Summary
The differential diagnosis of clear cell, papillary and chromophobe renal cell carcinoma is clinically important, because these tumor subtypes are associated with different pathobiology and clinical behavior. For cases in which histopathology is equivocal, immunohistochemistry and quantitative RT-PCR can assist in the differential diagnosis by measuring expression of subtype-specific biomarkers. Several renal tumor biomarkers have been discovered in expression microarray studies. However, due to heterogeneity of gene and protein expression, additional biomarkers are needed for reliable diagnostic classification. We developed novel bioinformatics systems to identify candidate renal tumor biomarkers from the microarray profiles of 45 clear cell, 16 papillary and 10 chromophobe renal cell carcinoma; the microarray data was derived from two independent published studies. The ArrayWiki biocomputing system merged the microarray datasets into a single file, so gene expression could be analyzed from a larger number of tumors. The caCORRECT system removed non-random sources of error from the microarray data, and the omniBioMarker system analyzed data with several gene-ranking algorithms, in order to identify algorithms effective at recognizing previously described renal tumor biomarkers. We predicted these algorithms would also be effective at identifying unknown biomarkers that could be verified by independent methods. We selected six novel candidate biomakers from the omniBioMarker analysis, and verified their differential expression in formalin-fixed paraffin-embedded tissues by quantitative RT-PCR and immunohistochemistry. The candidate biomarkers were carbonic anhydrase IX, ceruloplasmin, schwannomin-interacting protein 1, E74-like factor 3, cytochrome c oxidase subunit 5a and acetyl-CoA acetyltransferase 1. Quantitative RT-PCR was performed on 17 clear cell, 13 papillary and 7 chromophobe renal cell carcinoma. Carbonic anhydrase IX and ceruloplasmin were overexpressed in clear cell renal cell carcinoma; schwannomin-interacting protein 1 and E74-like factor 3 were overexpressed in papillary renal cell carcinoma; and cytochrome c oxidase subunit 5a and acetyl-CoA acetyltransferase 1 were overexpressed in chromophobe renal cell carcinoma. Immunohistochemistry was performed on tissue microarrays containing 66 clear cell, 16 papillary and 12 chromophobe renal cell carcinoma. Cytoplasmic carbonic anhydrase IX staining was significantly associated with clear cell renal cell carcinoma. Strong cytoplasmic schwannomin-interacting protein 1 and cytochrome c oxidase subunit 5a staining were significantly more frequent in papillary and chromophobe renal cell carcinoma, respectively. In summary, we developed a novel process for identifying candidate renal tumor biomarkers from microarray data, and verifying differential expression in independent assays. The tumor biomarkers have potential utility as a multiplex expression panel for classifying renal cell carcinoma with equivocal histology. Biomarker expression assays are increasingly important for renal cell carcinoma diagnosis, as needle core biopsies become more common and different therapies for tumor subtypes continue to be developed.
Keywords: Renal Cell Carcinoma, Microarrays, Quantitative RT-PCR, Immunohistochemistry, Bioinformatics
1. Introduction
Renal cell carcinoma (RCC) is the major adult malignancy of the kidney; it is subclassified into several subtypes including clear cell, papillary and chromophobe RCC. Renal oncocytoma is a relatively common benign tumor that may be related to chromophobe RCC [1]. Accurate classification is clinically important, because tumor subtypes are associated with different malignant potential, prognoses and optimal therapies [2]. In recent years, we and other groups have used cDNA and oligonucleotide microarrays to characterize gene expression profiles in renal tumor subtypes [3-6]. Based on unique expression patterns, several novel immunohistochemical markers have been identified for each RCC subtype. When used in conjunction with histopathology, these immunohistochemical markers are clinically useful for renal tumor diagnosis [7-10]. However, due to the heterogeneity of gene and protein expression in RCC, additional biomarkers are needed to develop clinically reliable immunohistochemical panels, with adequate diagnostic sensitivity and specificity for each RCC subtype. In this report, we describe the use of novel bioinformatics systems to identify candidate RCC biomarkers from previous microarray data [11]. The bioinformatics systems are designed to combine disparate datasets from independent microarray studies, remove non-random sources of error from the expression data, and analyze expression patterns in the context of pre-existing biological knowledge, in order to identify valid biomarkers more efficiently. Following identification of candidate biomarkers, we describe the verification of selected markers in formalin-fixed paraffin-embedded renal tumor tissues by quantitative RT-PCR and immunohistochemistry.
2. Materials and Methods
Acquisition of microarray data
Microarray datasets were obtained from previously published reports [3, 12]. Schuetz et al utilized Affymetrix HG Focus microarrays with over 8700 probe sets, in a study that included 13 clear cell, 5 papillary and 4 chromophobe RCC. The chromophobe carcinomas were combined with 3 additional renal oncocytomas to form a single class for biomarker discovery (n = 7). Jones et al. utilized Affymetrix HG-U133A microarrays with over 22000 probe sets, in a study that included 32 clear cell, 11 papillary, 6 chromophobe RCC and 12 oncocytomas. The HG-U133A microarray data were reduced to include only those probe sets shared with HG Focus data. The ArrayWiki biocomputing system [13] combined the microarray datasets into a single data file, in order to increase total sample size, while updating probe annotation with a knowledge management interface based on Wikipedia standards. The URL http://arraywiki.bme.gatech.edu/index.php/Andrew_Young contains visual representations of experiments described in this report. General information on ArrayWiki is available at the URL http://www.bio-miblab.org/arraywiki.
