Abstract
Heterogeneous nuclear ribonucleoproteins (hnRNPs) are multi-functional proteins that bind to newly synthesized mRNAs in the nucleus and participate in many subsequent steps of gene expression. A well-studied Saccharomyces cerevisiae hnRNP that has several nuclear functions is Npl3p. Here, we provide evidence that Npl3p also has a cytoplasmic role: it functions in translation termination fidelity. Yeast harboring the npl3-95 mutant allele have impaired ability to translate LacZ, enhanced sensitivity to cycloheximide and paromomycin, and increased ability to readthrough translation termination codons. Most of these defects are enhanced in yeast that also lack Upf1p, a factor crucial for the nonsense-mediated decay (NMD) RNA surveillance pathway that is also known to be involved in translation termination. We show that the npl3-95 mutant allele encodes an Npl3p that forms high molecular-weight complexes that co-fractionate with the poly(A)-binding protein Pab1p. Together, the data suggest that Npl3p plays a role in translation termination in yeast.
INTRODUCTION
The original notion that gene expression is a series of independent steps has been replaced with the view that gene expression is a highly coupled pathway linked by key integrator molecules 1; 2. For example, in the nucleus, the proteins required for mRNA capping are recruited to the transcription complex during transcription initiation, permitting mRNA capping to occur co-transcriptionally 3; 4. Because the 5'-cap protects mRNAs from decay, it is crucial that capping occurs soon after the 5′-end of the mRNA is transcribed. Later events, such as mRNA splicing, cleavage, and polyadenylation, often also occur while a mRNA is still being transcribed 5; 6. All these nuclear post-transcriptional events are coupled with transcription via the C-terminal domain (CTD) of the large subunit of RNA polymerase II (Pol II). The CTD achieves this by recruiting factors involved in all of these post-transcriptional events. Many other gene expression events in the nucleus are also coupled, including 5′-capping, splicing and polyadenylation 6; 7.
Following these nuclear processing events, transcripts must become competent for export across the nuclear pore 1; 4; 5; 8. This competence is accomplished by the recruitment of specific RNA export factors and hnRNPs to newly synthesized transcripts. For example, the transcription export (TREX) complex, which binds the mRNA during transcriptional elongation, serves to promote RNA export 9; 10. The Mex67p (TAP/NXF1 in vertebrates) protein is another important factor required for the export of mRNA to the cytoplasm 11. Mex67p forms a heterodimeric export receptor with the protein Mtr2, which together interact with nuclear pore proteins and other adaptor RNA binding proteins to facilitate RNA movement across the nuclear pore 12; 13; 14.
Once in the cytoplasm, mRNAs undergo additional coupled events. For example, translation and cytoplasmic mRNA decay are linked, such that translation sometimes impedes decay and in other cases it accelerates decay. A quality control mechanism that may link both nuclear and cytoplasmic events is nonsense-mediated mRNA decay (NMD). This RNA surveillance mechanism recognizes and rapidly degrades mRNAs harboring premature (nonsense) termination codons (PTCs) 15; 16; 17. NMD depends on several highly conserved factors, including Upf1, Upf2 and Upf318; 19; 20; 21; 22; 23. The Upf proteins are involved not only in RNA surveillance but they also act in general translation and translation termination fidelity19; 24; 25. As evidence for the latter, deletion of any one of the three UPF genes leads to partial suppression of nonsense codon recognition 26; 27. All together, during the process of gene expression a variety of factors associate with the mRNA to promote the translation of the encoded protein. Throughout this process, factors such as Mex67p associate during the mRNA export phase; whereas others, like the Upf proteins, bind to the mRNA upon export to the cytoplasm 16. This constant exchange of factors that results in a highly dynamic mRNA molecule is commonly referred to messenger ribonucleoprotein (mRNP) remodeling 4.
In this paper, we focus on Npl3p, a well-studied multi-functional hnRNP that participates in many steps of gene expression. Npl3p interacts with the CTD of Pol II and directly stimulates the elongation activity of RNA polymerase II 28. It also binds to pre-mRNA and promotes its processing by recruiting RNA splicing factors 29; 30; 31. Following mRNA processing, Npl3p remains bound to the mRNA and actively promotes its export 32. Recent evidence has suggested that Npl3p also functions in translation. Yeast cells harboring the npl3-27 allele are defective in releasing the mutant form of Npl3p from translating polyribosomes and demonstrate impaired translation 33. Furthermore, Npl3p copurifies with ribosomal proteins and the poly(A+) mRNA binding protein, Pab1, both key components of the translational apparatus 33; 34. Together, these results suggest that Npl3p is a good candidate to be a crucial factor that couples events in both the nucleus and the cytoplasm.
In this study, we address the role of Npl3p in translation. We provide evidence that a mutant form of Npl3p functions in translation termination fidelity. The translation termination defects observed in cells harboring a mutant allele of NPL3 are enhanced by loss of a factor previously known to have a role in translation termination fidelity: Upf1p. Together, our results suggest that Npl3p plays a role in translation termination in yeast.
RESULTS
Evidence that Npl3p and Upf1p cooperate to facilitate translation in S. cerevisiae
Previous studies have demonstrated that Npl3p is recruited early during transcription and plays a role in many subsequent events, including transcription termination, 3′ end formation, mRNP assembly, and RNA export 29; 31; 35. More recent studies have also linked Npl3p with translation, but a specific role for Npl3p in this process has not been elucidated 33. To address this issue, we used S. cerevisiae strains harboring the npl3-95, npl3-101 and npl3-1 mutant alleles. The npl3-95 mutant allele was originally identified as temperature-sensitive mutation that negatively silences the HMR locus 36; it has also been shown to be required for mRNA export in yeast 32. The npl3-101 and npl3-1 alleles were shown to mislocalize nuclear-bound proteins to other cellular compartments 32. The mutations in these three alleles are all located within RNA recognition motif 2 (RRM2) of Npl3p (Fig. 1A). The npl3 mutant alleles used in this study share several phenotypes including: a) temperature sensitivity (Tsm−) and b) defects in mRNA export. These phenotypes are only manifested at the restrictive temperature (≥28C°). Hence, the assays in this study were conducted at the permissive temperature (24C°) where no mRNA export defects are observed 32.
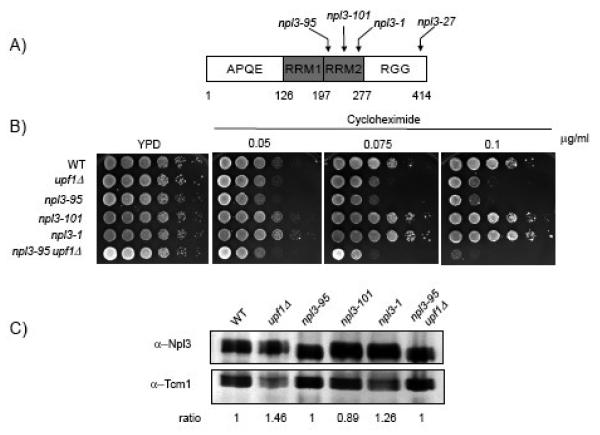
Figure 1. Npl3p plays a role in translation.
(A) Schematic diagram of Npl3p showing the two RNA binding motifs (RRMs) located in the central portion of the polypeptide, the APQE motif in the N-terminal end and the RGG repeats in the carboxyl end. The arrows show the location of the three mutant alleles (npl3-95, npl3-101, npl3-1 and npl3-27) in the RRM2 that were used in this study. B) Ten-fold serial dilutions of yeast cells spotted onto YPD or YPD media containing various concentrations (0.05 g/ml, 0.075 g/ml or 0.10 g/ml) of cycloheximide are shown. Plates were incubated for three days at 24°C. C) Western blot analysis was performed with lysates from WT, upf1Δ, npl3-95, npl3-101, npl3-1 and npl3-95/upf1Δ yeast strains. Ten micrograms of total protein extracts were immunoblotted with antibodies against Npl3p and Tcm1p (ribosomal protein control).
