Abstract
Eukaryotic translation initiation factor (eIF) 1 is a central mediator of start codon recognition. Dissociation of eIF1 from the pre-initiation complex allows release of phosphate from the G-protein factor eIF2, triggering downstream events in initiation. Mutations that weaken binding of eIF1 to the pre-initiation complex decrease the fidelity of start codon recognition (Sui− phenotype) by allowing increased eIF1 release at non-AUG codons. Consistent with this, over-expression of these mutant proteins suppresses their Sui− phenotypes. Here, we have examined mutations at the penultimate residue of eIF1, G107, that produce Sui− phenotypes without increasing the rate of eIF1 release. We provide evidence that, in addition to its role in gating phosphate release, dissociation of eIF1 triggers conversion from an open, scanning-competent state of the pre-initiation complex to a stable, closed one. We also show that eIF5 antagonizes binding of eIF1 to the complex and that key interactions of eIF1 with its partners are modulated by the charge at and around G107. Our data indicate that eIF1 plays multiple roles in start codon recognition and suggest that prior to AUG recognition it prevents eIF5 from binding to a key site in the pre-initiation complex required for triggering downstream events.
Keywords: initiation codon, eIF5, protein synthesis, ribosome, kinetics
INTRODUCTION
Eukaryotic translation initiation factor (eIF) 1 is a 12 kDa protein that plays a number of important roles in the initiation of translation in eukaryotes1,2. Together with eIF1A and other factors, eIF1 mediates the assembly of the 43S pre-initiation complex (PIC) by recruiting the eIF2•GTP•Met-tRNAi ternary complex (TC) to the 40S ribosomal subunit2–4. It also plays a key role in start codon recognition by acting as a negative regulator of phosphate (Pi) release from eIF2 within the PIC. Hydrolysis of GTP in the PIC can occur as it scans the mRNA leader from the 5′ m7G cap, and is dependent on the GTPase-activating factor eIF5. When the start codon in the mRNA is recognized, eIF1 is ejected from the PIC, which allows release of Pi from eIF2•GDP•Pi, converting eIF2 to its inactive, GDP-bound state5,6. eIF2•GDP can then be released from the PIC and other downstream events can proceed culminating in formation of a final initiation complex on the start codon of the message7,8.
Structural studies have indicated that eIF1 and eIF1A synergistically mediate a conformational change from a closed state of the 40S subunit to an open one that is more receptive to TC binding9. In addition, recent studies have indicated that formation of three base-pairs between the anticodon of tRNAi and the start codon in mRNA induces a conformational transition in the 43S PIC that stabilizes TC binding10. This conformational change is likely to be related to the transition from the open, scanning-competent conformation to the closed, scanning-arrested one upon start codon recognition5,11,12. This transition is also regulated by eIF1A, which binds more tightly to the PIC through a functional interaction with eIF5, and promotes release of eIF1, during the transition from open to closed conformational states13–15. However, the role of eIF1 in regulating this conformational transition is unclear.
Genetic analyses of yeast eIF1 identified mutations that affect TC loading and start codon recognition15–18. Mutations that slow TC loading onto the 40S subunit or otherwise destabilize PIC assembly have a Gcd− phenotype (general control derepressed): derepressed translation of GCN4 mRNA in a gcn2Δ strain19. Another set of mutations in eIF1 reduce the stringency of start codon recognition, a Sui− phenotype (suppressors of initiation codon mutation)16,17,20,21. These Sui− mutations restore translation of a mutant version of the HIS4 mRNA (his4-303), in which the AUG initiation codon is changed to AUU. In most cases, initiation from this mRNA in Sui− strains appears to occur at an in-frame UUG near the original start site19. Reporter assays with HIS4-lacZ fusions show increased initiation at UUG codons with the Sui− mutants as compared to WT eIF115,20.
In vitro analyses using a reconstituted yeast translation initiation system revealed that these Sui− mutants of eIF1 have significantly reduced affinities for the 40S ribosomal subunit and 43S PIC15. One mutation, ISQLG93–97ASQAA (hereafter, 93–97), was shown to increase the rate of dissociation of eIF1, and correspondingly of Pi, from the 43S PIC. Over-expression of each of these eIF1 mutants suppressed its Sui− phenotype, presumably by increasing occupancy of the factor within the PIC. Hence, the Sui− mutations in eIF1 examined thus far appear to work by increasing the ease with which eIF1 dissociates from the PIC at non-AUG codons, the step that commits the complex to downstream events that lead to formation of the 80S initiation complex.
A screen for yeast mutants with altered frame-shifting efficiency identified a mutation in eIF1 (mof2-1)22 that was later shown to also reduce the fidelity of start codon recognition23 This allele had a substitution from G to R at the penultimate residue in yeast eIF1, G107. In contrast to the “classical” Sui− eIF1 mutants, G107R binds the 40S subunit and 43S PIC with an affinity similar to WT and is actually released more slowly than WT in response to AUG recognition5, indicating that it reduces the fidelity of start codon recognition through a different mechanism than the classical Sui− mutations. We reasoned that by understanding this alternative mechanism we might shed light on additional roles of eIF1 in start codon recognition.
RESULTS
Changes in and around G107 produce Sui− and Gcd− phenotypes
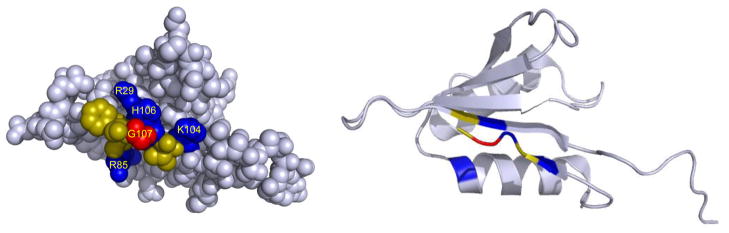
The NMR structures of mammalian and yeast eIF124,25 show that the penultimate G107 residue is in a pocket surrounded by basic and hydrophobic amino acids (Fig. 1), which are conserved between yeast and mammals. The G107R mutation increases the positive charge on this surface of the protein, and we reasoned this might play a role in the phenotypes associated with the change. To test the role of charge in this region of eIF1, we created five additional mutants: G107K, G107E, G107S, R85S, and R29S in a His6-tagged SUI1 allele.
Figure 1.
Space-filling (left) and ribbon (right) representations of yeast eIF1 (pdb id 2ogh;25) showing the pocket created by G107 (red) surrounded by basic (blue) and hydrophobic (yellow) residues. Only part of the unstructured N-terminus (bottom right in structure) is shown.
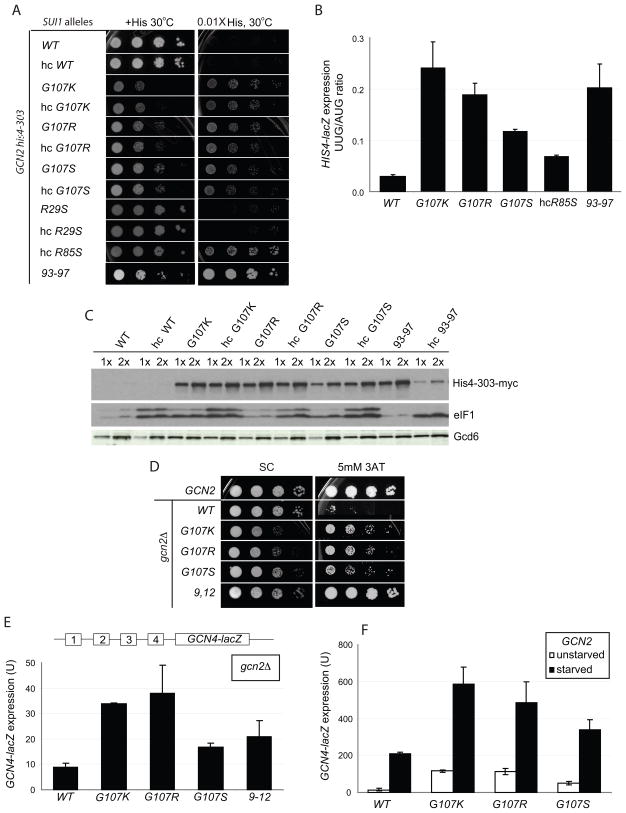
The phenotypes of sui1Δ yeast strains expressing these eIF1 mutants on low- (lc) and high-copy (hc) plasmids were assessed. The G107R, G107K and G107S mutations produce recessive slow growth phenotypes (Slg−) at 30 °C, while both hc and lc G107E and lc R85S are recessively lethal (Fig. 2A). Over-expression rescued growth of the R85S mutant. In all cases, the levels of expression of the mutant proteins were found to be at or above WT levels (Figs. 2C and S1).
Figure 2.