Quality assurance of microarray data
The RCC microarray datasets were analyzed with caCORRECT (chip artifact CORRECTion; http://www.bio-miblab.org), a web-based bioinformatics system that detected and removed localized array, or “chip”, artifacts [14]. For this purpose, artifacts were defined as spatially prominent data variances caused by chip manufacturing or lab processing errors For detection of artifacts, quantile normalization was used to align signal distributions from each microarray and remove global array biases within the set of experiments [15]. After quantile normalization, variance scores were calculated for each probe on each microarray chip, using a modified t-statistic calculated from other chips in a leave-one-out fashion. A sliding window image-processing algorithm was then run to identify high-variance probes that were geographically clustered on the array platform; regions of clustered high-variance probes represented potential artifacts, while geographically isolated high-variance probes were left alone. After this first round was complete, four additional rounds of artifact-omitting quantile normalization, and artifact-weighted artifact detection, were performed in order to identify subtle artifacts that may have been overshadowed in earlier rounds by larger defects. At this point, quality metrics were calculated to describe the artifact coverage and noise content of each chip and of the experiment as a whole. Probe data that were identified as artifacts were then replaced with the probe-specific median intensity of all other chips in the dataset. Completion of the caCORRECT process resulted in the following files: (i) heatmap images of probe variance score for all chips, with and without logical artifact masks; (ii) new versions of “clean” probe expression files with appropriate replacements; and (iii) gene expression value tables, calculated by R implementation of the Robust Microarray Averaging (RMA) algorithm [16] from data before and after caCORRECT.
Identification of candidate biomarkers
The omniBioMarker bioinformatics resource [17] was used to identify genes expressed differentially in RCC subtypes, using microarray data processed with caCORRECT. Processed gene expression value tables were combined into a master gene expression data file, which was assessed by hierarchical clustering [18] to ensure that combined data continued to classify RCC subtypes as in the original reports [3, 12]. omniBioMarker then analyzed the combined RCC data in an iterative fashion using support vector machine classifiers (SVM) to rank the genes individually by classification ability, as determined by bootstrapping [19]. In order to identify the optimal algorithm for subtype classification, omniBioMarker varied the SVMs by adjusting two parameters that control classifier complexity and generalization ability. The algorithm searched for the best set of parameters over a predefined parameter space. In the first iteration, omniBioMarker ranked the performance of each SVM classifier using control biomarkers, which were defined as biomarkers that had been verified in previous studies by RT-PCR or IHC [3, 20]. The optimal classifier generally ranked control biomarkers before non-control biomarkers. After identifying the optimal gene-ranking classifier for the combined RCC microarray data, the corresponding ranking results were used to identify additional candidate biomarkers with consistent differential expression in clear cell, papillary and chromophobe RCC. Candidates were interpreted with Gene Ontology analysis tools including GO-Miner [21], and selected for subsequent verification by quantitative RT-PCR and IHC.
Quantitative RT-PCR
Gene expression was assessed by quantitative RT-PCR, using total RNA from fixed tissues of 17 clear cell, 13 papillary and 7 chromophobe RCC. Duplicate experiments were performed according to published protocols with minor modifications [22]: Histological sections were deparaffinized with ethanol and xylene, and cells of interest were microdissected with a sterile scalpel. Tissues were digested in buffer containing proteinase K at 60°C overnight. RNA was extracted with phenol/chloroform, and genomic DNA was removed with DNase. RNA quality and quantity were assessed with a Bioanalyzer (Agilent Technologies). Up to 3 μg of RNA was used for first strand cDNA synthesis with Superscript III (Invitrogen). PCR was performed with a custom-designed Taqman Low Density Array (LDA, Applied Biosystems) in a 96-well microfluidic card format, using the ABI PRISM 7900HT Sequence Detection System (high-throughput real-time PCR system). Gene expression data were normalized relative to the geometric mean of two housekeeping genes (18S, ACTB). LDA runs were analyzed by using Relative Quantification (RQ) Manager (Applied Biosystems) software. The following test genes were analyzed: carbonic anhydrase IX (CA9, Assay ID: Hs00154208_m1, Applied Biosystems); ceruloplasmin (CP, Assay ID: Hs00236810_m1, Applied Biosystems); schwannomin-interacting protein 1 (SCHIP1, Assay ID: Hs00205829_m1, Applied Biosystems); E74-like factor 3 (ELF3, Assay ID: Hs00231786_m1, Applied Biosystems); cytochrome c oxidase subunit 5a (COX5A, Assay ID: Hs00362067_m1, Applied Biosystems); and acetyl-CoA acetyltransferase 1 (ACAT1, Assay ID: Hs00608002_m1, Applied Biosystems). Test gene expression was normalized to 18S ribosomal RNA and referenced to a normal kidney reference RNA specimen. Relative normalized gene expression was compared in renal tumor subtypes, with statistical significance assessed by two-tailed T-test.
Immunohistochemistry
Selected biomarkers were further verified by immunohistochemistry, performed on the KIC1501 tissue microarray (Clonagen), which included 66 clear cell, 16 papillary and 12 chromophobe RCC. Tissue sections were incubated with the following primary antibodies: anti-CA9 (rabbit polyclonal serum, Novus Biological), anti-SCHIP1 (rabbit polyclonal IgG, Sigma), and anti-COX5A (rabbit polyclonal IgG, Protein Tech Group). After washing unbound primary antibody, sections were treated with goat anti-rabbit immunoglobulin conjugated to a peroxidase-labeled polymer, according to the manufacturer's instructions (Envision kit; DAKO Corp., Carpinteria, CA). Immunohistochemical reactions were developed with diaminobenzidine as the chromogenic peroxidase substrate. Sections were counterstained with hematoxylin after immunohistochemistry. Specificity was verified by negative control reactions without primary antibody, as well as appropriate staining reactions in positive control tissues. The intensity of immunohistochemical staining in tumor cells was graded as negative (0), weak (1+), moderate (2+) and strong (3+); negative-to-weak staining was classified as low-level expression, whereas moderate-to-strong staining was classified as high-level expression. Frequency of cases with high-level expression was compared among renal tumor subtypes, with statistical significance assessed by chi-square analysis.
