Abstract
T helper type 17 (TH-17) cells, together with their effector cytokines including interleukin 17 (IL-17) family members, are emerging as key mediators of chronic inflammatory and autoimmune disorders. Here we present the crystal structure of a 1:2 complex of IL-17RA bound to IL-17F. The manner of complex formation is unique for cytokines, and involves two fibronectin-type domains of IL-17RA engaging IL-17 within a groove between the IL-17 homodimer interface in a knob-and-hole fashion. The first receptor-binding event to the IL-17 cytokines modulates the affinity and specificity of the second receptor-binding event, thereby promoting heterodimeric versus homodimeric complex formation. IL-17RA utilizes a common recognition strategy to bind to several IL-17 family members, allowing it to potentially act as a shared receptor within multiple different signaling complexes.
Introduction
Naïve T cells are stimulated to differentiate into specialized effector cells primarily through theactions of secreted cytokines. T helper (TH) cells have been typically considered to fall into one of two effector cell lineages; TH-1 and TH-2 cells modulating cellular and humoral T cell immunity, respectively, based on their cytokine expression profiles1. More recent work described TH-17 cells, a third lineage of effector TH cells distinct from, and in fact antagonized by products of the TH-1 and TH-2 lineages2,3. Named after their ‘signature’ cytokine interleukin 17 (IL-17), this subset of TH cells appear to have evolved as an arm of the adaptive immune system specialized for enhanced host protection against extracellular bacteria and some fungi, as these microbes may not be effectively controlled by TH-1 or TH-2 responses4,5. The varied tissue sources of cytokines that induce differentiation and regulate homeostasis of TH-17 cells, namely IL-23, IL-6, and transforming growth factor-β (TGF-β, together with the presence of IL-17 receptors on both hematopoietic and non-hematopoietic cells, have highlighted the complicated relationships that exist between adaptive and innate immune cells. While the full scope of TH-17 cell effector functions is still emerging, the strong inflammatory response promoted by TH-17 cells has been associated with the pathogenesis of a number of autoimmune and inflammatory disorders previously attributed to TH-1 or TH-2 cells including rheumatoid arthritis, multiple sclerosis and psoriasis4. As such, the targeting of TH-17 cells for treatment of autoimmune and inflammatory disorders, either directly through IL-17 blockade, or indirectly through inhibition of IL-23, is currently being pursued clinically. However, the structural uniqueness of the IL-17 system, combined with a dearth of biochemical and structural information on receptor interactions, is a current barrier to the development of mechanism or structure-based antagonists. The IL-17 family is composed of six cytokines and five receptors, and the ligand-receptor pairing is not completely worked out for all members6. On the basis of the crystal structure of IL-17F, the six structurally related IL-17 cytokines (IL-17A IL-17F) are predicted to form a homodimeric fold (or heterodimeric fold in the case of IL-17A-F) homologous to that of the cysteine-knot growth factors such as nerve growth factor (NGF)7,8. TH-17 cell-derived IL-17A and IL-17F share the greatest homology within the family (50%) and require both IL-17RA (http://www.signaling-gateway.org/molecule/query?afcsid=A001253) and IL-17RC for signaling9,10. While it has been shown that fibroblasts, epithelial and endothelial cells coexpress both IL-17RA and IL-17RC, T cells do not express IL-17RC, and only express IL-17RA11. Until recently, it was thought that lymphocytes are not responsive to IL-17; however, Flavell and coworkers reported that T cells indeed can directly respond to IL-1712.
The five IL-17 receptors (IL-17RA IL-17RE) are not homologous to any known receptors, and exhibit considerable sequence divergence between one another. All appear to contain extracellular domains composed of fibronectin type-III (FnIII) domains, and cytoplasmic SEF/IL-17R (SEFIR) domains that show loose homology to Toll/IL-1R (TLR) domains13,14. The IL-17 receptors mediate signaling events that are distinct from those triggered by the more widely known receptors for type I four helix cytokines15,16. Like TLR stimulation, IL-17 receptor stimulation results in activation of NF-κB and mitogen-activated protein kinases (MAPK). However, IL-17 receptor signaling does not utilize the same set of membrane proximal adaptor molecules as TLR signaling; IL-17R requires the adaptor Act1 which alsocontains a SEFIR domain17–19. These unique signaling properties of IL-17 receptors enable TH-17 cells to act as a bridge between innate and adaptive immune cells.
Mechanistically, fluorescence resonance energy transfer (FRET) studies have suggested that IL-17RA may exist as a preformed dimer on the cell surface that undergoes a conformational change upon IL-17 binding to form a heterodimeric signaling complex with IL-17RC. However, the molecular basis for how a homodimeric IL-17 cytokine would pair with two different receptors remains unknown14,20.
Results
Structure of IL-17RA bound to IL-17F
We determined the crystal structure of IL-17RA bound to IL-17F at 3.3 A resolution using single isomorphous replacement with anomalous scattering (SIRAS) phasing (Table 1). We expressed IL-17F from baculovirus, and the IL-17RA extracellular domain (ECD) using 293S GnTI- cells. To facilitate crystallization, the complex was methylated, and the heavily glycosylated receptor ECD was ‘shaved’ with endoglycosidase H prior to crystallization to improve homogeneity, leaving one GlcNAc residue at each of the Asn-linked glycosylation sites (Fig. 1). Biochemically the shaved and unshaved complexes behaved identically (data not shown). By gel filtration, mixtures of IL-17F or IL-17A with IL-17RA ECD resulted in co-elution of complexes with 2:2 (2 receptors + 1 IL-17 dimer) and 1:2 (1 receptor + 1 IL-17 dimer) stoichiometries, with the major species being the 1:2. The 2:2 was only detected at high protein concentrations, whereas at lower concentrations the 1:2 predominated even in the presence of excess IL-17RA. The crystals contained one IL-17RA bound to one IL-17F homodimer (Fig. 1). As discussed below, this ‘partial’ signaling complex may, in fact, be the biologically relevant form of the IL-17RA-IL-17F and IL-17RA-IL-17A complexes.
Table 1.