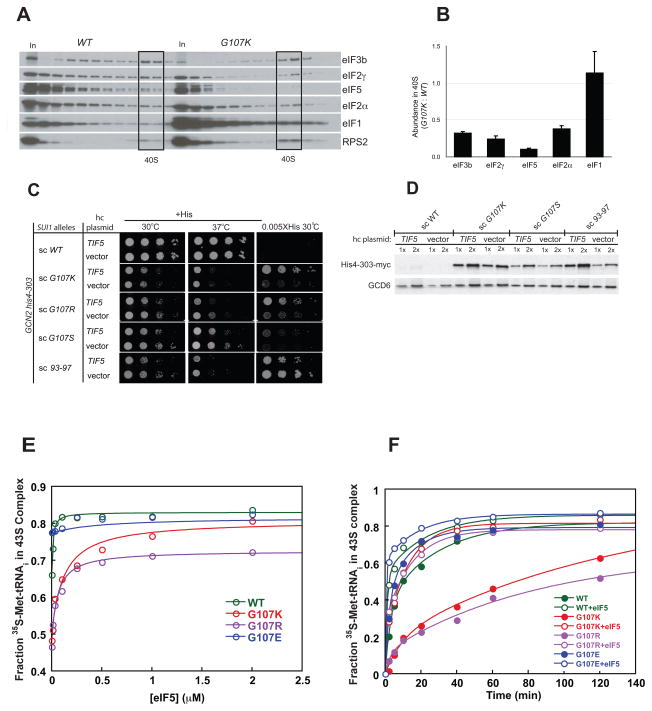
Figure 2. (A–C) Substitutions at G107 in eIF1 reduce stringent AUG selection in vivo. (A) Sui− phenotypes of G107 mutations. GCN2 his4-303 strains with the indicated WT or mutant His-SUI1 alleles on sc or hc plasmids were grown at 30 °C for 2d on SC medium lacking leucine (+His; 120 μM histidine) and for 7d on SD medium plus uracil, tryptophan and 1.2 μM histidine (0.01X His). (B) Quantification of Sui− phenotypes. Strains from (A) bearing plasmids p367 or p391 containing HIS4-lacZ fusions with an AUG or UUG start codon, respectively, were grown in SD with 0.3 mM His and Trp at 30 °C and β-galactosidase activities were measured in WCEs. Mean ratios of expression of UUG to AUG reporters are shown with standard errors (SE) from 3 experiments on 6 independent transformants. (C) Sui− mutants increase myc-his4-303 expression. his4-303-myc strains harboring the indicated WT or mutant SUI1 alleles were grown overnight in YPD and WCEs were subjected to Western analysis with anti-myc, anti-eIF1, and anti-GCD6 antibodies. We do not yet know the nature of the slower migrating eIF1 band. (D–F) G107 substitutions partially derepress GCN4 translation. (D) gcn2Δ strains harboring sc His-SUI1 alleles were grown on SC-L medium at 30 °C for 2d, and on SC-L lacking histidine and containing 5 mM 3-AT for 4d. (E) The gcn2Δ strains described in (D) harboring the GCN4-lacZ reporter (with all 4 uORFs) were grown in SC-L, and β-galactosidase activities (nmol o-nitrophenyl-β-D-galactopyranoside cleaved min−1 mg−1) were measured in WCEs. The means and SEs from 3 or more measurements on 6 independent transformants are shown. (F) The GCN2 his4-303 strains with sc His-SUI1 alleles from (A) harboring GCN4-lacZ plasmid p180 were grown in SC lacking uracil, leucine, isoleucine and valine for 6h with or without sulfometuron 5 mg/ml) and β-galactosidase activities were measured in WCEs.
Strong Sui− mutations allow growth on medium lacking histidine in the his4-303 background by permitting initiation at a non-AUG codon in his4-303. While none of the G107 mutations conferred this phenotype, G107R, G107K, G107S and hc R85S did allow some growth with only 0.01X of the normal histidine supplement, which cannot support growth of the his4-303 strain harboring WT eIF1 (Figure 2A). Thus, the Sui− phenotypes of these mutants were weaker than that conferred by the 93–97 mutation (Fig. 2A), which allows growth on medium lacking histidine and acts by decreasing the affinity of eIF1 for the PIC15. The Sui− phenotypes of these new mutants were confirmed in the HIS4-lacZ reporter assay. G107K, G107R, and G107S conferred 5–10-fold increases in the UUG/AUG initiation ratio, with G107K producing the largest effect and G107S the smallest (Fig. 2B). The effect of the 93–97 mutation was similar to that of G107K. Hc R85S conferred only a 2-fold increase in the UUG/AUG initiation ratio; however, we showed previously that over-expression of Sui− eIF1 mutants greatly reduces their Sui− phenotypes. Hence, R85S would likely confer a stronger Sui− phenotype if it were not being overexpressed from a hc plasmid to overcome the lethality of this substitution. In contrast to these results, the R29S mutation produced no detectable phenotypes. We previously analyzed the effects of the G107R and 93–97 mutations using this assay15 and the results reported here are in good agreement with that work.
It was surprising that in the his4-303 background the G107K and G107R mutations did not allow growth on medium lacking histidine, given that their effects in the reporter assay were similar to those of the 93–97 mutation, which produces a robust His+ phenotype15 (Fig. 2A, B). To investigate this discrepancy, we replaced the genomic copy of his4-303 with a C-terminally myc-tagged version and used western blotting with anti-myc antibodies to assess levels of his4-303 expression in the mutants (Fig. 2C). The data demonstrate that his4-303 expression is increased dramatically by the G107K, G107R and G107S mutations. The levels of increase are similar to that produced by 93–97 and the G31R Sui− mutation in eIF5 (data not shown). Thus the western measurements of his4-303 expression are in good agreement with the HIS4-lacZ reporter assays in revealing strong Sui− phenotypes for the G107 mutants. The inherent Slg− phenotypes of the G107 mutants likely contribute to their relatively weak His+ phenotypes.
We previously showed that the Sui− phenotypes of eIF1 mutants that decrease the factor’s affinity for the PIC could be suppressed by over-expression, which presumably increases occupancy of the mutant factor in the complex by mass action15. This effect can be clearly seen at the level of his4-303 expression for the 93–97 mutant (Fig. 2C, compare 93–97 to hc 93–97). In contrast, the effects of the G107 mutants are not suppressed by over-expression (Fig. 2A, C), further indicating that these mutations act via a different mechanism than the previously analyzed Sui− mutations in eIF1.
Next, we screened for the effect of the mutations on expression of GCN4 in a strain lacking the eIF2α kinase Gcn2. Translation of the GCN4 ORF is controlled by four short, upstream ORFs that repress GCN4 translation when TC levels are high. Amino acid starvation activates Gcn2, which phosphorylates eIF2α, reducing TC levels and allowing read-through of the upstream ORFs and increased translation of the GCN4 ORF19. Mutations that reduce the rate of TC binding to the 40S subunit can allow bypass of the upstream ORFs and expression of Gcn4 in the absence of Gcn2 during amino acid starvation, a phenotype called Gcd−. This phenotype can be measured as resistance of gcn2Δ strains to the histidine biosynthesis inhibitor 3-aminotriazole (3-AT), and by increased expression of a GCN4-lacZ reporter. Figure 2D shows that the G107K, -R and -S mutations all produce Gcd− phenotypes, allowing increased growth in the presence of 5 mM 3-AT relative to cells expressing WT eIF1, with the R and K substitutions giving the strongest effects (relative to their inherent Slg− phenotypes). The 3-AT resistance of the G107R and -S mutants was not suppressed by co-overexpression of the three subunits of eIF2 and the initiator tRNA (hc TC) while the effect of hc TC on G107K was difficult to assess because it exacerbated the inherent Slg− phenotype (data not shown). Consistent with the mutants’ Gcd− phenotypes, Gcn4-lacZ expression is increased 3–4-fold for G107K and G107R and ~2-fold for G107S in the gcn2Δ background. These effects are similar in magnitude to those observed previously for another Gcd− eIF1 mutant, FDPF9,12ADPA (hereafter, 9–12) (Fig. 2E)15. It is interesting that the 9,12 strain exhibits much stronger growth than the G107R and G107K mutants on 3AT medium (Fig. 2D) even though the latter exhibit higher levels of GCN4-lacZ expression in the gcn2Δ background (Fig. 2E). The inherent slow-growth phenotype of the G107R and G107K mutants (Fig. 2D, SC) is one factor contributing to this apparent discrepancy. It is also possible that the G107 mutations confer increased sensitivity to amino acid starvation by a mechanism unrelated to their effects on GCN4 mRNA.
The derepression of GCN4 translation caused by these mutations could also be seen in unstarved GCN2 cells (Fig. 2F), where eIF2α phosphorylation occurs at low basal levels. Starvation of these mutant strains increases production of Gcn4-lacZ to levels even higher than in WT starved cells, indicating that further derepression by eIF2α phosphorylation is still possible. Thus, the G107K, -R and -S mutations do not compromise starvation-induced derepression. The Gcd− phenotypes suggest that these mutations reduce the efficiency of TC binding to the 40S subunit.
In order to understand the mechanisms through which these mutations generate Gcd− and Sui− phenotypes, we carried out in vitro studies using a reconstituted S. cerevisiae initiation system4.
The charge in the region around G107 is important for tight binding to the 40S subunit and interaction with eIF1A
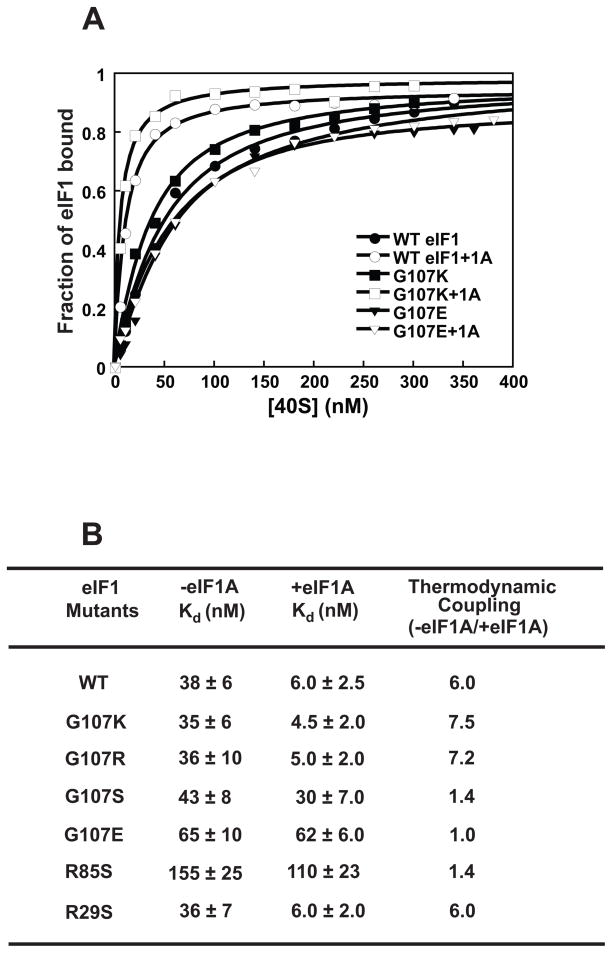
To determine the affinity of the eIF1 mutants for 40S ribosomal subunits we labeled the mutant factors with fluorescein at their C-termini using expressed protein ligation13,26. Increasing concentrations of 40S subunits were added to these fluorescently-derivatized factors and the change in fluorescence anisotropy measured at each point. Fluorescence anisotropy reflects the rate of rotational diffusion of the fluorescently labeled protein, which decreases when the labeled factor binds the 40S subunit, leading to an increase in observed anisotropy. Fig. 3A shows binding curves for WT eIF1 and the G107K and G107E mutants (closed symbols). We previously showed that binding of eIF1 and eIF1A to the 40S ribosomal subunit is cooperative13 and thus we also measured binding of the labeled eIF1 mutants in the presence of saturating eIF1A (Fig. 3A, open symbols). Binding data for all the mutant factors are given in Figure 3B.
