Abstract
A library of 5″-modified neomycin derivatives were synthesized for an antibacterial structure-activity optimization strategy. Two leads exhibited prominent activity against both methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE). Antibacterial activities were measured when combined with other clinically used antibiotics. Significant synergistic activities were observed which may lead to the development of novel therapeutic practices in the battle against infectious bacteria.
Introduction
Aminoglycosides are an important class of antibiotics used against infectious diseases. Their usefulness, however, has been significantly compromised by the emergence of resistant bacteria, especially those equipped with aminoglycoside-modifying enzymes (AME's).1-3 Structural modification of aminoglycosides remains to be an effective approach for reviving their antibacterial activities against resistant bacteria. The typical goal is to provide novel aminoglycosides with structural motifs that cannot be accommodated by AME's but are still being able to bind to the targeted site on rRNA.4,5 Most aminoglycosides exert their antibacterial activity by binding selectively to the A-site decoding region of the 16S rRNA and disrupting the functions that are vital to bacteria. While such modification strategies may lead to development of novel broad spectrum antibiotics, X-ray structural and enzymatic studies suggest that higher concentrations of AME's as compared to that of rRNA and the substrate promiscuity of AME's could limit the success of this approach.6
Recently, we discovered that structural modifications of aminoglycosides may alter the traditional mode of action of aminoglycosides and lead to revived activities against resistant bacteria.7 Encouraged by these findings and prompted by an article that reported a similar discovery,8 we conducted further syntheses of derivatives based on the leads to reveal structure-activity relationships (SAR). In an effort to gain more insight into possible modes of action, we also investigated the use of these new aminoglycosides in combination with other clinically used antibiotics with known modes of action.
Materials and Methods
General Experimental Procedures
General Procedure for Coupling of Compound 31 with Carboxylic Acids
To a solution of compound 3 (0.20 g, 0.14 mmol) and carboxylic acids (0.28 mmol) in DMF (10 mL) and Et3N (0.04 mL, 0.28 mmol), HOBt (0.030 g, 0.21 mmol) and EDC (0.040 g, 0.21 mmol) were added. The reaction was stirred at room temperature overnight. After completion of the reaction, the reaction was concentrated and diluted with EtOAc. The organic solution was washed with water, saturated NaHCO3(aq), brine and dried over anhydrous Na2SO4. After removal of the solvent followed by a fast gradient column chromatography (eluted from hexane/EtOAc = 1/1 to EtOAc/MeOH = 9:1), the product was usually obtained as a solid, which was subjected to hydrogenation without further purification.
General Procedure for Hydrogenation and Purification
The solids from acid/amine coupling reaction (0.1 – 0.2 mmol) were dissolved in degassed MeOH (9 mL) followed by the addition of 1 mL HOAc : H2O (1/4 ratio) degassed solution. Catalytic amount of Pd(OH)2/C powder was added and the system was well sealed and further degassed. The system was stirred under atmospheric H2 at room temperature for 10 hours. The reaction was then quenched by filtering through Celite and the residue was washed with H2O and the combined solutions were concentrated. The crude product was purified with Amberlite CG50 (NH4+) eluted with a gradient of NH4OH solution (0% – 20%). After collection of the desired fractions and removal of solvent, the product was re-dissolved in water and loaded to an ion-exchange column packed with Dowex 1X8-200 (Cl- form), and eluted with water. After removal of solvent, the product was obtained as white solid.
5″-Deoxy-5″-decanamidoneomycin B (4b)
1H NMR (D2O, 300 MHz) δ 5.87 (d, J = 5.0 Hz, 1H), 5.32 (d, J = 4.1 Hz, 1H), 5.18 (d, J = 1.7 Hz, 1H), 4.35 (t, J = 5.2 Hz, 1H), 4.1 – 4.2 (m, 4H), 3.9 – 4.0 (m, 3H), 3.6 – 3.7 (m, 2H), 3.2 – 3.6 (m, 12H), 2.40 (dt, J = 12.4 Hz, J = 4.1 Hz, 1H), 2.18 (t, J = 7.2 Hz, 2H), 1.8 (m, 1H), 1.5 (m, 2H), 1.1 (m, 12H), 0.72 (t, J = 6.5 Hz, 3H); 13C NMR (D2O, 75 MHz) δ 178.0, 109.1, 95.7, 94.9, 84.8, 80.7, 77.3, 75.0, 73.5, 72.3, 70.5, 70.2, 69.8, 68.1, 67.6, 67.4, 53.3, 50.9, 49.6, 48.6, 41.1, 40.6, 40.1, 36.0, 31.2, 28.7, 28.5 (2 carbons), 28.4, 28.0, 25.5, 22.1, 13.5; ESI/APCI Calcd for C33H66N7O13+ ([M+H]+) m/e 768.4713; measure m/e 768.4698.
