients of variation changed for the 4 sequences considered, in the order presented above, from (2.87%, 4%, 5.65% and 3.18%) for the “low” MM signals, to (3.1%, 4.3%, 5.6% and 3.2%) for the “high” MM signals. Note that this result is only valid if using a quantification approach that can accommodate unexpected (broad) resonances through a flexible baseline.
Third, simulations may also depend on metabolite and macromolecule T2’s. To verify that we have not accidentally favored the CPRESS sequences by using larger metabolite/MM T2’s than for the other sequences, the CPRESS 2 simulations were repeated, while using metabolite T2’s of 250ms, and MM T2’s of 35ms (the same as the ones used for the other pulse sequences, but lower than the published values for a CPRESS sequence 20). The results were comparable, resulting in Glu CRLB’s of 5%, and %SD’s of 3.2% for metabolite/MM T2 values of 250ms/75ms, vs. Glu CRLB’s of 5.4% and CV’s of 3.3% for metabolite/MM T2 values of 375ms/35ms (see Table 1). This result is not unexpected—as the metabolite and MM T2’s were changed in concert. While longer metabolite T2’s result in higher SNR, and increased repeatability, longer MM T2’s complicate the baseline and decrease repeatability.
The effects of Eddy currents were also not simulated in this work. While these effects are probably insignificant for medium to long echo time sequences mainly considered in this work, they might become significant for very short TE sequences. In such cases, the effects of eddy currents might need to be added to the simulated signals, while also insuring that the data fitting approach attempts to correct for these effects.
The simulations presented here offer a good measure of intra-volunteer CV’s to be encountered in vivo. It is possible for the inter-volunteer, inter-session CV’s to be higher in a cohort of subjects, due to factors which were not considered here, such as biological diversity, variable shimming, variable lipid/MM signals, neuropathology, functional status, etc. Although inclusion of such effects is, in principle, possible in the framework of the simulations presented here, existing literature reports (indicating variable contribution to the total variability from “between session” effects for different metabolites 28) render simulation of these effects questionable, and was omitted in this work.
Last, but not least, the program used for data quantification may influence the findings of this study. Based on a previous report 29, it is expected that frequency domain fitting methods using a flexible baseline (as LCModel) and time-domain fitting methods employing weighting of the first data points (as AMARES) will result in comparable results. It is possible, however, that the use of a peak height or peak integration method for data quantification to result in an acquisition such as JPRESS (that results in very simple, baseline-free spectra) offering both the most accurate and repeatable method for Glu detection.
Acknowledgments
Grant support: NIH 1R21NS054303-01A2
References
- 1.Kandel ER, Schwartz JH, Jessell TM. Neurotransmitters. In: Hill M, editor. Principles of neural science. 2000. pp. 280–298. [Google Scholar]
- 2.Srinivasan R, Sailasuta N, Hurd R, Nelson S, Pelletier D. Evidence of elevated glutamate in multiple sclerosis using magnetic resonance spectroscopy at 3 T. Brain. 2005;128(Pt 5):1016–1025. doi: 10.1093/brain/awh467. [DOI] [PubMed] [Google Scholar]
- 3.Purdon SE, Valiakalayil A, Hanstock CC, Seres P, Tibbo P. Elevated 3T proton MRS glutamate levels associated with poor Continuous Performance Test (CPT-0X) scores and genetic risk for schizophrenia. Schizophr Res. 2008;99(1–3):218–224. doi: 10.1016/j.schres.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 4.Sailasuta N, Shriner K, Ross B. Evidence of reduced glutamate in the frontal lobe of HIV-seropositive patients(dagger) NMR Biomed. 2008;33(3):326–331. doi: 10.1002/nbm.1329. [DOI] [PubMed] [Google Scholar]
- 5.Chamuleau RA, Bosman DK, Bovee WM, Luyten PR, den Hollander JA. What the clinician can learn from MR glutamine/glutamate assays. NMR Biomed. 1991;4(2):103–108. doi: 10.1002/nbm.1940040213. [DOI] [PubMed] [Google Scholar]
- 6.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13(3):129–153. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 7.Choi C, Coupland NJ, Bhardwaj PP, Malykhin N, Gheorghiu D, Allen PS. Measurement of brain glutamate and glutamine by spectrally-selective refocusing at 3 Tesla. Magn Reson Med. 2006;55(5):997–1005. doi: 10.1002/mrm.20875. [DOI] [PubMed] [Google Scholar]
- 8.Hurd R, Sailasuta N, Srinivasan R, Vigneron D, Pelletier D, Nelson S. Measurement of brain glutamate using TE-averaged PRESS at 3T. Magn Reson Med. 2004;51:435–440. doi: 10.1002/mrm.20007. [DOI] [PubMed] [Google Scholar]