Figure 3. The charge at and around position 107 affects eIF1’s affinity for the 40S subunit and its thermodynamic coupling with eIF1A.
(A) Binding of fluorescein-labeled eIF1 mutants was monitored using fluorescence anisotropy: (●) WT eIF1; (○) WT eIF1 + eIF1A; (■) G107K; (□) G107K + eIF1A; (▼) G107E; (∇) G107E + eIF1A. Each curve is the average of at least three experiments. (B) Kd values for eIF1 mutants binding to the 40S subunit. The ratio of Kds in the absence and presence of eIF1A is the fold thermodynamic coupling between the two factors. Errors given are mean deviations.
As previously reported15, G107R eIF1 binds the 40S subunit with similar affinity as the WT factor and has a similar degree of cooperativity (thermodynamic coupling) with eIF1A, ~6–7-fold. The G107K and R29S mutations behave similarly. The G107S substitution does not change the affinity of the factor for the 40S subunit in the absence of eIF1A, but decreases it ~5-fold in eIF1A’s presence relative to WT eIF1. This change amounts to a loss of cooperative binding between eIF1 and eIF1A. The lethal G107E change, which introduces a negative charge at this position, results in ~2-fold weaker binding of the factor to the 40S subunit in the absence of eIF1A and nearly 10-fold weaker binding in its presence, also resulting in a loss of cooperativity between the two factors. R85S, which is also lethal at normal cellular concentrations of eIF1 and also decreases the positive charge in this region of the protein, likewise decreases affinity of the factor in the presence and absence of eIF1A (4-fold and 20-fold, respectively) and eliminates cooperative binding.
The effects of the mutant proteins on the affinity of the ribosome for eIF1A was measured in separate experiments (not shown), and G107S, G107E and R85S were shown to reduce or eliminate cooperativity in this direction as well, as expected for a thermodynamic cycle. In addition, affinity of the eIF1 mutants for the 40S subunit was also measured by competitive binding of unlabeled mutant factor and fluorescein-labeled WT eIF113. The results of these experiments (data not shown) are consistent with all of the effects of the mutations observed in the direct binding assays, indicating that the C-terminal fluorophore does not alter the behavior of the mutant proteins.
Overall, the observations that two changes in this region of the protein that decrease positive charge (G107E and R85S) weaken binding of eIF1 to the 40S subunit and eliminate cooperativity with eIF1A, whereas mutations that increase the positive charge do not, suggest that the charge on this face of the protein is important for proper interactions with other components of the initiation machinery. The lethal mutations at G107 and R85 that eliminate coupled binding of eIF1 and eIF1A to the 40S subunit are the first to be found in either factor that have this effect.
Ternary complex loading onto the 40S subunit in vitro is modulated by the charge at positions 85 and 107
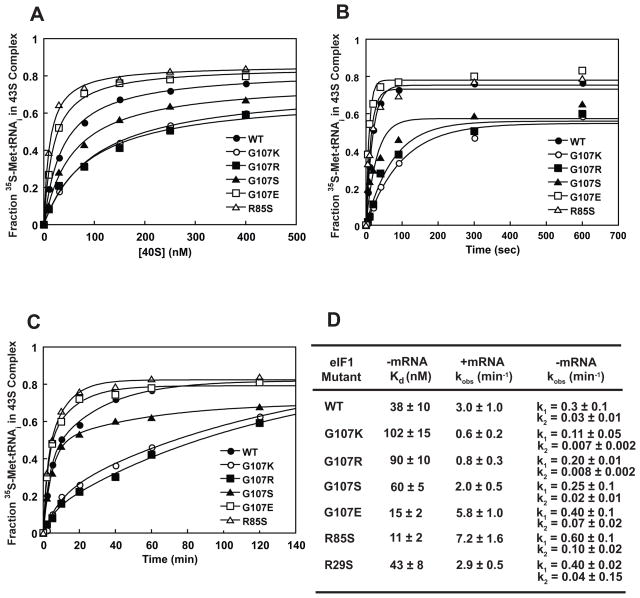
The Gcd− phenotypes of the G107K, -R and -S mutants suggested that these changes reduce the efficiency of TC binding to the 40S subunit. We therefore measured the kinetics and overall affinity of TC loading in the presence of saturating concentrations of eIF1A and each of the mutant versions of eIF1, both in the presence and absence of a model mRNA. TC binding was followed using 35S-Met-tRNAi and a native gel shift assay10. In the presence of model mRNA with an AUG codon, TC binds very tightly to the 40S subunit regardless of whether WT or a mutant eIF1 is used (Kd < 1 nM; data not shown), indicating that stable 43S•mRNA complex can be formed in all cases. In the absence of mRNA (Fig. 4A, D), TC binds to the 40S subunit with lower affinity (Kd = 38 nM with WT eIF1). The G107K and G107R mutants decreased affinity of TC for the 40S subunit ~3-fold and the G107S mutant decreased the affinity by ~2-fold. In contrast, TC binds to the 40S subunit with 2- and 4-fold higher affinities with the R85S and G107E mutants, respectively, than with WT eIF1. These results parallel those obtained for the effects of the substitutions on the factor’s affinity for the 40S subunit, with those that increase the positive charge in this region acting in one way (tight eIF1 binding and weakened TC binding) and those that decrease the positive charge acting in the opposite manner (weakened eIF1 binding and stronger TC binding). The R29S mutation has no effect on the affinities of eIF1 or TC for the 40S subunit, suggesting that it does not make up part of the same functional interface as G107 and R85.
Figure 4. TC recruitment to the 40S subunit is affected by the charge at and around position 107 in eIF1.
(A) The native gel-shift assay was used to measure TC affinity for the 40S subunit in the presence of saturating WT or mutant eIF1 and eIF1A in the absence of mRNA: (●) WT; (○) G107K; (■) G107R; (▲) G107S; (□) G107E; (△) R85S. (B – C) The kinetics of TC recruitment in the presence (B) and absence (C) of mRNA. The symbols are the same as in (A). The concentration of 40S subunits used was 10 nM in presence of mRNA and 200 nM in absence of mRNA. The curves shown are averages of three independent experiments. (D) Thermodynamic and kinetic constants for TC recruitment to the 40S•eIF1•eIF1A complex. TC binding in the absence of mRNA is biphasic indicating the existence of an additional step on the pathway10.
To better understand the effects of these mutations on PIC formation, we measured the kinetics of TC loading onto the 40S subunit in the presence and absence of mRNA (Fig. 4B and C). It was previously reported that the rate of TC binding to the 40S subunit in vitro was reduced by the Gcd− substitutions G107R and 9–1215. Consistent with these previous measurements, in the presence of mRNA the G107R substitution reduced the rate of TC loading 4-fold (Fig. 4B, D). G107K had a similar effect, reducing the rate constant for TC binding by 5-fold, and G107S had a smaller effect, reducing the rate constant 1.5-fold. The reduction in TC binding rate caused by these mutations is consistent with the Gcd− phenotypes they produce in vivo (Fig. 2D). In contrast, the G107E and R85S mutations actually increased the rate of TC loading by approximately 2-fold. Results consistent with these were obtained when TC recruitment was measured in the absence of mRNA (Fig. 4C, D).
The G107K, -R and -S substitutions affect binding of TC and other MFC components to the 40S subunit in vivo
To further investigate the effect of the eIF1 mutations on binding of TC and other factors to the 40S subunit, we performed in vivo cross-linking experiments. Cells expressing G107K, G107S or WT eIF1 were treated with formaldehyde to cross-link factors to ribosomes, and cell lysates were then fractionated on sucrose gradients. Fractions from these gradients were separated by SDS-PAGE and analyzed by western blotting for the levels of initiation components (Fig. 5A, B and Fig. S2). Consistent with the in vitro studies and the Gcd− phenotypes, the G107K and G107S substitutions both reduced the amount of eIF2 bound per 40S subunit (i.e., normalized to Rps2 signal), with G107K having a larger effect. G107K also reduced the levels of both eIF3 and eIF5 bound per 40S (Fig. 5A, B), whereas G107S (Fig. S2) reduced eIF5 binding but did not affect eIF3 binding. In both cases eIF1 binding per 40S was at or above WT levels. The degree of the effects for G107K and G107S seen in these experiments correlate very well with the strength of the Gcd− phenotype of each mutant in vivo (Fig. 2D,E) and the effect on TC binding in vitro (Fig. 4), with G107K (and G107R) always producing larger effects than G107S.
Figure 5. G107K impairs PIC assembly in vivo.
(A–B) Sucrose gradient analysis of formaldehyde cross-linked WCEs from SUI1 (JCY145) and SUI1-G107K (JCY645) strains. Gradient fractions and 1% of each WCE (In), were subjected to Western analysis with antibodies against the indicated eIFs or 40S subunit protein RPS2. Fractions containing free 40S subunits are boxed. (B) Initiation factor binding to 40S subunits in the experiment in (A) and two replicates were quantified by calculating the ratios of eIF to RPS2 intensities in the 40S and normalizing the resulting mutant ratio by the corresponding WT ratio. The means ± SEs are plotted. The direction of sedimentation is from left to right. The lack of an increase in abundance of unbound factors in the gradients with the G107K mutant, which might have been expected given the loss of these proteins from the PICs, is attributed to increased instability of free eIFs relative to those in MFC or PICS in vivo or during sedimentation14. (C–D) eIF5 over-expression enhances the Sui− phenotypes of eIF1 mutants. GCN2 his4-303 or GCN2 his4-303-myc strains described in Figs. 2A and C, respectively, were transformed with hc TIF5 plasmid (p4438) and analyzed for growth phenotypes (C) and His4-303-myc expression (D) as described in Fig. 2. (E–F). The G107K and -R mutations affect a functional interaction between eIF1 and eIF5. (E) The effect of eIF5 on TC binding to 40S subunits in the absence of mRNA with saturating eIF1 variants: WT, green; G107K, red; G107R, purple; G107E, blue. (F) The effect of saturating (2 μM) eIF5 on the kinetics of TC recruitment to the 40S subunit with eIF1 variants. The colors are the same as in E with open and closed symbols indicating the presence and absence of eIF5, respectively.