3. Results
We analyzed two RCC microarray datasets with a series of novel bioinformatics systems, in order to identify candidate diagnostic biomarkers. First, the ArrayWiki knowledge management system was used to combine and annotate the datasets in compatible formats. The caCORRECT quality assurance system was then used to identify non-random physical artifacts on the microarrays, and eliminate potentially confounding results from these defective regions. Examples of chip artifacts included scratches on the array surface and bubbles in the hybridization medium (Figure 1). Next, the omniBioMarker system was used to analyze the combined microarray dataset with a variety of support vector machine classifiers, in order to identify optimal algorithms for identifying candidate RCC biomarkers. In this step, the support vector machines were compared for performance in classifying RCC subtypes, using only biomarkers in the dataset that had been previously verified in independent studies by quantitative RT-PCR or immunohistochemistry. The strongest algorithm was then applied to the entire dataset, in order to identify additional gene products with differential expression in RCC subtypes. From this group of gene products, we selected candidate diagnostic biomarkers for subsequent verification by quantitative RT-PCR and immunohistochemistry.
Figure 1. caCORRECT analysis of microarray data.
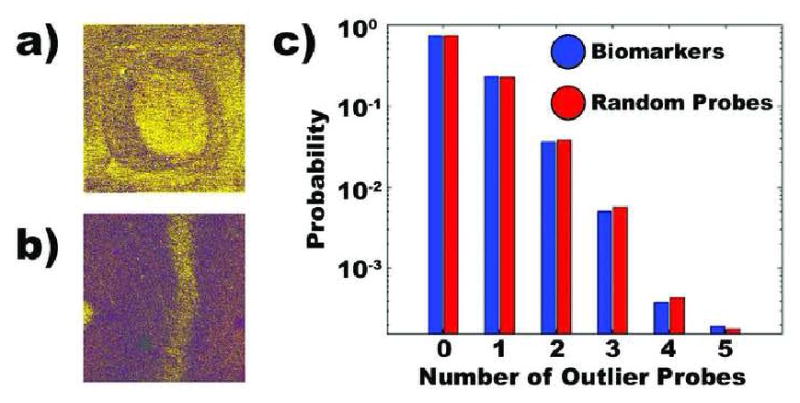
a) Detection of localized artifacts caused by an air bubble in hybridization medium. b) Detection of localized artifacts caused by a scratch on the microarray surface. c) Analysis of published microarray data from Schuetz et al. The biomarkers proposed in this study were randomly distributed with the microarray artifacts detected by caCORRECT. If biomarkers did colocalize with microarray artifacts, the biomarker bar (blue) would be larger than the random probe bar (red) at higher number of outlier probes.
Differential expression of six gene products was verified by quantitative RT-PCR, using formalin-fixed paraffin-embedded specimens from 17 clear cell, 13 papillary and 7 chromophobe RCC (Figure 2). CA9 and CP were overexpressed in clear cell RCC (p = 9.83 × 10-05 and 3.59 × 10-06, respectively); SCHIP1 and ELF3 were overexpressed in papillary RCC (p = 1.48 × 10-03 and 4.14 × 10-08, respectively); and COX5A and ACAT1 were overexpressed in chromophobe RCC (p = 1.32 × 10-05 and 1.40 × 10-07, respectively). Differential expression of CA9, SCHIP1 and COX5A was further verified by immunohistochemistry, using commercial primary antibodies and a formalin-fixed paraffin-embedded tissue microarray that included 66 clear cell, 16 papillary and 12 chromophobe RCC (Figure 3). By immunohistochemistry, CA9 was strongly overexpressed in the tumor cell cytoplasm of clear cell RCC (p < 0.001). Cytoplasmic SCHIP1 staining was seen in papillary and clear cell RCC, but 3+ intensity was significantly more frequent in tumor cells of papillary RCC (p < 0.001). Cytoplasmic COX5A staining was seen in all RCC subtypes; however, 3+ intensity was significantly more frequent in tumor cells of chromophobe RCC (p < 0.02). Immunohistochemical data are summarized in Table 1.
Figure 2. Quantitative RT-PCR in formalin-fixed paraffin-embedded renal tumors.