Data collection and refinement statistics
| Native 1 | K2PtCl4 | SeMet | Native 2 | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P41212 | P41212 | P41212 | P41212 |
| Cell dimensions | ||||
| a, b, c (Å) | 171.6, 171.6, 84.3 | 171.7, 171.7, 83.6 | 171.4, 171.4, 82.3 | 170.7, 170.7, 81.9 |
| Wavelength (Å) | 1.00 | 1.07 | 0.978 | 1.03 |
| Resolution (Å) | 3.4, 3.4, 3.9 | 4.5 | 3.8 | 3.3 |
| Completeness (%) | 100 (100) | 99.9 (100) | 100 (100) | 99.2 (99.9) |
| Redundancy | 7.8 (8.0) | 15.0 (15.5) | 15.4 (15.6) | 6.3 (6.4) |
| Rmerge | 0.10 (0.47) | 0.10 (0.39) | 0.12 (0.48) | 0.15 (0.45) |
| I/σI | 15.6 (3.8) | 19.7 (6.8) | 12.7 (7.0) | 7.6 (3.2) |
| Refinement statistics | ||||
| Resolution range (Å) | 40–3.3 | |||
| No. reflections (total/test) | 18479/948 | |||
| Rfactor/Rfree (%) | 22.7/25.3 | |||
| No. atoms | ||||
| Protein | 3779 | |||
| Carbohydrate | 98 | |||
| Calcium | 1 | |||
| R.M.S. deviations | ||||
| Bond lengths (Å) | 0.009 | |||
| Bond angles (°) | 1.36 | |||
| Mean B value (Å 2) | 111 | |||
| Ramachandran plot (most favored, allowed, generously allowed, disallowed) | 86.2, 13.6, 0.2, 0.0 | |||
Values in parentheses represent the highest resolution shell
K2PtCl4, heavy metal derivative
SeMet, selono-methionine-labeled IL-17RA
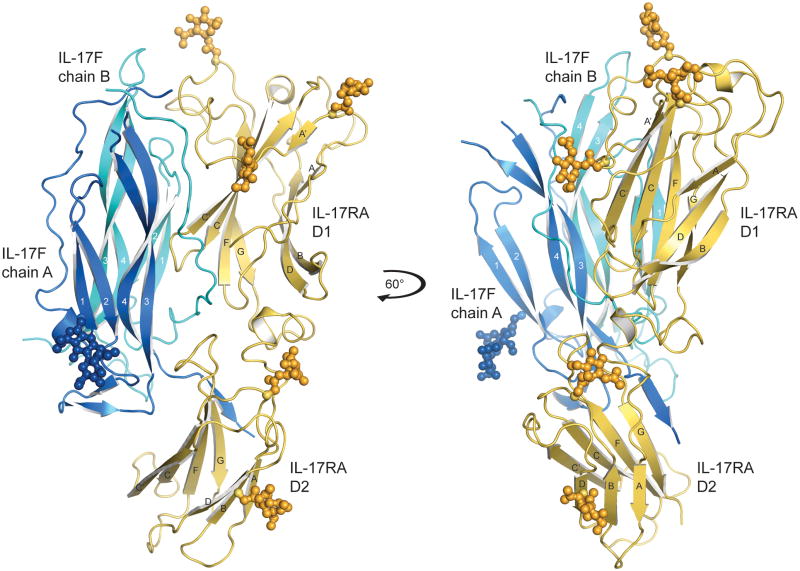
Figure 1.
Structure of the IL-17RA-IL-17F complex. Ribbon diagram of IL-17RA in yellow bound to IL-17F (chain A, blue; chain B, cyan), N-linked glycans are shown in ball-and-stick representation. IL-17RA is composed of two fibronectin type III domains (D1 and D2) joined by a short helical linker. The right-hand panel shows the complex rotated by 60° around the y-axis.
The IL-17RA ectodomain is composed of two unusual FnIII domain modules joined by an 18-amino acid linker (Fig. 1 and Supplementary Fig. 1). Although not apparent from the sequence, the IL-17RA structure is reminiscent of hematopoietic cytokine receptors in that it contains tandem β-sandwich domains; however, the domains themselves contain some substantial deviations from canonical FnIII folds, and the manner of ligand interaction is entirely distinct from other cytokine receptors. Residues 2–272 of the predicted 286 ectodomain residues (where residue 1 is the first amino acid of the mature peptide) were modeled into continuous electron density for the receptor chain and five of the potential seven N-linked glycans were clearly visualized (see Supplementary Fig. 2 for examples of electron density map quality). The first FnIII domain (D1) has an additional 40 amino acid N-terminal extension that forms a unique fold (Supplementary Fig. 1). The chain makes a hairpin-like turn bridged by a disulfide bond (Cys12–Cys19), and the second strand of the turn forms a β-strand (A′) that extends the FnIII β-sheet and then wraps around the face of the D1 domain, disulfide bonding with the C′ strand Cys95, before passing over the domain to start the A-strand of the FnIII domain. The interdomain linker region contains a short helix and is stabilized by an internal disulfide bond (Cys154–Cys165). The second FnIII domain (D2) has two atypical disulfide bonds, one linking the C–C′ loop (Cys214) to the D–F loop (Cys245) and a second within the F–G loop (Cys259–Cys263). We predict that a third disulfide bond exists between F–G loop (Cys246) and C-terminus of the G-strand (Cys272), similar to that observed in class-II cytokine receptors21, however this bond is not well defined in the current electron density map.
While the core structure of the IL-17RA-bound IL-17F molecule was essentially unchanged compared to that of the unliganded form of IL-17F7, peripheral strands and loops underwent structural accommodations to facilitate binding to IL-17RA. The conformation observed in the unliganded IL-17F structure could not be maintained in the IL-17RA-bound state, as it would generate steric clashes with the N-terminal coil region of the receptor. Each IL-17F monomer is composed of two pairs of anti-parallel β-sheets (strands 1–4) with the second and fourth strands connected by two disulfide bonds in a manner homologous to cysteine-knot family proteins. There is a 50 amino acid N-terminal extension of which residues 29–42 run parallel to strands 3 and 4 of the second IL-17F protomer. This coil region is stabilized by numerous interactions, including several hydrogen bonds with the adjacent strands. In the IL-17RA-bound IL-17F conformation this region (residues 33–42) moves out to open up the binding pocket and interact with the receptor (Fig. 2A). The first 24 amino acids of each IL-17F chain, and residues 105–109 from the 3–4 loop on one IL-17F protomer, could not be modeled. In the unliganded IL-17F structure Cys17 forms a disulfide bond with Cys107 at the tip of the 3–4 loop on the adjacent IL-17F chain. These interchain disulfide bonds were not modeled, but were present as our protein behaved as a disulfide-linked dimer on SDS-PAGE (data not shown).