The reduction in 40S binding of eIF2, eIF3 and eIF5 is consistent with a defect in TC binding because these factors associate with one another in the Multifactor Complex (MFC)27. Intriguingly, for both the G107K and G107S mutants the effect on eIF5 binding was larger than the effect on eIF2 binding, suggesting that the mutations may directly influence eIF5’s interaction with the PIC in addition to reducing binding of TC (and thus binding of eIF5, which is associated with it). These effects are very similar to those observed previously for G107R eIF115.
eIF5 promotes start codon selection by stimulating GTP hydrolysis by eIF2 and stabilizing the closed conformation of the PIC. Hence, we reasoned that the low eIF5 occupancy of native PICs in the G107K and –R mutants might be a factor limiting their Sui− phenotypes. Supporting this hypothesis, we found that over-expressing eIF5 (TIF5) from a hc plasmid enhanced the Sui− phenotypes of the G107K, -R and -S mutants, allowing growth on medium containing only 0.005X of the normal histidine supplement (Fig. 5C). Experiments monitoring expression of his4-303-myc directly using western blots also demonstrated that over-expression of eIF5 increases his4-303 production with the G107K and G107S mutants (Fig. 5D). Interestingly, over-expression of eIF5 also increased his4-303-myc levels in the 93–97 eIF1 mutant (which produces a synthetic slow-growth phenotype with hc TIF5 (Fig. 5C), making assessment of effects on the His+ phenotype difficult), indicating that the effect is not restricted to the G107 mutants. In contrast, eIF5 over-expression did not increase his4-303-myc expression in a WT eIF1 background. These findings suggest that increasing eIF5 occupancy in the PIC enhances selection of non-cognate start codons, which is opposite to the effect described previously for overexpressing WT eIF128. One explanation for these findings would be that increased eIF5 occupancy evokes more rapid dissociation of eIF1 from the PIC at near-cognate start codons, for which direct evidence is provided below.
Functional interactions between eIF1 and eIF5 within the 43S PIC
In vitro, with WT eIF1 and eIF1A, stable recruitment of TC to the 40S subunit in the absence of mRNA is enhanced slightly by the addition of eIF5 (Fig. 5E, green open circles). We used this effect to test for a functional interaction between eIF1 and eIF5 in TC recruitment and eIF5 binding to the PIC. Adding increasing concentrations of eIF5 to TC binding experiments done at 250 nM 40S subunits in the presence of either G107R or G107K eIF1 (and WT eIF1A) resulted in a nearly 2-fold increase in TC binding, in contrast to the only ~20 % increase observed with WT eIF1 (Fig. 5E, compare purple and red to green circles). The concentrations of eIF5 required to give half-maximal enhancement of TC binding (K1/2) were also higher with the G107K and -R mutant factors than with WT eIF1 (K1/2 = 120, 60 and 10 nM, respectively). Importantly, under the same conditions binding of TC to 40S was unaffected by eIF5 when the G107E mutant was used (Fig. 5E, blue open circles).
We also measured the rate of TC binding to 40S subunits under the same conditions at a saturating concentration of eIF5 (2 μM) to determine whether eIF5 could rescue the binding defect associated with the G107K and -R substitutions. As shown in Figure 5F, with the G107K and -R mutants, eIF5 increased the observed rate of TC binding to the 40S subunit in the absence of mRNA 10- and 5-fold, respectively, whereas it increased the rate only 1.5-fold for WT or G107E. Taken together, these data suggest that the positively charged substitutions of G107 impair TC recruitment in a manner that can be rescued by increased eIF5 occupancy in the PIC. G107E, by contrast, appears to mimic the stimulatory effect of eIF5 on TC recruitment, conferring stronger than WT binding of TC to the 40S subunit that is not elevated further by adding more eIF5. Considering that G107R dissociates more slowly than WT eIF1 from the PIC on AUG recognition6, eIF5 might be acting to increase the rate of dissociation of G107R and G107K eIF1, which in turn would lead to more stable binding of the TC. This interpretation is consistent with previous suggestions that eIF1 release from the PIC allows conversion from the open to the closed state9,15 as well as our data indicating that the rate of stable recruitment of TC to the PIC in vitro is determined by the rate of this conformational change10. To test this model, we set out to determine the effects of the eIF1 mutations and eIF5 on the rate of eIF1 release.
G107 substitutions alter rates of eIF1 dissociation and Pi release from the PIC in response to AUG and non-AUG codons
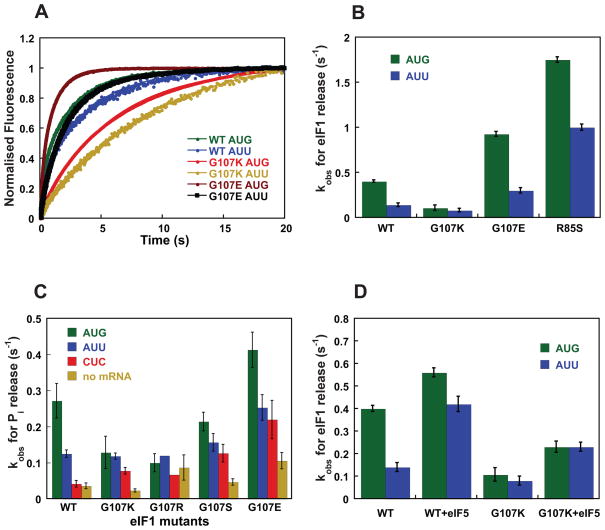
To determine the effects of the G107 substitutions on eIF1 dissociation from the 43S•mRNA complex, fluorescently-labeled mutant eIF1-Fl and WT eIF1A-TAMRA were preassembled in 43S complexes and then mixed with a model mRNA in a stopped-flow fluorometer. An increase in fluorescein fluorescence over time was observed due to loss of fluorescence resonance energy transfer (FRET) between the fluorescein and TAMRA dyes upon dissociation eIF1-Fl (the FRET donor)5. The data are summarized in Figure 6B and Table S3. When the model mRNA has an AUG start codon, dissociation of G107K eIF1 is 4-fold slower than WT (kobs = 0.1 and 0.4 s−1, respectively), similar to our previous findings for G107R6. With an AUU start codon, dissociation of WT eIF1 slows 3-fold (kobs = 0.13 s−1), whereas the rate of dissociation of G107K remains the same as it is for mRNA with an AUG codon (kobs = 0.08 s−1). Thus the G107K substitution makes the rate of eIF1 release independent of whether the start codon is an AUG or an AUU. The same result was obtained with an AUC start codon in the model mRNA (data not shown). In contrast, release of G107E eIF1 is 2-fold faster and release of R85S eIF1 ~5-fold faster than WT with an AUG or AUU start codon in the mRNA (Table S3). This behavior is similar to that of previously characterized Sui− mutants of eIF1 which promote recognition of non-AUG codons as initiation sites by increasing the ease with which eIF1 dissociates from the PIC15.
Figure 6. Kinetics of eIF1 and Pi release from the PIC are affected by the charge at and around position 107.
(A) eIF1 release was monitored by following the decrease in FRET (increase in fluorescein fluorescence) between eIF1-Fl and eIF1A-TAMRA in the PIC after addition of model mRNA with an AUG or AUU start codon. Curves were fit with double-exponential rate equations. The fast phase corresponds to a conformational change and the slow phase to eIF1 release. WT eIF1: AUG (green), AUU (blue); G107K: AUG (red), AUU (yellow); G107E: AUG (brown), AUU (black). (B) Comparison of rate constants for eIF1 release from 43S PICs with model mRNA with an AUG (green) or AUU (blue) start codon. (C) Rate constants for Pi release from PICs made with WT and mutant eIF1 in the absence and presence of model mRNAs with different start codons: AUG (green); AUU (blue); CUC (red); no mRNA (yellow). (D) The effect of eIF5 on the rate of release of WT and G107K eIF1 from 43S PICs with model mRNA with an AUG (green) or AUU (blue) start codon.
We next measured the effect of the mutations on the rate of Pi release from eIF2 in the PIC in response to recognition of cognate (AUG), near-cognate (AUU) and non-cognate (CUC) start codons. To this end, 43S complex was made with TC containing γ-32P-GTP, WT or mutant eIF1, and eIF1A. GTP hydrolysis was initiated by rapidly mixing the 43S complex with saturating amounts of eIF5 (3 μM) and mRNA (10 μM) in a rapid quench device. The reaction was stopped at different times in 100 mM EDTA and the amount of Pi produced over time quantified by gel electrophoresis and phosphorimager analysis. We previously showed that the kinetics of GTP hydrolysis in this experiment are biphasic, with the first phase corresponding to GTP hydrolysis (or a step limiting hydrolysis) and the second phase corresponding to Pi release6.
The rate constants for Pi release are shown in Fig. 6C and are in good agreement with the trends observed in the eIF1 release experiments described above. With WT eIF1 the rate constant for Pi release is ~2.5-fold lower with an AUU codon than with an AUG codon (0.12 s−1 vs. 0.27 s−1, respectively), and ~10-fold lower with a CUC codon or in the absence of mRNA. In contrast, for the G107K and G107R mutants there is no difference in rate constants for Pi release from the complexes with an AUG or AUU codon. This normalization of Pi release rates by the mutations results from a decrease in the rate of release on an AUG start codon without a corresponding decrease in the rate on an AUU codon (0.27 and 0.12 s−1 for AUG and AUU with WT eIF1 vs. ~ 0.1 s−1 on both codons with G107R/K eIF1; Fig. 6C). The reduction in the rates of both eIF1 and Pi release caused by these mutations in the presence of an AUG codon is consistent with the previously described role for eIF1 as an inhibitor of Pi release, and with our previous results with G107R eIF16,15. The fact that the G107R and -K mutations do not also reduce the rate of eIF1 dissociation and subsequent Pi release when the potential start codon is an AUU, however, was unexpected and suggests that the mutations disrupt an interaction that allows preferential release of eIF1 in response to AUG codons relative to non-AUG codons.