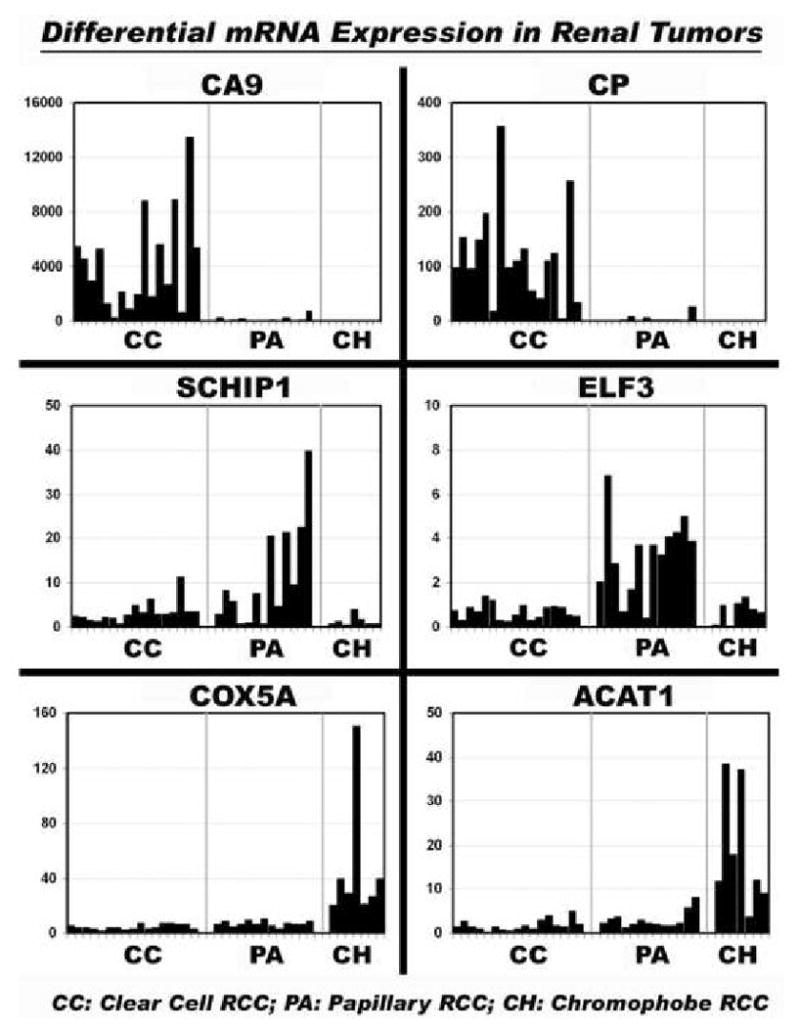
In each panel, bars indicate relative mRNA expression in a single tumor specimen. Tumor subtypes are designated as CC (clear cell RCC), PA (papillary RCC) and CH (chromophobe RCC). Top left) Carbonic anhydrase IX (CA9). Top right) Ceruloplasmin (CP). Middle left) Schwannomin-interacting protein 1 (SCHIP1). Middle right) E74-like factor 3 (ELF3). Bottom left) Cytochrome c oxidase subunit 5A (COX5A). Bottom right) Acetyl-CoA acetyltransferase 1 (ACAT1). CA9 and CP were significantly overexpressed in clear cell RCC; SCHIP1 and ELF3 were significantly overexpressed in papillary RCC; COX5A and ACAT1 were significantly overexpressed in chromophobe RCC.
Figure 3. Immunohistochemistry in formalin-fixed paraffin-embedded renal tumors.
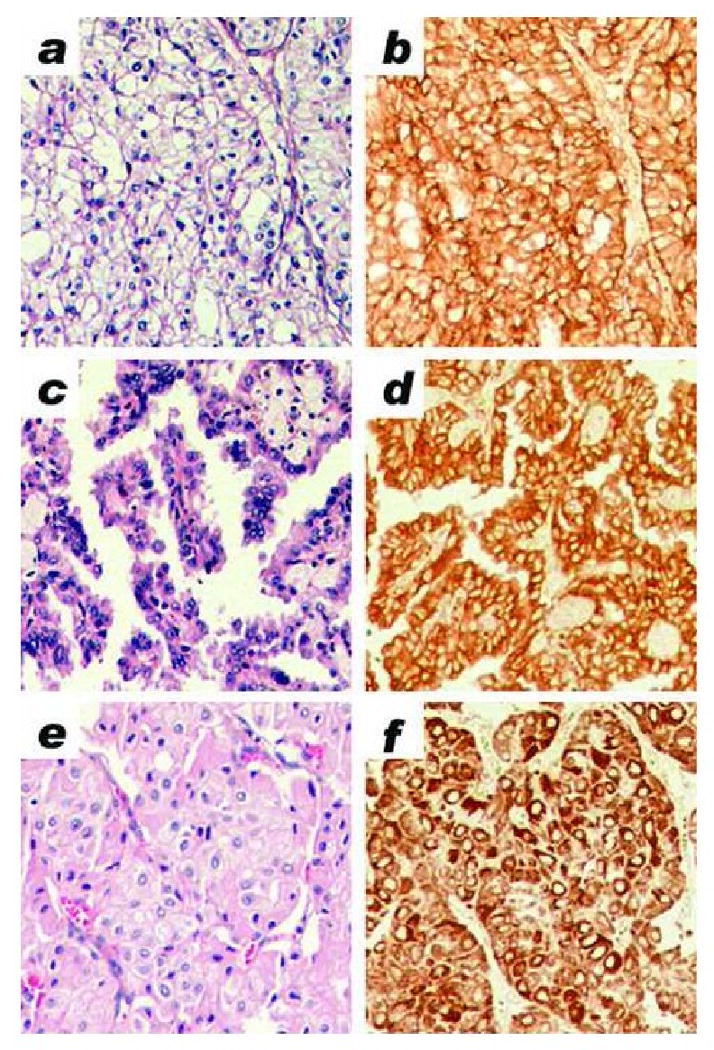
All images were taken at 100× magnification a) Clear cell RCC, hematoxylin & eosin stain. b) Clear cell RCC, CA9 immunohistochemical stain (3+ positive). c) Papillary RCC, hematoxylin & eosin stain. d) Papillary RCC, SCHIP1 immunohistochemical stain (3+ positive). e) Chromophobe RCC, hematoxylin & eosin stain. f) Chromophobe RCC, COX5A immunohistochemical stain (3+ positive).
Table 1. Immunohistochemical Analysis of Candidate Biomarkers in RCC Subtypes.