5″-Deoxy-5″-dodecanamidoneomycin B (4c)
1H NMR (D2O, 300 MHz) δ 5.86 (d, J = 4.1 Hz, 1H), 5.31 (d, J = 4.1 Hz, 1H), 5.18 (d, J = 1.4 Hz, 1H), 4.34 (t, J = 4.8 Hz, 1H), 4.1 – 4.2 (m, 4H), 3.9 – 4.0 (m, 3H), 3.6 – 3.7 (m, 2H), 3.2 – 3.6 (m, 12H), 2.40 (dt, J = 8.2 Hz, J = 4.1 Hz, 1H), 2.16 (t, J = 7.2 Hz, 2H), 1.9 (m, 1H), 1.5 (m, 2H), 1.1 (m, 16H), 0.72 (t, J = 6.5 Hz, 3H); 13C NMR (D2O, 75 MHz) δ 177.9, 109.2, 95.7, 94.9, 84.8, 80.7, 77.3, 75.0, 73.5, 72.3, 70.5, 70.2, 69.8, 68.1, 67.6, 67.4, 53.3, 50.8, 49.6, 48.6, 41.1, 40.5, 40.1, 36.0, 31.2, 28.8 (2 carbons), 28.7, 28.6, 28.5, 28.4, 28.0, 25.5, 22.1, 13.5; ESI/APCI Calcd for C35H70N7O13+ ([M+H]+) m/e 796.5026; measure m/e 796.5016.
5″-Deoxy-5″-tetradecanamidoneomycin B (4d)
1H NMR (D2O, 300 MHz) δ 5.86 (d, J = 3.8 Hz, 1H), 5.31 (d, J = 4.1 Hz, 1H), 5.18 (d, J = 1.7 Hz, 1H), 4.34 (t, J = 5.2 Hz, 1H), 4.1 – 4.2 (m, 4H), 3.9 – 4.0 (m, 3H), 3.6 – 3.7 (m, 2H), 3.2 – 3.6 (m, 12H), 2.40 (dt, J = 12.4 Hz, J = 4.1 Hz, 1H), 2.17 (t, J = 7.2 Hz, 2H), 1.8 (m, 1H), 1.5 (m, 2H), 1.1 (m, 20H), 0.72 (t, J = 6.5 Hz, 3H); 13C NMR (D2O, 75 MHz) δ 178.0, 109.1, 95.7, 94.9, 84.8, 80.7, 77.3, 75.0, 73.5, 72.3, 70.5, 70.2, 69.8, 68.1, 67.6, 67.4, 53.3, 50.8, 49.6, 48.6, 41.1, 40.6, 40.1, 36.0, 31.2, 28.9 (2 carbons), 28.8 (2 carbons), 28.7, 28.6, 28.5, 28.4, 28.0, 25.5, 22.1, 13.5; ESI/APCI Calcd for C37H74N7O13+ ([M+H]+) m/e 824.5339; measure m/e 824.5333.
Minimum Inhibitory Concentration (MIC) Determinations9
A solution of selected bacteria was inoculated into trypticase soy broth at 35°C for 1 - 2hrs. The absorbance at 625 nm was measured, and diluted with broth, if necessary, to an absorbance of 0.08 to 0.1. The adjusted inoculated medium (100 μL) was diluted with 10 mL broth, and then applied to a 96-well microtilter plate (50 μL). A series of solutions (50 μL each in 2-fold dilution) of the tested compounds was added to the testing wells. The 96-well plate was incubated at 35°C for 12 - 18 hrs. The minimum inhibitory concentration (MIC) is defined as the lowest concentration of compound needed to inhibit the growth of bacteria. The determinations were repeated at least three times.
Quantitative Structure–Activity Relationship (QSAR) Analysis
MIC's presented in Table 1 have been recalculated to molar concentrations (μM). MIC's represented as ranges in Table 1 (e.g. “16-32”) were calculated as arithmetic means of these values. Log P (logarithm of the partition coefficient octanol/water) values for neomycin B and each of the new compounds were calculated using the HyperChem™ 7.0 package, with atomic charges calculated using the AM1 semi-empirical method.
Table 1.
