Abstract
Understanding tissue architecture and physical and chemical reciprocity between cells and their microenvironment provides vital insight into key events in cancer metastasis, such as cell migration through three-dimensional extracellular matrices. Yet many mechanistic details associated with metastasis remain elusive due to difficulty studying cancer cells in relevant three-dimensional microenvironments. Recently optical imaging has facilitated direct observation of single cells undertaking fundamental steps in the metastatic processes. As such, optical imaging is providing novel “optical biomarkers” with diagnostic potential that may be linked to cell motility pathways associated with metastasis, and can help guide new approaches in cancer diagnosis and therapy. Herein, we present recent advances in one subclass of optical imaging of particular promise for cellular imaging, multiphoton microscopy, that can be used to improve detection of malignant cells as well as advance our understanding of the cell biology of cancer metastasis.
Introduction
The spread of cancer to nearby and distant sites involves several steps, termed the metastatic cascade: local invasion, entrance into the vasculature (intravasation), exit from the vasculature (extravasation), and growth in a distant tissue1,2. Despite advances in our understanding of cancer progression, key steps in the metastatic process are still poorly understood and seldom directly observed in vivo. We have now reached an understanding of several biochemical changes and gene profiles that accompany tumor progression. However, our understanding of the basic cell biology of metastasis will remain incomplete until we take this information forward to image molecular events within tumors. To accomplish this, advanced techniques are needed to visualize tumor cells in their relevant environment, and capture their behavior and molecular changes as they navigate the stages of metastasis.
Advanced imaging approaches are not only important for understanding the biology of metastasis on a fundamental level, but also are likely to impact patient outcome. Timely detection and accurate staging of human cancer is critical because the selected course of treatment is directly dependent on the location and pathological characterization of the tumor mass. Therefore, methods to accurately detect early stage cancer, and reliably diagnose and stage progressing cancers, can have a profound impact on treatment courses that may increase patient survival. In particular, imaging techniques to distinguish early hyperplastic lesions from normal tissue and malignant versus benign tumors are decidedly necessary. As such, optical techniques that identify changes in cellular behavior and/or the extracellular matrix (ECM) to produce consistent and quantifiable markers of cellular and molecular changes associated with cancer, i.e. “optical biomarkers”, are desired. Such optical biomarkers, when linked to molecular pathways, have the potential to provide guidance for specifically targeted therapeutic intervention. Several recent advances in optical imaging of tumor progression (reviewed below) are beginning to make this potential achievable.
The need to see cancer at the cellular scale
To date, most clinical tools focus on identifying a tumor mass that is large enough to be detected with whole body/organ imaging technologies, such as ultrasound, magnetic resonance imaging, computed tomography, fluorescence-mediated tomography, and positron emission tomography (PET)3,4. PET in particular has promise due to its ability to selectively image tumors based on metabolism, proliferation, or specifically labeled receptor or antigen biomarkers5. Yet, while these tools are extremely valuable in clinical oncology, and are the primary means for non-invasively detecting a tumor mass, they are not capable of subcellular resolution3,5–7. As such, they are unlikely to provide a highly accurate indication of disease state by themselves since comprehensively understanding cancer progression requires the ability to visualize single cell phenotypes that are linked to defined molecular pathways. Therefore a great deal of work has been directed toward imaging single cells and identifying optical biomarkers that are reliably indicative of early cell transformation, such as changes in cell metabolism8–14 or stromal extracellular matrix (ECM) microenvironment architecture8,12,13,15,16. However, at the time of detection, it is common for tumors to have already progressed to an invasive stage. As such, information that helps stop or limit the spread of primary tumor cells to new sites in the body, or indicates treatment for secondary tumor risk, is of importance. Yet, detecting optical biomarkers that specify an invasive tumor is more complex than tumor mass detection. Moreover, in order for optical biomarkers associated with metastasis to reach their full potential, a detailed understanding of their coupled molecular mechanisms need to be determined in the context of the cell phenotype, initially in animal studies, and ultimately in patients.
Recent insights into the mechanisms of cell motility events and characterization of optical biomarkers associated with metastatic processes have been gained using multiphoton laser-scanning microscopy (MPLSM), allowing visualization of invasive cells within 3D microenvironments in vivo and in vitro. Here, we highlight these novel findings obtained from MPLSM imaging of cancerous cells within their native environments, with particular emphasis placed on the potential to link molecular mechanisms of motility during local invasion through the collagenous stroma with optical biomarkers indicative of disease state. Current limitations of optical imaging as a diagnostic tool mean that emerging technologies will increase the diagnostic potential of MPLSM.
Of course, while MPLSM provides particular advantages over many of the more conventional optical imaging technologies and is uniquely suited for high resolution imaging of the collagen matrix, it is important to note that recent advances in other imaging modalities - such as optical coherence tomography17–19, spinning disk confocal microscopy20,21, and photoacoustic tomography22,23 - are also capable of providing insight into single cancer cell behavior. However, discussion of these imaging modalities is beyond the scope of the current review and the reader is encouraged to examine the listed references for additional information. Moreover, since technologies such as these can often be combined with MPLSM for multimodal imaging approaches (e.g.17,24) they can provide an even greater depth of information to aid our understanding of tumor biology.
Nonlinear optical imaging for cell biology
Multiphoton laser-scanning microscopy is an optical sectioning technique where the fluorescence emission is proportional to the molecular cross-section and has a quadratic dependence on the excitation laser power25–27 (Box 1; for detailed reviews of MPLSM and second harmonic generation see refs.7,27–30). Due to this quadratic dependence on excitation intensity, higher order processes are unlikely to occur outside the focal volume (i.e. the probability of two or more photons simultaneously exciting a fluorophore outside the focal volume is extremely low)26. As a result, improved signal to noise and background discrimination is achieved without the need for a confocal aperture, which allows the majority of emitted photons to be collected while not photobleaching the out-of-focus volumes of the tissue. Moreover, mode-locked lasers used for MPLSM excite at longer wavelengths (typically 650–1050 nm excitation; this broad tunability also has practical advantages for being able to excite a wide range of fluorophores) than conventional fluorescence imaging allowing for deeper penetration into tissues since longer wavelengths are more immune to light scattering and a more efficient collection of scattered photons emitted following deep tissue imaging is utilized with MPLSM. In addition, using pulsed lasers such that the mean power at the sample is moderate minimizes photo-damage. It has been demonstrated that for many cells and tissues the viability is improved when longer wavelengths are used due to circumventing some of the know mechanisms of phototoxicity (including cycle checkpoints that are UV light sensitive)31,32. Hence, MPLSM26 can image deeper26,33 into 3D structures, with improved signal-to-noise ratio26,29,33 and viability32, than more conventional optical imaging techniques, such as confocal microscopy33.
Box 1. Multiphoton Laser-Scanning Microscopy (MPLSM).
Multiphoton microscopy possesses several advantages over more conventional microscopy techniques for imaging cells within 3D matrices:
- Short pulses of longer wavelength light (typically 650–1050 nm) are used such that two or more photons (i.e. twice the normal, single-photon, excitation wavelength for 2-photon excitation) are absorbed to excite a fluorophore resulting in reduced scattering and permitting much deeper tissue imaging 26,33. The longer excitation wavelength does not decrease in resolution relative to other optical imaging techniques 102.
- Excitation remains restricted to the plane of focus (optical sectioning) because the probability of two or more photons simultaneously exciting a fluorophore outside the focal volume is extremely low 26,27. Therefore, an aperture (used with confocal microscopy 103–105) to reduce out-of-plane fluorescence is not needed, permitting a much greater emission signal collection efficiency and the use of non-descanned (direct) detection to capture more light, particularly during deep tissue imaging 7,106.
- The pulsed lasers used with multiphoton microscopy use a high peak-power (which also significantly enhances the signal 29), but average power delivered to the sample is quite moderate since the excitation is not continuous; and the long wavelengths used have been shown to be less damaging to tissue. When combined with the lack of fluorescence excitation outside of the focal plane, MPLSM results in significantly less photo-toxicity 32, which is very important for long term cell biology studies, imaging precious live tissue biopsies, or when performing live cell intravital imaging in vivo.