| Tumor Subtype | N | CA9 | SCHIP1 | COX5A | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0+ | 1+ | 2+ | 3+ | p | 0+ | 1+ | 2+ | 3+ | p | 0+ | 1+ | 2+ | 3+ | p | ||
| CC-RCC | 66 | 2 | 3 | 16 | 45 | 23 | 26 | 15 | 2 | 0 | 33 | 32 | 1 | |||
| PA-RCC | 16 | 0 | 12 | 3 | 1 | < 0.001* | 1 | 1 | 5 | 9 | < 0.001** | 0 | 0 | 10 | 6 | < 0.02*** |
| CH-RCC | 12 | 10 | 2 | 0 | 0 | 8 | 2 | 2 | 0 | 0 | 0 | 2 | 10 | |||
Abbreviations: CC- Clear Cell; PA - Papillary; CH - Chromophobe
CC-RCC vs other RCC subtypes
3+ staining intensity in PA-RCC vs other RCC subtypes
3+ staining intensity in CH-RCC vs other RCC subtypes
4. Discussion
Gene expression profiling is an important approach to discover molecular markers for diagnostic pathology. Microarrays are used to identify complex expression profiles, which are screened to identify large numbers of differentially expressed genes. These differential expression profiles provide a list of candidate diagnostic biomarkers for clinical pathology laboratories, using assays such as immunohistochemistry and quantitative RT-PCR [3, 4, 6-10, 23-25]. While this approach has been applied effectively, it remains limited because the number of genes in most microarray studies exceeds the number of experimental samples by several orders of magnitude. Therefore, differential expression profiles tend to contain numerous false positives (candidate biomarkers that are not verified when different samples and analytical methods are tested) and false negatives (true biomarkers that are not differentially expressed among the small number of samples in the microarray study). In order to maximize the potential contribution of microarray technology, new information tools are needed to identify candidate biomarkers with the greatest likelihood of validity. In this report, we describe an integrated series of biocomputation systems, called ArrayWiki, caCORRECT and omniBioMarker, which we developed to make the process of biomarker discovery effective and efficient [11, 13, 14, 17]. The bioinformatics systems are designed to maximize the experimental sample size, remove systematic error from the microarray data, and empirically identify optimal algorithms to identify differentially expressed biomarkers. Our data are presented publicly at the ArrayWiki Internet site (see Materials & Methods). Data in ArrayWiki are open to community contribution, comment, and modification, using syntax and structure common to Wikipedia and similar resources [26, 27]. Ongoing annotation from the community could enhance the value of this knowledge base for future biomarker discovery experiments.
We selected six candidate RCC biomarkers for verification by RT-PCR and immunohistochemistry; each biomarker has potential relevance for renal tumor pathobiology and clinical management. Carbonic anhydrase IX (CA9) and ceruloplasmin (CP) were verified as biomarkers for clear cell RCC. CA9, a hypoxia-inducible protein, is well-established as a clear cell RCC biomarker [20, 28]. It is overexpressed in clear cell tumors compared to benign lesions and other RCC subtypes, and thus may be useful for diagnostic classification. Several studies also suggest that comparatively low CA9 expression in clear cell RCC is a negative prognostic indicator [29, 30]. In addition, CA9 is a potentially important therapeutic biomarker, since it is the protein target for G250 monoclonal antibody-based immunotherapy and vaccines against clear cell RCC [31]. Along with CA9, the acute phase reactant CP was overexpressed in clear cell RCC in our previous microarray experiments. This pattern was seen with many other genes related to inflammation and the acute phase response [3, 6]. Similarly, other groups have identified CP as a clear cell RCC biomarker by suppression subtractive hybridization [32, 33]. In addition, serum CP protein levels are elevated in patients with RCC and other malignancies compared to healthy controls [34-38]. CP has antioxidant properties that may be involved with the host response to neoplasia [39].
Schwannomin-interacting protein 1 (SCHIP1) and E74-like factor 3 (ELF3) were verified as biomarkers for papillary RCC. Neither gene product has been described in RCC previously. SCHIP1 was first discovered as a protein that interacts specifically with spliced isoforms and naturally occurring mutants of neurofibromatosis type 2 (NF2) tumor suppressor protein, also known as schwannomin or merlin [40]. Specific interactions of NF2 with a variety of proteins, including SCHIP1, have been associated experimentally with the PI3-kinase, MAP kinase and small GTPase signaling pathways, which may represent therapeutic targets for inhibiting tumor proliferation [41]. ELF3 (also termed ESX and ESE-1) encodes an ETS-family nuclear transcription factor that is expressed specifically in epithelial cells [42, 43]. Microarray studies have shown that ELF3 is overexpressed in several types of carcinoma and sarcomas with epithelial differentiation [43-45]. Transfection of ELF3 into breast epithelial cell lines results in malignant transformation [46, 47]. ELF3 expression may be involved in feedback regulatory pathways with transforming growth factor beta type II receptor and erbB2 receptor, and thus may be a potential therapeutic target [48-50].
Cytochrome c oxidase subunit 5a (COX5A) and acetyl-CoA acetyltransferase 1 (ACAT1) were verified as biomarkers for chromophobe RCC. The COX5A gene product is localized to the mitochondrion and is critical for oxidative phosphorylation [51]. Proteomic studies have shown that COX5A is expressed differentially in gastric carcinoma [52]. In addition, we have shown that chromophobe RCC overexpresses many mitochondrial proteins and other gene products related to energy pathways, electron transport, and oxidative phosphorylation [3, 6], a molecular signature that may reflect the abundant mitochondria in neoplastic cells of these tumors [53]. Previous research on renal tumors has correlated high content of oxidative phosphorylation complexes with a slow growing, noninvasive phenotype [54]. ACAT1 is an integral membrane protein that localizes to the endoplasmic reticulum. It controls cholesterol ester formation in kidney and other organs [55]. Interferon gamma and STAT1 regulate ACAT1 expression in prostate cancer cells [56, 57], but the regulation of ACAT1 in renal cancer is still unknown.