G107S eIF1 also reduces the rate of Pi release on AUG codons relative to AUU and CUC codons, but to a lesser extent than the R and K mutants (Fig. 6C). In contrast, the G107E mutation increases the rate of Pi release in all cases, consistent with its effects on the rate of eIF1 dissociation (Fig. 6A, B).
eIF5 promotes faster eIF1 release from the PIC
To test the hypothesis that eIF5 acts to antagonize eIF1 binding to the PIC, we measured the effect of eIF5 on the rate constants for eIF1 release in response to either an AUG or AUU codon (Fig. 6D). Consistent with previous results5 and the small effect of eIF5 on TC binding with WT eIF1 (Fig. 5E), addition of eIF5 had only a modest effect on the rate of release of WT eIF1 with an AUG codon (kobs = 0.4 and 0.56 s−1, without and with eIF5, respectively). With an AUU codon, however, addition of eIF5 increased the rate constant 3-fold, from 0.14 to 0.4 s−1. These data support the proposal that eIF5 promotes eIF1 release from the PIC and can enhance selection of near-cognate start codons. It is possible that in the presence of an AUG codon the rate of release is maximal even in the absence of eIF5 and thus eIF5 cannot stimulate it further.
Because the data described above indicated that the G107K mutation perturbed the interaction between eIF1 and eIF5 in the PIC, we also measured the effect of this amino acid change on eIF5’s ability to enhance eIF1 release. Fig. 6D shows that 2 μM eIF5 increases the rate of eIF1 release 2–3-fold from PICs made with G107K eIF1, regardless of whether the start codon is an AUG or AUU. A possible explanation for these data is that eIF1 and part of eIF5 compete (directly or indirectly) for binding to the same site in the PIC and that the G107K substitution, which stabilizes eIF1 binding to the PIC, requires the competitive action of eIF5 to achieve full displacement in vitro. These results are consistent with the effect of eIF5 overexpression on the Sui− phenotypes of the G107K and R mutants observed in vivo.
The fidelity of start codon recognition during translation in cell-free extracts is affected by the G107 substitutions
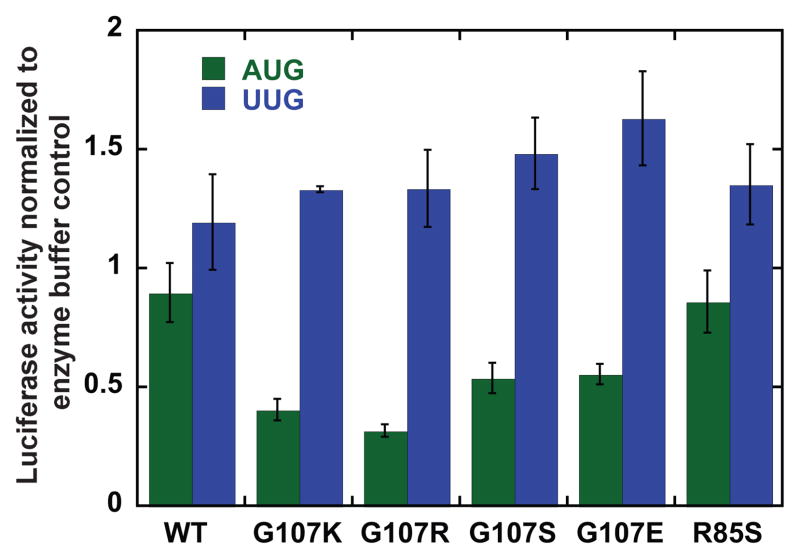
To help connect the results in the reconstituted system to those from experiments performed in vivo, we examined the effects of the eIF1 mutants on the fidelity of start codon recognition in yeast cell extracts29. We used extracts from WT cells supplemented with 2.8 μM purified WT or mutant eIF1. The extracts were programmed with renilla luciferase mRNA with either an AUG or UUG start codon. Replacing the AUG start codon with a UUG resulted in a 10-fold decrease in efficiency of protein production. In each case, luciferase expression after 15 min (linear range) was normalized to that of a control with the same mRNA and eIF1 storage buffer added to the extract instead of eIF1. Similar to what was observed for the rates of eIF1 and Pi release, in the extracts the G107K and -R mutants decrease expression from an mRNA with an AUG start codon but do not significantly alter expression from the mRNA with a UUG start codon (Fig. 7). The G107S and -E mutants also diminish expression from the mRNA with an AUG start codon, but to a smaller extent. In addition, G107E reproducibly increases expression from the mRNA with a UUG codon relative to the case with WT eIF1 (p = 0.017 by t-test), consistent with its faster release from the PIC in the reconstituted system. The R85S mutant has little effect on expression from an AUG or UUG start codon, perhaps because its binding defect is too severe to effectively compete with the endogenous WT protein for incorporation into 43S complexes in the extracts.
Figure 7. Substitutions at G107 and R85 in eIF1 affect the fidelity of start codon recognition during translation in cell-free extracts.
Renilla luciferase mRNA with either an AUG or UUG start codon was translated in WT yeast extracts supplemented with 2.8 μM WT or mutant eIF1. The mRNA with the UUG start codon is translated 10-fold less efficiently than the mRNA with the AUG codon in extracts with only WT eIF1. Luciferase activity was measured after 15 minutes (linear range). The reported values are normalized to the value for that mRNA with eIF1 storage buffer added to the extract instead of the additional eIF1; no change caused by the added WT or mutant eIF1 will give a normalized value of 1.0.
DISCUSSION
In this study we have presented data indicating that the charge at and around position 107 in eIF1 plays an important role in the factor’s interactions with other components of the initiation machinery, impacting TC recruitment and start codon recognition. Changing G107 to R or K, which increases the positive charge density, slows release of eIF1 in response to recognition of AUG codons, which in turn slows Pi release from eIF2. These mutations also diminish the stability of eIF5 binding to the PIC in vivo and reduce the rate and stability of TC recruitment to the 40S subunit in vitro and in vivo. In contrast, changing G107 to an E, which decreases the positive charge density, has the opposite effects on most of these steps and interactions, increasing the rates of eIF1 and Pi release from the PIC and the rate and affinity of TC binding to 40S. Likewise, mutation of the neighboring R85 to S, which also decreases positive charge, increases the rate of TC recruitment and the stability of the resulting 43S complex, and accelerates eIF1 release. In addition to their effects on TC binding, eIF1 dissociation and Pi release, the G107E and R85S mutations weaken binding of eIF1 to the 40S subunit and eliminate the ~7-fold cooperative enhancement of this binding when eIF1A is also bound. The G107K and R mutations have no effect on eIF1 binding to the 40S subunit alone or on cooperativity with eIF1A. The G107S mutation has weaker effects than the other mutations, both in vivo and in vitro, consistent with its smaller perturbation of charge density in this region of the protein. Its effect could be due to its ability to form hydrogen bonds, its inhibition of the free rotation at this position afforded by the wild-type G residue, or both. In general, however, it is striking how the effects of the mutations on the steps and interactions we can observe correlate with increased or decreased positive charge in and around position 107.
Evidence that conversion of the 43S complex to a stable, closed state involves eIF1 release
Our previous kinetic and thermodynamic analysis of the role of base pairing between the start codon and initiator tRNA anticodon provided evidence that initial binding of TC to the 40S subunit produces a complex that is unstable during native gel electrophoresis and that this complex rapidly converts to a stable state when start codon:anticodon base pairing occurs10. A single mistmatch in this three base-pair duplex reduces the rate constant for this induced-fit step by over three orders of magnitude. The data indicated that the observed rate of TC binding in our experiments predominantly reflects this conformational transition rather than the initial encounter of TC with the 40S subunit, which appears to be fast. It seems likely that this conformational change is related to the previously proposed rearrangement from an “open” complex, which can scan the mRNA, to a “closed” one that is arrested on the mRNA and is strongly stabilized by recognition of the start codon11,14,30,31. As the 43S complex scans the mRNA it will encounter many potential near-cognate start codons (CUG, AUU, etc.) but it generally should not stop at them. Consistent with this, the rate of conversion to the stable state observed in our TC binding experiments is very slow on these codons, which would allow the complex to continue scanning the mRNA rather than stop. When an AUG codon is encountered the rate of conversion to the stable state is increased by over three orders of magnitude, which would cause the complex to arrest on the start codon immediately10.
One question that was not addressed in our previous work was the role of eIF1 release from the complex in the conversion to the stable, presumably closed, state. Cryo-EM reconstructions of the yeast 40S subunit alone and bound to eIF1, eIF1A, or both demonstrated that the two factors together cause a conformational change in the subunit from a closed state (40S alone) to an open one (40S•eIF1•eIF1A) that is receptive to TC binding and proposed to be competent for scanning9. The stable conversion from the closed state to the open one requires both factors and, in fact, the 40S eIF1A complex appears even more closed than the naked 40S subunit. These data suggested that release of eIF1 from the PIC, which leaves eIF1A behind32, might be a key trigger of conversion from an open, scanning-competent state to a closed, scanning-arrested one7,11. Moreover, although eIF1 stimulates the rate of TC binding, TC is more stably associated with the PIC in eIF1’s absence9. Thus, it could be predicted that eIF1 dissociation stabilizes the closed, scanning-arrested state that binds TC more tightly.
The data presented here provide strong support for this proposal. The G107R and –K substitutions in eIF1 slow its release from the PIC and also slow the observed rate of TC binding, whereas G107E and R85S increase the rates of both eIF1 release and TC binding. Consistent with this trend, we previously showed that the 93–97 eIF1 mutant, which causes a Sui− phenotype by increasing the rate of eIF1 release from the PIC, also enhances the rate of TC binding, although at the time we did not note the connection between eIF1 release and TC binding rates15. Thus our data support the proposal that eIF1 release is a key trigger of the conformational transition to the stable, closed form of the PIC with tightly bound TC (Fig. 8).