- Another advantage of MPLSM is that it readily generates harmonic signals 107, which differ from fluorescent signals in that harmonic emission signal is coherent (e.g. 2-photon excitation at 900nm for second harmonic generation, SHG, results in a perfect 450nm emission signal). As such, in theory, harmonic generation does not release energy into the sample 107, and is therefore optical noninvasive, allowing long term imaging of biological structures. Strong harmonophores, based on biomolecule arrangement and structure, relevant to cell biology include fibrillar collagen 48–52, myosin, and tubulin 30,48,53. Harmonic signals from these proteins can be easily separated from fluorescent signals with standard filtering technologies as well as measurements of the fluorophore lifetime (since harmonic signals do not undergo fluorescence intensity decay over time).
- Emission signals from multiphoton excitation can be used to determine changes in fluorescence anisotropy or lifetime which can indicate changes in molecular conformation, biochemical microenvironment, and binding state 108. Moreover, fluorescence lifetime imaging microscopy (FLIM 109,110) is particularly useful for conducting fluorescence resonance energy transfer (FRET) experiments since a change in donor or acceptor fluorescence lifetime can be used to indicate FRET independent of the molar concentration of each fluorophore 111–113. Fluorescence lifetime analysis of multiphoton emission signals allows acquisition of fundamental signaling information in cells within 3D microenvironments in vitro or in vivo.
It should be noted, however, that the discussion of deep tissue imaging herein is in the context of optical imaging, where MPLSM provides an effective imaging depth that greatly exceeds more conventional optical imaging approaches, but is still significantly less than current clinical tools that can image through the human body. Standard MPLSM can effectively image to depths of 400–1000 m, depending on the tissue structure25,29, which currently precludes imaging through the skin into human organs, but does not exclude MPLSM as a potential diagnostic technology (see Box 2) or its utility for studying fundamental cell biology in context. With MPLSM, cell biology studies can now be performed on cells within relevant 3D matrices34–36 as well as in vivo with intravital imaging37–40. This allows detailed analysis of cell phenotype, and a quantitative description of cell behavior (i.e. migration speed, direction etc.) within biologically relevant environments. Moreover, MPLSM is particularly well suited to image quantum dots or cells expressing fluorescent protein(s) (while a detailed discussion of fluorescent proteins and biosensors is beyond the scope of this review, the reader is encouraged to examine recent reviews describing this rapidly growing and exciting field, such as41–47 and Lidke and Wilson in this issue). In addition, utilization of fluorescent protein technologies for localization studies or multiple fluorescent labels for protein-protein interaction/activation studies with Förster resonance energy transfer (FRET)42–44 provide novel insight into signal transduction in vivo (see review by Balla, this issue).
Box 2. Diagnostic potential of MPLSM.
Although MPLSM has emerged as a powerful technology to study cell phenotype and associated signal transduction in 3D microenvironments, the clinical capability of MPLSM currently remains limited since the imaging depth is still not adequate for deep organ imaging through the skin. Yet, MPLSM still holds great promise as a diagnostic technology.
- Current work is focusing on imaging live tissue tumor biopsies and stained and unstained histology slides as a way to obtain additional quantitative information to supplement the standard pathology process8,11,12,15,114,115. It is feasible for live tumor biopsies or resected tumors to be rapidly imaged immediately following harvest as a means of initial diagnostic examination before standard processing for histology. Histology samples can then be examined further for quantitative metrics to assist the pathological assessment.
- MPLSM has the potential to be utilized as a diagnostic tool in vivo through multiphoton fluorescence endoscopy116–120, which has sub-cellular resolution118,120, or by imaging skin directly121. Furthermore, recent advances in microlenses may further extend the applicability of these approaches122,123. In particular, gradient refractive index (GRIN) lenses, which can be several centimeters in length and ~1–2 mm in diameter, hold particular promise for extending in vivo imaging with MPLSM 120,122,123.
- MPLSM has been used to effectively detect circulating tumor cells in the small vessels of the peripheral vasculature with multiphoton intravital flow cytometry124. If this technology can be effectively coupled with fluorescent markers of the circulating tumor cells in humans or other markers of cancerous cells in the vasculature it may hold the potential to detect circulating tumor cells through thin regions of skin in human patients.
- Ongoing advances in adaptive optics125 may considerably increase the effective imaging depth of MPLSM by correcting for aberrations that arise due to inhomogeneities in the refractive index within tissues 125–128. Correction for such aberrations will greatly increase the effective imaging depth for MPLSM. When combined with the technologies discussed above, MPLSM may in fact prove to be a very effective tool for imaging thick extracted tumor specimens and in vivo imaging in human patients.
MPLSM is additionally advantageous because it generates harmonic signals, which are not fluorescent, but rather a polarization process with an emission wavelength equal to exactly half of the excitation wavelength48–51, and therefore can be easily separated from fluorescence signals. In the context of cell biology, this results in second and third harmonic generation (SHG and THG, respectively) signals from biological components, such as fibrillar collagen48–52, myosin, and tubulin30,48,53. This proves to be a very useful aspect that can be exploited, since biological materials such as collagen are strong harmonophores; providing a powerful means to simultaneously image collagen and selected fluorophores. This approach thus allows imaging of cells (and relevant intracellular components) within complex 3D collagen matrices to define molecular pathways representative of the in vivo condition, and can also provide diagnostic markers of the ECM that are linked to tumor progression15,34.
Imaging tumor cell motility in 3D microenvironments in vivo and in vitro
Multiphoton laser-scanning microscopy has been used to study cancer cell motility and local invasion in vivo and within 3D microenvironments in vitro to provide novel information about these processes. The studies presented here, while not a comprehensive review of the regulation of these processes in metastasis, demonstrate the link particular cell phenotypes with molecular pathways, providing a potential link between the fundamental cell biology and optical biomarkers.
Tumor cell phenotypes during local invasion and intravasation
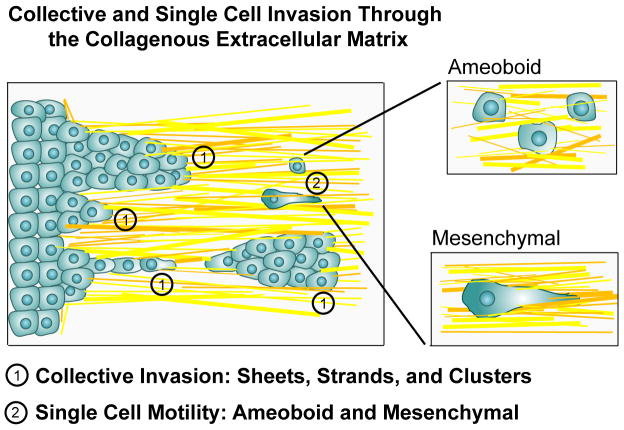
Three primary cell phenotypes have been reported for cells migration through the ECM during local invasion: 1) amoeboid, 2) mesenchymal (also referred to as fibroblastoid), and 3) collective migration depending on tumor type, stage, and location relative to the primary tumor mass or vasculature (Figure 1). Fundamental work by Condeelis and colleagues54 using intravital multiphoton microscopy identified key differences between well-described motility phenotypes on 2D substrates and the mechanisms of cell motility during metastasis in vivo. Notably, they observed migrating cancer cells moving along collagen fibers toward and into blood vessels with rapid shape changes and no cell polarity, characterized as amoeboid motility37,54, which is distinct from fibroblastic migration where cells are elongated and form a well defined lamellapodia37. Furthermore, MPLSM imaging of live tumors and carcinoma cells in 3D culture show features of collective invasion (sheets, strands, and tubes) in regions of local invasion, as well as single cell mesenchymal and amoeboid phenotypes, in contact with aligned collagen fibers 8,15,34,36,55–58. Each of these phenotypes has been linked to regulation of specific signal transduction networks. A brief description of these findings follows.
Figure 1. Cell phenotypes associated with invasion through the stromal matrix.
In vivo MPLSM imaging of tumor cells in mice, as well as MPLSM of tumor cells in relevant 3D matrices, have identified three distinct cell phenotypes: collective invasion, single cell amoeboid migration, and 3D migration of cells with a mesenchymal phenotype. The presence or absence of these phenotypes largely depends on tumor type, the state of the stromal ECM, and the cellular composition of the microenvironment. Collective migration (1), where cells remain physically connected to each other and have a multicellular polarity 58, can present as sheets of cells with a leading edge, lines of cells or groups of cells, or as a collective group that has detached from the primary tumor. These groups of tumor cells have focalized 3D-matrix adhesions, generate traction force and have an active proteolytic program to modify the ECM. Amoeboid motility (2a) is characterizes by a rounded or ellipsoid morphology, a lack of cell polarity, and non-focalized 3D-matrix adhesions to facilitate high velocity movement through the matrix. Mesenchymal migration (2b) is distinguished by an elongated morphology, focalized 3D-matrix adhesions and an active proteolytic program. Each of these phenotypes are regulated by unique molecular programs. Furthermore, it is clear that invasive cells have multiple compensatory mechanisms to switch between these phenotype in order to efficiently migrate through collagenous matrices. For in depth discussions of these phenotypes the readers in encouraged to examined references 37,58,137.