In summary, we describe several candidate biomarkers for RCC classification, derived from microarray data analyzed with a series of novel biocomputational tools. These biomarkers have potential clinical utility as a multiplex expression panel for classification of RCC with equivocal histology, when combined with H&E morphology and other gene expression or cytogenetic studies. For many tumor types, multiplex biomarker panels are more sensitive and specific than assays for individual markers [10, 20, 58]. In addition, multiplex immunohistology platforms will be important to develop the emerging class of immunoassays based on nanoparticle optical detection tags [59, 60]. Therefore, CA9, SCHIP1 and COX5A could be combined with other immunohistochemical markers for renal tumor classification, such as glutathione S-transferase (GSTA) and adipophilin (ADFP) for clear cell RCC [23, 25]; alpha methyacyl CoA racemase (AMACR) for papillary RCC [9]; and parvalbumin (PVALB), beta defensin-1 (DEFB1), claudin 7 (CLDN7) and claudin 8 (CLDN8) for chromophobe RCC [7, 10]. Multiplex immunohistochemical profiling of RCC is likely to become more important for classification, as the use of diagnostic needle core biopsies grows, and differential therapies for primary and metastatic RCC subtypes continue to be developed [2, 61, 62].
Acknowledgments
This work was supported by a grant from the National Cancer Institute Centers for Cancer Nanotechnology Excellence Program (U54CA119338).
Footnotes
Disclosure/Conflict of Interest: The authors declare no conflicts of interest related to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Amin MB, Amin MB, Tamboli P, et al. Prognostic impact of histologic subtyping of adult renal epithelial neoplasms: an experience of 405 cases. Am J Surg Pathol. 2002;26:281–91. doi: 10.1097/00000478-200203000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Beck SD, Patel MI, Snyder ME, et al. Effect of papillary and chromophobe cell type on disease-free survival after nephrectomy for renal cell carcinoma. Ann Surg Oncol. 2004;11:71–7. doi: 10.1007/BF02524349. [DOI] [PubMed] [Google Scholar]
- 3.Schuetz AN, Yin-Goen Q, Amin MB, et al. Molecular Classification of Renal Tumors by Gene Expression Profiling. J Mol Diagn. 2005;7:206–18. doi: 10.1016/S1525-1578(10)60547-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi M, Yang XJ, Sugimura J, et al. Molecular subclassification of kidney tumors and the discovery of new diagnostic markers. Oncogene. 2003;22:6810–8. doi: 10.1038/sj.onc.1206869. [DOI] [PubMed] [Google Scholar]
- 5.Yang XJ, Sugimura J, Schafernak KT, et al. Classification of renal neoplasms based on molecular signatures. J Urol. 2006;175:2302–6. doi: 10.1016/S0022-5347(06)00255-2. [DOI] [PubMed] [Google Scholar]
- 6.Young AN, Amin MB, Moreno CS, et al. Expression profiling of renal epithelial neoplasms: a method for tumor classification and discovery of diagnostic molecular markers. Am J Pathol. 2001;158:1639–51. doi: 10.1016/S0002-9440(10)64120-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Osunkoya AO, Cohen C, Lawson D, et al. Claudin-7 and claudin-8: immunohistochemical markers for the differential diagnosis of chromophobe renal cell carcinoma and renal oncocytoma. Hum Pathol. 2008 doi: 10.1016/j.humpath.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Pan CC, Chen PC, Chiang H. Overexpression of KIT (CD117) in chromophobe renal cell carcinoma and renal oncocytoma. Am J Clin Pathol. 2004;121:878–83. doi: 10.1309/A7M2-XTMJ-QK0K-PQER. [DOI] [PubMed] [Google Scholar]
- 9.Tretiakova MS, Sahoo S, Takahashi M, et al. Expression of alpha-methylacyl-CoA racemase in papillary renal cell carcinoma. Am J Surg Pathol. 2004;28:69–76. doi: 10.1097/00000478-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 10.Young AN, de Oliveira Salles PG, Lim SD, et al. Beta defensin-1, parvalbumin, and vimentin: a panel of diagnostic immunohistochemical markers for renal tumors derived from gene expression profiling studies using cDNA microarrays. Am J Surg Pathol. 2003;27:199–205. doi: 10.1097/00000478-200302000-00008. [DOI] [PubMed] [Google Scholar]
- 11.Stokes TH, Phan J, Quo CF, et al. Bio-nano-informatics: an integrated information management system for personalized oncology. Conf Proc IEEE Eng Med Biol Soc. 2006;1:3325–8. doi: 10.1109/IEMBS.2006.259752. [DOI] [PubMed] [Google Scholar]
- 12.Jones J, Otu H, Spentzos D, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11:5730–9. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 13.Stokes TH, Torrance JT, Li H, et al. ArrayWiki: an enabling technology for sharing public microarray data repositories and meta-analyses. BMC Bioinformatics. 2008;9 6:S18. doi: 10.1186/1471-2105-9-S6-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stokes TH, Moffitt RA, Phan JH, et al. chip artifact CORRECTion (caCORRECT): a bioinformatics system for quality assurance of genomics and proteomics array data. Ann Biomed Eng. 2007;35:1068–80. doi: 10.1007/s10439-007-9313-y. [DOI] [PubMed] [Google Scholar]
- 15.Steinhoff C, Vingron M. Normalization and quantification of differential expression in gene expression microarrays. Brief Bioinform. 2006;7:166–77. doi: 10.1093/bib/bbl002. [DOI] [PubMed] [Google Scholar]