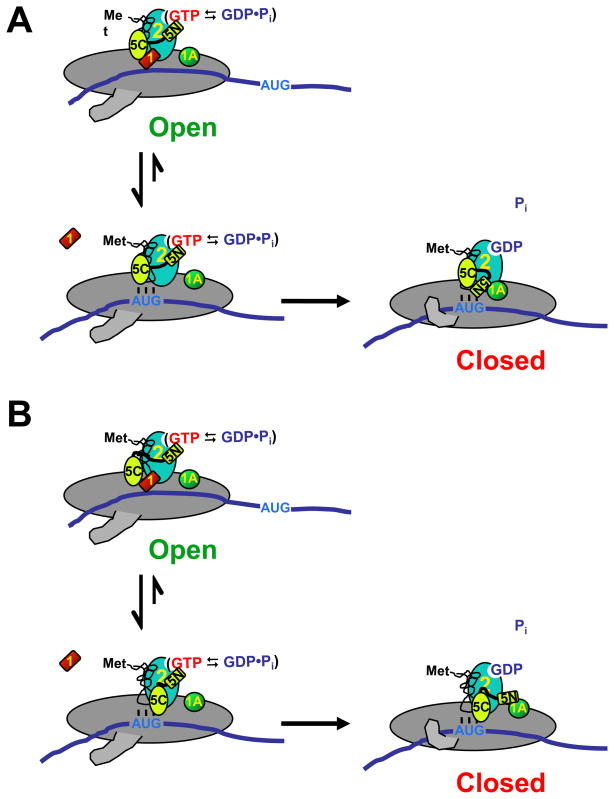
Figure 8. Possible models of events surrounding start codon recognition.
Prior to start codon recognition, the PIC is in an open conformation with relatively unstable TC binding. eIF1 interacts with the CTD of eIF5 and the NTD of eIF5 interacts with eIF2, activating it for GTP hydrolysis. When the start codon is encountered, codon-anticodon base pairing induces release of eIF1, which causes the PIC to undergo a conformational change into the closed state. eIF1 release stabilizes TC binding. In (A), release of eIF1 allows the NTD of eIF5 to enter the eIF1 binding site. In (B), eIF1 release allows the CTD of eIF5 to interact with eIF2β, which in turn causes movement of eIF5’s NTD. In either case, movement of eIF5’s NTD and/or a conformational change in eIF2 allows Pi release from eIF2 and establishment of the strong interaction between eIF5 and eIF1A. For simplicity, eIF2 is shown as a single protein instead of a trimer and the acceptor end of tRNAi is shown free instead of bound to eIF2.
The fact that the Gcd− phenotype of the G107R mutant (and potentially that of G107K as well) is not suppressed by hc TC is consistent with the above interpretation. Our data indicate that a first-order step–eIF1 release–limits the rate of TC binding with these mutant versions of eIF1, and increasing the concentration of TC would not be expected to speed this step. In contrast, Gcd− mutations that are suppressed by hc TC (e.g., 9–12) presumably affect the initial TC binding event, which is second-order, rather than a first-order step that follows it.
eIF1-eIF5 antagonism and start codon recognition
Previous studies established that eIF1 and the CTD of eIF5 can interact in solution and mapped residues on the surface of eIF1 whose NMR chemical shifts change when eIF5 binds25. Included in this interaction surface was a basic region of the protein termed “KH” because it contains lysine and histidine residues: K100, K101, K104 and H106. Changing all of these residues at once to glutamines was recessively lethal and produced a weak dominant Sui− phenotype. These changes reduced binding of eIF1 to the 40S subunit/43S complex in vivo and to eIF5 and the β subunit of eIF2 in vitro. G107, the penultimate residue in eIF1, borders the KH region. Our in vivo and in vitro data indicate that changing G107 to R or K reduces eIF5 and TC binding to the PIC in vivo (Fig. 5A)15, and impairs TC recruitment in vitro in a manner rescued by higher concentrations of eIF5 (Fig. 5E, F). In contrast, the G107E mutant elevates TC recruitment and is refractory to additional eIF5. Given that the G107R and -K substitutions retard eIF1 dissociation, while G107E accelerates it, our results can be explained by a model (Fig. 8) in which eIF1 and a part of eIF5 (e.g., its N-terminal domain) compete for binding to a site in the PIC (e.g., the 40S subunit or eIF2). When eIF1 is in the complex it prevents binding of eIF5 to this site and destabilizes TC binding, but ejection of eIF1 upon start codon recognition allows eIF5 to enter the site. This proposal can explain the antagonism between eIF1 and eIF5 in binding to the PIC (Fig. 6D) and the stimulatory effect of eIF5 on stable TC recruitment (Fig. 5E, F), which requires release of eIF1. It can also explain our finding that selection of UUG start codons in vivo is enhanced by overexpressing eIF5 (Fig. 5C) but suppressed by overexpressing eIF118. The charge at G107 is important for the strength of eIF1’s interaction with this site, perhaps because it alters the KH region’s affinity for the 40S subunit or eIF2β. Positive charge strengthens the interaction, thus further inhibiting eIF5’s ability to enter the site, whereas negative charge weakens the interaction, reducing the effect of eIF5. This can, at least partially, explain the reduced occupancies of eIF5 and TC in native PICs in G107K and –R cells, and why adding eIF5 restores TC loading in the presence of G107K and -R but has virtually no effect with G107E present (Fig. 5E, F). An additional component of the dramatic effect of the G107K and –R mutations on eIF5 association with the PIC in vivo could be effects of these mutations on MFC assembly. For example, the mutations might perturb the ability of the KH region in eIF1 to mediate interaction with eIF5.
The NMR structures of eIF1 and the NTD of eIF5 revealed that the two have similar folds24,25,33. Based on this, it was suggested that eIF1 and the NTD of eIF5 might compete for the same site within the PIC, possibly the γ-subunit of eIF233. This hypothesis is consistent with our proposal (Fig. 8A). Alternatively, although previous work suggested that eIF1 and the CTD of eIF5 can interact simultaneously in solution with the NTD of eIF2β34, it is possible that in the context of the PIC these interactions are mutually exclusive. If this were the case, eIF5’s CTD might be prevented from interacting with the NTD of eIF2β until eIF1 is released from the complex upon start codon recognition (Fig. 8B).
It was previously shown that a direct or indirect interaction between eIF1A and eIF5 occurs upon start codon recognition and that Sui− mutations in these two factors perturb this interaction30. Both versions of our proposal (Figs. 8A and B) could explain these observations if the exclusion of eIF5 from the putative binding site in the PIC by eIF1 prevents the interaction between eIF5 and eIF1A. In this case, release of eIF1 upon start codon recognition would allow the eIF1A-eIF5 interaction to occur. It is possible that the change in eIF5’s interaction partners when eIF1 is released from the PIC is the event that allows Pi to be released from eIF2, perhaps via movement of the N-terminal domain of eIF5 relative to eIF2’s GTPase active site or via a conformational change in eIF2 itself.
Mutations at positions 107 and 85 of eIF1 reduce the fidelity of start codon recognition
Our results indicate that reducing the positive charge at and around position 107 of eIF1–either by changing G107 to E or R85 to S–decreases the affinity of eIF1 for the 40S subunit and increases its rate of release from the 43S complex in response to both AUG and non-AUG codons. Both of these mutations are lethal when expressed as the sole copy of eIF1 on a low-copy plasmid. When the R85S mutant is over-expressed, the lethality is suppressed but the cells have a Sui− phenotype. It is possible that the lethal phenotype associated with these mutations is the result of a strong reduction in the fidelity of start codon recognition due to weakened binding of the mutant factors to the PIC and that, in the case of R85S, this effect is sufficiently suppressed by over-expression that the cells can grow again, albeit with a Sui− phenotype. However, the previously characterized D83G (sui1-1) and Q84P (sui1-17) mutants bind even more weakly to 40S subunits in vitro15 yet allow growth of yeast when expressed as the sole source of eIF1. This suggests that other defects must contribute to the lethal phenotypes of G107E and R85S, for example the loss of thermodynamic coupling between eIF1 and eIF1A in 40S binding, which may affect PIC assembly or other events in initiation. The G107S mutation reduces the affinity of the factor for the 40S•eIF1A complex by 5-fold, suggesting its weak Sui− phenotype might arise from increased spurious eIF1 release from the PIC, as do the previously characterized eIF1 mutants (93–97, D83G and Q84P). Thus the data suggest that G107E, G107S and R85S all reduce the fidelity of start codon recognition by lowering eIF1’s affinity for the small ribosomal subunit.
The G107R and G107K mutants, on the other hand, slow release of eIF1 from the PIC in response to recognition of an AUG codon, making their Sui− phenotypes harder to explain a priori. Our data demonstrate that although these mutations slow eIF1 and Pi release on AUG codons they do not slow these steps on non-AUG codons. The repression of response on an AUG codon but lack of repression on a near-cognate codon is observed not only in the assays in the reconstituted system but also in the in vitro translation experiments in yeast cell extracts, a system that contains all of the initiation factors and is dependent on both the 5′-cap and 3′-poly(A) tail on the mRNA. These observations also are consistent with the in vivo effects of these mutations in elevating the ratio of UUG:AUG initiation with the HIS4-lacZ reporters (Fig. 2B), which would be expected if release of eIF1 and Pi are slowed on AUG relative to near-cognate codons. However, unlike the classical eIF1 Sui− mutants, G107R and K do not increase the rate of these steps with near-cognate start codons and yet they produce an increase in the cellular level of the his4-303 product, which requires initiation at a UUG or AUU start codon (Fig. 2C).