MIC of the 5″-Modified Neomycin Derivativesa
| Compounds | Antibiotic susceptible strains | Antibiotic resistant strains | ||||||
|---|---|---|---|---|---|---|---|---|
| E. coli ATCC 25922 | S. aureus ATCC 25923 | K. pneumoniae ATCC13883 | S. aureus ATCC 33591 (MRSA) | K. pneumoniae ATCC 700603 | P. aeruginosa ATCC 27853 | E. faecalis ATCC 29212 | E. faecalis ATCC51299 (VRE) | |
| Neomycin B | 4 | 1 | 4 | 125 | 16-32 | 64 | 64-125 | ≥ 250 |
| Amikacin | 1 | 1 | 1 | 8-16 | 0.5 | 0.5-1 | 32-64 | ≥ 250 |
| Vancomycin | 125-250 | 0.5-1 | ≥ 250 | 1 | ND | ≥ 250 | 1-2 | 125 |
| 4a | 16 | 2-4 | 16-32 | 125 | 32-64 | ≥ 250 | 32-64 | ≥ 250 |
| 4b | 32 | 16 | 64-125 | 125-250 | 125 | 16-32 | 32-64 | 64-125 |
| 4c | 16-32 | 8-16 | 64-125 | 16-32 | 64-125 | 16 | 8-16 | 64-125 |
| 4d | 8-16 | 2-4 | 64 | 4-8 | 32-64 | 8 | 2-4 | 8-16 |
| 4e | 4-8 | 1-2 | 8-16 | 2-4 | 16-32 | 4 | 2-4 | 4 |
| 4f | 4-8 | 2-4 | 8-16 | 2-4 | 32 | 8-16 | 4-8 | 8-16 |
Unit: μg/mL, ND: Not Determined.
Combinational Study
A block of 6×6 wells on a 96-well microtiter plate was created with 2-fold serial dilution of 4e and the selected antibiotic to locate the optimal range of inhibitory concentrations. Then a block of 8×8 wells on a 96-well microtitter plate with 2-fold serial dilution of 4e and the selected antibiotic was created to study the combinational studies. For each compound, there was a row or column with only one compound so that the MIC could be determined. The fractional inhibitory concentration (FIC) index was calculated following the equation: FIC = [A]/MICA + [B]/MICB. The combinational effect was defined as: synergism: FIC ≤ 0.5; addition: FIC = 0.5–1.0; indifference: FIC = 1-4; antagonism: FIC ≥ 4. The procedure for every combination of compound 4e and selected antibiotic was repeated 2-4 times.
Hemolytic Activity
Hemolytic activity was determined using methods described by Dartois et al.10 and Sorensen et al.11 with modification. Sheep erythrocytes were used to test hemolytic activities of 4e and 5f. Sheep red blood cells (RBCs) were obtained by centrifuging whole blood at 1,000 × g, washed four times with phosphate-buffered saline (PBS), and resuspended in PBS to a final concentration of 108 erythrocytes per mL. The RBC suspension (80 μL) was added to each well containing different concentrations of 4e and 5f (20 μL). The plate was incubated at 37°C for 60 min. Wells with added deionized water and Triton X-100 (1%, w/v) served as negative (blank) and positive controls, respectively. The percent of hemolysis was calculated using the following equation: % hemolysis = [(absorbance of sample) – (absorbance of blank)]×100/ (absorbance of positive control).
Results
Chemistry
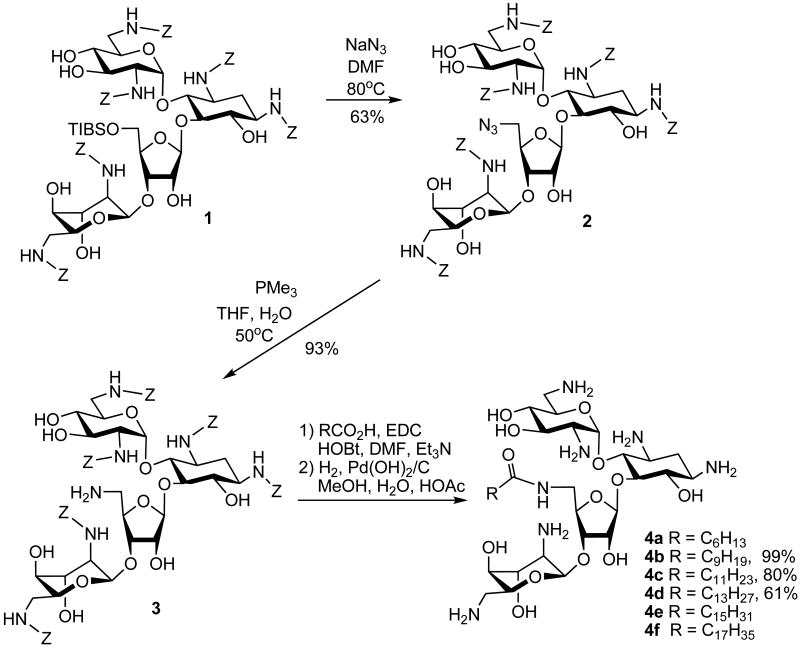
We have demonstrated that neomycin derivatives bearing linear acyl groups at the 5″ position display unusual antibacterial activities. Three additional derivatives with acyl groups of various chain lengths (C7, C16 and C18) were synthesized (Scheme 1). While the derivative with a heptanoyl group (4a)7 maintains its traditional antibacterial profile, derivatives with a hexadecanoyl group from palmitic acid (C16, 4e)7 and an octadecanoyl group from stearic acid (C18, 4f)7 manifest unexpected activity against methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE). MRSA strains harbor genes that encode APH(3′), ANT(4′), and AAC(6′)/APH(2″), which render the bacteria resistant to various aminoglycosides, such as gentamicin and tobramycin.13 VRE contains vanB, ant(6)-I, and aac(6′)-aph(2″) resistance genes with high levels of resistance to aminoglycosides and vancomycin.12,15 The vanB gene and other genes encode for proteins that produce abnormal d-Ala- d-Lac terminal ends of peptidoglycan precursors.16 Vancomycin binds to d-Ala- d-Lac with much lower affinity as compared to the normal d-Ala- d-Ala allowing the bacteria to become resistant to vancomycin. Since both MRSA and VRE are known to possess high levels of resistance against traditional aminoglycosides,12-14 we decided to further explore the SAR regarding the linear acyl groups by synthesizing 4b, 4c and 4d with C10 (decanoyl), C12 (dodecanoyl) and C14 (tetradecanoyl) chain length, respectively. The synthesis of these 5″-acylated neomycin derivatives proceeded via the reported method using neomycin B as the starting material (Scheme 1).7
Scheme 1.
Synthesis of Neomycin Derivatives
Following the protection of amino groups of neomycin B with a carbobenzyloxy (Cbz or Z) group, the 5″-OH was selectively substituted via compound 117 with azide forming compound 2. After the Staudinger reduction of azide, compound 37 with 5″-NH2 was employed and coupled with desired carboxylic acids leading to the preparation of desired neomycin derivatives.
Biological Testing
The synthesized aminoglycosides were assayed against both Gram-positive (G+) and Gram-negative (G-) susceptible and resistant bacterial strains using neomycin B, amikacin and vancomycin as controls. Aminoglycoside susceptible Escherichia coli (G-, ATCC 25922) and S. aureus (G+, ATCC 25923) were used as standard reference strains. Also used were Klebsiella. pneumoniae (G-, ATCC 13883) resistant to ampicillin but susceptible to aminoglycosides, Pseudomonas. aeruginosa (G-, ATCC 27853) which expresses APH(3′)-IIb and manifests modest resistance toward aminoglycosides,18 MRSA (ATCC 33591), Enterococcus faecalis (G+, ATCC51299, VRE), and a E. faecalis strain (ATCC 29212) which is susceptible to vancomycin but moderately resistance to aminoglycosides. The minimum inhibitory concentrations (MIC's) are summarized in Table 1.
From the MIC values, the 5″-acylated neomycin derivatives are generally more active against G+ bacteria than G- bacteria. When comparing the antibacterial activity of each compound, compound 4a displayed a similar MIC profile as neomycin although it was 4-8 fold less active than neomycin against aminoglycoside susceptible strains while inactive against aminoglycoside resistant strains. Increasing the acyl chain length from heptanoyl (4a) to decanoyl (4b) led to a decrease in antibacterial activity: 8-16 fold less active than neomycin against aminoglycoside susceptible strains and inactive against aminoglycoside resistant strains. However, the MIC profile of 4b remained similar to that of 4a and neomycin. As the acyl chain length was extended to C14, C16 and C18 (designated 4d, 4e and 4f, respectively), the activities against aminoglycoside resistant strains increased which implicates different modes of antibacterial action.