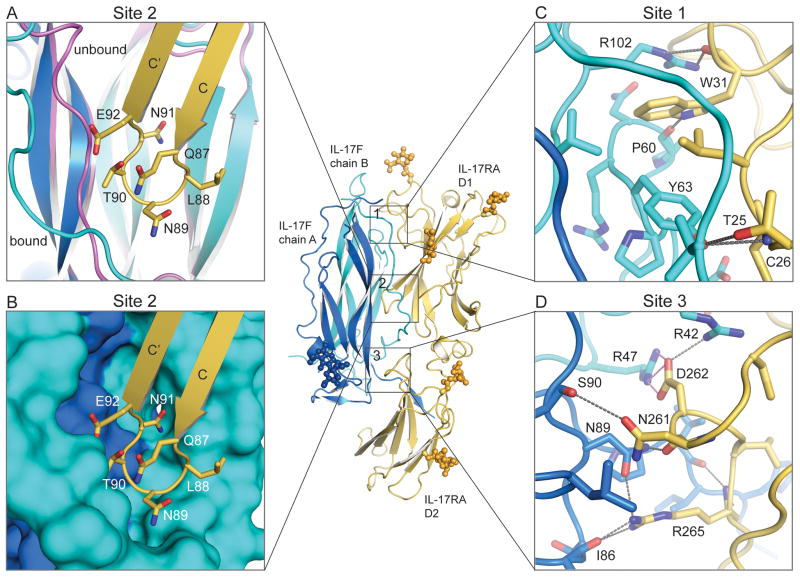
Figure 2.
IL-17F binding to IL-17RA is mediated by three distinct interfaces. (A) Site 2, the IL-17RA D1 C–C’ loop (yellow) inserts between the N-terminal coil region and strands 1 and 2 of the IL-17F chain B (cyan). The N-terminal coil undergoes a conformational change between the unbound (magenta) and bound (cyan) conformations. (B) Site 2, surface representation of the knob-in-holes IL-17F binding pocket complementarity. (C) Site 1, the IL-17RA D1 N-terminal binding site. (D) Site 3, the IL-17RA D2 binding site. Contact residues are shown as stick models. Gray dotted lines represent hydrogen bonds and pink dotted lines represent salt-bridges.
IL-17RA-IL-17F binding interface
The overall binding mode of IL-17F to IL-17RA, in which both receptor FnIII domains bind in a ‘side-on’ orientation and use edge strands to insert into a crevasse formed at the dimeric interface of the ligand, is unlike other cytokine or growth factor receptor complexes. IL-17RA forms an extensive binding interface with IL-17F, burying ~2200 A2 of surface area; ~70% of this buried surface area is mediated by the IL-17RA D1 domain. There are three major interaction sites at the binding interface (Fig. 2). Site 1 is formed between the N-terminal extension of IL-17RA (Thr25–Trp31) and the 1–2 loop (Pro60–Tyr63) plus the C-terminal region of strand 3 (Val100, Arg102) of IL-17F chain B; this interaction buries ~330 A2 (Fig. 2C). Trp31 of the receptor is buried in the center of this binding site; the main-chain O forms hydrogen bonds with Arg102 and the side chain forms hydrogen bonds with Pro60. Two additional hydrogen bonds are formed between IL-17RA Thr25 and Cys26 and IL-17F Tyr63. Site 2 is the most prominent interface feature of the complex, and is composed of the IL-17RA D1 C′ C loop (Leu86–Arg93) which slots into a deep binding-pocket flanked by the N-terminal extension and strand 2 of IL-17F chain B and strand 3 of IL-17F chain A; this interaction buries almost 550 A2 (Fig. 2A,B). This 8-amino acid IL-17RA loop forms extensive hydrophobic and polar interactions with both chains of IL-17F including a potential salt bridge between IL-17RA Glu92 and IL-17F chain B Arg37, and a hydrogen bond between the main-chain O of IL-17RA Asn89 and IL-17F chain A Asn95. Site 3, which encompasses ~410 Å2 of buried surface area (BSA), is formed between the IL-17RA D2 F G loop (Cys259–Arg265) and the C-terminal regions of stands 3 and 4 of IL-17F chain A, and the N-terminal extension of IL-17F chain B (Fig. 2D). Site 3 is rich in charged interactions with nine potential hydrogen bonds and a salt bridge between IL-17RA Asp262 and IL-17F chain B Arg47. Overall the interface is extensive and is composed of numerous specific contacts. It is envisaged that an analogous binding mode will be used by other IL-17 receptor-cytokine pairs, given the sequence conservation of contact residues (discussed below). However, a greater bond-network and/or shape complementarity may be employed in the higher affinity complexes.
Heterodimeric receptor complex formation
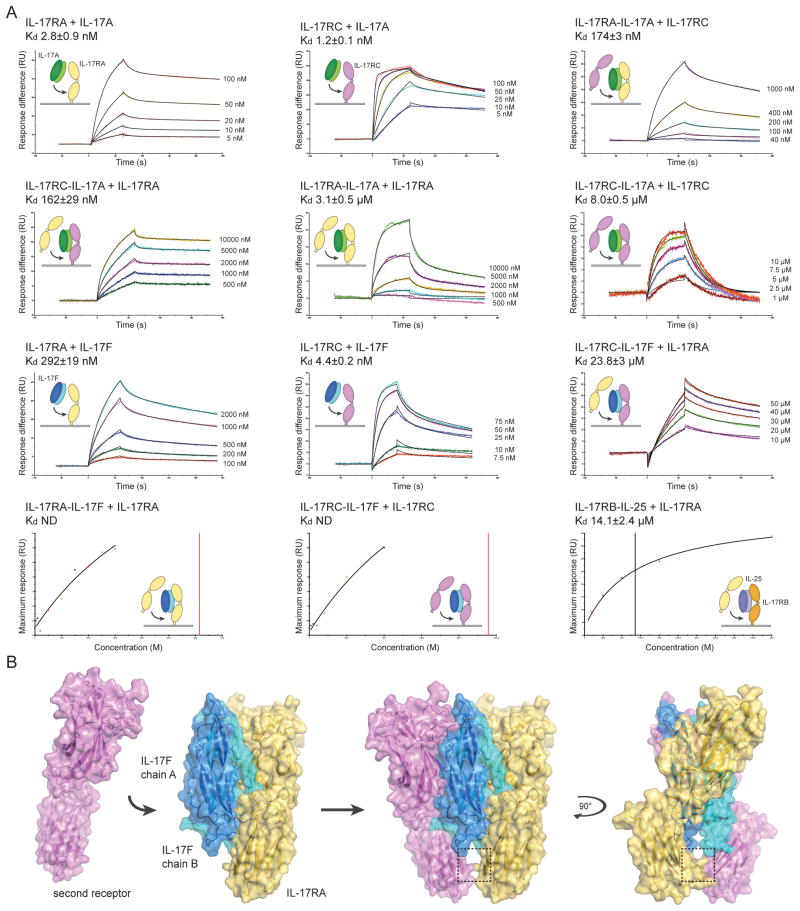
The stoichiometries of the receptor complexes remain to be fully elucidated6, but the asymmetric IL-17RA-IL-17F complex hints at a preference for heterodimerization with a second, different receptor. We therefore investigated the mechanism by which a homodimeric cytokine could possibly coordinate two different receptors. Both IL-17RA and IL-17RC can bind independently to IL-17A and IL-17F, but both receptors are necessary for signaling9,10,22. To further understand how the signaling complex is formed we devised a surface plasmon resonance (SPR) strategy using soluble proteins to measure the affinities of both the homodimeric and heteromeric receptor complexes for cytokine in vitro. Whilst others have reported the binding affinities of IL-17RA and IL-17RC for IL-17A and IL-17F7,22, we considered it pertinent to assess the binding affinity of the second receptor-binding site. The strategy was to immobilize one receptor on the SPR chip at a low coupling density in order to minimize possible homo-dimerization (e.g. cross-linking) of the receptors on the chip. The dimeric IL-17 cytokine was then captured by this receptor so that each receptor would be bound to one dimeric IL-17 ligand, leaving an exposed and accessible second receptor-binding site. The second receptor was subsequently passed over the preformed receptor-cytokine complexes to measure the affinity of the second receptor-binding event. In this fashion, the complex was assembled in a stepwise manner and each of the binding affinities was measured (Fig. 3). IL-17A bound to both IL-17RA (2.8±0.9 nM) and IL-17RC (1.2±0.1 nM) with high affinity. Once IL-17A was bound by one IL-17RA molecule, the binding affinity for a second IL-17RA was reduced to 3.1±0.5 μM whereas the IL-17RC affinity for this second binding site was 174±3 nM. If the IL-17A was originally captured by IL-17RC, a second IL-17RA bound to the existing IL-17RC-IL-17A complex with 162±29 nM affinity; the binding affinity of a second IL-17RC to existing IL-17RC-IL-17A complex was only 8.0±0.5 μM.