Elucidating the influence of extracellular proteases with MPLSM
Interestingly, amoeboid motility phenotypes were also identified during migration within 3D matrices in vitro in a protease-dependent manner56,59. In 3D collagen matrices, broadly inhibiting multiple protease families results in a switch from mesenchymal motility, which is characterized by strong β1-integrin localization at the cell-matrix interface56,60 co-localized with active MT1-MMP (MMP14)56, to an effective amoeboid-like migration, termed a mesenchymal-amoeboid transition56. Intravital multiphoton microscopy of fibrosarcoma cells treated with protease inhibitors and then injected into the mouse dermis supports conclusions from in vitro 3D matrix experiments, suggesting a mechanism for protease-independent amoeboid movement in vivo56. Moreover, MPLSM imaging of collagen fibrils in 3D reconstituted collagen matrices demonstrated that broad inhibition of proteases did not inhibit the ability of the cell to reorganize the matrix or invade into the matrix34,36. However, it is generally accepted that in vivo, before invasive cells can break free of the primary tumor mass and invade into the stroma, the physical ECM boundary at the tumor-stroma interface must be broken down and/or reorganized61,62. A need for protease activity in vivo makes intuitive sense when one observes the tumor boundary using SHG imaging, which shows that non-invasive regions of tumors are confined by collagen fibers that are arranged parallel to the tumor boundary8,15 (which is also an optical biomarker used when inspecting for the presence or absence of local invasion; see Box 3 and Figure 2). Thus, while multiple studies have presented strong evidence demonstrating that tumor cells can migrate through the collagen matrix in the absence of protease activity34,36,56,59, most of this work has been performed in 3D culture in vitro or on cells injected into mouse models.
Box 3. Optical Biomarkers.
Optical biomarkers, the detection of changes in cellular behavior, phenotype, or physical-chemical properties, and/or changes in the extracellular matrix composition and architecture by optical imaging technologies, that can consistently quantify cellular and molecular changes associated with cancer have diagnostic potential.
Metabolic Markers:
- Due to changes in metabolic state associated with increased glycolysis in tumors, detecting changes in fluorescence intensity and/or fluorescence lifetime for metabolic intermediates such as NADH and FAD, and their redox ratio can provide information indicating cellular transformation or subpopulations of tumor cells with metastatic potential 8–13
Stromal Collagen Markers:
Deposition of collagen surrounding tumors has been noted by pathologists, yet only recently has the functional significance of desmoplasia and collagen architecture been investigated in vivo. Significant changes in the collagen matrix occur with tumor initiation and progression, which can be used as optical biomarkers:
- Tumor-Associated Collagen Signatures (TACS) 8,15: To date three TACS have been defined related to collagen architecture:
- TACS-1 is the presence of locally dense collagen within the globally increased collagen concentration surrounding tumors, identified by increased signal intensity at a region near the tumor, which serves as a reliable hallmark for locating small tumor regions.
- TACS-2 is straightened collagen fibers stretched around the tumor (i.e. tumor expansion that strains the fibers resulting in the loss of the wavy collagen crimp pattern). The collagen is organized parallel to the tumor boundary and is associated with non-invasive regions of the tumor mass.
- TACS-3 is collagen that has been re-organized so that it is radially aligned (perpendicular) to the tumor-stroma boundary and is characteristic of regions of local invasion.
- Changes in collagen structure due to degradation (resulting from extracellular protease activity), altered fibril composition, and/or defective collagen fibril formation and the associated changes in SHG signal scattering may also be indicative of changes in the stroma that point to tumor initiation or tumor progression. Because the SHG signal is sensitive to fibril organization and structure, changes in the scatter properties of the SHG signal can be used to determine changes in collagen structure. As an example of this, collagen structure is abnormal in connective tissue disorders such as osteogenesis imperfecta (OI). Dermis from a mouse model of OI demonstrates significant differences in forward/backward scatter of the collagen SHG signal compared to wild-type tissue129. Thus, quantitative analysis of scattering can be used as an optical biomarker, particularly since changes in collagen structure can influence invasion of cancer cells 130.
- Intermolecular cross-links of collagen are fluorescent and can be imaged with MPLSM131. Tissue transglutaminase and lysyl oxidase, both of which cross-link collagen, are altered in tumors132,133, and collagen crosslinking may influence cancer cell invasion61. Thus, autofluorescence signals from collagen cross-links can be readily detected with MPLSM and therefore represents another potential optical biomarker for tumor progression.
Additional Optical Biomarkers:
While collagen and the metabolites NADH and FAD have been the best characterized intrinsic fluorophores that are of relevance to cancer, there are many other intrinsic fluorophores that warrant further study as being of benefit to cancer research (and are perhaps future biomarkers) such as Protoporphyrin IX134,135 (component of the Heme pathway), lipofuscin91,136, elastin91, and tryptophan91,93.
Figure 2. Optical biomarkers detected with MPLSM.
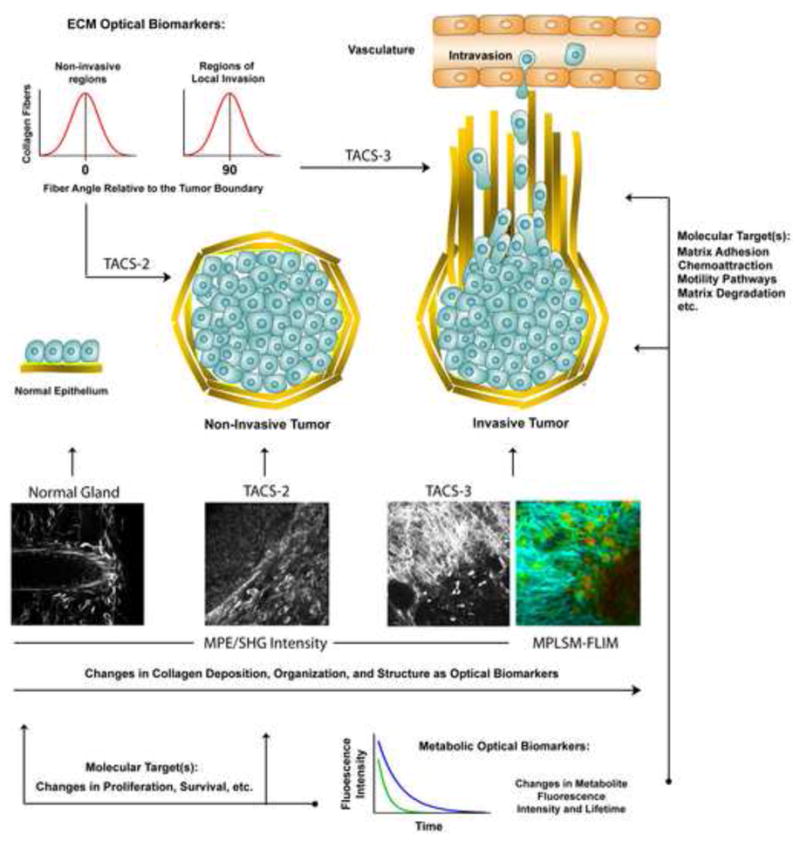
Multiphoton microscopy allows the study of single cell behaviors relevant to metastasis in 3D microenvironments in vitro and in vivo. MPLSM imaging live tumor cells has not only provided new information about the cell biology of metastasis, but has also provided novel optical biomarkers that have diagnostic potential. For instance, changes in the collagen matrix architecture, termed tumor associated collagen signatures (TACS), have been described that help distinguish invasive from non-invasive tumor regions (i.e. TACS-2 vs. TACS-3). Shown are MPLSM micrographs (multiphoton excitation of endogenous fluorescence from FAD with simultaneous SHG of the collagen matrix) of live, intact, mammary gland (i), non-invasive regions of mammary tumor (ii; TACS-2), and invasive regions of mammary tumor (iii; TACS-3; See Box 3). As highlighted in the associated cartoons, the collagen fiber angle distributions associated with these matrix morphologies can be quantified (i.e. fiber angle distribution diagrams shown in the upper left corner), and used to quantitatively distinguish regions of invasion from non-invasive regions of the tumor. Furthermore, changes in cell metabolism that are associated with transformation and tumor progression can be detected in single tumor cells and provide an optical biomarker for disease state (e.g. the MPLSM-FLIM image of TACS-3 showing differences in the fluorescence lifetime of FAD in invading cells versus cells in the primary tumor mass; note the difference in heat map color and intensity in cells in the stroma versus the primary tumor mass). When linked to defined signal transduction pathways, optical biomarkers such as these may help guide therapeutic intervention by indicating whether targeted therapy against a particular pathway is relevant.. Collagen is depicting in yellow and basement membrane in the normal gland is illustrated in light green. MPLSM and MPLSM-FLIM images are adapted and reproduced from Provenzano et al., 15, with permission.