As a first test of whether Npl3p has a role in translation, we examined the sensitivity of these strains to the translation inhibitor cycloheximide. Previous studies have shown that yeast strains with translational defects are hypersensitive to the growth-inhibitory effects of this compound 37; 38. As shown in Fig. 1B, the npl3-95 strain was hypersensitive to cycloheximide, while the npl3-101 and npl3-1 strains were resistant to this translational inhibitor (Fig. 1B). The Npl3-95p was expressed at levels comparable to that of the wild type Npl3p (Fig. 1C), suggesting that the defect observed was a result of a functional deficiency in translation and not due to reduced protein levels.
To further investigate the role of Npl3p in translation, we tested whether Npl3p genetically interacts with Upf1p, a central component of the NMD RNA surveillance pathway that has a role in translation and is known to physically interact with the hnRNP family member Hrp1p 39. In agreement with previous reports, we found that a strain harboring a deletion of the UPF1 gene (upf1Δ) was sensitive to cycloheximide (Fig. 1B) 40. The upf1Δ strain showed less sensitivity to this drug than the npl3-95 strain. When the two mutations were combined, this elicited an enhanced growth defect in response to cycloheximide treatment relative to either the upf1Δ or npl3-95 single-mutant strains (Fig. 1B). The synthetic enhancement of the cycloheximide sensitivity phenotype in the double-mutant (npl3-95/upf1Δ) strain supports the notion for a role of the Npl3-95p mutant in translation 41.
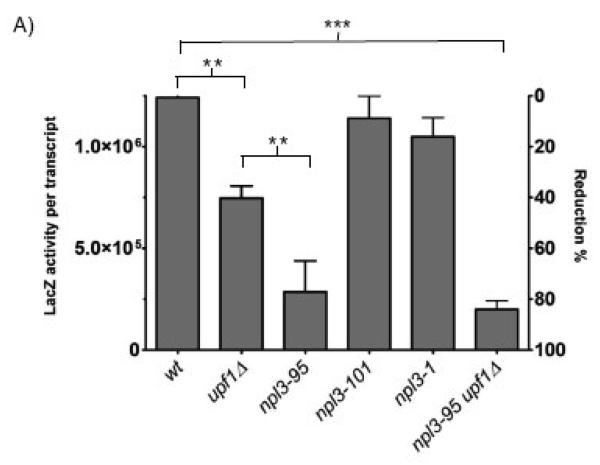
As an independent assay for detecting defects in protein synthesis, we transformed npl3-95, npl3-101 and npl3-1 strains with a β-galactosidase reporter and measured translation rates. The total activity for each strain was normalized to lacZ mRNA and expressed as β-galactosidase activity (data not shown). As shown in Fig. 2, the npl3-95 strain exhibited a ~70% reduction in total lacZ activity compared to wild-type cells. This translation defect was allele specific, as cells harboring the npl3-101 or npl3-1 alleles did not exhibit reduced lacZ activity. The upf1Δ strain exhibited a less marked (~40%) reduction in total lacZ activity, while the double npl3-95/upf1Δ mutant strain exhibited a strong (~85%) reduction in total lacZ activity. These results suggest defects in translation of the lacZ mRNA in these strains and are consistent with the cycloheximide sensitivity phenotypes observed in Fig. 1B.
Figure 2. General translation rates of the npl3 mutant strains.
A) Wild-type control (WT), upf1Δ, npl3-95, npl3-101 and npl3-1 strains were transformed with a lacZ reporter construct and β-galactosidase assays were performed. LacZ activity is expressed in β-galactosidase units per transcript for each strain used in the analysis. Total lacZ activity was normalized to total lacZ mRNA levels (activity per transcript). The LacZ/mRNA activity obtained for the wild-type strain was set as 100% and used as a reference point for comparison. n = 3 independent experiments. Error bars represent 1 SEM above and below the mean. * p< 0.05, **p< 0.001, ***p< 0.0001.
Npl3p and Upf1p are required for accurate translation termination
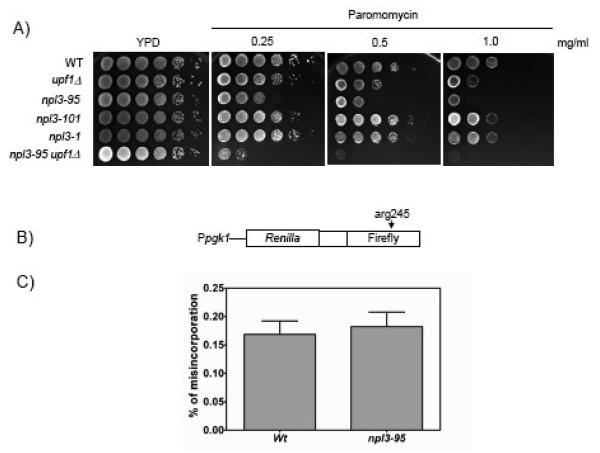
To determine whether Npl3p functions to promote translation by increasing its fidelity, we examined the effect of paromomycin on the growth phenotypes of the npl3 mutant strains. This aminoglycoside increases the error rates of translating ribosomes; hence strains compromised for translational fidelity are hypersensitive to paromomycin 37; 42. We found that the npl3-95 strain was sensitive to paromomycin, indicative of a defect in translational fidelity in these cells (Fig. 3A). In contrast, the npl3-101 and npl3-1 strains did not show sensitivity to this translational inhibitor (Fig. 3A). In agreement with previous reports, we found that the growth of the upf1Δ strain was also sensitive to paromomycin; albeit less so (Fig. 3A). Analysis of the double-mutant (npl3-95/upf1Δ) strain revealed a synthetically enhanced sensitivity to paromomycin relative to the single-mutant strains. These results are consistent with the assay performed in the presence of cycloheximide (Fig. 1B) and the translational rates of lacZ reporters (Fig. 2), providing further evidence for a role of Npl3p in translation fidelity.
Figure 3. The npl3-95 mutant allele is sensitive to paromomycin.
A) Ten-fold serial dilutions of various yeast strains spotted on YPD or YPD media containing 0.25 mg/ml, 0.50 mg/ml or 1.0 mg/ml of the translational inhibitor paromomycin. Cells were incubated for three days at 24°C. B) Schematic diagram of the dual luciferase reporter chimera used to measure translation elongation fidelity. C) The percentage of elongation misincorporation was expressed as wild-type or mutant Firefly/Renilla luciferase activity. No significant difference on translation elongation fidelity was observed between the wild type NPL3 and mutant npl3-95 strains (P = 0.7108).
To determine whether Npl3p promotes translation fidelity during the elongating phase of translation, we used a dual-luciferase reporter construct previously developed to measure translation elongation fidelity 42. This reporter construct contains Renilla and firefly luciferase coding sequences in tandem, separated by a linker region (Fig. 3B). The firefly luciferase gene in this construct harbors a histidine-toarginine mutation (Arg245) within a catalytically important residue that, under normal conditions, generates an inactive form of firefly luciferase 42. Cells with reduced translation elongation accuracy incorporate histidine at this position instead of arginine, resulting in the production of an active form of firefly luciferase. The misincorporation of histidine instead of arginine at this position occurs at relatively high frequency because the histidine-tRNA is the near-cognate tRNA for arginine. This system allows for an accurate measure of translation elongation fidelity by determining the activity of the firefly luciferase enzyme relative to the upstream Renilla luciferase activity 42. Using this assay, we found that both npl3-95 and wild-type strains expressed similar levels of firefly luciferase activity suggesting that npl3-95 cells are not defective in translation elongation fidelity (Fig. 3C).