Figure 3.
Assembly and model of the heterodimeric IL-17 signaling complex. (A) IL-17 receptor-cytokine affinity was measured by surface plasmon resonance (SPR). IL-17RA, IL-17RB and IL-17RC were immobilized on the SPR chip surface, and the binding affinity of IL-17A, IL-17F or IL-25 was measured. Where indicated, the affinity of a second receptor binding to the pre-assembled receptor-cytokine complex on the chip was then measured. For kinetic experiments (top 3 rows), representative SPR sensorgrams are shown as colored lines and the curve-fit as a black line. Time in seconds (s) is plotted against response (RU, resonance units). The injected concentrations are to the right of the sensorgrams. For equilibrium experiments (fourth row), the injected concentration (M) is plotted against the maximum response (RU) for a representative experiment; the curve fit is shown as a black line and the dissociation constant (Kd) is marked as a vertical line. The insets show cartoon representations of the binding event. IL-17RA is colored yellow, IL-17RB in orange, IL-17RC in magenta, IL-17A in dark and light green, IL-17F in dark and light blue and IL-25 in dark and light purple. The Kd is reported as the mean of at least two independent experiments ± the standard error of the mean. (B) Model of heterodimeric signaling complex formation. The second receptor (magenta) was modeled assuming that both receptors bind to IL-17F in the same orientation. The C-terminal domains (D2) of the receptors come into close proximity as highlighted by the box (see Supplementary Fig. 3 for more details).
A similar pattern was observed for IL-17F, which has a higher affinity for IL-17RC (4.4±0.2 nM) compared to IL-17RA (292±19 nM). Given the divergent affinities it seems likely that IL-17F would be initially captured by IL-17RC; once bound, the affinity of IL-17RA for the IL-17RC-IL-17F complex was 23.8±3 μM. In contrast, the binding affinity of IL-17RA and IL-17RC for preformed IL-17RA-IL-17F and IL-17RC-IL-17F complexes, respectively, was so weak that it could not be accurately calculated over the concentration range used for these experiments. Thus, these findings clearly show that engagement of IL-17RA or IL-17RC by IL-17A or IL-17F encourages a preference for the second receptor-binding site to engage a different receptor and thereby to form the heterodimeric receptor complex.
IL-17RA has been implicated in IL-17E (also known as IL-25) signaling together with IL-17RB23. IL-25 promotes Th2 inflammatory responses and shares approximately ~20% identity with IL-17A and IL-17F. Binding experiments have demonstrated that whilst IL-25 binds to IL-17RB with high affinity, it has no apparent affinity for IL-17RA23–25. We hypothesized that IL-17RA may only bind IL-25 once IL-25 is captured by IL-17RB. To test this hypothesis, we immobilized IL-17RB on an SPR chip, captured IL-25 and measured the affinity of IL-17RA for the IL-17RB-IL-25 complex. Supporting our hypothesis, IL-17RA bound to the IL-17RB-IL-25 complex with 14.1±2.4 μM affinity (Fig. 3). At concentrations up to 50 μM, no interaction could be observed between IL-17RA and IL-25, or between the IL-17RB-IL-25 complex and a second IL-17RB molecule. Together with the IL-17A and IL-17F binding data, these results indicate that the formation of the heteromeric complex may be mediated by allostery and/or an interaction between the receptors.
To further address this concept we modeled a second IL-17RA molecule to form the hypothetical 2:2 receptor-cytokine complex (Fig. 3B). Assuming that the second receptor binds in an identical fashion to the first, the base of IL-17RA D2 would come into very close proximity with the D2 of the second IL-17RA (Fig. 3B, dashed box). In the case of two IL-17RA molecules bound to IL-17F, His212 on the C–C′ loop of one IL-17RA would clash with the second IL-17RA His212 (Fig. S3). This potential interaction site may allow the receptors to regulate their pairing. Steric clashes may cause reduced affinity for a second identical receptor, or favorable receptor-receptor interactions may stabilize heteromeric receptor complexes. We do not rule out the possibility that homodimeric receptor complexes could form on cells under certain conditions, however, our data argues that receptor heterodimers will likely be the predominant signaling species (see Discussion).