In vivo, a defined matrix architecture that results from normal development is in place, and becomes stretched by the growing tumor, and is later reorganized and aligned, facilitating local invasion into the stroma15. In live tumors, MPLSM imaging reveals that this transition occurs over time (weeks to months in rapidly progressing mouse tumor models; which may indicate changes over months to years in human patients). Therefore, it seems likely that a remodeling of the collagen matrix may be required61,63,64 to aid matrix re-alignment that facilitates invasion. Moreover, recent work by Sabeh and co-workers61 suggests that part of the discrepancy between protease-dependent and protease-independent 3D migration may be influenced by the preparation of the collagen matrix; in particular, whether or not the ECM is crosslinked, and whether or not a path for the cell has already been created55,61,65, suggesting that more work needs to be performed in native environments in vivo. Combined, these reports advocate studies to understand proteolytic degradation of the ECM in vivo that will likely require imaging single cells at the invasion boundary, along with the tumor-associated cells of the microenvironment, and the collagen fibers adjacent to the invading cells in endogenous tumors over time in order to understand the process. However, in vivo studies also provide strong evidence that an amoeboid phenotype exists in tumors36,37,54,56, and suggests that the invading cells may not be susceptible to protease inhibition therapy. As such, the detection of ameoboid phenotypes in biopsy section may guide decisions regarding the therapeutic regime.
Molecular pathways that regulate motility: Therapeutic Targets?
Sahai and co-workers identified a link between the mechanism for migration through 3D matrices and the Rho/ROCK signaling pathway59. In rounded, dynamically blebbing cells analogous to amoeboid phenotypes, blockade of Rho/ROCK signaling is significantly more effective at reducing 3D migration than in cell lines with elongated phenotypes59; and knockdown of Smurf1, a ubiquitin ligase that can target RhoA for degradation, increases RhoA activity and promotes tumor cell motility through a mesenchymal-amoeboid transition66, suggesting that in contrast to protease therapy, cells with amoeboid phenotypes may be susceptible to targeted inhibition of the Rho pathway. Importantly, it has recently been established using MPLSM that Rho/ROCK signaling and resulting acto-myosin regulated contractile force are necessary for local collagen matrix deformation/reorganization near the cell boundary34,36. This matrix reorganization near the cell boundary provides physical contact guidance cues that strongly influence cell migration through 3D matrices34,67,68. Interestingly, when the collagen matrix is engineered to mimic matrix architecture that most efficiently promotes 3D migration, Rho and ROCK become unnecessary for migration of invasive breast carcinoma cells through the collagen matrix34, suggesting that the role of Rho-mediated intracellular contractile force during 3D migration may be more related to organizing the microenvironment than the movement of single motile cells.
In regions of local invasion, MPLSM imaging has revealed that collective invasion takes place at regions of matrix re-alignment along with single motile cells progressing deep into the aligned stroma8,15,34. This may be especially true for a subpopulation of individual tumor cells that may be migrating particularly fast if they have been strongly stimulated by paracrine signaling from tumor-associated macrophages37,69–71 or provided with a migration promoting ECM architecture or paracrine stimulation by tumor-associated fibroblasts65,72. Chemotactic gradients can enhance the invasion process by stimulating intracellular signaling networks associated with cell motility. Microarray analysis of invasive MTLn3 rat adenocarcinoma cells in vivo (compared to the non-metastatic MTC cell line), which were used to identify amoeboid movement with MPLSM for cells migrating along collagen toward blood vessels54, identified the growth factor regulated (EGF and CSF-1) ‘minimum motility pathway’54,73,74. This pathway is characterized by regulation of the actin cytoskeleton and in particular three end-stage effectors, Arp2/3 complex, capping protein, and cofilin, that influence actin dynamics associated with cell motility73,75,76, and directly link an in vivo cell phenotype identified by MPLSM with molecular targets. Furthermore, Mena, an Ena/Vasp protein that antagonizes actin capping, is upregulated54,73,74, and intravital multiphoton imaging experiments showed that cells overexpressing EGFP-Mena in orthotopic xenografts are more motile77. Interestingly an isoform of Mena (termed MenaINV) enhances sensitivity EGF stimulation and promotes EGF-induced cell motility and invasion, suggesting that these cells are more likely to metastasize even in the presence of basal levels of EGF77. Hence, it is clear that invasive cells have multiple compensatory mechanisms to efficiently migrate through collagenous matrices, highlighting the need for a detailed understanding of the link between cell phenotype and active signal transduction for identifying cell populations that may or may not be susceptible to a particular therapeutic intervention.
The need to image 3D-matrix adhesions in vivo
In addition to chemoattraction, integrin-mediated adhesion is known to play a role in tumor progression to metastasis78. While matrix adhesions in motile cells in 3D environments are not well understood largely due to a current lack of high-resolution imaging studies of these structures relative to the ECM microenvironment in vivo, and are likely less developed than large focal adhesions that form on stiff 2D substrates, current evidence strongly suggests a role for integrins and focal adhesion proteins in regulating metastasis-associated motility in vivo. For instance, focal adhesion kinase (FAK) is a well-described regulator of focal adhesions and cell motility79, is overexpressed in numerous human cancers80, and its loss has been shown to block malignant conversion81,82. In addition, FAK has been shown to regulate invasion by influencing urokinase plasminogen activator (uPA) activity83, which may result in changes to the ECM that are detectable with MPLSM. Moreover, FAK deletion represses a number of transcripts associated with adhesion, motility, and the cytoskeleton (many of which are ‘minimum motility pathway’ genes identified by MPLSM imaging and simultaneous collection of invasive cells in vivo54,73), which suggests FAK regulates actin dynamics and cell protrusion and motility81. In addition, FAK-null cells injected into the vasculature cannot efficiently form protrusions across the vessel wall that are necessary for them to extravasate84. Hence, adhesion complexes and regulators of the actin cytoskeleton are emerging as viable targets for metastasis therapy. Yet, while FAK has been associated with regulating a subset of genes that are predictive of human patient outcome81, and a number of gene signatures have been compiled to predict patient outcome85–88, specific quantitative information linking cell and/or matrix phenotype to specific cells bearing these signatures is currently lacking. Therefore, studies that can identify optical biomarkers and signal transduction pathways/gene signatures within invading cells are of great value not only in increasing our basic understanding of tumor biology, but also providing information that can be used in a diagnostic capacity to guide therapeutic intervention.
Optical Biomarkers
In addition to the use of exogenous fluorescent markers, such as GFP, Quantum dots, etc., to track molecular events in cells, a set of “built-in” endogenous fluorophores exists that can be studied to further our understanding of normal and pathologic processes. These molecules have the advantage of being naturally present in cells and tissues of interest, are involved in key biological processes, and therefore can be imaged in live, unstained tissue. Not only are these endogenous fluorophores of great use in various cell culture and animal models, but will ultimately be of use as biomarkers of disease state in human samples where fluorescently-labeled proteins are not available.
Metabolic intermediates
Although it has long been known that glycolysis is increased in tumors89,90, specific metabolic intermediates are emerging as intrinsic optical biomarkers of these changes. In particular, the metabolites nicotinamide adenine dinucleotide (NADH; reduced form) and flavin adenine dinucleotide (FAD; oxidized form) have characteristic fluorescent properties and spectral emissions that have been well characterized by spectroscopy91–93, and are detectable in live, unstained cells10,11. Moreover, changes in the redox state of the cell can be determined by imaging the relative intensity of NADH and FAD12,94, which correlates linearly with glucose uptake in tumor models95.