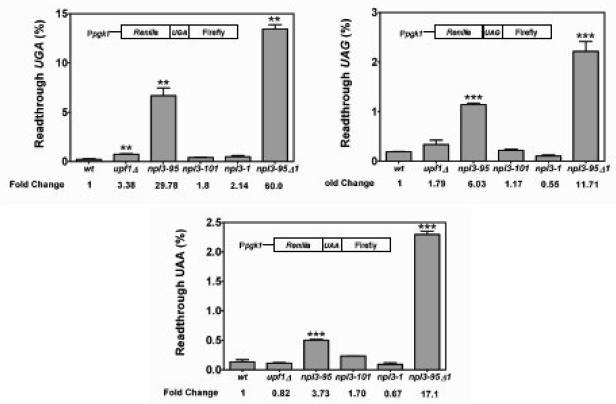
Given that Npl3p did not appear to function in translation elongation fidelity, we determined its role in translation termination. To test this, we used a dual-luciferase assay system previously developed to quantify translation termination efficiency 26. This assay consists of a bicistronic reporter mRNA that contains the Renilla and the firefly luciferase in tandem, separated by a linker sequence that harbors a stop codon (Fig. 4). In yeast strains defective in translation termination, higher levels of firefly luciferase are synthesized due to a higher frequency of ribosomes readthrough of the stop codon. Therefore, an increase in activity of the firefly luciferase relative to the Renilla activity is directly proportional to defects in translation termination. Using this dual-luciferase assay, we quantitated the readthrough activity at all three stop codons. The upf1Δ strain expressed modestly higher levels of luciferase than the wild-type strain (by 3- and 2-fold for the UGA and UAG constructs, respectively; Fig. 4), in agreement with previous reports 26. Under similar conditions, we were not able to observe translational readthrough activity for the upf1Δ strain harboring the UAA stop codon construct. Strikingly, npl3-95 cells exhibited much more elevated readthrough activity (by 30-, 6-, and 4-fold for the UGA, UAG, and UAA constructs, respectively; Fig. 4) than upf1Δ cells, indicating that defects in Npl3p cause a much more profound defect in translation termination fidelity than loss of Upf1p. As was the case for cycloheximide, translational rates and paromomycin sensitivity (Figs. 1B, 2 & 3A, respectively), this effect was specific for the npl3-95 allele, as the npl3-1 and npl3-101 strains did not have significantly higher luciferase activity than wild-type cells (Fig. 4). Double-mutant (npl3-95/upf1Δ) cells had higher luciferase activity than either single-mutant (npl3-95 or upf1Δ) strain (Fig. 4). The increased readthrough activity was greater than additive in the double-mutant cells as compared to the single-mutant cells at all three stop codons, implicating Npl3p in translation termination fidelity. Together with the data showing that the double-mutant strain had increased sensitivity to translation inhibitors (Fig. 1B & 3A), these results suggest that Npl3 is required to promote accurate translation termination.
Figure 4. npl3-95 cells display defects in translation fidelity.
A) Dual luciferase assays were conducted to determine the efficiency of stop codon recognition (UAA, UAG, or UGA). The percentage of translational readthrough for each strain was calculated as the Firefly/Renilla luciferase activity (nonsense) divided by the Firefly/Renilla luciferase activity (sense) multiplied by 100. Two tailed t-tests were used for statistical testing. n = 3 for all experiments shown (the asterisk indicates a statistically significant result when compared to the wild type strain).
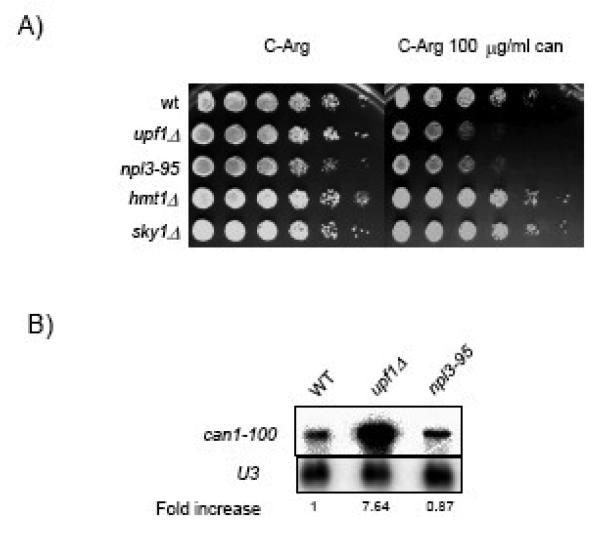
Hmt1p and Sky1p do not play a role in translation termination
Npl3p, like other hnRNP proteins, is subjected to several post-translational modifications including methylation and phosphorylation 11; 43; 44. The nuclear arginine methyl-transferase, Hmt1p, is involved in the methylation of the RGG domain of Npl3p. The methylation of these amino acid residues promotes nuclear-cytoplasmic shuttling of Npl3p 43; 45. Deletion of Hmt1p is lethal in combination with several npl3 mutant alleles, including npl3-95, a phenotype that may reflect an effect in a critical process such as translational accuracy 46. Npl3p is also subjected to phosphorylation by the serine arginine (SR)-rich protein kinase Sky1p. This phosphorylation and dephosphorylation cycle allows Npl3p to disengage from the mRNA and shuttle back into the nuclear compartment 47; 48. To determine whether Hmt1p or Sky1p are required for proper translation termination, we assessed the nonsense suppression phenotype of the can1-100 allele (which contains an UAA premature termination codon) in these mutant strains. In yeast strains defective in translation termination, nonsense codon readthrough produces a functional form of the arginine permease Can1p that is capable of importing canavanine, a toxic arginine analog. Therefore, decreased growth rates correlate with enhanced nonsense suppression activity. 49; 50. As shown in Fig. 5, hmt1Δ or sky1Δ strains did not display nonsense suppression phenotypes. In contrast, the npl3-95 and the upf1Δ mutant strains grew much more poorly (Cans) in the presence of canavanine (Fig. 5), in agreement with the defects in translation termination observed in both strains (Figs. 3 & 4). The upf1Δ strain showed more sensitivity to canavanine than npl3-95 due to the mRNA stabilizing effect on the can1-100 transcript that occurs in upf1Δ strain (Fig. 5B). The stabilization of the can1-100 NMD substrate provides for enhanced substrate availability and worse growth conditions for the upf1Δ strain. Therefore, these results are consistent with Npl3-95p mutant having a strong defect in translation termination. The results also indicate that Hmt1p or Sky1p do not appear to play a role in translation termination in yeast. Hence, the role of Npl3p in translation termination is independent of the post-translational modifications mediated by Hmt1p and/or Sky1p.
Figure 5. hmt1Δ or sky1Δ strains do not display nonsense suppression phenotypes.
A) Ten-fold serial dilutions of yeast suspensions with similar cell numbers from the indicated strains were plated on C-Arg (left panel) and C-Arg containing 100 g/ml of canavanine (right panel). Cells were incubated for three days at 24°C. B) Northern blot analysis of the can1-100 nonsense-containing mRNA measured in wild type, upf1Δ and npl3-95 strains.