QSAR Analysis
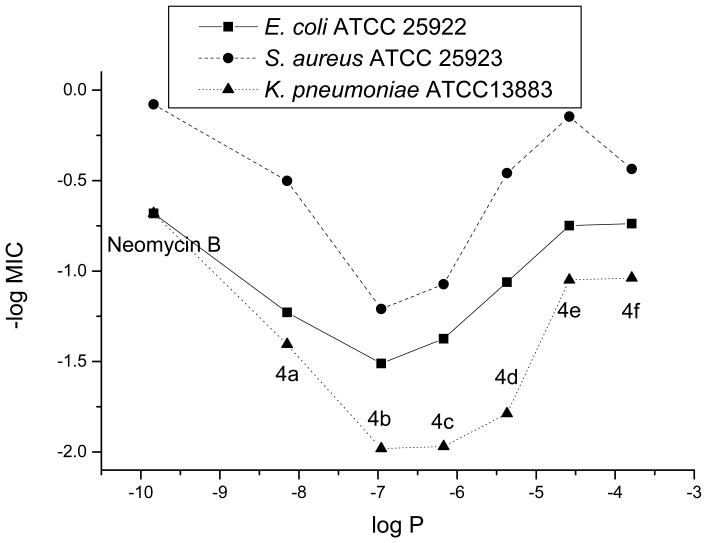
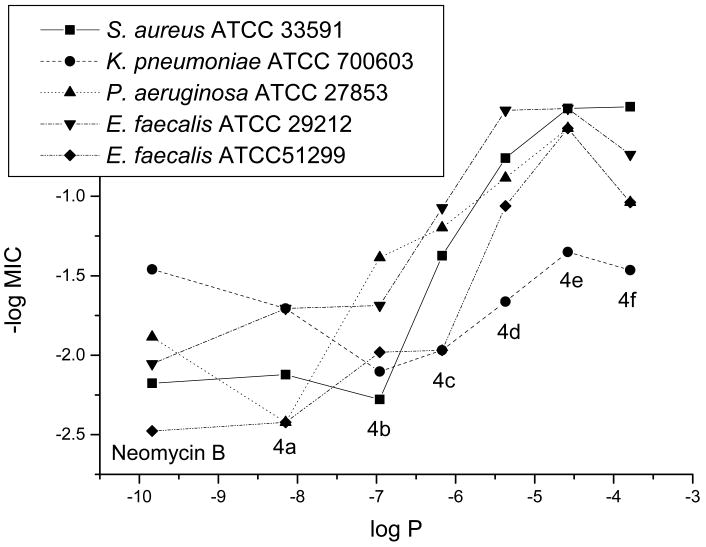
To provide support for our hypothesis of different modes of antibacterial action, we conducted a simple, one-descriptor (logP) QSAR analysis. The data set was divided into two groups: results from aminoglycoside susceptible and results from aminoglycoside resistant strains (Figures 1 and 2). From the results against susceptible strains, a “V-shape” relation was obtained instead of a linear, or at least monotonous relationship. The latter was found in the cases of the resistant strains.
Figure 1.
QSAR Analysis for MIC's from Aminoglycoside Susceptible Strains
Figure 2.
QSAR Analysis for MIC's from Aminoglycoside Resistant Strains
Hemolysis Studies
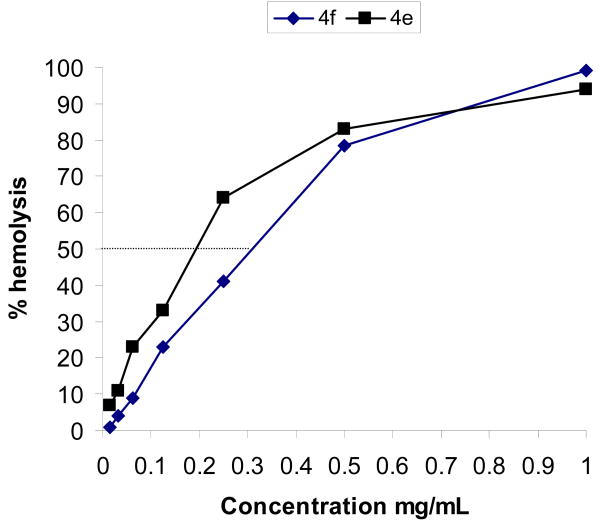
G+ bacteria differ from G- bacteria by the absence of an outer membrane found in the latter with abundant and structurally diverse lipopolysaccharides. Judging from the length of acyl groups and the activity profile of compounds 4d, 4e and 4f with lower MICs against G+ bacteria, we postulate new modes of antibacterial action involving interaction with bacterial inner membranes. Therefore, one of the concerns of employing these newly synthesized compounds as an antibacterial is their potential toxicity to mammalian cells. Thus, we decided to conduct the hemolysis study by using compounds 4e and 4f with longer linear acyl chains. From the hemolysis data, compounds 4e and 4f cause an estimated 50% hemolysis at 0.2 and 0.3 mg/mL, respectively (Figure 3).
Figure 3.