In addition to intensity changes, fluorescent lifetime microscopy (FLIM; Box 1) can be exploited to detect changes in metabolic state that accompany tumor progression. The fluorescence lifetime of protein-bound vs. free NADH or of FAD differ, and can be distinguished by FLIM. This approach has been used to discern precancerous lesions from normal tissue11, and invasive mammary carcinoma cells from non-invasive cells8. Specifically, tumor cells have a higher NADH and FAD fluorescent intensity and a longer fluorescent lifetime than normal cells11. Moreover, the fluorescence lifetime for FAD is longer in invasive cells than cells in the primary tumor mass, suggesting changes that accompany carcinoma progression8. Unexpectedly, these fluorophores are preserved in fixed, paraffin-embedded tissue, where the fluorescence intensity and lifetime of NADH and FAD discerns tumor from normal mammary epithelium in mouse samples11. Thus, application of MPLSM/MPLSM-FLIM to human pathological samples or fresh biopsy tissues may allow the readout of metabolic states, and may help further differentiate subsets of patients, or suggest more targeted therapies. For example, as recently reviewed by Thompson and co-workers90 the phosphoinositide 3-kinase (PI3K) pathway (Akt, the tumor suppressor PTEN etc..), which is important in many human cancers and a target for therapeutic intervention96, can regulate cell proliferation and motility, as well as glucose metabolism. AKT, a downstream kinase of PI3K, can regulate glucose transporter expression and capture, even in cells that are not insulin dependent90,97. Findings such as these suggest that optical detection of changes in fluorescent lifetimes of endogenous fluorophores such as NADH and FAD, which indicate altered glycolysis, may indicate whether targeted therapy against the PI3K pathway is relevant. Moreover, oncogenes such as Myc and Ras, and tumor suppressors such as p53 either cause or are sensitive to changes in glucose or glutamine metabolism90. Hence, a real-time understanding of metabolic state in single cells can not only be an indicator of transformation or metastatic potential, but optical biomarkers of metabolism, particularly when combined with information about single cell phenotype, may be useful for identifying particular oncogenic signaling pathways important in human cancer.
Collagen changes in pathology
For many years, the deposition of collagen surrounding tumors, termed “desmoplasia,” has been noted by pathologists, yet only recently has the functional significance of desmoplasia been investigated. Importantly, collagen is perhaps the most readily imaged endogenous molecule, as fibrillar collagen is a strong harmonophore48–52, and therefore changes in collagen within mammary15,98,99 and ovarian12 tumors with SHG can be detected.
As described above, visualization of collagen surrounding mammary tumors demonstrates a characteristic sequence in the intensity, organization and alignment of collagen that accompanies tumor progression15 (see Figure 2, Box 3). These “tumor associated collagen signatures,” or TACS, are potentially relevant optical biomarkers, which is consistent with general collagen changes in human breast cancer detected by SHG99. Because aligned collagen fibers in animal models and in 3D cell culture are associated with and facilitate local invasion, these studies link collagen organization to cellular behavior and specific signal transduction pathways, such as Rho/Rock-dependent matrix reorganization34; and suggest that inhibition of intracellular contractility in a non-invasive primary tumor mass may help block conversion to an invasive tumor. Moreover, the observation that locally invasive regions near the tumor mass have a collective migration phenotype suggests contractile and proteolytically active behavior15,34,55,60,61 that could be targeted with a treatment regime specific to the mesenchymal phenotype. Furthermore, single cells invading away from the primary tumor mass can show mesenchymal or amoeboid phenotypes that may depend on the matrix architecture61 and the mechanism of chemoattraction37,69, and therefore understanding the matrix architecture and the cell phenotype could direct therapeutic intervention as well (i.e. amoeboid motility may be targeted for pieces of the ‘minimum motility pathway’54,73,74,77). Hence, it is becoming clear that a real-time understanding of matrix architecture and the corresponding cell behavior in vivo or in situ with optical imaging is of great utility in cancer biology. Moreover, as our understanding of the molecular basis of invasion into 3D matrices increases, future therapies targeted at these events will hopefully emerge, and would be of particular benefit to patients with an “invasive” optical biomarker.
The further development of collagen as a biomarker of tumor progression, will benefit from additional visualization tools to make measurements of intensity and alignment readily quantified, such as algorithms to identify collagen structures in SHG images and color map their angle or scatter properties. In a broader sense, because collagen is the most abundant protein in the body of most vertebrates100, the potential to use collagen as a means to depict tissue architecture in the context of a wide variety of cell biological, developmental, and pathological investigations in model organisms is great.
Emerging and Future Applications
Because optical biomarkers, such as NADH, FAD, and collagen, have relevance to carcinoma progression and potential to be linked to relevant signal transduction cascades they are potential read outs for high-throughput screens. For example, because collagen matrix alignment facilitates invasion8,15, and can be recapitulated in 3D culture34, SHG imaging and analysis of collagen alignment by cells can be used to find small molecules that may prevent this process. Already, we know that this is a Rho-mediated contractility event34, and molecules that inhibit the Rho pathway may be effective in preventing carcinoma progression. Similarly, if the tumor-associated switch to glycolysis is recapitulated in cells or tissue explants in culture, then imaging of NADH and FAD may be a useful end-point for screens of small molecules to prevent the glycolytic switch.
Quantitative techniques and computational tools to standardize and quantify complex datasets from 4D (x-y-z-time) imaging101, will greatly aid our ability to understand fundamental questions in cancer biology, but will also be extremely important for accurately identifying optical biomarkers associated with disease (see Swedlow & Eliceiri, this issue). Once an optical biomarker (or a consensus panel of biomarkers) is established for a particular human cancer, the link between biomarker and molecular mechanism may be elucidated, and with the appropriate detection technology, analytical tools, and drug screens, a personalized tumor-phenotype-specific treatment regime may be in hand. Therefore, the information we gain from optical imaging studies in animal and culture models of human cancer will not only provide insight into the mechanisms of disease, but hold great promise for improving disease outcome.
Because of its advantages for imaging in tissue, the use of MPLSM as a tool for the cell biologist to advance our understanding of cancer progression in animal models is likely to expand, as additional analysis approaches, instrumentation, and expertise become more commonly available. Already, MPLSM has assisted in our understanding of cell invasion and metastasis, and captured single cells as they invade in vivo. In addition to cancer biology, these imaging approaches are likely to illuminate many normal developmental and pathological processes in vivo. Moreover, MPSLM has great potential to become of use as a clinical diagnostic tool (see Box 2) to take advantage of optical biomarkers in fresh tissue biopsies even as a patient is undergoing surgery, or in classic histopathology samples.