To help reconcile these data, it is useful to recall that derepression of translation of the GCN4 transcription factor provoked by G107R and G107K (Gcd− phenotype) will elicit increased transcription of his4-30319. However, although induction of his4-303 transcription contributes to the Sui− phenotype35, it is not sufficient to explain the large increase in his4-303-myc expression we observed in these mutants (Fig. 2C). Thus, we speculate that the decreased initiation at AUG codons will result in an increase in free 43S complexes, which could in turn increase the initiation frequency at non-AUG codons (at which initiation has not been compromised by the mutations) by mass action. This effect might be pronounced in vivo (Fig. 2C) where free 43S complex concentration is usually low and likely limits initiation frequency, but less pronounced in the cell-free extract system (Fig. 7) where 43S complex is more abundant relative to initiating mRNAs. Our finding that the G107 mutations confer much weaker His+ phenotypes but comparable levels of his4-303-myc expression compared to 93–97, might indicate that the reduced efficiency of AUG initiation in the G107 mutants compromises the expression of one or more WT histidine biosynthetic gene.
Finally, the fact that the G107R and -K mutations decrease the rate of eIF1 release in response to AUG codons but leave the rate unchanged on near cognate codons suggests that the mutations disrupt a function of the factor required for specific recognition of AUG codons rather than compromising a more general aspect of eIF1’s function, as the classical eIF1 Sui− mutations do. How this region of the factor mediates response to AUG but not near-cognate codons remains to be elucidated but the data suggest that eIF1 plays a more active role in start codon recognition than might have been previously thought. Thus, in addition to its roles in suppressing Pi release and conversion to the stable, closed state of the 43S complex, our data suggest that eIF1 helps the complex distinguish between different potential start codons.
MATERIALS AND METHODS
Buffers and Reagents
Reaction (Recon) buffer: 30 mM Hepes-KOH, pH 7.4, 100 mM KOAc, pH 7.4, 3 mM Mg(OAc)2, and 2 mM DTT. Enzyme buffer: 20 mM Hepes-KOH, pH 7.4, 100 mM KOAc, pH 7.4, 2 mM DTT and 10 % Glycerol. Purification of all components was performed as described previously36. The model mRNAs used were of the sequence GGAA[UC]7UNNN[CU]10 C, where NNN was either AUG, AUU, AUC or CUC. The use of the unstructured model mRNA obviates the need for mRNA recruitment and remodeling factors (e.g., eIF3 and eIF4F) as well as a 5′-cap and 3′-poly(A) tail.
Fluorescent labeling of mutant initiation factors eIF1 and eIF1A
eIF1 and its mutants and eIF1A were labeled at their C-termini with cysteine-lysine-fluorescein or TAMRA dipeptide conjugate, respectively, using the expressed protein ligation system as previously described13.
Fluorescence Anisotropy experiments
Fluorescence anisotropy experiments were performed using a T-format Spex Fluorolog-3 (J.Y. Horiba) as previously described13. The excitation and emission wavelengths were 497 nm and 520 nm, respectively. Experiments were carried out in 1X recon buffer. Data were fit with the quadratic solution of an equation describing the binding of fluorescently labeled eIF1 mutants to 40S subunits to give Kd values as described13.
Ternary complex binding and kinetic experiments
Measurement of the affinity and kinetics of TC binding to 40S was carried out using a native gel assay as described36. Component concentrations were 0.5–1 nM 35S-Met-tRNAi, 10 μM mRNA (AUG), 200 nM eIF2, 1μM mutant or WT eIF1 and 1μM eIF1A. With all mutants, the concentration of eIF1 was saturating, thus effects on TC recruitment cannot be attributed to reduced eIF1 binding to 40S. eIF5 was 3 μM when present.
TC affinity measurements
0.2 μM eIF2, 0.5–1 nM 35S-Met-tRNAi and 1 mM GDPNP were incubated for 15 minutes to form TC and then added to a mixture of 40S, 1, 1A, and mRNA at 26 °C. The fraction of 35S-Met-tRNAi bound after 2 hrs was determined by electrophoresis on a native polyacrylamide gel and plotted against [40S]. The data were fit with a hyperbolic binding curve to determine Kd values10.
TC binding kinetics
Determination of the kinetics of 43S complex formation was carried out essentially as described 9,10. TC made with 35S-Met-tRNAi was mixed with 40S subunits, eIF1 (1 μM), eIF1A (1 μM) and mRNA (10 μM). At different times, binding of labeled TC was stopped by adding a chase of excess (at least 300-fold) unlabeled TC. The fraction of labeled Met-tRNAi bound to 40S at each time point was determined by native gel electrophoresis and kobs values calculated by fitting the data with a single exponential equation. In experiments where kinetics of 43S complex formation was studied in the absence of mRNA the reaction was stopped by loading time points directly on a running native gel (25 W, constant power); when the complexes enter the gel, no further binding or dissociation takes place. In this case the data were fit with a double exponential equation10.
eIF1 Dissociation Kinetics
The kinetics of eIF1 dissociation was performed on an SX.180MV-R stopped flow fluorometer (Applied Photophysics) as described previously5 experiments. Briefly, 43S complex was made with 50 nM fluorescein-labeled mutant or WT eIF1 (donor), 60 nM TAMRA-labeled eIF1A (acceptor), 100 nM 40S and 450 nM TC (made with GDPNP). This was mixed rapidly with an equal volume of 20 μM model mRNA and excess of unlabeled eIF1 (3 μM). When eIF5 was used it was included with the mRNA and had a final concentration of 1 μM. Loss of FRET between the two factors when eIF1 dissociates is observed as an increase in fluorescein fluorescence. The data were fit with a double exponential equation, with the first phase corresponding to a conformational change and the second to eIF1 dissociation5.
Pi Release Kinetics
The kinetics of phosphate release from 43S•mRNA complexes was measured by carrying out GTP hydrolysis experiments in a rapid quench (Kintek) as described previously6. TC was formed at 4X concentration: 3.2 μM eIF2, 3.2 μM Met-tRNAi and 250 pM GTPγ[32P] were incubated in 1X recon buffer for 15 minutes at 26 °C. Ribosomal complex was also made at 4X concentration in 1X recon buffer using 800 nM 40S, 3.2 μM eIF1 and 3.2 μM eIF1A. Equal volumes of TC and ribosomal complex were mixed and incubated at 26 °C for 1 hr to form 2X 43S complex. To initiate reaction, 43S complex was mixed with 1.6 μM eIF5, 20 μM mRNA in the rapid quench. Reactions were quenched at different times with 100 mM EDTA. The experiments were also done in the absence of mRNA. Electrophoresis on 15 % polyacrylamide -TBE gels followed by PhosphorImager analysis was used to quantify the fraction of GTP hydrolyzed over time. The data were fit with the sum of two exponentials. The fast first phase corresponds to GTP hydrolysis and the slow second phase corresponds to Pi release, which drives hydrolysis forward6.
In vitro translation assay
In vitro translation was performed with the eIF1 mutants as described previously 29 with some modifications. WT yeast lysate was prepared from yeast strain BY4741. Renilla luciferase mRNA with either an AUG or UUG start codon was transcribed and capped (mMESSAGE mMACHINE, Ambion) and a poly (A) tail added (Poly(A) Tailing Kit, Ambion). 1 and 10 ng of the mRNA with an AUG or UUG start codon, respectively, were used per 15 μL translation reaction. 2.8 μM WT or mutant eIF1 was added to each reaction prior to adding mRNA. For controls an equal volume of eIF1 storage buffer was added instead of eIF1. After 15 min, 5 μL of each reaction was added to 50 μL of Passive Lysis Buffer (Promega) and luciferase activity measured as described15 in a Turner Biosystems luminometer. Data with each mRNA were normalized to the control with only eIF1 storage buffer added for that mRNA. No change from this control would yield a value of 1 for either mRNA.
Yeast strains and plasmids
The yeast strains and plasmids employed in this work are listed in Supplementary Tables S1 and S2, respectively. Strain JCY04 containing chromosomal his4-303-myc10::kanMX6 was constructed as follows. A PCR fragment containing a portion of the his4-303-myc10::kanMX6 allele, beginning 207 nt upstream of the myc10 coding sequences and ending 176 nt downstream of the his4-303 stop codon, was amplified from genomic DNA of H3374 using primers CHA209 (5′ –CCAACCTATGGTTACGCTAGGC-3′) and CHA210 (5′ –CGCTATTTGATACCCACTCTTGC-3′). The PCR product was used to transform strain JCY03 to G418 resistance, replacing his4-303 with his4-303-myc10::kanMX6. The gene replacement was verified by PCR analysis of chromosomal DNA using a primer corresponding to sequences at the 3′ end of the his4-303 ORF, CHA211 (5′ –GATCGTTCTTTGTGACGGTTA- 3′), and sequences downstream of the myc10 coding sequence, HQ505 (5′ –AGTGGCGCGAATTCACTAGTG- 3′). The PCR product was sequenced with primer CHA209.
Plasmid p4438 was constructed by inserting the 2.2 kb EcoRI-Sal I fragment containing the TIF5 promoter, ORF and terminator into YEplac112 cut with the same restriction enzymes.
Derivatives of strains CHY01 (HIS4 gcn2Δ sui1Δ::hisG), JCY103 (his4-303GCN2 sui1Δ::hisG), or JCY04 (his4-303-myc10::kanMX6 GCN2 sui1Δ::hisG) harboring mutant SUI1-His alleles on LEU2 plasmids were constructed by plasmid-shuffling using growth on 5-FOA medium to evict the WT SUI1 URA3 plasmid p1200 resident in these strains.
The following GCN2 his4-303 sui1Δ::hisG strains harboring sc His-SUI1 alleles were examined in Fig. 2A: sc WT (JCY145), hc WT (JCY149), sc G107K (JCY645), hc G107K (JCY649), sc G107R (JCY203), hc G107R (JCY197), sc G107S (JCY653), hc G107S (JCY657), sc R29S (JCY661), hc R29S (JCY665), hc R85S (JCY671), 93–97 (JCY189).
The following gcn2Δ HIS4 sui1Δ::hisG strains harboring sc His-SUI1 alleles were analyzed in Fig. 2D: WT (JCY13), G107K (JCY607), G107R (JCY209), G107S (JCY615), and 9–12 (JCY115).