Hemolysis of 4e and 4f
Combinational Studies
Employing a combination of antibiotics is a common practice in the treatment of bacterial infection.19 Such a practice has the advantages of potentially enhancing the efficacy of treatment as in the case of synergism, lowering the possibility of inducing drug resistance from microbes, and reducing the dose of antibiotics. An example of such synergism is the use of vancomycin (bacterial cell membrane action) in combination with gentamicin, an aminoglycoside.20 Therefore, we explored the possible use of the new neomycin derivatives in combination with clinically used antibiotics. Since 4e is the most active neomycin derivative, we selected 4e along with amikacin, neomycin B and vancomycin for combinational antibacterial studies.
We employed a checkerboard assay for the antibiotic combinational study.21 The selected antibiotics were used in combination with the lead, 4e. The results are summarized in Table 2. The combinational effect can be evaluated based on the fractional inhibitory concentration (FIC) index, which can be calculated based on the following equation:
Table 2.
FIC from Combinational Studies of 4e
| Strains of Bacteria | Neomycin | Amikacin | Vancomycin |
|---|---|---|---|
| E. coli ATCC 25922 | 0.27 – 0.56 (Synergism) a | 0.38 - 0.53 (Synergism) | ND |
| S. aureus ATCC 25923 | ND | 1.0 (Addition or Indifference) | 0.53 – 1.5 (Addition or Indifference) |
| K. pneumoniae ATCC 13883 | ND | 0.08 - 0.16 (Synergism) | ND |
| K. pneumoniae ATCC 700603 | ND | 0.38 - 0.53 (Synergism) | ND |
| S. aureus ATCC 33591 (MRSA) | ND | 0.28 – 0.53 (Synergism) | 0.53 – 0.56 (Addition) |
| P. aeruginosa ATCC 27853 | ND | 0.38 – 0.50 (Synergism) | ND |
| E. faecalis ATCC 29212 | ND | 0.53 – 0.56 (Addition) | 0.28 – 0.53 (Synergism) |
FIC = [A]/MICA + [B]/MICB, Synergism: FIC≤0.5; Addition: FIC = 0.5–1.0; Indifference: FIC = 1-4; Antagonism: FIC ≥ 4, ND: Not Determined.
FIC = [A]/MICA + [B]/MICB, Synergism: FIC≤0.5; Addition: FIC = 0.5–1.0; Indifference: FIC = 1-4; Antagonism: FIC ≥ 4.
Discussion
From the relationship between acyl chain length and MIC profile, it appears that when a shorter chain length was incorporated, the derivatives maintain the original mode of antibacterial action as the parent neomycin (i.e. binding to the A-site decoding region of 16S rRNA). The added acyl chain, however, reduces the activity of 4a, likely due to the steric interference of the acyl group on the binding of the neomycin derivative to rRNA. As the chain length increases, the steric hindrance will increase resulting in further decrease in the antibacterial activity. Nevertheless, when even longer acyl chains were incorporated, the derivatives displayed significant antibacterial activity with the SAR in the order of C16 ≈ C18 > C14 against both susceptible and resistant strains. Since elevated steric hindrance from these long acyl groups are expected, it is unlikely that compounds 4d, 4e and 4f regain their antibacterial activity by exerting the same mode of action. Our previous enzymatic and molecular modeling studies also revealed that AME's can inactivate aminoglycosides with diverse structural motifs.7 Thus, it is probable that these three neomycin derivatives have different modes of antibacterial action. The activity of 4d, 4e and 4f against enterococci, which are known to be intrinsically resistant against traditional aminoglycosides, also supports this hypothesis.
The results from QSAR analysis also support our hypothesis that different modes of actions are likely for compounds 4d, 4e and 4f due to the increase in lipophilicity. The compounds presented in the plot of Figure 1 can be roughly divided into two groups: neomycin, 4a (C8) and 4b (C10) as the first group and compounds 4c (C12), 4d (C14), 4e (C16) and 4f (C18) as the second group. The linear relationship of the first group suggests that when the sizes of acyl groups are relatively small, compounds 4a and 4b still exert the same antibacterial mode of action as neomycin. As the sizes of acyl groups increase, the second group shows a different linear relationship with a reverse dependence of the activity on the lipophilicity unlike the first group. These two different linear relationships are consistent with our speculation that compounds 4d, 4e and 4f regain their antibacterial activity by exerting different modes of action. The results from Figure 2 also manifest two roughly linear relationships from two groups of compounds as described above. Since traditional aminoglycosides are inactive against these resistant bacteria, it is expected to obtain a rather leveling line for the first group. Once again, for the second group, the linear relationship implies a strong connection between the increased lipophilicity and the antibacterial activity.