IL-17RA as a common receptor
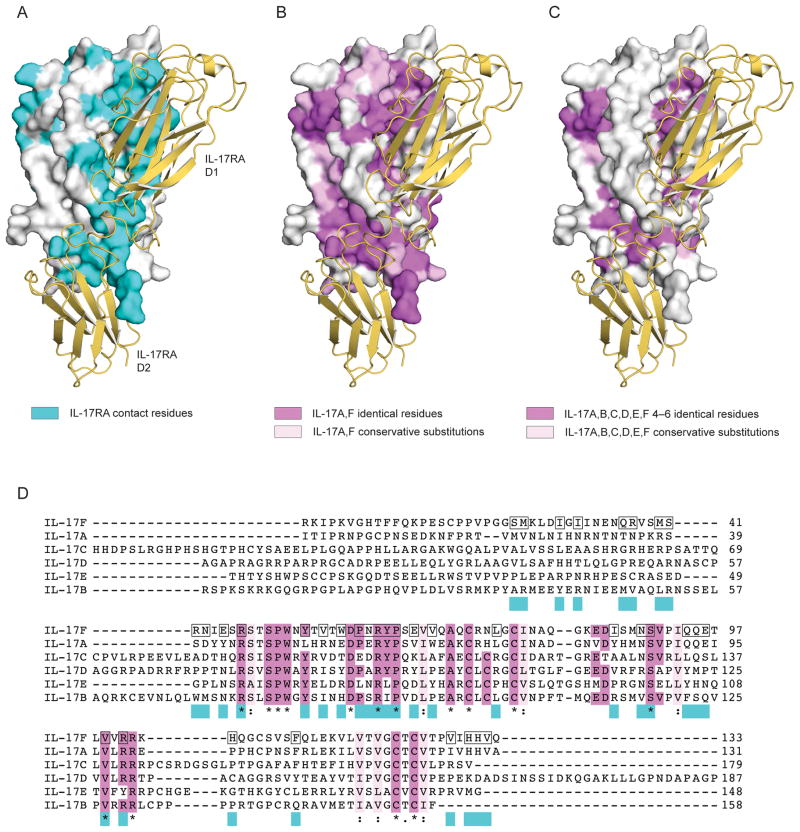
IL-17RA binds to IL-17A with ~100-fold higher affinity than IL-17F. IL-17A and IL-17F share ~50% identity, and mapping the conserved residues onto the structure of IL-17F reveals a horseshoe-shaped ring of variable residues around the receptor-binding pocket (Fig. 4). The majority of the IL-17RA C′–C loop interactions are formed with residues that differ between the IL-17A and IL-17F molecules whereas the N-terminal region and IL-17RA D2 F–G loop interactions involve predominately conserved residues. We reported here that the extracellular region of IL-17RA can also bind to the IL-17RB-IL-25 complex, and it was recently shown that IL-17RD can interact with IL-17RA to mediate IL-17A signaling26. Given this association of IL-17RA with diverse IL-17 family members we speculate that IL-17RA may act as a shared receptor analogous to those utilized in class I cytokine receptor complexes27. To investigate this possibility, we mapped the residues conserved among all IL-17 family members onto the IL-17F surface. Analyzing the location of these residues in the IL-17RA-IL-17F complex, it seems plausible that IL-17RA contacts these conserved residues with the N-terminal region of the D1 domain and the F–G loop of the D2 domain (Figure 4C). In contrast, IL-17RA may modulate specificity for each cytokine by contacting non-conserved cytokine residues with the C–C′ loop (Figure 4C). Collectively, then, IL-17RA appears to use a strategy of cross-reactivity based on a subset of conserved contacts, amongst a background of distinct contacts, with several different IL-17 cytokines. This is similar to the strategy utilized by the shared p75 receptor for recognition of different neurotrophin ligands28, and stands in contrast to the mechanism used for cross-reactivity by, for example, gp130 and γc chain, which form largely disparate molecular interactions with different four-helix cytokines27.
Figure 4.
Binding interface and conserved IL-17 residues. Surface representation of IL-17F in white with IL-17RA in ribbon format colored yellow. (A) IL-17RA-IL-17F contact residues highlighted in cyan. (B) Residues conserved among IL-17A and IL-17F are mapped onto the IL-17F structure; identical residues are colored magenta and conservative substitutions in light pink. (C) Residues identical among 4, 5 or 6 IL-17 cytokine family members are colored magenta and conservative substitutions across all six cytokines in light pink. (D) Alignment of human IL-17 cytokines. Residues that form contacts in the IL-17RA-IL-17F structure are highlighted by a black box on the IL-17F sequence and in cyan underneath the alignment. Residues that are identical in 4, 5 or 6 cytokines are highlighted in magenta; those identical in all 6 cytokines are also marked with ‘*’; conserved groups are colored light pink and marked with ‘:’.
Receptor binding modes of cysteine-knot growth factors
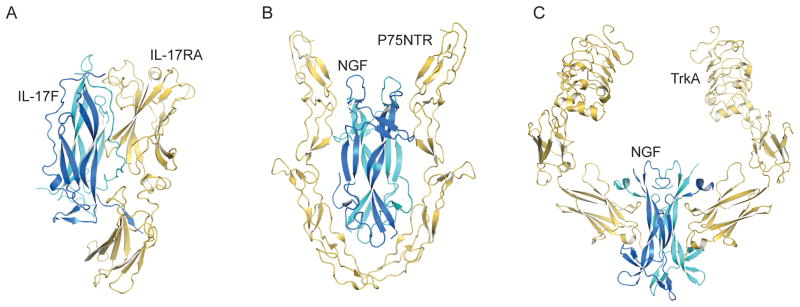
Several crystal structures for receptor-cysteine-knot growth factor ligand complexes, such as nerve growth factor (NGF)28–30, vascular endothelial growth factor (VEGF)31, two glial cell-derived neurotrophic factor (GDNF) family members32, and others; these structures can serve as instructive comparisons with the mode of ligand engagement mediated by IL-17RA (Fig. 5). In the complex of NGF bound to the p75 neurotrophin receptor (p75NTR, a death receptor family member)28,30, the receptor bears no structural similarity to IL-17RA; however, like IL-17RA, p75NTR engages NGF within a concave groove at the ligand dimer interface (Fig. 5B). In the TrkA complex with NGF29,33, an immunoglobulin (Ig)-domain in TrkA, which is structurally related to the FnIII domains of IL-17RA, is used for ligand binding. However, the Ig-domain of TrkA binds end-on to a flat face in the ‘saddle’ of NGF formed by the NGF β-sheets; thus the mode of binding is distinct (Fig. 5C). Interestingly, the NGF-p75NTR complex has been reported as both 1:2 and 2:2 complexes that may represent partial and complete forms of a homodimeric p75 signaling complex, respectively28,30. However, in that case, homodimeric NGF ligand engages two identical p75 molecules, and thus does not require a structural mechanism for the symmetric dimeric ligand to heterodimerize two different receptors.
Figure 5.