- 9.Yahya A, Madler B, Fallone BG. Exploiting the chemical shift displacement effect in the detection of glutamate and glutamine (Glx) with PRESS. J Magn Reson. 2008;191(1):120–127. doi: 10.1016/j.jmr.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Yang S, Hu J, Kou Z, Yang Y. Spectral simplification for resolved glutamate and glutamine measurement using a standard STEAM sequence with optimized timing parameters at 3, 4, 4.7, 7, and 9.4T. Magn Reson Med. 2008;59(2):236–244. doi: 10.1002/mrm.21463. [DOI] [PubMed] [Google Scholar]
- 11.Bartha R. Effect of signal-to-noise ratio and spectral linewidth on metabolite quantification at 4 T. NMR Biomed. 2007;20(5):512–521. doi: 10.1002/nbm.1122. [DOI] [PubMed] [Google Scholar]
- 12.Kim H, Wild JM, Allen PS. Strategy for the spectral filtering of myo-inositol and other strongly coupled spins. Magn Reson Med. 2004;51(2):263–272. doi: 10.1002/mrm.10697. [DOI] [PubMed] [Google Scholar]
- 13.Hancu I. Which pulse sequence is optimal for myo-inositol detection at 3T? NMR Biomed. 2009;22(4):426–435. doi: 10.1002/nbm.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith S, Levante T, Meier B, Ernst R. Computer simulations in magnetic resonance. An object-oriented programming approach. J Magn Reson Series A. 1994;106(1):75–105. [Google Scholar]
- 15.Young K, Matson GB, Govindaraju V, Maudsley AA. Spectral simulations incorporating gradient coherence selection. J Magn Reson. 1999;140(1):146–152. doi: 10.1006/jmre.1999.1809. [DOI] [PubMed] [Google Scholar]
- 16.Mullins PG, Chen H, Xu J, Caprihan A, Gasparovic C. Comparative reliability of proton spectroscopy techniques designed to improve detection of J-coupled metabolites. Magn Reson Med. 2008;60(4):964–969. doi: 10.1002/mrm.21696. [DOI] [PubMed] [Google Scholar]
- 17.Schubert F, Gallinat J, Seifert F, Rinneberg H. Glutamate concentrations in human brain using single voxel proton magnetic resonance spectroscopy at 3 Tesla. Neuroimage. 2004;21(4):1762–1771. doi: 10.1016/j.neuroimage.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Seeger U, Klose U, Seitz D, Nagele T, Lutz O, Grodd W. Proton spectroscopy of human brain with very short echo time using high gradient amplitudes. Magn Reson Imaging. 1998;16(1):55–62. doi: 10.1016/s0730-725x(97)00239-7. [DOI] [PubMed] [Google Scholar]
- 19.Hennig J, Thiel T, Speck O. Improved sensitivity to overlapping multiplet signals in in vivo proton spectroscopy using a multiecho volume selective (CPRESS) experiment. Magn Reson Med. 1997;37(6):816–820. doi: 10.1002/mrm.1910370603. [DOI] [PubMed] [Google Scholar]
- 20.Soher BJ, Pattany PM, Matson GB, Maudsley AA. Observation of coupled 1H metabolite resonances at long TE. Magn Reson Med. 2005;53(6):1283–1287. doi: 10.1002/mrm.20491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sailasuta N, Hurd R, Cunningham C, Vigneron DB, Nelson SJ, Pauly J. A novel approach for reducing chemical shift registration error at 3T: short TE CPRESS. Kyoto; Proc Int Soc Magn Res Med 12-th Meeting. 2004. p. 2297. [Google Scholar]
- 22.Mlynarik V, Gruber S, Moser E. Proton T (1) and T (2) relaxation times of human brain metabolites at 3 Tesla. NMR Biomed. 2001;14(5):325–331. doi: 10.1002/nbm.713. [DOI] [PubMed] [Google Scholar]
- 23.Traber F, Block W, Lamerichs R, Gieseke J, Schild HH. 1H metabolite relaxation times at 3.0 tesla: Measurements of T1 and T2 values in normal brain and determination of regional differences in transverse relaxation. J Magn Reson Imaging. 2004;19(5):537–545. doi: 10.1002/jmri.20053. [DOI] [PubMed] [Google Scholar]
- 24.Gottschalk M, Lamalle L, Segebarth C. Short-TE localised 1H MRS of the human brain at 3 T: quantification of the metabolite signals using two approaches to account for macromolecular signal contributions. NMR Biomed. 2008;21(5):507–517. doi: 10.1002/nbm.1219. [DOI] [PubMed] [Google Scholar]
- 25.Zhong K, Ernst T. Localized in vivo human 1H MRS at very short echo times. Magn Reson Med. 2004;52(4):898–901. doi: 10.1002/mrm.20201. [DOI] [PubMed] [Google Scholar]
- 26.Cavassila S, Deval S, Huegen C, van Ormondt D, Graveron-Demilly D. Cramer-Rao bounds: an evaluation tool for quantitation. NMR Biomed. 2001;14(4):278–283. doi: 10.1002/nbm.701. [DOI] [PubMed] [Google Scholar]
- 27.Maudsley AA, Govindaraju V, Young K, et al. Numerical simulation of PRESS localized MR spectroscopy. J Magn Reson. 2005;173(1):54–63. doi: 10.1016/j.jmr.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 28.Brooks WM, Friedman SD, Stidley CA. Reproducibility of 1H-MRS in vivo. Magn Reson Med. 1999;41(1):193–197. doi: 10.1002/(sici)1522-2594(199901)41:1<193::aid-mrm27>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 29.Kanowski M, Kaufmann J, Braun J, Bernarding J, Tempelmann C. Quantitation of simulated short echo time 1H human brain spectra by LCModel and AMARES. Magn Reson Med. 2004;51(5):904–912. doi: 10.1002/mrm.20063. [DOI] [PubMed] [Google Scholar]