Based on the results of hemolysis studies, the concentrations for 90% hemolysis for both compounds are expected to be greater than 1 mg/mL. Compared to the MIC values of these two compounds (ranging from 2 to 16 μg/mL) the concentrations that give significant hemolysis are 50 – 500 fold higher. Thus, these neomycin derivatives are not hemolytic at their effective antibacterial concentrations.
According to the fractional inhibitory concentration (FIC) index, compound 4e displays strong synergistic effect with amikacin and/or neomycin against G- bacteria including E. coli, K. pneumoniae, and P. aeruginosa. Since compound 4e alone is less active against G- bacteria, one possible reason could be that 4e causes damage to the bacterial membrane and facilitates the entrance of amikacin into bacteria. The synergistic effect with 4e and amikacin was consistently observed with all G- bacterial strains tested. With G+ bacteria, the synergistic effects with 4e were inconsistent with the outcomes ranging from synergism to indifference with amikacin and vancomycin. The synergism of amikacin and 4e against MRSA is of interest. Since MRSA is equipped with various AME's which should drastically reduce the activity of aminoglycosides including amikacin, the observed synergism is unexpected. The synergism of vancomycin and 4e against E. faecali is difficult to explain. It is possible that the action of 4e may not interfere with the action of vancomycin, and unlike the combinational effect against G-bacteria, it may dependent on the individual bacterial strain. The difference in the combinational effect of compound 4e against G+ and G- could possibly reflect the action of compound 4e towards inner and outer membranes, respectively.
In conclusion, we have completed the structural optimization of the 5″-acylated neomycin derivatives. Interesting antibiotic combinational effects have been revealed. The lead, 4e, alone has a prominent antibacterial activity against G+ bacteria like MRSA and VRE. Facing the emergence of vancomycin-resistant S. aureus (VRSA),22,23 we believe that compound 4e represents a possible countermeasure against this formidable pathogen. Judging from the data of QSAR analysis, it is evident that the lipophilicity is responsible for the observed antibacterial activity of the lead.
Based on the combinational study, compound 4e will be more effective when used alone against G+ bacteria. Nevertheless, we have shown that it is possible to use 4e in combination with aminoglycosides against G- bacteria. This finding is consistent with a novel mode of antibacterial action of 4e. The synergism between 4e and amikacin against E. coli and other G- bacteria may provide potential application in counteracting food-borne bacterial outbreaks such as those caused by E coli O157:H7, Salmonella spp. and other G- pathogens.
Supplementary Material
Acknowledgments
We acknowledge National Institutes of Health (AI053138) for financial support.
Footnotes
Supplementary data: Spectroscopic information for the synthesized compounds can be found in the online version.
References
- 1.Umezawa S, Tsuchiya T. In: Aminoglycoside Antibiotics. Umezawa H, Hooper IR, editors. Springer-Verlag; New York: 1982. pp. 37–110. [Google Scholar]
- 2.Haddad J, Kotra LP, Mobashery S. In: Glycochemistry Principles, Synthesis, and Applications. Wang PG, Bertozzi CR, editors. Marcel Dekker, Inc.; New York/Basel: 2001. pp. 353–424. [Google Scholar]
- 3.Wang J, Chang CWT. In: Aminoglycoside Antibiotics. Arya DP, editor. John Wiley & Sons, Inc.; 2007. pp. 141–180. [Google Scholar]
- 4.Wang J, Li J, Chen HN, Chang H, Tanifum CT, Liu HH, Czyryca PG, Chang CWT. Glycodiversification for optimization of the kanamycin class aminoglycosides. J Med Chem. 2005;48:6271–6285. doi: 10.1021/jm050368c. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Chiang FI, Chen HN, Chang CWT. Investigation of the regioselectivity for Staudinger reaction and its application for the synthesis of aminoglycosides with N-1 modification. J Org Chem. 2007;72:4055–4066. doi: 10.1021/jo062588j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fong DH, Berghuis AM. Substrate promiscuity of an aminoglycoside antibiotic resistance enzyme via target mimicry. EMBO J. 2002;21:2323–2331. doi: 10.1093/emboj/21.10.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Chiang FI, Wu L, Czyryca PG, Li D, Chang CWT. Surprising alteration of antibacterial activity of 5″-modified neomycin against resistant bacteria. J Med Chem. 2008;51:7563–7573. doi: 10.1021/jm800997s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bera S, Zhanel GG, Schweizer F. Design, synthesis, and antibacterial activities of neomycin-lipid conjugates: polycationic lipids with potent Gram-positive activity. J Med Chem. 2008;51:6160–6164. doi: 10.1021/jm800345u. [DOI] [PubMed] [Google Scholar]
- 9.The procedure was modified from Methods for Dilution Antimicrobial Susceptibility Testing for Bacteria that Grow Aerobically. Approved standard M7-A5, and Performance Standards for Antimicrobial Disk Susceptibility Tests Approved standard M2-A7, National Committee for Clinical Laboratory Standards, Wayne, PA.