The translation-deficient mutant Npl3p forms high molecular-weight complexes associated with Pab1p
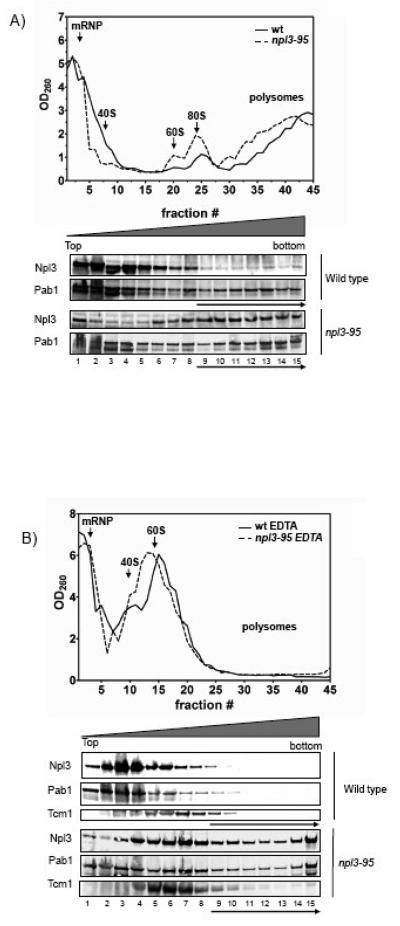
To determine whether the translational defects observed in cells harboring the npl3-95 mutant are due to abnormalities in polyribosome formation, we compared the polysome profile of this mutant strain with the wild-type strain using sucrose density sedimentation. We found that the npl3-95 strain exhibited a polysome profile very similar to that of wild-type cells (Fig. 6A). The finding that polysome formation was not significantly perturbed by the mutation in the npl3-95 allele suggested that Npl3p does not function in steps that lead up to polysome formation (e.g., translation initiation) and was consistent with our data showing that this mutation did not affect translation elongation fidelity (Fig. 3C) but rather affected translation termination (Fig. 4).
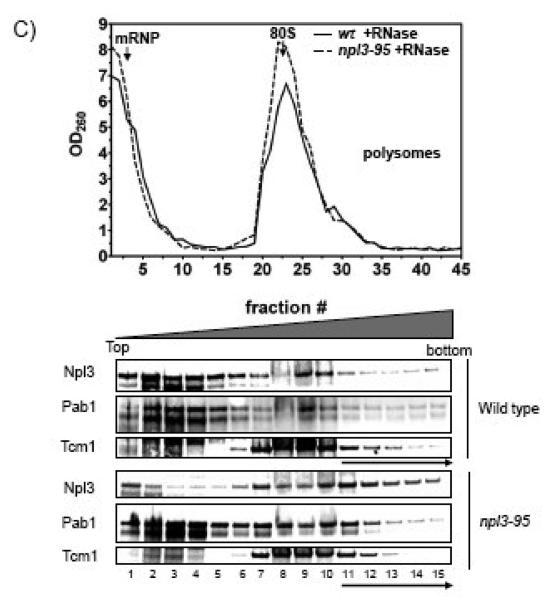
Figure 6. Cells harboring the npl3-95 allele form heavy mRNP arrangements.
The OD260 plot shows the UV absorption pattern from the sucrose gradient for wild type and npl3-95. The lower panels show Western blot analysis of the pooled fractions with antibodies against Npl3p and Pab1p. High density fractions refer to fractions 9 and above (except for the RNaseA experiment in which we the high density fractions refer to fractions 11 and above), these fractions are indicated with an arrow. Each protein lane represents three fractions from the UV plots that were pooled together. A) Distribution of Npl3p and Pab1p on sucrose density gradients. B) Polysome profile of wild type and npl3-95 extracts treated with 15 mM EDTA. C) Polysome profile of wild type and npl3-95 extracts treated with RNaseA (0.1 g/ml). Similar results were obtained in at least two independent experiments.
Because a role in translation termination could affect Npl3p's association with polysomes, we next determined the distribution of Npl3p in sucrose gradients. We found that ~25% of wild-type Npl3p associated with the high-density fractions in sucrose gradients (lane 9 and above), consistent with a protein involved in translation (Fig. 6A). To determine whether the presence of Npl3p in such high-density fractions of the gradient was indicative of its association with polysomes, we monitored Npl3p distribution in the presence of EDTA. Previous studies have demonstrated that treatment of whole cell extracts with EDTA dissociates polysomes by disrupting the interaction between the 40S and 60S ribosomal subunits 19. We found that treatment of lysates from wild-type cells with EDTA elicited a shift in Npl3p from the high-density fractions to lower fractions (lanes 1-8) such that only ~5% of it remained in the high-density fractions (Fig. 6B). Likewise, control proteins (Pab1p and the ribosomal protein Tcm1p) shifted towards the lighter fractions and were largely absent from the high-density fractions. Together, this data demonstrated that EDTA effectively disrupted the polysomes and that a significant fraction of Npl3p is normally associated with polysomes. This data also agreed with a previous study from Krebber and colleagues who found that ~30% of Npl3p associates with actively translating ribosomes 33.
The inability of the Npl3-95 mutant to efficiently promote translation termination suggested that this mutant protein might have an altered association with polysomes. Consistent with this, we found that the npl3-95 mutant strain expressed a form of Npl3p that was much more abundant in the high-density fractions than was wild-type Npl3p (Fig. 6A). More than 50% of the Npl3-95p mutant was found in the high-density fractions. To determine whether this was the result of an association with polysomes, we assayed whether EDTA would disrupt the association. Surprisingly, we found that the mutant Npl3p did not significantly shift from the high-density fractions when the lysates were treated with EDTA (Fig. 6B). Likewise, ~40% of Pab1p remained in the high-density fraction of EDTA-treated lysates prepared from the mutant npl3-95 strain (Fig. 6B). In marked contrast, only ~5% of Pab1p was in the high-density fraction of EDTA-treated lysates from cells harboring wild-type Npl3p. As a control, we examined the ribosomal protein Tcm1 and found that it shifted from the high-density fractions to the low-density fraction in response to EDTA treatment of both mutant and wild-type Npl3p lysates. Together, these results indicated that the Npl3-95p mutant co-sediments with high molecular-weight fractions that contain Pab1p but lack actively translating ribosomes.
To determine whether the Npl3p mutant and Pab1p co-fractionation depends on the presence of RNA, we treated the lysates with RNase A before sucrose gradient centrifugation. RNase treatment reduced the amount of Pab1p in the high-density fractions, whereas it had little or no effect on the amount of the Npl3p mutant in such fractions (Fig. 6C). This indicated that mutant Npl3p is probably an integral component of the high molecular-weight complexes, whereas Pab1p probably depends on interactions with the poly(A) tail of mRNA to be present in such complexes. In the Discussion, we speculate as to the nature of these high molecular-weight complexes.
DISCUSSION
In this study, we report that two proteins, Npl3p and Upf1p, previously shown to be involved at different steps of gene expression have a common function in translation termination. The mRNA-binding protein Npl3p functions in mRNA processing, transcription termination, and RNA export 28; 29; 31; 35, while the RNA helicase Upf1p is a central component of the NMD RNA surveillance pathway 51; 52; 53. We provide several lines of evidence that Npl3p and Upf1p function in translation termination in yeast. First, cells harboring mutations either in Npl3p or Upf1p were hypersensitive to the translational inhibitors cycloheximide and paromomycin (Figs. 1B and 3A). Second, the general translation rates were reduced in both npl3-95 and upf1Δ strains (Fig. 2). Third, a double npl3-95/upf1Δ mutant strain exhibited enhanced sensitivity to both of these translational inhibitors relative to the single-mutant (npl3-95 or upf1Δ) strains. This effect was also observed in the general translation rates as measured by lacZ reporters (Fig. 2). Finally, a quantitative read-through assay using a dual-luciferase assay system42 revealed an enhancement in translational readthrough activity (ranging from 12- to 60-fold) at all stop signals for the npl3-95/upf1Δ strain relative to the single-mutant strains (Fig. 4). Together, the results outlined above support the notion that Npl3p and Upf1p act to maintain accurate translation termination.