Acknowledgments
We apologize to those authors whose work we were unable to cite because of space and date of publication limitations. We thank members of the Keely laboratory and the Laboratory for Optical and Computational Instrumentation (LOCI) for helpful discussions regarding this work. This work was supported by a NIH postdoctoral training grant (T32CA009681) to PPP, and grants from the DOD: W81XWH-04-1-042 (PPP), the Mary Kay Ash Foundation (PJK), DOD CDMRP BC074970 (PJK), Am. Cancer Society RSG-00-339CSM (PJK), NIH CA076537 (PJK), and NIH EB000184 (KWE).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sahai E. Illuminating the metastatic process. Nat Rev Cancer. 2007;7 (10):737–749. doi: 10.1038/nrc2229. [DOI] [PubMed] [Google Scholar]
- 2.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127 (4):679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17 (5):545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 4.Weissleder R. Scaling down imaging: molecular mapping of cancer in mice. Nat Rev Cancer. 2002;2 (1):11–18. doi: 10.1038/nrc701. [DOI] [PubMed] [Google Scholar]
- 5.Gambhir SS. Molecular imaging of cancer with positron emission tomography. Nat Rev Cancer. 2002;2 (9):683–693. doi: 10.1038/nrc882. [DOI] [PubMed] [Google Scholar]
- 6.Weissleder R, Ntziachristos V. Shedding light onto live molecular targets. Nat Med. 2003;9 (1):123–128. doi: 10.1038/nm0103-123. [DOI] [PubMed] [Google Scholar]
- 7.Provenzano PP, et al. Multiphoton microscopy and fluorescence lifetime imaging microscopy (FLIM) to monitor metastasis and the tumor microenvironment. Clin Exp Metastasis. 2009;26 (4):357–370. doi: 10.1007/s10585-008-9204-0. [DOI] [PubMed] [Google Scholar]
- 8.Provenzano PP, et al. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008;6 (1):11. doi: 10.1186/1741-7015-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skala MC, et al. In vivo multiphoton fluorescence lifetime imaging of protein-bound and free nicotinamide adenine dinucleotide in normal and precancerous epithelia. J Biomed Opt. 2007;12 (2):024014. doi: 10.1117/1.2717503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skala MC, et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc Natl Acad Sci U S A. 2007;104 (49):19494–19499. doi: 10.1073/pnas.0708425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conklin MW, et al. Fluorescence lifetime imaging of endogenous fluorophores in histopathology sections reveals differences between normal and tumor epithelium in carcinoma in situ of the breast. Cell Biochem Biophys. 2009;53 (3):145–157. doi: 10.1007/s12013-009-9046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirkpatrick ND, et al. Endogenous optical biomarkers of ovarian cancer evaluated with multiphoton microscopy. Cancer Epidemiol Biomarkers Prev. 2007;16 (10):2048–2057. doi: 10.1158/1055-9965.EPI-07-0009. [DOI] [PubMed] [Google Scholar]
- 13.Georgakoudi I, et al. NAD(P)H and collagen as in vivo quantitative fluorescent biomarkers of epithelial precancerous changes. Cancer Res. 2002;62 (3):682–687. [PubMed] [Google Scholar]
- 14.Ramanujam N. Fluorescence spectroscopy of neoplastic and non-neoplastic tissues. Neoplasia. 2000;2 (1–2):89–117. doi: 10.1038/sj.neo.7900077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Provenzano PP, et al. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006;4 (1):38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hompland T, et al. Second-harmonic generation in collagen as a potential cancer diagnostic parameter. J Biomed Opt. 2008;13 (5):054050. doi: 10.1117/1.2983664. [DOI] [PubMed] [Google Scholar]
- 17.Tang S, et al. Combined multiphoton microscopy and optical coherence tomography using a 12-fs broadband source. J Biomed Opt. 2006;11 (2):020502. doi: 10.1117/1.2193428. [DOI] [PubMed] [Google Scholar]
- 18.Anandasabapathy S. Endoscopic imaging: emerging optical techniques for the detection of colorectal neoplasia. Curr Opin Gastroenterol. 2008;24 (1):64–69. doi: 10.1097/MOG.0b013e3282f2df8d. [DOI] [PubMed] [Google Scholar]
- 19.Wilder-Smith P, et al. Noninvasive imaging of oral premalignancy and malignancy. J Biomed Opt. 2005;10 (5):051601. doi: 10.1117/1.2098930. [DOI] [PubMed] [Google Scholar]
- 20.Egeblad M, et al. Visualizing stromal cell dynamics in different tumor microenvironments by spinning disk confocal microscopy. Dis Model Mech. 2008;1(2–3):155–167. doi: 10.1242/dmm.000596. discussion 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graf R, et al. Live cell spinning disk microscopy. Adv Biochem Eng Biotechnol. 2005;95:57–75. doi: 10.1007/b102210. [DOI] [PubMed] [Google Scholar]
- 22.Wang LV. Prospects of photoacoustic tomography. Med Phys. 2008;35 (12):5758–5767. doi: 10.1118/1.3013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang HF, et al. Functional photoacoustic microscopy for high-resolution and noninvasive in vivo imaging. Nat Biotechnol. 2006;24 (7):848–851. doi: 10.1038/nbt1220. [DOI] [PubMed] [Google Scholar]
- 24.Jhan JW, et al. Integrated multiple multi-photon imaging and Raman spectroscopy for characterizing structure-constituent correlation of tissues. Opt Express. 2008;16 (21):16431–16441. doi: 10.1364/oe.16.016431. [DOI] [PubMed] [Google Scholar]
- 25.Helmchen F, Denk W. New developments in multiphoton microscopy. Curr Opin Neurobiol. 2002;12 (5):593–601. doi: 10.1016/s0959-4388(02)00362-8. [DOI] [PubMed] [Google Scholar]
- 26.Denk W, et al. Two-photon laser scanning fluorescence microscopy. Science. 1990;248 (4951):73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 27.Diaspro A, Sheppard CJR. Two-Photon Excitation Fluorescence Microscopy. In: Diaspro A, editor. Confocal and Two-Photon Microscopy: Foundations, Applications, and Advances. Wiley-Liss, Inc; 2002. pp. 39–73. [Google Scholar]
- 28.Zipfel WR, et al. Nonlinear magic: multiphoton microscopy in the biosciences. Nat Biotechnol. 2003;21 (11):1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 29.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Methods. 2005;2 (12):932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 30.Mohler W, et al. Second harmonic generation imaging of endogenous structural proteins. Methods. 2003;29 (1):97–109. doi: 10.1016/s1046-2023(02)00292-x. [DOI] [PubMed] [Google Scholar]
- 31.Hockberger PE, et al. Activation of flavin-containing oxidases underlies light-induced production of H2O2 in mammalian cells. Proc Natl Acad Sci U S A. 1999;96 (11):6255–6260. doi: 10.1073/pnas.96.11.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Squirrell JM, et al. Long-term two-photon fluorescence imaging of mammalian embryos without compromising viability. Nat Biotechnol. 1999;17 (8):763–767. doi: 10.1038/11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Centonze VE, White JG. Multiphoton excitation provides optical sections from deeper within scattering specimens than confocal imaging. Biophys J. 1998;75 (4):2015–2024. doi: 10.1016/S0006-3495(98)77643-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Provenzano PP, et al. Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization. Biophys J. 2008;95 (11):5374–5384. doi: 10.1529/biophysj.108.133116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee PF, et al. Nonlinear optical microscopy reveals invading endothelial cells anisotropically alter three-dimensional collagen matrices. Exp Cell Res. 2009;315 (3):396–410. doi: 10.1016/j.yexcr.2008.10.040. [DOI] [PubMed] [Google Scholar]
- 36.Wyckoff JB, et al. ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr Biol. 2006;16 (15):1515–1523. doi: 10.1016/j.cub.2006.05.065. [DOI] [PubMed] [Google Scholar]
- 37.Condeelis J, Segall JE. Intravital imaging of cell movement in tumours. Nat Rev Cancer. 2003;3 (12):921–930. doi: 10.1038/nrc1231. [DOI] [PubMed] [Google Scholar]
- 38.Brown EB, et al. In vivo measurement of gene expression, angiogenesis and physiological function in tumors using multiphoton laser scanning microscopy. Nat Med. 2001;7 (7):864–868. doi: 10.1038/89997. [DOI] [PubMed] [Google Scholar]
- 39.Jain RK, et al. Dissecting tumour pathophysiology using intravital microscopy. Nat Rev Cancer. 2002;2 (4):266–276. doi: 10.1038/nrc778. [DOI] [PubMed] [Google Scholar]
- 40.Ahmed F, et al. GFP expression in the mammary gland for imaging of mammary tumor cells in transgenic mice. Cancer Res. 2002;62 (24):7166–7169. [PubMed] [Google Scholar]