Cell spotting assay
Growth phenotypes of yeast strains were determined by inoculating 2 ml of the appropriate synthetic complete (SC) medium37 lacking the appropriate nutrient to maintain selection for the relevant plasmid, and grown at 30 °C overnight or until saturation. 20 μl of the resulting culture was mixed with 180 μl of sterile distilled water and 10-fold serial dilutions were spotted on the appropriate selection plates and incubated at 30 °C, as indicated in the figure legends.
Biochemical assays using yeast extracts
For Western analysis of eIF1 and His4-303-myc expression in yeast, WCEs were prepared by trichloroacetic acid (TCA) extraction as previously described38. Briefly, cells cultured to an OD600 of ~1 were harvested, washed with 20 % TCA and resuspended in 200 μl of 20 % TCA in a 2 ml eppendorf tube at room temperature. A 1:1 ratio (v/v) of glass beads was added and vortexed for 2 min, and the lysate was transferred to a clean eppendorf tube. The beads were washed twice with 200 μl of 5 % TCA and the washes were combined with the lysate, which was centrifuged at 3,000 rpm at room temperature in a microfuge for 10 min. The supernatant was discarded and the pellet was resuspended in 100 μl of 2X SDS loading buffer and neutralized by adding 80 μl of 1M Tris-base. The samples were boiled for 3 min and centrifuged at 3,000 rpm at 4 °C for 10 min. The supernatant was collected and stored at −80°C. 3 μl and 6 μl samples were separated on 4–20 % Tris-Glycine polyacrylamide gels (Criterion, BioRad). After electroblotting to nitrocellulose, membranes were probed with rabbit polyclonal antibodies against eIF118 or GCD639 or mouse monoclonal antibodies against the myc epitope (Sigma). Secondary antibodies were HRP-conjugated donkey anti-rabbit or sheep anti-mouse. Detection was performed using ECL.
Assays of β-galactosidase activity in WCEs were performed as described previously 40. Analyses of polysome profiles and fractionation of native PICs in WCEs from HCHO cross-linked were conducted as already described15 using antibodies against eIF3b, eIF5, eIF2γ41, eIF2α42, eIF118 and RPS2 (kindly provided by Jon Warner).
Supplementary Material
Acknowledgments
We thank members of our labs and Tom Dever for comments on the manuscript. This work was funded by a grant from the NIH to JRL (GM62128) and by the Intramural Research Program of the NIH, NICHD (for AGH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mitchell SF, Lorsch JR. Should I stay or should I go? Eukaryotic translation initiation factors 1 and 1A control start codon recognition. J Biol Chem. 2008;283:27345–9. doi: 10.1074/jbc.R800031200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benne R, Amesz H, Hershey JW, Voorma HO. The activity of eukaryotic initiation factor eIF-2 in ternary complex formation with GTP and Met-tRNA. J Biol Chem. 1979;254:3201–5. [PubMed] [Google Scholar]
- 3.Chaudhuri J, Chowdhury D, Maitra U. Distinct functions of eukaryotic translation initiation factors eIF1A and eIF3 in the formation of the 40S ribosomal preinitiation complex. J Biol Chem. 1999;273:17975–17980. doi: 10.1074/jbc.274.25.17975. [DOI] [PubMed] [Google Scholar]
- 4.Algire MA, Maag D, Savio P, Acker MG, Tarun SZ, Jr, Sachs AB, Asano K, Nielsen KH, Olsen DS, Phan L, Hinnebusch AG, Lorsch JR. Development and characterization of a reconstituted yeast translation initiation system. RNA. 2002;8:382–97. doi: 10.1017/s1355838202029527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maag D, Fekete CA, Gryczynski Z, Lorsch JR. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol Cell. 2005;17:265–75. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 6.Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell. 2005;20:251–62. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV. Specific functional interactions of nucleotides at key -3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20:624–36. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das S, Maitra U. Functional significance and mechanism of eIF5-promoted GTP hydrolysis in eukaryotic translation initiation. Prog Nucleic Acid Res Mol Biol. 2001;70:207–31. doi: 10.1016/s0079-6603(01)70018-9. [DOI] [PubMed] [Google Scholar]
- 9.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 10.Kolitz SE, Takacs JE, Lorsch JR. Kinetic and thermodynamic analysis of the role of start codon/anticodon base pairing during eukaryotic translation initiation. RNA. 2009;15:138–52. doi: 10.1261/rna.1318509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–22. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pestova TV, Borukhov SI, Hellen CU. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–9. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 13.Maag D, Lorsch JR. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J Mol Biol. 2003;330:917–24. doi: 10.1016/s0022-2836(03)00665-x. [DOI] [PubMed] [Google Scholar]
- 14.Fekete CA, Mitchell SF, Cherkasova VA, Applefield D, Algire MA, Maag D, Saini AK, Lorsch JR, Hinnebusch AG. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO J. 2007;26:1602–14. doi: 10.1038/sj.emboj.7601613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung YN, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 2007;21:1217–30. doi: 10.1101/gad.1528307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castilho-Valavicius B, Yoon H, Donahue TF. Genetic characterization of the Saccharomyces cerevisiae translational initiation suppressors sui1, sui2 and SUI3 and their effects on HIS4 expression. Genetics. 1990;124:483–95. doi: 10.1093/genetics/124.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoon H, Donahue TF. Control of translation initiation in Saccharomyces cerevisiae. Mol Microbiol. 1992;6:1413–9. doi: 10.1111/j.1365-2958.1992.tb00861.x. [DOI] [PubMed] [Google Scholar]
- 18.Valasek L, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol Cell Biol. 2004;24:9437–55. doi: 10.1128/MCB.24.21.9437-9455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–50. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 20.Cigan AM, Pabich EK, Donahue TF. Mutational analysis of the HIS4 translational initiator region in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2964–75. doi: 10.1128/mcb.8.7.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donahue TF, Cigan AM. Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol Cell Biol. 1988;8:2955–63. doi: 10.1128/mcb.8.7.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dinman JD, Wickner RB. Translational maintenance of frame: mutants of Saccharomyces cerevisiae with altered -1 ribosomal frameshifting efficiencies. Genetics. 1994;136:75–86. doi: 10.1093/genetics/136.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui Y, Dinman JD, Kinzy TG, Peltz SW. The Mof2/Sui1 protein is a general monitor of translational accuracy. Mol Cell Biol. 1998;18:1506–16. doi: 10.1128/mcb.18.3.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fletcher CM, Pestova TV, Hellen CU, Wagner G. Structure and interactions of the translation initiation factor eIF1. EMBO J. 1999;18:2631–2637. doi: 10.1093/emboj/18.9.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reibarkh M, Yamamoto Y, Singh CR, del Rio F, Fahmy A, Lee B, Luna RE, Ii M, Wagner G, Asano K. Eukaryotic initiation factor (eIF) 1 carries two distinct eIF5-binding faces important for multifactor assembly and AUG selection. J Biol Chem. 2008;283:1094–103. doi: 10.1074/jbc.M708155200. [DOI] [PubMed] [Google Scholar]
- 26.Muir TW, Dolan S, Cole PA. Expressed protein ligation: A general method for protein engineering. Proc Natl Acad Sci USA. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asano K, Clayton J, Shalev A, Hinnebusch AG. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes Dev. 2000;14:2534–46. doi: 10.1101/gad.831800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valasek L, Mathew AA, Shin BS, Nielsen KH, Szamecz B, Hinnebusch AG. The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev. 2003;17:786–99. doi: 10.1101/gad.1065403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarun SZJ, Sachs AB. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 30.Maag D, Algire MA, Lorsch JR. Communication between Eukaryotic Translation Initiation Factors 5 and 1A within the Ribosomal Preinitiation Complex Plays a Role in Start Site Selection. J Mol Biol. 2006;356:724–37. doi: 10.1016/j.jmb.2005.11.083. [DOI] [PubMed] [Google Scholar]
- 31.Pestova TV, Lorsch JR, Hellen CU. The Mechanism of Translation Initiation in Eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 87–128. [Google Scholar]
- 32.Acker MG, Shin BS, Nanda JS, Saini AK, Dever TE, Lorsch JR. Kinetic Analysis of Late Steps of Eukaryotic Translation Initiation. J Mol Biol. 2009;385:491–506. doi: 10.1016/j.jmb.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conte MR, Kelly G, Babon J, Sanfelice D, Youell J, Smerdon SJ, Proud CG. Structure of the eukaryotic initiation factor (eIF) 5 reveals a fold common to several translation factors. Biochemistry. 2006;45:4550–8. doi: 10.1021/bi052387u. [DOI] [PubMed] [Google Scholar]
- 34.Singh CR, He H, Ii M, Yamamoto Y, Asano K. Efficient incorporation of eukaryotic initiation factor 1 into the multifactor complex is critical for formation of functional ribosomal preinitiation complexes in vivo. J Biol Chem. 2004;279:31910–20. doi: 10.1074/jbc.M313940200. [DOI] [PubMed] [Google Scholar]
- 35.Yoon HJ, Donahue TF. The suil suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNA(iMet) recognition of the start codon. Mol Cell Biol. 1992;12:248–60. doi: 10.1128/mcb.12.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–45. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- 37.Sherman F, Fink GR, Lawrence CW. Methods of yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1974. [Google Scholar]
- 38.Reid GA, Schatz G. Import of proteins into mitochondria. Yeast cells grown in the presence of carbonyl cyanide m-chlorophenylhydrazone accumulate massive amounts of some mitochondrial precursor polypeptides. J Biol Chem. 1982;257:13056–61. [PubMed] [Google Scholar]
- 39.Cigan AM, Pabich EK, Feng L, Donahue TF. Yeast translation initiation suppressor sui2 encodes the alpha subunit of eukaryotic initiation factor 2 and shares sequence identity with the human alpha subunit. Proc Natl Acad Sci U S A. 1989;86:2784–8. doi: 10.1073/pnas.86.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moehle CM, Hinnebusch AG. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phan L, Zhang X, Asano K, Anderson J, Vornlocher HP, Greenberg JR, Qin J, Hinnebusch AG. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol Cell Biol. 1998;18:4935–4946. doi: 10.1128/mcb.18.8.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dever TE, Feng L, Wek RC, Cigan AM, Donahue TD, Hinnebusch AG. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.