Notably, the translational defects observed in the npl3-95 strain are allele specific in nature. The npl3 alleles used in this study are located within the second RNA recognition motif (RRM2) of Npl3p (Fig. 1A). The npl3-95 mutant allele encodes a leucine to tyrosine mutation at position 216 within RRM2 of Npl3p, found within the α-helix one of the RRM2 domain of Npl3p 32. According to the available structural data, this helix packs against the back of the β-sheet which forms the RNA recognition surface 35. However, it is not localized within the RNA binding surface itself, thus making the disruption of accurate protein-RNA interactions an unlikely scenario 35. Interestingly, the residues that form the α-helix one are part of a heptapeptide repeat highly conserved among higher eukaryotic SR proteins such as SF2 and Drosophila SRp55 32. Mutational analysis of this region in SF2 have previously demonstrated a negative regulatory role of splicing in vitro 54. However, no significant inhibitory activity on splicing has been demonstrated for Npl3p. In contrast, the npl3-101 mutant allele is found within the β-sheet that forms the RNA binding surface of the RRM2 changing a glycine residue at position 241 to aspartate 32. This glycine at position 241 is predicted to be crucial for mRNA binding and a change to a negatively charged residue might be repulsive to the negatively charged mRNA molecule 55. The npl3-1 allele is located within α-helix 2 of the RRM2; this mutant represents a change in alanine 254 to a valine residue, conserving the hydrophobic nature at this location 32. Both mutations (npl3-101 and npl3-1) do not show any defects in general translation or translation termination, underscoring the importance of the conserved heptapetide sequence found within the RRM2. The RRM motif is known to mediate multiple types of interactions including protein-RNA, RRM-RRM and protein-protein interactions55; 56. Therefore, the npl3-95 mutant protein could be disrupting crucial protein-protein interactions that are necessary for accurate translational termination (e.g. aberrant Pab1p distribution observed in Fig. 6). This might explain the npl3-95 allele specific translational defects observed in this study.
Previous reports have suggested that both Npl3p and Upf1p play a role in translation 19; 33. In the case of Npl3p, it was previously shown to be physically associated with polysomes and proteins involved in translation, including Pab1p 9; 33. Moreover, cells harboring a mutant form of Npl3p (npl3-27) are defective in releasing the mutant form of Npl3p from translating polyribosomes and demonstrate impaired mRNA translation 33. Interestingly, the npl3-27 allele resembles the npl3-95 with regards to the defects observed in general translation and a robust presence in polyribosomal fractions. In contrast, the npl3-27 mutation is located at the C-terminal region of the protein within the RGG domain (Fig. 1A) 33. This region is crucial for the shuttling function of SR proteins, hence it is the region targeted for phosphorylation by the Sky1 protein kinase and methylation by the Hmt1 methylase 47. However, our results do not suggest a role for these modifications in the process of translation termination (Fig. 5). Together, these results suggest that the RGG and RRM2 domains of Npl3p might play important roles in the process of translation. The data in our paper clarifies the role of Npl3p by providing strong evidence that it functions in translation termination fidelity (Figs.1, 2, 4). These results might indicate a potential defect for the npl3-27 allele in nonsense suppression. However, dual luciferase assays fail to detect such effects (data not shown). This further underscores the allele specificity effects of npl3-95 in translation termination.
We also confirmed that Upf1p has a role in translation termination (Figs. 1, 2, 4, 5) 19; 57. Recent studies have suggested that human UPF1 also has role in translation: phosphorylated UPF1 interacts with eIF3, a translation initiation factor required for the recycling of post-termination complexes, and triggers repression during NMD 58. It remains to be determined precisely how Npl3p and Upf1p function in translation termination fidelity. We found that the combination of mutations in both genes leads to a synthetic enhancement of the translational defects observed in either mutation alone (Figs. 1B, 2, 3A, 4, 5). The synthetic enhancement of the npl3-95 translational defects by the deletion of upf1Δ further supports a role for Npl3p in translation termination.
We found that the translation-deficient mutant form of Npl3p (Npl3-95p) is associated with high molecular-weight fractions in sucrose density gradients raising the possibility that it may associate with high molecular-weight complexes (Fig. 6A). These complexes are unlikely to contain polysomes, as EDTA treatment had little effect on them (Fig. 6B). Strikingly, a significant fraction (40%) of Pab1p cosedimented with these high molecular-weight fractions in a RNA-dependent manner (Figs. 6B & 6C), suggesting that the npl3-95 mutant strain harbors a form of Npl3p that might sequester Pab1p in large RNPs. Based on these observations, we propose that the Npl3-95p mutant protein promotes an aberrant mRNP conformation that can sequester Pab1p from the actively translating mRNA pool, thus compromising translation termination fidelity. Consistent with this idea, Pab1p has been previously implicated in the regulation of translation termination and is an important component of the translation machinery 59; 60. Other proteins, including Npl3p-interacting proteins (e.g. Gbp2p, Hrb1p), might also be present in these high molecular-weight complexes and thereby contribute to the translation termination defect.
Our model also implies that a correct mRNP environment is a prerequisite for proper translation termination to occur 4; 61; 62. Upf1p may have a role in creating this environment, as previous studies have suggested that Upf1p plays a role in mRNP remodeling 62. We further hypothesize that the mRNP composition at the translation termination event requires the integrity of the machinery for proper termination to occur. The improper rearrangement of the mRNP due to shortage of Pab1p (titrated by the mutant Npl3-95p) at the termination event may provoke a delay in the change of the ribosome conformation(s) required for polypeptide chain release 63. This, in turn, might open a window for a near cognate tRNA to enter the ribosomal-A site and promote translational readthrough 63.
Our studies suggest that Npl3p has a role in translation that is independent of its function in mRNA export 33, transcription and/or nuclear protein import 28; 29. We hypothesize that other multi-functional proteins with similar roles to Npl3p might participate in this mRNP remodeling event. For example, recent studies have linked the RNA helicase, Dbp5 to mRNP remodeling and this factor is also required for translational fidelity 37; 64. Therefore, factors such as TAP1/NXF1, Gle1 and Mex67 are potential candidates that might be involved in this nucleocytoplasmic coupling of mRNA export, translation and turnover 61; 64; 65. In summary, our work establishes Npl3p as a multi-functional RNA-binding protein with functions that extend beyond the nucleus. Future studies will be required to determine precisely how Npl3p promotes translation termination fidelity in the cytoplasm.
METHODS
Yeast strains, growth media and plasmids
All strains used in this study are derived from the W303 genetic background. The genotypes of all yeast strains used in this study are listed in Table 1. Yeast strains were routinely grown at 24°C on either standard yeast extract/peptone (YP) medium or synthetic medium supplemented with appropriate amino acids and 2% dextrose as carbon source 66. Genetic manipulations were performed via PCR-mediated gene disruption and correct integration confirmed by PCR 67. The presence of the can1-100 allele was confirmed by PCR amplification of the CAN1 locus and DNA sequencing. Canavanine (250 μg/ml), cycloheximide (0.05 μg/ml, 0.075 μg/ml and 0.1 μg/ml) and paromomycin (0.25 mg/ml to 1 mg/ml) were added to the autoclaved YPD medium. Yeast transformations were performed by the lithium acetate procedure and plasmids were maintained by growth in selective media 66; 68. Plasmids used in this study are listed in Table 2.