IL-17RA-IL-17F receptor complex compared to homodimeric cysteine-knot growth factor receptor complexes. (A) IL-17RA-IL-17F, (B) P75NTR-NGF and (C) TrkA-NGF are shown as ribbon models with the receptors in yellow and the cytokines and growth factors in blue and cyan.
Discussion
IL-17 family cytokines are central mediators of chronic inflammatory and autoimmune conditions. Here we reported the crystal structure of IL-17RA bound to IL-17F, illuminating both the receptor binding interface and a potential mechanism by which the IL-17 family of homodimeric cytokines can coordinate two different receptors. Our structure revealed one IL-17RA bound to the dimeric IL-17F cytokine, leaving the second potential receptor-binding interface free to engage a second receptor. Biochemical studies demonstrated that the heteromeric complex can be reconstituted in vitro and that the binding of the first receptor modulates the affinity of the second.
Although we show here that IL-17F forms heterodimeric signaling complexes using IL-17RA as a shared receptor, the possibility remains that, in principle, homodimeric receptor complexes could form under certain conditions34,35. The differential receptor affinities shown by our binding data suggests that under conditions where one type of receptor would be highly overexpressed relative to another, homodimeric signaling pairs could be formed. In this manner, cells may be able to tune their responsiveness to IL-17 through heterodimeric versus homodimeric receptor complexes using a mass-action based mechanism based on modulations in relative receptor expression, as has been shown for the IL-4 and IL-13 system36. However, the generality of homodimeric complexes in vivo is debatable given that cells from IL-17RA-deficient or IL-17RC-deficient mice are not responsive to IL-17A or IL-17F9,10. FRET-based experiments have previously established that IL-17RA chains exist as preformed dimers on the surface of cells20. The addition of cytokine causes a decrease in FRET efficiency suggesting that the IL-17RA chains either undergo a conformational change separating the intracellular domains or dissociate upon binding. Overexpression of both IL-17RA and IL-17RC in the absence of cytokine is not sufficient to induce signaling, consistent with the notion that the cytokine elicits a change necessary for signaling9. Given that both IL-17RA and IL-17RC are required for signaling it seems plausible that cytokine binding imposes the necessary receptor pairing. The wide range of affinities between IL-17 receptors and cytokines further exacerbates this need to modulate receptor pairing. Our data indicates that pairing may be dictated by an interaction between the membrane proximal domains (D2) of the receptors. For like receptors, this may represent a repulsive force in the bound conformation reducing the affinity for the second like-receptor. Gaffen and colleagues reported that the murine IL-17RA D2 actually mediates homotypic interactions14. We have not observed any dimer formation with our extracellular IL-17RA protein (data not shown) despite a high degree of conservation between the murine and human IL-17RA extracellular regions. As the SEFIR domains in the intracellular region of IL-17 receptors and the signaling adaptor Act1 are predicted to dimerize13, it seems plausible that the intracellular domains may mediate the preassembly of receptor chains whilst cytokine binding imposes the correct receptor pairing for signaling.
IL-17RA is essential for IL-17A, IL-17F and IL-25 signaling9,23. More recently, IL-17RA has also been implicated in IL-17RD signaling, although the ligand for this interaction remains unknown26. Given this emerging trend, along with our observation that IL-17RA contact residues in site 1 and site 3 binding interfaces are somewhat conserved amongst the IL-17 cytokine family members, we propose that IL-17RA may act as a shared receptor for the IL-17 family. Shared receptors such as gp130 and the γc chain are the hallmark of many cytokine-receptor signaling complexes27. These common receptors bind to an array of multimeric receptor-cytokine complexes, often in a manner mediated by their affinity for the cytokine bound α-receptor complex rather than a direct binding affinity for the cytokine itself. IL-17RA may represent the newest member of the shared receptor family, serving in effect as a ‘γc chain’ of the IL-17R family, and adding a new twist to the paradigm in which the shared-receptor binding site utilizes conserved ‘anchor points’ to engage variant ligands.
Methods
Protein expression and purification
The native signal peptide and extracellular region of human IL-17RA (residues 1–286) was cloned into the BacMam expression vector pVLAD637. Recombinant protein was transiently expressed in suspension 293 GnTI- cells grown in Pro293 media (Lonza) supplemented with 1% fetal calf serum (FBS) and 10 mM Na butyrate at 37° C. Full length IL-17F with a C-terminal hexa-His tag was cloned into the pAcGP67-A expression vector (BD Biosciences) and the protein secreted by High Five insect cells grown in Insect Xpress media (Lonza) at 27° C. The supernatants containing the IL-17RA and IL-17F proteins were mixed and concentrated before Ni-affinity purification. The IL-17RA protein was deglycosylated via endoglycosidase-H treatment and the IL-17RA and IL-17F purification tags cleaved using 3C-protease and carboxypeptidase A (Sigma-Aldrich). The protein complex was subjected to reductive lysine methylation using dimethylamine-borane complex and formaldehyde as described by Walter et al.38. The IL-17RA-IL-17F complex was further purified using a Superdex 200 size exclusion column (GE Healthcare) equilibrated in 10 mM Hepes pH 7.4 and 150 mM NaCl. Fractions containing the IL-17RA-IL-17F complex were concentrated to ~15 mg/ml for crystallization trials.
Selono-methione (SeMet) labeled IL-17RA protein was prepared as described with the following modifications39. Untransfected adherent 293 GnTI- cells were cultivated in FBS-supplemented DMEM media (Invitrogen). The media was exchanged after a single phosphate-buffered saline wash, for Met and Cys-free DMEM (Invitrogen) supplemented with 40 mg/l L-Cys, 45 mg/l selenon-L-Met, 2% FBS, L-glutamate, Na pyruvate, IL-17RA BacMam virus and 10 mM Na butyrate. Expression was allowed to proceed for 72 hours. IL-17RA-SeMet protein supernatants were mixed with IL-17F and purified as described above. For binding experiments, proteins were expressed and purified essentially as described above. The IL-17RA, IL-17RB and IL-17RC extracellular domains were expressed by 293s GnTI- cells with and without a C-terminal BirA ligase tag. IL-17RC was expressed with an additional C-terminal Fc tag that was cleaved by 3C-protease prior to size exclusion chromatography. IL-17A, IL-17F and IL-25 cytokines were expressed by High Five cells with C-terminal hexa-His tags. Proteins were enymatically biotinylated using BirA ligase and purified via size exclusion chromatography.