- 10.Dartois V, Sanchez-Quesada J, Cabezas E, Chi E, Dubbelde C, Dunn C, Granja J, Gritzen C, Weinberger D, Ghadiri MR, Parr TR., Jr Systemic Antibacterial Activity of Novel Synthetic Cyclic Peptides. Antimicrob Agents Chemother. 2005;49:3302–3310. doi: 10.1128/AAC.49.8.3302-3310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorensen KN, Kim KH, Takemoto JY. In vitro antifungal and fungicidal activities and erythrocyte toxicities of cyclic lipodepsinonapeptides produced by Pseudomonas syringae pv. Syringae. Antimicrob Agents Chemother. 1996;40:2710–2713. doi: 10.1128/aac.40.12.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swenson JM, Clark NC, Sahm DF, Ferraro MJ, Doern G, Hindler J, Jorgensen JH, Pfaller MA, Reller LB, Weinstein MP, Zabransky RJ, Tenover FC. Molecular characterization and multilaboratory evaluation of Enterococcus faecalis ATCC 51299 for quality control of screening tests for vancomycin and high-level aminoglycoside resistance in Enterococci. J Clin Microbiol. 1995;33:3019–3021. doi: 10.1128/jcm.33.11.3019-3021.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ida T, Okamoto R, Shimauchi C, Okubo T, Kuga A, Inoue M. Identification of aminoglycoside-modifying enzymes by susceptibility testing: epidemiology of methicillin-resistant Staphylococcus aureus in Japan. J Clin Microbiol. 2001;39:3115–3121. doi: 10.1128/JCM.39.9.3115-3121.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fridkin SK, Lawton R, Edwards JR, Tenover FC, McGowan JE, Jr, Gaynes RP. Monitoring antimicrobial use and resistance: comparison with a national benchmark on reducing vancomycin use and vancomycin-resistant enterococci. Emerg Infect Dis. 2002;8:702–707. doi: 10.3201/eid0807.010465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cetinkaya Y, Falk P, Mayhall CG. Vancomycin-Resistant Enterococci. Clin Microbiol Rev. 2000;13:686–707. doi: 10.1128/cmr.13.4.686-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barna JCJ, Williams DH. The Structure and Mode of Action of Glycopeptide Antibiotics of the Vancomycin Group. Ann Rev Microbiol. 1984;38:339–357. doi: 10.1146/annurev.mi.38.100184.002011. [DOI] [PubMed] [Google Scholar]
- 17.Kling D, Hesek D, Shi Q, Mobashery S. Design and synthesis of a structurally constrained aminoglycoside. J Org Chem. 2007;72:5450–5453. doi: 10.1021/jo0707636. [DOI] [PubMed] [Google Scholar]
- 18.Hachiler H, Santanam P, Kayser FH. Sequence and characterization of a novel chromosomal aminoglycoside phosphotransferase gene, aph (3′)-IIb, in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:1254–1256. doi: 10.1128/aac.40.5.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neu HC, Gootz TD. In: Medical Microbiology. 4th. Baron S, editor. The University of Texas Medical Branch; Galvaston: 1996. pp. 183–185. [PubMed] [Google Scholar]
- 20.Cottagnoud P, Cottagnoud M, Täuber MG. Vancomycin Acts Synergistically with Gentamicin against Penicillin-Resistant Pneumococci by Increasing the Intracellular Penetration of Gentamicin. Antimicrob Agents Chemother. 2003;40:144–147. doi: 10.1128/AAC.47.1.144-147.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorian V. Antibiotics in Laboratory Medicine. New York: Williams & Wikins; 1996. pp. 330–396. [Google Scholar]
- 22.Zhu W, Clark NC, McDougal LK, Hageman J, McDonald LC, Pate JB. Vancomycin-Resistant Staphylococcus aureus Isolates Associated with Inc18-Like vanA Plasmids in Michigan. Antimicrob Agents Chemother. 2008;52:452–457. doi: 10.1128/AAC.00908-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenover FC, Weigel LM, Appelbaum PC, McDougal LK, Chaitram J, McAllister S, Clark N, Killgore G, O'Hara CM, Jevitt L, Patel JB, Bozdogan B. Vancomycin-Resistant Staphylococcus aureus Isolate from a Patient in Pennsylvania. Antimicrob Agents Chemother. 2004;48:275–280. doi: 10.1128/AAC.48.1.275-280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.