Table 1.
Yeast strains used in this study.
| Strains | source | Genotype |
|---|---|---|
| YCG9 | This study | MATa leu2-3 can1-100 ura3-1 ade2-1 his3-11,15 trp1-1 |
| YCG11 | Henry et. al. | Matα leu2-3 can1-100 ura3-1 ade2-1 his3-11,15 trp1-1 npl3-101 |
| YCG12 | Henry et. al. | MATa leu2-3 can1-100 ura3-1 ade2-1 his3-11,15 trp1-1 npl3-95 |
| YCG16 | Henry et. al. | Matα his3-11 ade2-1 leu2-1 ura3-1 npl3-1 |
| YCG25 | Henry et. al. |
MATa ade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 upf1::HIS3 |
| YCG35 | This study |
MATa ade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 upf1::his5 npl3-95 |
| YCG46 | This study |
MATa ade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 sky1::HIS3 |
| YCG47 | This study |
MATa ade2-1 his3-11, 15 leu2-3, 112 trp1-1 ura3-1 can1-100 hmt1::HIS3 |
Table 2.
Plasmids used in this study.
| Plasmids | source | Plasmid description |
|---|---|---|
| pCG95 |
Salas-Marco et.al. |
URA3 vector CAAA control for pCG96. |
| pCG96 |
Salas-Marco et.al |
URA3 vector UAAA stop codon. |
| pCG97 |
Salas-Marco et.al |
URA3 vector CAGC control for pCG98. |
| pCG98 |
Salas-Marco et.al |
URA3 vector UAGC stop codon. |
| pCG99 |
Salas-Marco et.al |
URA3 vector CGAC control for pCG100. |
| pCG100 |
Salas-Marco et.al |
URA3 vector UGAC stop codon. |
| pCG106 | Pappas et. al. | PGK1 (TATA)-lacZ URA3. |
| pCG107 |
Salas-Marco et.al |
Dual luciferase assay reporter for missincorporation. Firefly luciferase H245 (CAC to CGC). |
RNA isolation and Northern blotting
Total RNA was isolated via the hot acid phenol method and mRNA abundance was determined by Northern blot analysis as described previously 39; 69. Briefly, 50 ml of yeast cells were grown at 24°C to OD600 of 0.6-0.8. Cells were centrifuged, washed twice in RNA extraction buffer (0.3M Sodium acetate, 50mM Tris pH 8.0, 5mM EDTA, 1% SDS) and resuspended in 400μl of RNA extraction buffer and equal volume of hot saturated phenol (phenol pH 6.8). The mixtures were incubated at 65°C for 5 minutes with vortex every minute, followed by centrifugation (15,682 g) at 4°C. This procedure was repeated twice and following centrifugation, the aqueous phase was extracted with RNA phenol (phenol-chloroform-isoamyl alcohol 125:24:1, pH 4.3). The supernatant was collected and one ml of ice cold 100% ethanol was added to the supernatant to precipitate the RNA. RNA samples were then centrifuged for 30 minutes at 15,682 g at 4°C. Pellets were washed with 70% ethanol and vacuum dried before resuspension in 50 μl of 10 mM Tris pH 7.5. Random-primed probes were prepared from PCR-amplified fragments of CAN1, LacZ, and U3 to high specific activity with 32P-dCTP following the recommendations from the manufacturer (Roche). The results from these studies were quantified using a Molecular Imager FX (BioRad, Hercules, CA). The values shown represent the average of at least three independent experiments. Statistical analysis was performed using GraphPad Prism 5 software.
β-galactosidase assays
Yeast cultures (50 ml) were grown to OD600 of 0.6-0.8 at 24°C. One-half of the culture was used for total RNA isolation as described above. The second half of the culture was used for protein isolation. Cells were lysed with glass beads (0.5 μm in diameter) in breaking buffer (100 mM Tris-HCl [pH 8], 1 mM dithiothreitol, 20% glycerol; 2 mM phenylmethylsulfonyl fluoride and 1X protease inhibitor cocktail). Extracts were cleared by centrifugation (15,682 g) for ten minutes and protein concentrations were determined using the Bradford method. β-galactosidase liquid assays were performed by assaying each extract twice on Z buffer (using 200 μl of 4 mg/ml ONPG substrate. After thirty minutes, the reactions were stopped by adding 500 μl 1M Na2CO3. β-galactosidase activities shown represent at least three independent transformants per yeast strain and are expressed according to the following formula: (1.7 ml × OD420 units)/(0.0045 × cell extract volume [milliliters] × reaction time [minutes] × protein concentration [milligrams per milliliter]) 66; 70. Statistical analysis was performed using GraphPad Prism 5 software.
Dual luciferase assays
Dual luciferase assays were performed with the dual luciferase reporter assay system (Promega Corp., Madison, WI) as described previously with minor modifications 42; 71. Cells were grown to mid-logarithmic phase (OD600 0.8) in SC-Ura medium. Ten microliters of cells were removed directly from the culture and transferred to 100 μl of 1x passive lysis buffer. After allowing lysis for 15 seconds, 10 μls were used for luminescence measurements using a TD-20/20 luminometer (Tuner Designs, Sunnyville, CA.). The following steps were used for luminescence measurements: 10 μl of the firefly luciferase reagent (LARII) were added to the sample with a 2 seconds equilibration time and measurement of luminescence with a 10 seconds integration time, followed by addition of 10 μl of the Renilla luciferase reagent and firefly quenching (Stop & Glo), 2 seconds equilibration time, and measurement of luminescence with a 10 seconds integration time. The values shown represent the ratio of the firefly luciferase activity to the Renilla activity of the stop codon-containing constructs divided by the ratio of the firefly luciferase activity to the Renilla activity of the sense codon-containing construct multiplied by 100. For misincorporation assays, cells were treated and processed in a similar manner. For each strain at least three independent transformants were assayed and each individual extract was scored for luminescence activity in triplicates. Statistical analysis was performed using GraphPad Prism 5 software.
Whole cell extract preparation
Yeast cell cultures (50ml) were grown at 24°C to mid-logarithmic phase and harvested by centrifugation. Cell pellets were washed twice in lysis buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM EDTA, 10% glycerol, 1% Triton X-100) containing 1 mM PMSF and 1X protease inhibitor cocktail (Sigma) and transferred to microcentrifuge tubes. Cells were then resuspended in 250 μl of lysis buffer and 1/3 volume of 0.5 μm glass beads were added for mechanical lysis. Cells were vortexed six times for 30s at maximum speed with 2 minutes intervals on ice. Cell lysates were collected and cleared by centrifugation at 15,682 g for 10 minutes at 4°C. The cleared lysates were quantified using standard Bradford protein assay (Bio-Rad).
Western blot analysis
Yeast whole cell extracts were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Briefly, protein samples were mixed with appropriate amounts of 6X SDS-loading buffer and boiled for 5 minutes before loading onto a 10% SDS-PAGE gel and electrophoresed at 100V for 1.5 hours. The gel was then transferred to a nitrocellulose membrane (Bio-Rad) and blocked in 10% non-fat milk for 1 hour. The antibodies utilized were α-Npl3 (1:3000)45, α-Tcm1 (1:2500) 72 (kindly provided by Dr. Michael F. Henry and Dr. Allan Jacobson) and α-Pab1 (1:5000). Their corresponding secondary antibodies were used in 1:10,000 dilutions. Membranes were developed using the Western Dura chemiluminescense kit (Pierce).