- 16.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 17.Phan JH, Young AN, Wang MD. Selecting clinically-driven biomarkers for cancer nanotechnology. Conf Proc IEEE Eng Med Biol Soc. 2006;1:3317–20. doi: 10.1109/IEMBS.2006.259746. [DOI] [PubMed] [Google Scholar]
- 18.Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huynh KN, Phan JH, Vo TM, et al. Improved bolstering error estimation for gene ranking. Conf Proc IEEE Eng Med Biol Soc. 2007;2007:4633–6. doi: 10.1109/IEMBS.2007.4353372. [DOI] [PubMed] [Google Scholar]
- 20.Chen YT, Tu JJ, Kao J, et al. Messenger RNA expression ratios among four genes predict subtypes of renal cell carcinoma and distinguish oncocytoma from carcinoma. Clin Cancer Res. 2005;11:6558–66. doi: 10.1158/1078-0432.CCR-05-0647. [DOI] [PubMed] [Google Scholar]
- 21.Zeeberg BR, Feng W, Wang G, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Specht K, Richter T, Muller U, et al. Quantitative gene expression analysis in microdissected archival formalin-fixed and paraffin-embedded tumor tissue. Am J Pathol. 2001;158:419–29. doi: 10.1016/S0002-9440(10)63985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chuang ST, Chu P, Sugimura J, et al. Overexpression of glutathione s-transferase alpha in clear cell renal cell carcinoma. Am J Clin Pathol. 2005;123:421–9. doi: 10.1309/AQXR-6B2Q-PUGD-638C. [DOI] [PubMed] [Google Scholar]
- 24.Higgins JP, Shinghal R, Gill H, et al. Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray. Am J Pathol. 2003;162:925–32. doi: 10.1016/S0002-9440(10)63887-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao M, Tabuchi H, Nagashima Y, et al. Gene expression analysis of renal carcinoma: adipose differentiation-related protein as a potential diagnostic and prognostic biomarker for clear-cell renal carcinoma. J Pathol. 2005;205:377–87. doi: 10.1002/path.1693. [DOI] [PubMed] [Google Scholar]
- 26.Salzberg SL. Genome re-annotation: a wiki solution? Genome Biol. 2007;8:102. doi: 10.1186/gb-2007-8-1-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giles J. Key biology databases go wiki. Nature. 2007;445:691. doi: 10.1038/445691a. [DOI] [PubMed] [Google Scholar]
- 28.Dorai T, Sawczuk IS, Pastorek J, et al. The role of carbonic anhydrase IX overexpression in kidney cancer. Eur J Cancer. 2005;41:2935–47. doi: 10.1016/j.ejca.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Bui MH, Seligson D, Han KR, et al. Carbonic anhydrase IX is an independent predictor of survival in advanced renal clear cell carcinoma: implications for prognosis and therapy. Clin Cancer Res. 2003;9:802–11. [PubMed] [Google Scholar]
- 30.Patard JJ, Fergelot P, Karakiewicz PI, et al. Low CAIX expression and absence of VHL gene mutation are associated with tumor aggressiveness and poor survival of clear cell renal cell carcinoma. Int J Cancer. 2008;123:395–400. doi: 10.1002/ijc.23496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shuch B, Li Z, Belldegrun AS. Carbonic anhydrase IX and renal cell carcinoma: prognosis, response to systemic therapy, and future vaccine strategies. BJU Int. 2008;101 4:25–30. doi: 10.1111/j.1464-410X.2008.07645.x. [DOI] [PubMed] [Google Scholar]
- 32.Stassar MJ, Devitt G, Brosius M, et al. Identification of human renal cell carcinoma associated genes by suppression subtractive hybridization. Br J Cancer. 2001;85:1372–82. doi: 10.1054/bjoc.2001.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishie A, Masuda K, Otsubo M, et al. High expression of the Cap43 gene in infiltrating macrophages of human renal cell carcinomas. Clin Cancer Res. 2001;7:2145–51. [PubMed] [Google Scholar]
- 34.Pejovic M, Djordjevic V, Ignjatovic I, et al. Serum levels of some acute phase proteins in kidney and urinary tract urothelial cancers. Int Urol Nephrol. 1997;29:427–32. doi: 10.1007/BF02551108. [DOI] [PubMed] [Google Scholar]
- 35.Senra Varela A, Lopez Saez JJ, Quintela Senra D. Serum ceruloplasmin as a diagnostic marker of cancer. Cancer Lett. 1997;121:139–45. doi: 10.1016/s0304-3835(97)00340-6. [DOI] [PubMed] [Google Scholar]
- 36.Nayak SB, Bhat VR, Upadhyay D, et al. Copper and ceruloplasmin status in serum of prostate and colon cancer patients. Indian J Physiol Pharmacol. 2003;47:108–10. [PubMed] [Google Scholar]
- 37.Nayak SB, Bhat VR, Mayya SS. Serum copper, ceruloplasmin and thiobarbituric acid reactive substance status in patients with ovarian cancer. Indian J Physiol Pharmacol. 2004;48:486–8. [PubMed] [Google Scholar]
- 38.Boz A, Evliyaoglu O, Yildirim M, et al. The value of serum zinc, copper, ceruloplasmin levels in patients with gastrointestinal tract cancers. Turk J Gastroenterol. 2005;16:81–4. [PubMed] [Google Scholar]
- 39.Zowczak M, Iskra M, Paszkowski J, et al. Oxidase activity of ceruloplasmin and concentrations of copper and zinc in serum of cancer patients. J Trace Elem Med Biol. 2001;15:193–6. doi: 10.1016/S0946-672X(01)80066-3. [DOI] [PubMed] [Google Scholar]