Crystallization and x-ray data collection
IL-17RA-IL-17F complexes were initially grown via hanging-drop vapor diffusion in 10% PEG6000 and 0.1 M bicine pH 9.0. Optimized native and SeMet protein complex crystals were grown in PEG6000 (4–14%) and 0.1 M CAPSO buffer (pH 9.1–9.3) with 20 mM CaCl2 or 10 mM CaCl2 and 1.5% w/v trimethylamine N-oxide dihydrate added directly to the protein-precipitant drop. Heavy metal derivatives were prepared by soaking the crystals in well solution supplemented with 0.5 mM K2PtCl4 and 2% ethylene glycol for 6 hours. Crystals were cryo-protected prior to data collection in the well solution plus 20–25% ethylene glycol and cooled to 100 K. The crystals belong to the space group P41212 and have unit cells dimensions of ~171, 171, 83 Å. The initial native data set was collected at Stanford Synchrotron Radiation Lightsource (SSRL) beamline 9-2 (Stanford, CA). The Pt-derivative and SeMet datasets were collected at SSRL beamline 11-1. The higher resolution native dataset was collected at the Advanced Photon Source (APS) beamline ID-23D (Argonne, IL). All data was indexed and integrated using the program Mosflm40 and scaled with SCALA from the CCP4 suite41. The diffraction is anisotropic and the initial native dataset was also subjected to ellipsoidal truncation and anisotropic scaling using the diffraction anisotropy server 42 rendering a data set scaled to 3.4, 3.4 and 3.9 Å.
Structure determination and refinement
A molecular replacement solution for a single IL-17F homodimer was determined using the program Phaser43 with the previously determined 2.85 Å IL-17F structure as a model (PDB ID 1JPY)7. The initial maps showed additional density on one side of the IL-17F dimer illuminating the binding site for IL-17RA. Phases were calculated using a K2PtCl4 derivative via single isomorphous replacement with anomalous scattering (SIRAS) in the program Sharp44. Density modified maps were calculated assuming 71% solvent and including the partial model from the IL-17F molecular replacement for 10 out of 20 rounds. A partial model of the IL-17RA main chain was manually built into this map using the program Coot45.
The position of the IL-17RA Met residues was calculated via fast Fourier transform (FFT) to generate an anomalous difference map using the program FFT in the CCP4 suite. As the SeMet dataset was not isomorphous with the native dataset and the signal too weak to locate the sites via single anomalous difference (SAD) phasing methods, the partially built model was used as a molecular replacement model for the SeMet dataset and the calculated phases used to find the selenonium peaks. Three of a potential six SeMet residues were located, corresponding to IL-17RA Met159, Met166 and Met218. These Met positions, in addition to the predicted Asn-linked glycosylation sites and disulfide bonds were used to register the polypeptide in the density and complete building the initial IL-17RA model. Iterative rounds of coordinate and B-factor refinement were performed using the program Phenix46 intersected with manual model building in Coot. Initial rounds of model building utilized B-factor sharpened σA-weighted phased- combined maps calculated by the program CNS47. The final model was refined to 3.3 Å with an Rfactor and Rfree of 22.7% and 25.3% respectively. There is one IL-17RA-IL-17F complex in the asymmetric unit. The model includes a dimethyl-lysine at position 43 of the IL-17RA chain, five single N-Acetylglucosamine (GlcNAc) sites on the IL-17RA chain, one site with two GlcNAc residues on the IL-17F chain B and a calcium ion. The programs PROCHECK48 and WHAT_CHECK49 were used to assess the geometry of the final model. The CCP4 suite programs Contact and Areaimol were used to determined the interface contacts and buried surface area respectively. All structural figures were generated using the program Pymol50.
Affinity measurements
Binding affinities were calculated via surface plasmon resonance (SPR) on a Biacore T100 (GE Healthcare). C-terminally biotinylated IL-17 receptors were coupled to immobilized streptavidin on either an SA or CM4-sensor chip (GE Healthcare). An irrelevant, biotinylated protein was captured at equivalent immobilization densities to control flow cells. To measure the second receptor binding interaction, the cytokine was first captured to the immobilized receptor, followed by the second receptor injection. Low coupling densities (200–400 RU) and excess cytokine concentrations were used to optimize the number of cytokine homodimers bound to a single receptor. The surface was regenerated using 3 M MgCl2 between each cycle. For kinetic experiments, a flow rate of 50 μl/min was used. Data was analyzed using Biacore T100 evaluation software Version 2.0 (GE Healthcare). Affinities are reported as the mean of at least two independent experiments ± the standard error of the mean (s.e.m.).
Supplementary Material
Acknowledgments
We thank S. Juo for assistance with data collection and structure determination, E. Ozkan for helpful discussion and D. Gorman for cDNA reagents. We thank the beamline staff at the Stanford Synchrotron Radiation Lightsource, Advanced Light Source and Advanced Photon Source for assistance with data collection. L.K.E. is a CJ Martin fellow supported by the National Health and Medical Research Council of Australia. K.C.G. is supported by the NIH (AI51321) and Howard Hughes Medical Institute.
Footnotes
Accession numbers
Coordinates and structure factors have been deposited in the Protein Data Bank (www.rcsb.org) under the accession number 3JVF.
Author contributions
L.K.E and K.C.G. designed experiments, L.K.E. and S.F. performed experiments, L.K.E. and K.C.G. analyzed data and wrote manuscript.
Conflict of interest
K.C.G. declares a competing financial interest, and plans to file a patent to use information gained from this crystal structure to design IL-17 antagonists.