Sucrose density gradient fractionations
Sucrose density gradients were performed essentially as previously described 18. Yeast cells (100 ml) were grown at 24°C in YPD medium to OD600 of 0.6. Cycloheximide was added to 100 μg/ml and incubated on ice for 10 minutes with occasional shaking 18. Cells were harvested at 835 g for five minutes and washed with 50 ml of polysomes lysis buffer (10 mM Tris-Cl pH 7.5, 100 mM NaCl, 30 mM MgCl2). After a second centrifugation, cells were resuspended in 10 mL of polysomes lysis buffer, centrifuged and the cell pellet was resuspended in one ml of lysis buffer. Glass beads (2/3 of the cell volume) were added to the samples and cells were lysed by vortexing 8 times for 15 seconds with a 30 seconds cool down period on ice. Lysates were centrifuged at 5,938 g for 5 minutes at 4°C; supernatants were collected and centrifuged at 11,228 g for 5 minutes at 4°C. Twenty OD260 units were loaded on 9ml 10-50% sucrose gradients. These gradients were centrifuged at 209,678 g for 3 hours at 4°C in an Optima L-100 XP ultracentrifuge (Beckman) using a SW 41 Ti rotor. Two- hundred microliter fractions were collected from the top of the sucrose gradients and their OD260nm monitored via spectrophotometry. Fractions were pooled and precipitated with acetone and resuspended in 1X-SDS protein sample buffer. Samples were incubated for 10 min at 22°. For the experiments in the presence of EDTA, the lysis buffer and the sucrose gradient contained EDTA to a final concentration of 15 mM. For the RNaseA studies, the extract was pre-incubated with RNaseA to a final concentration of 0.1 mg/ml at 24°C for 20 minutes before loading to the sucrose gradient. The fractions were resolved in a 10-20% Criterion™ precast gel (Bio-Rad), transferred to nitrocellulose membranes and analyzed by Western Blotting as described above. The percentage of ribosomal association was calculated by quantifying the total amount of protein distributed in Western blot lanes 9 thru 15 divided by the total amount of the protein in the fifteen lanes multiplied by 100.
ACKNOWLEDGEMENTS
We are very grateful to Dr. David Bedwell for the gift of plasmids and strains, Dr. Allan Jacobson for the anti-Tcm1 antibody, Dr. Michael F. Henry for Npl3 plasmids, anti-Npl3 antibody, yeast strains and helpful discussions. We also thank members of the González laboratory for helpful comments. We also thank the Biostatistics Core of the U54 grant for their assistance. This work was supported by grants from the National Institutes of Health to C.I.G. (GM008102-3052, KO1 HL-04355-05, U54 CA96297, FIPI), M.F.W. (GM058595, U54 CA96297). This publication was made possible by Grant P20 RR 016174 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
GLOSSARY
- mRNP
messenger ribonucleoprotein
- NMD
nonsense mediated mRNA decay
- RRM
RNA recognition motif
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Stewart M. Ratcheting mRNA out of the nucleus. Mol Cell. 2007;25:327–30. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 2.Cole CN, Scarcelli JJ. Transport of messenger RNA from the nucleus to the cytoplasm. Curr Opin Cell Biol. 2006;18:299–306. doi: 10.1016/j.ceb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Ares M, Jr, Proudfoot NJ. The spanish connection: transcription and mRNA processing get even closer. Cell. 2005;120:163–6. doi: 10.1016/j.cell.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Aguilera A. Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr Opin Cell Biol. 2005;17:242–50. doi: 10.1016/j.ceb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Goldstrohm AC, Greenleaf AL, Garcia-Blanco MA. Co-transcriptional splicing of pre-messenger RNAs: considerations for the mechanism of alternative splicing. Gene. 2001;277:31–47. doi: 10.1016/s0378-1119(01)00695-3. [DOI] [PubMed] [Google Scholar]
- 7.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. Embo J. 2000;19:6860–9. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole CN, Scarcelli JJ. Unravelling mRNA export. Nat Cell Biol. 2006;8:645–7. doi: 10.1038/ncb0706-645. [DOI] [PubMed] [Google Scholar]
- 9.Hurt E, Luo MJ, Rother S, Reed R, Strasser K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc Natl Acad Sci U S A. 2004;101:1858–62. doi: 10.1073/pnas.0308663100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katahira J, Yoneda Y. Roles of the TREX complex in nuclear export of mRNA. RNA Biol. 2009;6 doi: 10.4161/rna.6.2.8046. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert W, Guthrie C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell. 2004;13:201–12. doi: 10.1016/s1097-2765(04)00030-9. [DOI] [PubMed] [Google Scholar]
- 12.Yao W, Lutzmann M, Hurt E. A versatile interaction platform on the Mex67-Mtr2 receptor creates an overlap between mRNA and ribosome export. EMBO J. 2008;27:6–16. doi: 10.1038/sj.emboj.7601947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao W, Roser D, Kohler A, Bradatsch B, Bassler J, Hurt E. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol Cell. 2007;26:51–62. doi: 10.1016/j.molcel.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 14.Senay C, Ferrari P, Rocher C, Rieger KJ, Winter J, Platel D, Bourne Y. The Mtr2-Mex67 NTF2-like domain complex. Structural insights into a dual role of Mtr2 for yeast nuclear export. J Biol Chem. 2003;278:48395–403. doi: 10.1074/jbc.M308275200. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez CI, Bhattacharya A, Wang W, Peltz SW. Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene. 2001;274:15–25. doi: 10.1016/s0378-1119(01)00552-2. [DOI] [PubMed] [Google Scholar]
- 16.Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 17.Maquat LE. Nonsense-mediated mRNA decay in mammals. J Cell Sci. 2005;118:1773–6. doi: 10.1242/jcs.01701. [DOI] [PubMed] [Google Scholar]
- 18.Atkin AL, Schenkman LR, Eastham M, Dahlseid JN, Lelivelt MJ, Culbertson MR. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J Biol Chem. 1997;272:22163–72. doi: 10.1074/jbc.272.35.22163. [DOI] [PubMed] [Google Scholar]
- 19.Atkin AL, Altamura N, Leeds P, Culbertson MR. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol Biol Cell. 1995;6:611–25. doi: 10.1091/mbc.6.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shirley RL, Ford AS, Richards MR, Albertini M, Culbertson MR. Nuclear import of Upf3p is mediated by importin-alpha/-beta and export to the cytoplasm is required for a functional nonsense-mediated mRNA decay pathway in yeast. Genetics. 2002;161:1465–82. doi: 10.1093/genetics/161.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maderazo AB, Belk JP, He F, Jacobson A. Nonsense-containing mRNAs that accumulate in the absence of a functional nonsense-mediated mRNA decay pathway are destabilized rapidly upon its restitution. Mol Cell Biol. 2003;23:842–51. doi: 10.1128/MCB.23.3.842-851.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He F, Jacobson A. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol Cell Biol. 2001;21:1515–30. doi: 10.1128/MCB.21.5.1515-1530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhalla AD, Gudikote JP, Wang J, Chan WK, Chang YF, Olivas OR, Wilkinson MF. Nonsense codons trigger an RNA partitioning shift. J Biol Chem. 2009;284:4062–72. doi: 10.1074/jbc.M805193200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W, Czaplinski K, Rao Y, Peltz SW. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. Embo J. 2001;20:880–90. doi: 10.1093/emboj/20.4.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–22. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keeling KM, Lanier J, Du M, Salas-Marco J, Gao L, Kaenjak-Angeletti A, Bedwell DM. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. Rna. 2004;10:691–703. doi: 10.1261/rna.5147804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weng Y, Czaplinski K, Peltz SW. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol. 1996;16:5491–506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS ONE. 2008;3:e3273. doi: 10.1371/journal.pone.0003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bucheli ME, Buratowski S. Npl3 is an antagonist of mRNA 3' end formation by RNA polymerase II. EMBO J. 2005;24:2150–60. doi: 10.1038/sj.emboj.7600687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bucheli ME, He X, Kaplan CD, Moore CL, Buratowski S. Polyadenylation site choice in yeast is affected by competition between Npl3 and polyadenylation factor CFI. RNA. 2007;13:1756–64. doi: 10.1261/rna.607207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kress TL, Krogan NJ, Guthrie C. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol Cell. 2008;32:727–34. doi: 10.1016/j.molcel.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henry M, Borland CZ, Bossie M, Silver PA. Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics. 1996;142:103–15. doi: 10.1093/genetics/142.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Windgassen M, Sturm D, Cajigas IJ, Gonzalez CI, Seedorf M, Bastians H, Krebber H. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol Cell Biol. 2004;24:10479–91. doi: 10.1128/MCB.24.23.10479-10491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sachs AB, Varani G. Eukaryotic translation initiation: there are (at least) two sides to every story. Nat Struct Biol. 2000;7:356–61. doi: 10.1038/75120. [DOI] [PubMed] [Google Scholar]