- 41.Shaner NC, et al. A guide to choosing fluorescent proteins. Nat Methods. 2005;2 (12):905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 42.Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science. 2003;300 (5616):87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 43.Lippincott-Schwartz J, et al. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2 (6):444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- 44.Giepmans BN, et al. The fluorescent toolbox for assessing protein location and function. Science. 2006;312 (5771):217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 45.Sabouri-Ghomi M, et al. Visualizing and quantifying adhesive signals. Curr Opin Cell Biol. 2008;20 (5):541–550. doi: 10.1016/j.ceb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsien RY. Building and breeding molecules to spy on cells and tumors. FEBS Lett. 2005;579 (4):927–932. doi: 10.1016/j.febslet.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 47.Smith AM, et al. Bioconjugated quantum dots for in vivo molecular and cellular imaging. Adv Drug Deliv Rev. 2008;60 (11):1226–1240. doi: 10.1016/j.addr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campagnola PJ, et al. Three-dimensional high-resolution second-harmonic generation imaging of endogenous structural proteins in biological tissues. Biophys J. 2002;82 (1 Pt 1):493–508. doi: 10.1016/S0006-3495(02)75414-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cox G, et al. 3-dimensional imaging of collagen using second harmonic generation. J Struct Biol. 2003;141 (1):53–62. doi: 10.1016/s1047-8477(02)00576-2. [DOI] [PubMed] [Google Scholar]
- 50.Williams RM, et al. Interpreting second-harmonic generation images of collagen I fibrils. Biophys J. 2005;88 (2):1377–1386. doi: 10.1529/biophysj.104.047308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoller P, et al. Polarization-dependent optical second-harmonic imaging of a rat-tail tendon. J Biomed Opt. 2002;7 (2):205–214. doi: 10.1117/1.1431967. [DOI] [PubMed] [Google Scholar]
- 52.Zoumi A, et al. Imaging cells and extracellular matrix in vivo by using second-harmonic generation and two-photon excited fluorescence. Proc Natl Acad Sci U S A. 2002;99 (17):11014–11019. doi: 10.1073/pnas.172368799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plotnikov SV, et al. Characterization of the myosin-based source for second-harmonic generation from muscle sarcomeres. Biophys J. 2006;90 (2):693–703. doi: 10.1529/biophysj.105.071555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang W, et al. Single cell behavior in metastatic primary mammary tumors correlated with gene expression patterns revealed by molecular profiling. Cancer Res. 2002;62 (21):6278–6288. [PubMed] [Google Scholar]
- 55.Wolf K, et al. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nature Cell Biology. 2007;9 (8):893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 56.Wolf K, et al. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160 (2):267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sahai E, et al. Simultaneous imaging of GFP, CFP and collagen in tumors in vivo using multiphoton microscopy. BMC Biotechnol. 2005;5:14. doi: 10.1186/1472-6750-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10 (7):445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 59.Sahai E, Marshall CJ. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol. 2003;5 (8):711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- 60.Hegerfeldt Y, et al. Collective Cell Movement in Primary Melanoma Explants: Plasticity of Cell-Cell Interaction, {beta}1-Integrin Function, and Migration Strategies. Cancer Res. 2002;62 (7):2125–2130. [PubMed] [Google Scholar]
- 61.Sabeh F, et al. Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J Cell Biol. 2009;185 (1):11–19. doi: 10.1083/jcb.200807195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rowe RG, Weiss SJ. Breaching the basement membrane: who, when and how? Trends Cell Biol. 2008;18 (11):560–574. doi: 10.1016/j.tcb.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 63.Hotary KB, et al. Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell. 2003;114 (1):33–45. doi: 10.1016/s0092-8674(03)00513-0. [DOI] [PubMed] [Google Scholar]
- 64.Sabeh F, et al. Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J Cell Biol. 2004;167 (4):769–781. doi: 10.1083/jcb.200408028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gaggioli C, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9 (12):1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 66.Sahai E, et al. Smurf1 regulates tumor cell plasticity and motility through degradation of RhoA leading to localized inhibition of contractility. J Cell Biol. 2007;176 (1):35–42. doi: 10.1083/jcb.200605135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guido S, Tranquillo RT. A methodology for the systematic and quantitative study of cell contact guidance in oriented collagen gels. Correlation of fibroblast orientation and gel birefringence. J Cell Sci. 1993;105 (Pt 2):317–331. doi: 10.1242/jcs.105.2.317. [DOI] [PubMed] [Google Scholar]
- 68.Dickinson RB, et al. Biased cell migration of fibroblasts exhibiting contact guidance in oriented collagen gels. Ann Biomed Eng. 1994;22 (4):342–356. doi: 10.1007/BF02368241. [DOI] [PubMed] [Google Scholar]
- 69.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124 (2):263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 70.Goswami S, et al. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 2005;65 (12):5278–5283. doi: 10.1158/0008-5472.CAN-04-1853. [DOI] [PubMed] [Google Scholar]
- 71.Wyckoff J, et al. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64 (19):7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 72.Bhowmick NA, et al. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432 (7015):332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang W, et al. Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Res. 2007;67 (8):3505–3511. doi: 10.1158/0008-5472.CAN-06-3714. [DOI] [PubMed] [Google Scholar]
- 74.Wang W, et al. Tumor cells caught in the act of invading: their strategy for enhanced cell motility. Trends Cell Biol. 2005;15 (3):138–145. doi: 10.1016/j.tcb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 75.Wang W, et al. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J Cell Biol. 2006;173 (3):395–404. doi: 10.1083/jcb.200510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghosh M, et al. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304 (5671):743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- 77.Philippar U, et al. A Mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis. Dev Cell. 2008;15 (6):813–828. doi: 10.1016/j.devcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Guo W, Giancotti FG. Integrin signalling during tumour progression. Nat Rev Mol Cell Biol. 2004;5 (10):816–826. doi: 10.1038/nrm1490. [DOI] [PubMed] [Google Scholar]
- 79.Mitra SK, et al. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6 (1):56–68. doi: 10.1038/nrm1549. [DOI] [PubMed] [Google Scholar]
- 80.Golubovskaya VM, et al. Focal adhesion kinase and cancer. Histol Histopathol. 2009;24 (4):503–510. doi: 10.14670/HH-24.503. [DOI] [PubMed] [Google Scholar]
- 81.Provenzano PP, et al. Mammary epithelial-specific disruption of focal adhesion kinase retards tumor formation and metastasis in a transgenic mouse model of human breast cancer. Am J Pathol. 2008;173 (5):1551–1565. doi: 10.2353/ajpath.2008.080308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McLean GW, et al. Specific deletion of focal adhesion kinase suppresses tumor formation and blocks malignant progression. Genes Dev. 2004;18 (24):2998–3003. doi: 10.1101/gad.316304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mitra SK, et al. Intrinsic focal adhesion kinase activity controls orthotopic breast carcinoma metastasis via the regulation of urokinase plasminogen activator expression in a syngeneic tumor model. Oncogene. 2006;25 (32):4429–4440. doi: 10.1038/sj.onc.1209482. [DOI] [PubMed] [Google Scholar]
- 84.Pylayeva Y, et al. Ras- and PI3K-dependent breast tumorigenesis in mice and humans requires focal adhesion kinase signaling. J Clin Invest. 2009;119 (2):252–266. doi: 10.1172/JCI37160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van‘t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415 (6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 86.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347 (25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 87.Chang HY, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102 (10):3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ramaswamy S, et al. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33 (1):49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 89.Warburg O. The Metabolism of Tumors. Arnold Constable; London: 1930. [Google Scholar]
- 90.Vander Heiden MG, et al. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324 (5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zipfel WR, et al. Live tissue intrinsic emission microscopy using multiphoton-excited native fluorescence and second harmonic generation. Proc Natl Acad Sci U S A. 2003;100 (12):7075–7080. doi: 10.1073/pnas.0832308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang S, et al. Two-photon fluorescence spectroscopy and microscopy of NAD(P)H and flavoprotein. Biophys J. 2002;82 (5):2811–2825. doi: 10.1016/S0006-3495(02)75621-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Palmer GM, et al. Autofluorescence spectroscopy of normal and malignant human breast cell lines. Photochem Photobiol. 2003;78 (5):462–469. doi: 10.1562/0031-8655(2003)078<0462:asonam>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 94.Chance B, et al. Intracellular oxidation-reduction states in vivo. Science. 1962;137:499–508. doi: 10.1126/science.137.3529.499. [DOI] [PubMed] [Google Scholar]