References
- 1.Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 2.Harrington LE, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 3.Park H, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 5.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 6.Gaffen SL. Structure and signalling in the IL-17 receptor family. Nat Rev Immunol. 2009 doi: 10.1038/nri2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hymowitz SG, et al. IL-17s adopt a cystine knot fold: structure and activity of a novel cytokine, IL-17F, and implications for receptor binding. EMBO J. 2001;20:5332–5341. doi: 10.1093/emboj/20.19.5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright JF, et al. Identification of an interleukin 17F/17A heterodimer in activated human CD4+ T cells. J Biol Chem. 2007;282:13447–13455. doi: 10.1074/jbc.M700499200. [DOI] [PubMed] [Google Scholar]
- 9.Toy D, et al. Cutting edge: interleukin 17 signals through a heteromeric receptor complex. J Immunol. 2006;177:36–39. doi: 10.4049/jimmunol.177.1.36. [DOI] [PubMed] [Google Scholar]
- 10.Zheng Y, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 11.Ishigame H, et al. Differential roles of interleukin-17A and -17F in host defense against mucoepithelial bacterial infection and allergic responses. Immunity. 2009;30:108–119. doi: 10.1016/j.immuni.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 12.O’Connor W, Jr, et al. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat Immunol. 2009;10:603–609. doi: 10.1038/ni.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novatchkova M, Leibbrandt A, Werzowa J, Neubuser A, Eisenhaber F. The STIR-domain superfamily in signal transduction, development and immunity. Trends Biochem Sci. 2003;28:226–229. doi: 10.1016/S0968-0004(03)00067-7. [DOI] [PubMed] [Google Scholar]
- 14.Kramer JM, et al. Cutting edge: identification of a pre-ligand assembly domain (PLAD) and ligand binding site in the IL-17 receptor. J Immunol. 2007;179:6379–6383. doi: 10.4049/jimmunol.179.10.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maitra A, et al. Distinct functional motifs within the IL-17 receptor regulate signal transduction and target gene expression. Proc Natl Acad Sci U S A. 2007;104:7506–7511. doi: 10.1073/pnas.0611589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol. 2009;9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claudio E, et al. The adaptor protein CIKS/Act1 is essential for IL-25-mediated allergic airway inflammation. J Immunol. 2009;182:1617–1630. doi: 10.4049/jimmunol.182.3.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang SH, Park H, Dong C. Act1 adaptor protein is an immediate and essential signaling component of interleukin-17 receptor. J Biol Chem. 2006;281:35603–35607. doi: 10.1074/jbc.C600256200. [DOI] [PubMed] [Google Scholar]
- 19.Qian Y, et al. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat Immunol. 2007;8:247–256. doi: 10.1038/ni1439. [DOI] [PubMed] [Google Scholar]
- 20.Kramer JM, et al. Evidence for ligand-independent multimerization of the IL-17 receptor. J Immunol. 2006;176:711–715. doi: 10.4049/jimmunol.176.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bazan JF. Structural design and molecular evolution of a cytokine receptor superfamily. Proc Natl Acad Sci U S A. 1990;87:6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright JF, et al. The human IL-17F/IL-17A heterodimeric cytokine signals through the IL-17RA/IL-17RC receptor complex. J Immunol. 2008;181:2799–2805. doi: 10.4049/jimmunol.181.4.2799. [DOI] [PubMed] [Google Scholar]
- 23.Rickel EA, et al. Identification of functional roles for both IL-17RB and IL-17RA in mediating IL-25-induced activities. J Immunol. 2008;181:4299–4310. doi: 10.4049/jimmunol.181.6.4299. [DOI] [PubMed] [Google Scholar]
- 24.Shi Y, et al. A novel cytokine receptor-ligand pair. Identification, molecular characterization, and in vivo immunomodulatory activity. J Biol Chem. 2000;275:19167–19176. doi: 10.1074/jbc.M910228199. [DOI] [PubMed] [Google Scholar]
- 25.Lee J, et al. IL-17E, a novel proinflammatory ligand for the IL-17 receptor homolog IL-17Rh1. J Biol Chem. 2001;276:1660–1664. doi: 10.1074/jbc.M008289200. [DOI] [PubMed] [Google Scholar]
- 26.Rong Z, et al. IL-17RD (Sef or IL-17RLM) interacts with IL-17 receptor and mediates IL-17 signaling. Cell Res. 2009;19:208–215. doi: 10.1038/cr.2008.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Lupardus P, Laporte SL, Garcia KC. Structural biology of shared cytokine receptors. Annu Rev Immunol. 2009;27:29–60. doi: 10.1146/annurev.immunol.24.021605.090616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He XL, Garcia KC. Structure of nerve growth factor complexed with the shared neurotrophin receptor p75. Science. 2004;304:870–875. doi: 10.1126/science.1095190. [DOI] [PubMed] [Google Scholar]
- 29.Wehrman T, et al. Structural and mechanistic insights into nerve growth factor interactions with the TrkA and p75 receptors. Neuron. 2007;53:25–38. doi: 10.1016/j.neuron.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 30.Gong Y, Cao P, Yu HJ, Jiang T. Crystal structure of the neurotrophin-3 and p75NTR symmetrical complex. Nature. 2008;454:789–793. doi: 10.1038/nature07089. [DOI] [PubMed] [Google Scholar]
- 31.Wiesmann C, et al. Crystal structure at 1.7 A resolution of VEGF in complex with domain 2 of the Flt-1 receptor. Cell. 1997;91:695–704. doi: 10.1016/s0092-8674(00)80456-0. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Baloh RH, Milbrandt J, Garcia KC. Structure of artemin complexed with its receptor GFRalpha3: convergent recognition of glial cell line-derived neurotrophic factors. Structure. 2006;14:1083–1092. doi: 10.1016/j.str.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 33.Wiesmann C, Ultsch MH, Bass SH, de Vos AM. Crystal structure of nerve growth factor in complex with the ligand-binding domain of the TrkA receptor. Nature. 1999;401:184–188. doi: 10.1038/43705. [DOI] [PubMed] [Google Scholar]
- 34.You Z, et al. Interleukin-17 receptor-like gene is a novel antiapoptotic gene highly expressed in androgen-independent prostate cancer. Cancer Res. 2006;66:175–183. doi: 10.1158/0008-5472.CAN-05-1130. [DOI] [PubMed] [Google Scholar]
- 35.Li TS, Li XN, Chang ZJ, Fu XY, Liu L. Identification and functional characterization of a novel interleukin 17 receptor: a possible mitogenic activation through ras/mitogen-activated protein kinase signaling pathway. Cell Signal. 2006;18:1287–1298. doi: 10.1016/j.cellsig.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 36.LaPorte SL, et al. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008;132:259–272. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dukkipati A, Park HH, Waghray D, Fischer S, Garcia KC. BacMam system for high-level expression of recombinant soluble and membrane glycoproteins for structural studies. Protein Expr Purif. 2008;62:160–170. doi: 10.1016/j.pep.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter TS, et al. Lysine methylation as a routine rescue strategy for protein crystallization. Structure. 2006;14:1617–1622. doi: 10.1016/j.str.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barton WA, Tzvetkova-Robev D, Erdjument-Bromage H, Tempst P, Nikolov DB. Highly efficient selenomethionine labeling of recombinant proteins produced in mammalian cells. Protein Sci. 2006;15:2008–2013. doi: 10.1110/ps.062244206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;26 [Google Scholar]
- 41.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 42.Strong M, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de La Fortelle E, Bricogne G. SHARP: A Maximum-Likelihood Heavy-Atom Parameter Refinement Program for the MIR and MAD Methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Afonine PV, Grosse-Kunstleve RW, Adams PD. The Phenix refinement framework. CCP4 Newsletter. 2005;42 [Google Scholar]
- 47.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 48.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 49.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 50.DeLano WL. DeLano Scientific. San Carlos; CA, USA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.