- 35.Deka P, Bucheli ME, Moore C, Buratowski S, Varani G. Structure of the yeast SR protein Npl3 and Interaction with mRNA 3'-end processing signals. J Mol Biol. 2008;375:136–50. doi: 10.1016/j.jmb.2007.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loo S, Laurenson P, Foss M, Dillin A, Rine J. Roles of ABF1, NPL3, and YCL54 in silencing in Saccharomyces cerevisiae. Genetics. 1995;141:889–902. doi: 10.1093/genetics/141.3.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli JJ, Seedorf M, Cole CN, Krebber H. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315:646–9. doi: 10.1126/science.1134641. [DOI] [PubMed] [Google Scholar]
- 38.Salas-Marco J, Bedwell DM. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol Cell Biol. 2004;24:7769–78. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez CI, Ruiz-Echevarria MJ, Vasudevan S, Henry MF, Peltz SW. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol Cell. 2000;5:489–99. doi: 10.1016/s1097-2765(00)80443-8. [DOI] [PubMed] [Google Scholar]
- 40.Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–77. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guarente L. Synthetic enhancement in gene interaction: a genetic tool come of age. Trends Genet. 1993;9:362–6. doi: 10.1016/0168-9525(93)90042-g. [DOI] [PubMed] [Google Scholar]
- 42.Salas-Marco J, Bedwell DM. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J Mol Biol. 2005;348:801–15. doi: 10.1016/j.jmb.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 43.McBride AE, Cook JT, Stemmler EA, Rutledge KL, McGrath KA, Rubens JA. Arginine methylation of yeast mRNA-binding protein Npl3 directly affects its function, nuclear export, and intranuclear protein interactions. J Biol Chem. 2005;280:30888–98. doi: 10.1074/jbc.M505831200. [DOI] [PubMed] [Google Scholar]
- 44.Shen EC, Henry MF, Weiss VH, Valentini SR, Silver PA, Lee MS. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 1998;12:679–91. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu C, Henry MF. Nuclear export of hnRNP Hrp1p and nuclear export of hnRNP Npl3p are linked and influenced by the methylation state of Npl3p. Mol Cell Biol. 2004;24:10742–56. doi: 10.1128/MCB.24.24.10742-10756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen EC, Stage-Zimmermann T, Chui P, Silver PA. 7The yeast mRNA-binding protein Npl3p interacts with the cap-binding complex. J Biol Chem. 2000;275:23718–24. doi: 10.1074/jbc.M002312200. [DOI] [PubMed] [Google Scholar]
- 47.Gilbert W, Siebel CW, Guthrie C. Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. Rna. 2001;7:302–13. doi: 10.1017/s1355838201002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yun CY, Fu XD. Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J Cell Biol. 2000;150:707–18. doi: 10.1083/jcb.150.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maderazo AB, He F, Mangus DA, Jacobson A. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol Cell Biol. 2000;20:4591–603. doi: 10.1128/mcb.20.13.4591-4603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ono BI, Ishino Y, Shinoda S. Nonsense mutations in the can1 locus of Saccharomyces cerevisiae. J Bacteriol. 1983;154:1476–9. doi: 10.1128/jb.154.3.1476-1479.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cui Y, Hagan KW, Zhang S, Peltz SW. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–36. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 52.Ruiz-Echevarria MJ, Czaplinski K, Peltz SW. Making sense of nonsense in yeast. Trends Biochem Sci. 1996;21:433–8. doi: 10.1016/s0968-0004(96)10055-4. [DOI] [PubMed] [Google Scholar]
- 53.He F, Brown AH, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1997;17:1580–94. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–16. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. Febs J. 2005;272:2118–31. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 56.Kielkopf CL, Lucke S, Green MR. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 2004;18:1513–26. doi: 10.1101/gad.1206204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Czaplinski K, Weng Y, Hagan KW, Peltz SW. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. Rna. 1995;1:610–23. [PMC free article] [PubMed] [Google Scholar]
- 58.Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–27. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cosson B, Couturier A, Chabelskaya S, Kiktev D, Inge-Vechtomov S, Philippe M, Zhouravleva G. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol Cell Biol. 2002;22:3301–15. doi: 10.1128/MCB.22.10.3301-3315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brune C, Munchel SE, Fischer N, Podtelejnikov AV, Weis K. Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. Rna. 2005;11:517–31. doi: 10.1261/rna.7291205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kutay U, Panse VG. Gle1 does double duty. Cell. 2008;134:564–6. doi: 10.1016/j.cell.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 62.Hilleren P, Parker R. mRNA surveillance in eukaryotes: kinetic proofreading of proper translation termination as assessed by mRNP domain organization? Rna. 1999;5:711–9. doi: 10.1017/s1355838299990519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pape T, Wintermeyer W, Rodnina MV. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat Struct Biol. 2000;7:104–7. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- 64.Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol. 2006;8:668–76. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- 65.Alcazar-Roman AR, Tran EJ, Guo S, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol. 2006;8:711–6. doi: 10.1038/ncb1427. [DOI] [PubMed] [Google Scholar]
- 66.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 67.Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. (3rd) 1998;14:953–61. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 68.Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:31–4. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 69.Wang W, Cajigas IJ, Peltz SW, Wilkenson MF, Gonzalez CI. Role of Upf2 phosphorylation in Saccharomyces cerevisiae Nonsence-Mediated Decay. MCB. 2006 doi: 10.1128/MCB.26.9.3390-3400.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pappas DL, Jr, Hampsey M. Functional interaction between Ssu72 and the Rpb2 subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:8343–51. doi: 10.1128/mcb.20.22.8343-8351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McNabb DS, Reed R, Marciniak RA. Dual luciferase assay system for rapid assessment of gene expression in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1539–49. doi: 10.1128/EC.4.9.1539-1549.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mangus DA, Jacobson A. Linking mRNA turnover and translation: assessing the polyribosomal association of mRNA decay factors and degradative intermediates. Methods. 1999;17:28–37. doi: 10.1006/meth.1998.0704. [DOI] [PubMed] [Google Scholar]