- 95.Zhang Z, et al. Metabolic imaging of tumors using intrinsic and extrinsic fluorescent markers. Biosens Bioelectron. 2004;20 (3):643–650. doi: 10.1016/j.bios.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 96.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27 (41):5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.DeBerardinis RJ, et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7 (1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 98.Brown E, et al. Dynamic imaging of collagen and its modulation in tumors in vivo using second-harmonic generation. Nat Med. 2003;9 (6):796–800. doi: 10.1038/nm879. [DOI] [PubMed] [Google Scholar]
- 99.Falzon G, et al. Analysis of collagen fibre shape changes in breast cancer. Phys Med Biol. 2008;53 (23):6641–6652. doi: 10.1088/0031-9155/53/23/001. [DOI] [PubMed] [Google Scholar]
- 100.Birk DE, Linsenmayer TF. Collagen fibril assembly, deposition, and organization into tissue specific matrices. In: Yurchenco PD, et al., editors. Extracellular matrix assembly and structure. Academic Press; 1994. pp. 91–128. [Google Scholar]
- 101.Megason SG, Fraser SE. Imaging in systems biology. Cell. 2007;130 (5):784–795. doi: 10.1016/j.cell.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 102.Cox G, Sheppard CJ. Practical limits of resolution in confocal and non-linear microscopy. Microsc Res Tech. 2004;63 (1):18–22. doi: 10.1002/jemt.10423. [DOI] [PubMed] [Google Scholar]
- 103.Brakenhoff GJ, et al. Three-dimensional chromatin distribution in neuroblastoma nuclei shown by confocal scanning laser microscopy. Nature. 1985;317 (6039):748–749. doi: 10.1038/317748a0. [DOI] [PubMed] [Google Scholar]
- 104.White JG, et al. An evaluation of confocal versus conventional imaging of biological structures by fluorescence light microscopy. J Cell Biol. 1987;105 (1):41–48. doi: 10.1083/jcb.105.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pawley JB. Handbook of Biological Confocal Microscopy. Springer; 2006. [Google Scholar]
- 106.Wokosin DL, et al. An optical workstation with concurrent, independent multiphoton imaging and experimental laser microbeam capabilities. Review of Scientific Instruments. 2003;74 (1):193–201. doi: 10.1063/1.1524716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sun CK. Advances in Biochemical Engineering/Biotechnology. Vol. 95. Springer; Berlin/Heidelberg: 2005. Higher harmonic generation microscopy; pp. 17–56. [DOI] [PubMed] [Google Scholar]
- 108.Lakowicz JR, et al. Fluorescence lifetime imaging. Anal Biochem. 1992;202 (2):316–330. doi: 10.1016/0003-2697(92)90112-k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.van Munster EB, Gadella TW. Fluorescence lifetime imaging microscopy (FLIM) Adv Biochem Eng Biotechnol. 2005;95:143–175. doi: 10.1007/b102213. [DOI] [PubMed] [Google Scholar]
- 110.Becker W, et al. Fluorescence lifetime imaging by time-correlated single-photon counting. Microsc Res Tech. 2004;63 (1):58–66. doi: 10.1002/jemt.10421. [DOI] [PubMed] [Google Scholar]
- 111.Peter M, et al. Multiphoton-FLIM quantification of the EGFP-mRFP1 FRET pair for localization of membrane receptor-kinase interactions. Biophys J. 2005;88 (2):1224–1237. doi: 10.1529/biophysj.104.050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Parsons M, et al. Spatially distinct binding of Cdc42 to PAK1 and N-WASP in breast carcinoma cells. Mol Cell Biol. 2005;25 (5):1680–1695. doi: 10.1128/MCB.25.5.1680-1695.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Majoul I, et al. Practical Fluorescence Resonance Energy Transfer or Molecular Nanobioscopy of Living Cells. In: Pawley JB, editor. Handbook of Biological Confocal Microscopy. Springer; 2006. pp. 788–808. [Google Scholar]
- 114.Kantelhardt SR, et al. Imaging of brain and brain tumor specimens by time-resolved multiphoton excitation microscopy ex vivo. Neuro Oncol. 2007;9 (2):103–112. doi: 10.1215/15228517-2006-034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dimitrow E, et al. Sensitivity and specificity of multiphoton laser tomography for in vivo and ex vivo diagnosis of malignant melanoma. J Invest Dermatol. 2009;129 (7):1752–1758. doi: 10.1038/jid.2008.439. [DOI] [PubMed] [Google Scholar]
- 116.Bird D, Gu M. Fibre-optic two-photon scanning fluorescence microscopy. J Microsc. 2002;208 (Pt 1):35–48. doi: 10.1046/j.1365-2818.2002.01059.x. [DOI] [PubMed] [Google Scholar]
- 117.Bird D, Gu M. Resolution improvement in two-photon fluorescence microscopy with a single-mode fiber. Appl Opt. 2002;41 (10):1852–1857. doi: 10.1364/ao.41.001852. [DOI] [PubMed] [Google Scholar]
- 118.Bird D, Gu M. Two-photon fluorescence endoscopy with a micro-optic scanning head. Opt Lett. 2003;28 (17):1552–1554. doi: 10.1364/ol.28.001552. [DOI] [PubMed] [Google Scholar]
- 119.Flusberg BA, et al. Fiber-optic fluorescence imaging. Nat Methods. 2005;2 (12):941–950. doi: 10.1038/nmeth820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jung JC, Schnitzer MJ. Multiphoton endoscopy. Opt Lett. 2003;28 (11):902–904. doi: 10.1364/ol.28.000902. [DOI] [PubMed] [Google Scholar]
- 121.Konig K. Clinical multiphoton tomography. J Biophotonics. 2008;1 (1):13–23. doi: 10.1002/jbio.200710022. [DOI] [PubMed] [Google Scholar]
- 122.Barretto RP, et al. In vivo fluorescence imaging with high-resolution microlenses. Nat Methods. 2009;6 (7):511–512. doi: 10.1038/nmeth.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Levene MJ, et al. In vivo multiphoton microscopy of deep brain tissue. J Neurophysiol. 2004;91 (4):1908–1912. doi: 10.1152/jn.01007.2003. [DOI] [PubMed] [Google Scholar]
- 124.He W, et al. In vivo quantitation of rare circulating tumor cells by multiphoton intravital flow cytometry. Proc Natl Acad Sci U S A. 2007;104 (28):11760–11765. doi: 10.1073/pnas.0703875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marsh P, et al. Practical implementation of adaptive optics in multiphoton microscopy. Opt Express. 2003;11:1123–1130. doi: 10.1364/oe.11.001123. [DOI] [PubMed] [Google Scholar]
- 126.Rueckel M, et al. Adaptive wavefront correction in two-photon microscopy using coherence-gated wavefront sensing. Proc Natl Acad Sci U S A. 2006;103 (46):17137–17142. doi: 10.1073/pnas.0604791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sherman L, et al. Adaptive correction of depth-induced aberrations in multiphoton scanning microscopy using a deformable mirror. J Microsc. 2002;206 (Pt 1):65–71. doi: 10.1046/j.1365-2818.2002.01004.x. [DOI] [PubMed] [Google Scholar]
- 128.Neil MA, et al. Adaptive aberration correction in a two-photon microscope. J Microsc. 2000;200 (Pt 2):105–108. doi: 10.1046/j.1365-2818.2000.00770.x. [DOI] [PubMed] [Google Scholar]
- 129.Lacomb R, et al. Quantitative second harmonic generation imaging of the diseased state osteogenesis imperfecta: experiment and simulation. Biophys J. 2008;94 (11):4504–4514. doi: 10.1529/biophysj.107.114405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Demou ZN, et al. Lack of telopeptides in fibrillar collagen I promotes the invasion of a metastatic breast tumor cell line. Cancer Res. 2005;65 (13):5674–5682. doi: 10.1158/0008-5472.CAN-04-1682. [DOI] [PubMed] [Google Scholar]
- 131.Raub CB, et al. Noninvasive assessment of collagen gel microstructure and mechanics using multiphoton microscopy. Biophys J. 2007;92 (6):2212–2222. doi: 10.1529/biophysj.106.097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chhabra A, et al. Tissue transglutaminase promotes or suppresses tumors depending on cell context. Anticancer Res. 2009;29 (6):1909–1919. [PubMed] [Google Scholar]
- 133.Payne SL, et al. Paradoxical roles for lysyl oxidases in cancer--a prospect. J Cell Biochem. 2007;101 (6):1338–1354. doi: 10.1002/jcb.21371. [DOI] [PubMed] [Google Scholar]
- 134.Kantelhardt SR, et al. Multiphoton excitation fluorescence microscopy of 5-aminolevulinic acid induced fluorescence in experimental gliomas. Lasers Surg Med. 2008;40 (4):273–281. doi: 10.1002/lsm.20623. [DOI] [PubMed] [Google Scholar]
- 135.Major AL, et al. In vivo fluorescence detection of ovarian cancer in the NuTu-19 epithelial ovarian cancer animal model using 5-aminolevulinic acid (ALA) Gynecol Oncol. 1997;66 (1):122–132. doi: 10.1006/gyno.1996.4502. [DOI] [PubMed] [Google Scholar]
- 136.Rogart JN, et al. Multiphoton imaging can be used for microscopic examination of intact human gastrointestinal mucosa ex vivo. Clin Gastroenterol Hepatol. 2008;6 (1):95–101. doi: 10.1016/j.cgh.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3 (5):362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]