- 40.Goutebroze L, Brault E, Muchardt C, et al. Cloning and characterization of SCHIP-1, a novel protein interacting specifically with spliced isoforms and naturally occurring mutant NF2 proteins. Mol Cell Biol. 2000;20:1699–712. doi: 10.1128/mcb.20.5.1699-1712.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scoles DR. The merlin interacting proteins reveal multiple targets for NF2 therapy. Biochim Biophys Acta. 2008;1785:32–54. doi: 10.1016/j.bbcan.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Brembeck FH, Opitz OG, Libermann TA, et al. Dual function of the epithelial specific ets transcription factor, ELF3, in modulating differentiation. Oncogene. 2000;19:1941–9. doi: 10.1038/sj.onc.1203441. [DOI] [PubMed] [Google Scholar]
- 43.Tymms MJ, Ng AY, Thomas RS, et al. A novel epithelial-expressed ETS gene, ELF3: human and murine cDNA sequences, murine genomic organization, human mapping to 1q32.2 and expression in tissues and cancer. Oncogene. 1997;15:2449–62. doi: 10.1038/sj.onc.1201427. [DOI] [PubMed] [Google Scholar]
- 44.He J, Pan Y, Hu J, et al. Profile of Ets gene expression in human breast carcinoma. Cancer Biol Ther. 2007;6:76–82. doi: 10.4161/cbt.6.1.3551. [DOI] [PubMed] [Google Scholar]
- 45.Tschoep K, Kohlmann A, Schlemmer M, et al. Gene expression profiling in sarcomas. Crit Rev Oncol Hematol. 2007;63:111–24. doi: 10.1016/j.critrevonc.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 46.Manavathi B, Rayala SK, Kumar R. Phosphorylation-dependent regulation of stability and transforming potential of ETS transcriptional factor ESE-1 by p21-activated kinase 1. J Biol Chem. 2007;282:19820–30. doi: 10.1074/jbc.M702309200. [DOI] [PubMed] [Google Scholar]
- 47.Prescott JD, Koto KS, Singh M, et al. The ETS transcription factor ESE-1 transforms MCF-12A human mammary epithelial cells via a novel cytoplasmic mechanism. Mol Cell Biol. 2004;24:5548–64. doi: 10.1128/MCB.24.12.5548-5564.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang J, Lee C, Hahm KB, et al. Over-expression of ERT(ESX/ESE-1/ELF3), an ets-related transcription factor, induces endogenous TGF-beta type II receptor expression and restores the TGF-beta signaling pathway in Hs578t human breast cancer cells. Oncogene. 2000;19:151–4. doi: 10.1038/sj.onc.1203252. [DOI] [PubMed] [Google Scholar]
- 49.Neve RM, Ylstra B, Chang CH, et al. ErbB2 activation of ESX gene expression. Oncogene. 2002;21:3934–8. doi: 10.1038/sj.onc.1205503. [DOI] [PubMed] [Google Scholar]
- 50.Scott GK, Chang CH, Erny KM, et al. Ets regulation of the erbB2 promoter. Oncogene. 2000;19:6490–502. doi: 10.1038/sj.onc.1204041. [DOI] [PubMed] [Google Scholar]
- 51.Uddin M, Opazo JC, Wildman DE, et al. Molecular evolution of the cytochrome c oxidase subunit 5A gene in primates. BMC Evol Biol. 2008;8:8. doi: 10.1186/1471-2148-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishigaki R, Osaki M, Hiratsuka M, et al. Proteomic identification of differentially-expressed genes in human gastric carcinomas. Proteomics. 2005;5:3205–13. doi: 10.1002/pmic.200401307. [DOI] [PubMed] [Google Scholar]
- 53.Tickoo SK, Lee MW, Eble JN, et al. Ultrastructural observations on mitochondria and microvesicles in renal oncocytoma, chromophobe renal cell carcinoma, and eosinophilic variant of conventional (clear cell) renal cell carcinoma. Am J Surg Pathol. 2000;24:1247–56. doi: 10.1097/00000478-200009000-00008. [DOI] [PubMed] [Google Scholar]
- 54.Simonnet H, Alazard N, Pfeiffer K, et al. Low mitochondrial respiratory chain content correlates with tumor aggressiveness in renal cell carcinoma. Carcinogenesis. 2002;23:759–68. doi: 10.1093/carcin/23.5.759. [DOI] [PubMed] [Google Scholar]
- 55.Chang TY, Chang CC, Ohgami N, et al. Cholesterol sensing, trafficking, and esterification. Annu Rev Cell Dev Biol. 2006;22:129–57. doi: 10.1146/annurev.cellbio.22.010305.104656. [DOI] [PubMed] [Google Scholar]
- 56.Yang JB, Duan ZJ, Yao W, et al. Synergistic transcriptional activation of human Acyl-coenzyme A: cholesterol acyltransterase-1 gene by interferon-gamma and all-trans-retinoic acid THP-1 cells. J Biol Chem. 2001;276:20989–98. doi: 10.1074/jbc.M011488200. [DOI] [PubMed] [Google Scholar]
- 57.Locke JA, Wasan KM, Nelson CC, et al. Androgen-mediated cholesterol metabolism in LNCaP and PC-3 cell lines is regulated through two different isoforms of acyl-coenzyme A:Cholesterol Acyltransferase (ACAT) Prostate. 2008;68:20–33. doi: 10.1002/pros.20674. [DOI] [PubMed] [Google Scholar]
- 58.Oliva E, Young RH, Amin MB, et al. An immunohistochemical analysis of endometrial stromal and smooth muscle tumors of the uterus: a study of 54 cases emphasizing the importance of using a panel because of overlap in immunoreactivity for individual antibodies. Am J Surg Pathol. 2002;26:403–12. doi: 10.1097/00000478-200204000-00001. [DOI] [PubMed] [Google Scholar]
- 59.Qian X, Peng XH, Ansari DO, et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 60.Xing Y, Chaudry Q, Shen C, et al. Bioconjugated quantum dots for multiplexed and quantitative immunohistochemistry. Nat Protoc. 2007;2:1152–65. doi: 10.1038/nprot.2007.107. [DOI] [PubMed] [Google Scholar]
- 61.Kummerlin I, ten Kate F, Smedts F, et al. Core biopsies of renal tumors: a study on diagnostic accuracy, interobserver, and intraobserver variability. Eur Urol. 2008;53:1219–25. doi: 10.1016/j.eururo.2007.11.054. [DOI] [PubMed] [Google Scholar]
- 62.Motzer RJ, Bacik J, Mariani T, et al. Treatment outcome and survival associated with metastatic renal cell carcinoma of non-clear-cell histology. J Clin Oncol. 2002;20:2376–81. doi: 10.1200/JCO.2002.11.123. [DOI] [PubMed] [Google Scholar]


